RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans (original) (raw)
Abstract
Introduction of exogenous double-stranded RNA (dsRNA) into Caenorhabditis elegans has been shown to specifically and potently disrupt the activity of genes containing homologous sequences. In this study we present evidence that the primary interference effects of dsRNA are post-transcriptional. First, we examined the primary DNA sequence after dsRNA-mediated interference and found no evidence for alterations. Second, we found that dsRNA-mediated interference with the upstream gene in a polar operon had no effect on the activity of the downstream gene; this finding argues against an effect on initiation or elongation of transcription. Third, we observed by in situ hybridization that dsRNA-mediated interference produced a substantial, although not complete, reduction in accumulation of nascent transcripts in the nucleus, while cytoplasmic accumulation of transcripts was virtually eliminated. These results indicate that the endogenous mRNA is the target for interference and suggest a mechanism that degrades the targeted RNA before translation can occur. This mechanism is not dependent on the SMG system, an mRNA surveillance system in C. elegans responsible for targeting and destroying aberrant messages. We suggest a model of how dsRNA might function in a catalytic mechanism to target homologous mRNAs for degradation.
Eukaryotic organisms have a variety of responses to double-stranded RNA (dsRNA), some of which may be viewed as attempts to ward off the threat of a viral invasion. Mammalian cells unleash a global panic response that results in cessation of translation of all mRNAs (1, 2). Other organisms have a more specific “tactical” approach. In the nematode Caenorhabditis elegans, dsRNA induces a homology-dependent and highly effective decrease in the activity of the corresponding homologous gene, with no evident global effect (3). A similar response has been proposed (4) to underlie certain RNA-mediated cosuppression processes in plants (see ref. 5 for a general review of cosuppression effects in plants). On a technical level, researchers who study C. elegans and plants are now harnessing these responses as an effective means by which to selectively disrupt a specific gene’s activity. The technical application of these processes is referred to as RNA interference, or RNAi.
In addition to the remarkable degree of specificity for the homologous locus, the potency of dsRNA effects in C. elegans has surprised us; in certain cases only a few molecules of dsRNA per cell are required to achieve effective knock-out (3). Standard antisense models of RNA-based interference involve base pairing between antisense and a complementary endogenous sense strand, thereby sequestering the mRNA from the translational machinery and/or targeting it for destruction (6). The substoichiometric activity of dsRNA indicates that RNAi in C. elegans cannot work by a traditional antisense mechanism: there is not enough antisense RNA in the injected material to bind stoichiometrically to the endogenous mRNA. How can dsRNA so effectively target and disrupt a specific gene’s activity? To gain some insight into the mechanism by which dsRNA mediates interference, we have undertaken a series of experiments designed to identify the exact nature of the primary target (i.e., gene locus, nascent transcript, or mature RNA).
MATERIALS AND METHODS
Strains and Alleles.
Standard methods were used for culturing the nematode C. elegans (7). We used the wild-type strain N2 and the following mutant or transgenic strains. TR 1327: unc-54 (r293)I; smg-3 (r867)IV. PD 9556: ccIs9556. ccIs9556 is a chromosomally integrated array containing a pes-10_∷_lacZ translational fusion (pGS15.24) (G. Seydoux, personal communication) and rol-6 (su1006dom) (pRF4) (8) as a selectable marker. JH 103: axIs36[pJH1.16(pes-10_∷_gfp);pMH86(dpy-20)]X. This is a transgenic line with a chromosomally integrated array containing a pes-10_∷_gfp translational fusion (pJH1.16), kindly provided by M. Wallenfang and G. Seydoux.
Clones.
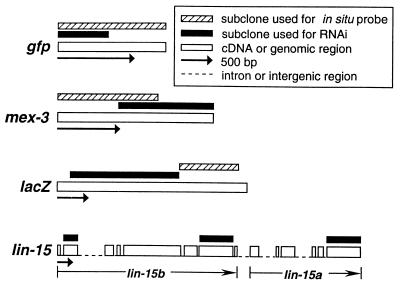
The following plasmids were used to generate dsRNA for RNAi experiments and gene-specific probes for in situ hybridization (see Fig. 1). gfp: pPD79.44, a derivative of pTU#65 (9), was used to make full-length probes for in situ hybridization. pPD128.28 is a construct that contains the 5′ half of gfp and a segment of unc-22 cDNA; dsRNA made with this construct effectively disrupts both unc-22 function and gfp expression. lacZ: pPD124.15 contains an _Fsp_I to _Sac_I fragment from lacZ and was used for RNAi. pPD123.102 and pPD123.103 contain a nonoverlapping _Sac_I to Bst1107I fragment from lacZ and were used to generate probes for in situ hybridization. mex-3: A 772-bp _Eco_RI fragment from a mex-3 cDNA clone (pJP656, ref. 10) was ligated into pBluescript KS+ (Stratagene) to create pMM719, a clone used to make RNA for interference. An 815-bp _Xba_I to _Sac_I fragment from pJP656 was subcloned into pBluescript KS+ to create pMM721, which was used to generate antisense probes for in situ hybridization. cey-2: pMC422 is a cDNA clone (11) and was used to generate antisense probes for in situ hybridization.
Figure 1.
Structure of clones used to generate dsRNA for RNAi and probes for in situ hybridization.
The following lin-15 clones were made by using PCR amplification of C. elegans genomic DNA; the primers were designed with either _Pst_I or _Eco_RI linkers to facilitate cloning into pBluescript KS+. pMM771 contains sequence corresponding to exon 2 of lin-15b; primers used were mm-174 (AACTGCAGAAGCGCAGCAGCAATTCAAAAG) and mm-175 (AACTGCAGCGGTAAGTAGCAATTTCCGCAG). pMM772 contains sequence corresponding to exon 7 of lin-15b; primers used were mm-176 (AACTGCAGAATTGAAGAAACTTGGACAAC) and mm-177 (AACTGCAGTCTTCCAGCAATCTTGGGCTTT). pMM775 contains sequence corresponding to exon 6 of lin-15a; primers used were mm-180 (GGAATTCGTGCCTCCATCGACGAATCTAA) and mm-181 (GGAATTCGTTAAAAAATTGGCTCAGGCTT).
RNA Synthesis and Microinjection.
DNA plasmid constructs that were to serve as templates were linearized with appropriate blunt or 5′-overhang restriction enzymes; sense and antisense RNAs were synthesized in vitro by using T3 and T7 polymerases. DNA templates were then removed with a 15-min DNase treatment. RNAs were extracted with phenol/chloroform and chloroform, precipitated in ethanol, and resuspended in 5 mM Tris⋅HCl, pH 7.9. Sense/antisense RNA strands were heated to 80°C for 3 min and allowed to anneal in injection buffer (2% polyethylene glycol 8000/20 mM potassium phosphate/3 mM potassium citrate, with the mixture adjusted to pH 7.5 with KOH) at 37°C for 30 min prior to injection into the gonads of adult hermaphrodites. C. elegans hermaphrodites possess two gonad arms. For most experiments dsRNA was injected into only one gonad arm; previous experiments have demonstrated that this is sufficient to target endogenous RNAs synthesized in both gonad arms and indeed throughout most cells and tissues of the animal (3).
Amplification of unc-22 Sequences After RNAi.
Adult wild-type C. elegans (N2) were injected with dsRNA produced from a cDNA clone corresponding to bases 17896–17686 from the published genomic sequence of unc-22 (12, 13). DNA was isolated from affected F1 animals (all of which showed a strong twitching phenotype) and was amplified by PCR using the following primer combinations. An error-correcting DNA polymerase (Pfu; Stratagene) was used for amplification. Nucleotides in lowercase represent restriction site linkers used for cloning into standard plasmid vectors.
zf229+zf232: zf229 [CCACGGAGCACAAAYCGGGYC(C)] is an antisense sequencing primer within the interference region. To critically test the model that adenosine deaminase activity might modify the injected dsRNA and eventually produce covalent changes at the DNA level (ref. 14; see also Results and Discussion), the primer was designed so that A → I transitions in the coding strand would have no effect on the ability of the oligonucleotide to prime PCR synthesis.
zf232 (aaactgcagATGTCTTTGAAGATAATCTGAACC) is a sense sequencing primer upstream of the interference region.
zf224+zf233: zf224 (AACAAGGCCGGACCGGGAGAGGCC) is a primer within the interference sequence. This primer was designed so that A → I transitions in the noncoding strand would have no effect on the ability of the oligonucleotide to prime PCR synthesis.
zf233 (ggggtaccAATTCCGGCTTGTCAACTTTTCC) is a primer downstream of the coding sequence.
After amplification, 17 individual clones (13 from zf229+zf232 and 4 from zf224+zf233) were sequenced. In each case, approximately 300 bp of well-defined sequence from each end of the PCR product was analyzed for identity with the expected genomic sequence. All differences from the expected genomic sequence in initial analysis (<1%) were subsequently found to be due to uncertainty in base identification by the automated sequence software. Manual examination of electropherograms indicated these all to be the expected sequence.
In Situ Hybridization.
Whole-mount in situ hybridizations were performed essentially as described by Seydoux and Fire (15, 16), with the following modifications. Adult hermaphrodites were squashed to extrude gonads and embryos prior to freeze cracking. A commercially available fixative, Streck’s tissue fixative (Streck Laboratories, Omaha, NE), was used to fix the animals overnight rather than a 20-min formaldehyde fixation described in the original protocol (17). Digoxigenin (DIG)-labeled single-stranded DNA probes were synthesized by multiple cycles of primer extension in the presence of DIG-dUTP as described (16, 18), using subclones depicted in Fig. 1 as templates. An anti-digoxigenin antibody conjugated to alkaline phosphatase was used to visualize the probes (15, 16).
RESULTS AND DISCUSSION
RNA-Mediated Interference Leaves the Primary DNA Sequence Unchanged.
Certain gene silencing processes in the fungus Neurospora crassa (named “RIP” effects) have been shown to involve covalent changes in DNA sequence (for review see ref. 19). Although this type of mechanism has not been demonstrated in metazoans, Wagner and Sun (14) have proposed a model in which changes in primary DNA sequence might be a component of RNA-mediated interference in C. elegans. We looked for changes in DNA sequence by two different means: first by direct sequencing of genes from affected animals and second by examining the inheritance properties of the interference state.
Following injection of dsRNA for a segment of the unc-22 gene, we obtained a population of F1 animals that exhibited a strong twitching phenotype (see ref. 3 for details of plasmid constructs used). PCR was then used to amplify a genomic DNA segment surrounding and containing the region corresponding to the injected dsRNA. Adenosine deaminases that specifically target and modify dsRNAs by converting adenosines to inosines have been characterized for a variety of organisms (20). Wagner and Sun (14) postulated that such deaminase activity on the injected dsRNA in C. elegans could subsequently lead to changes at the DNA level, effectively mutating the locus such that a loss-of-function phenotype would result. Thus, primers were designed so that A → I deamination reactions would not interfere with the ability to amplify the PCR product (see Materials and Methods). A total of 7.4 kb from 18 individual clones was analyzed; 4.4 kb of this was from sequences in common between the dsRNA and genomic DNA, whereas the remaining 3 kb was from introns and transcribed sequences immediately downstream. The presence of unmodified introns in the PCR amplification product indicates that the amplification material was indeed the genomic DNA template (and not the cDNA clone used to make the injected dsRNA). No modifications in DNA sequence were observed in any of the sequences analyzed from affected animals.
If RNAi induced changes at the DNA sequence level, we would have expected to generate genetic variants with heritable differences in phenotype. We examined animals that had been subjected to interference with the endogenous genes unc-54, unc-22, fem-1 or with transgene-driven gfp and lacZ. In each case, the progeny of injected animals (F1) showed strong interference effects (3), whereas 100% of the next generation (F2) reverted to the wild-type phenotype. We examined >104 F2 animals for unc-22 and unc-54, >5 × 103 animals for gfp and lacZ, and >103 animals for fem-1 (the F1 females from injection of fem-1 dsRNA were not self-fertile, and so needed to be individually outcrossed with wild-type males to analyze further generations; the presence of females in the resulting populations was then analyzed over the next two generations). In these experiments, we used a careful dsRNA preparation protocol that removes all traces of contaminating DNA (3). This precaution was important in that contaminating DNA might conceivably form heritable transgene arrays that might be capable of continuously generating interfering RNA products from spurious transcripts; such a mechanism is likely responsible for the generation of heritable Unc-22 animals after injection of cloned DNA fragments of the unc-22 gene (21).
The case of fem-1 is particularly convincing in ruling out DNA sequence changes as a source of the RNAi effect: fem-1 is thought to act autonomously in the germ line (22), with loss of function in the germ line generating the feminization phenotype seen in fem-1 null mutations (23, 24) and after RNAi (3). The fact that Fem-1 (F1 RNAi) animals produce fem-1+/fem-1+ progeny in the following generation (F2) is a strong indication that RNAi has not been accompanied by any changes in DNA sequence.
Although the above evidence argues strongly against an alteration of the chromosome at the primary sequence level, it was still conceivable that DNA modification or alteration of the chromatin template might play a key role in the interference effect. RNA-mediated modification of DNA has been suggested for certain cases of gene silencing in plants (25). Alternatively, a co-transcriptional or post-transcriptional process could be affected. We further addressed the question of what is the primary target of RNAi by analyzing the ability of dsRNA to disrupt gene activity at different stages of RNA processing.
Is Initiation of Transcription Affected?
C. elegans is an unusual eukaryote in that some of its genes (approximately 25%) are clustered in operons and transcribed as polycistronic units (26). If RNAi worked by blocking initiation or elongation of transcription, then we would expect that all genes in an operon would be knocked out simply by targeting the most upstream gene in the group. This is not the case. The genes lin-15b and lin-15a, which together form a standard operon, do not have any phenotype when either is mutated alone; however, when activity of both genes is disrupted a multivulva (MUV) phenotype results (27, 28). By injecting dsRNAs against the two genes of the lin-15 operon either separately or simultaneously, we demonstrated that both genes need to be targeted to produce MUV animals; RNAi against only one gene produced little or no phenotypic consequence (Table 1). This result was unaffected by the number of exons or relative size of the targeted region (i.e., total number of base pairs). For example, MUV phenotypes were produced when exon 2 of lin-15b and exon 6 of lin-15a were jointly targeted (total of 1,539 bp), whereas no MUV phenotypes resulted when exons 2 and 7 of lin-15b were jointly targeted (total of 1,533 bp) (Table 1).
Table 1.
Effect of RNAi on the _lin_-15 operon
| Segment targeted by dsRNA injections | Progeny with MUV phenotype | |
|---|---|---|
| % | MUV/total | |
| lin_-15_b exon 7 | 2 | 6/255 |
| lin_-15_b exon 7 and lin_-15_b exon 2 | 0 | 0/173 |
| lin_-15_a exon 6 | 0 | 0/114 |
| lin_-15_b exon 7 and lin_-15_a exon 6 | 52 | 88/168 |
| lin_-15_b exon 2 and lin_-15_a exon 6 | 47 | 98/208 |
lin-15 provides a particularly good case in which to assess the sensitivity of genes in an operon. For this experiment to be meaningful, the gene cluster should function as a true operon in that activity of the downstream gene should depend on transcripts that come through the upstream gene. Thus, blocking transcription of the upstream coding region would also affect the downstream region. This is apparently the case for lin-15, in that deletions around the 5′ end of the upstream gene lead to a loss of function of both genes (27, 28). The demonstrated genetic polarity in lin-15 expression, combined with the lack of polarity in the RNAi effect, strongly argues that the primary target of RNAi is not the initiation or elongation of transcription. Additional results describing the effects of injecting specific single-stranded RNA preparations on several operon-like clusters have recently been reported (29; B. Meyer and D. Pasqualone, personal communication; T. Blumenthal, personal communication). Although it is not clear in these cases whether transcription is naturally polar (i.e., that expression of the downstream coding region depends on promoter-proximal sequences in the upstream gene), it is of interest to note that in each case reported RNAi effects are specific for the gene homologous to the injected RNA sequences (i.e., the upstream gene can be affected by RNAi without affecting the function of the downstream gene).
The ability to separately interfere with individual genes in an operon, combined with previous observations that dsRNA segments corresponding to intron and promoter sequences are ineffective in causing RNAi (3), argue that the targets of RNAi are most likely nascent or processed RNA transcripts. To address this issue further, we directly followed the effects of RNAi on products of two transgenes with well characterized transcription and mRNA distribution patterns in the early embryo.
Are Nascent Transcripts Affected?
Seydoux and Fire (15) observed that transcripts from reporter genes with a low density of introns tend to accumulate in the nucleus. When examined by in situ hybridization, these transcripts first appear as “double dots” that are presumably sites of new transcription. Transgenic lines expressing such intron-poor reporters could form the basis of an assay to examine the effect of RNAi on nascent transcripts. We chose to use a pes-10_∷_lac-Z translational fusion. The pes-10 mRNA is among the earliest products of the zygotic genome, with the promoter active in all of the somatic blastomeres of the early embryo, beginning at the 4-cell stage. Transcripts in each somatic blastomere first appear as double dots; subsequently the RNA appears uniform within the nucleus, and then a small fraction is transported to the cytoplasm (15). A pes-10_∷_gfp fusion (kindly provided by M. Wallenfang and G. Seydoux) was also analyzed. This intron-rich gfp construct expresses in a cellular pattern similar to that of the pes-10_∷_lacZ construct, with the major difference being greater accumulation of transcripts in the cytoplasm.
We used in situ hybridization to examine the effects of dsRNA injection on the quantity and subcellular distribution of RNA products from the two reporter transgenes. For these experiments, 20–50 adult hermaphrodites were injected with dsRNA, allowed to recover, and then processed for in situ hybridization the following day. Probes used for in situ hybridization corresponded to a segment of the RNA having at most a partial overlap with the segment targeted by injected dsRNA (see Fig. 1); in this way we could be sure that lack of signal was because of loss (or extensive modification) of the endogenous RNA and not because of “masking” of the endogenous RNA by hybridization to the introduced dsRNA.
We found that injection of lacZ and gfp dsRNAs completely blocked the accumulation of β-galactosidase and green fluorescent protein, respectively. At the RNA level, transcripts from the target gene still appeared in the nucleus of embryonic blastomeres, although these transcripts appeared only transiently and at lower levels compared with the embryos of uninjected controls (Fig. 2). Interestingly, a strong nuclear signal was often detected in the EMS blastomere of the 4-cell stage after RNAi; this strong signal frequently corresponded with evidence that EMS was undergoing the metaphase stage of mitosis at the time of fixation, indicating that targeted transcripts can persist for at least a short time after being made (Fig. 2 B and C). Although a nuclear signal was seen in many early somatic blastomeres, a cytoplasmic signal was never detected. These results, in conjunction with our lin-15 operon results, indicate that transcription is unaffected and suggest that endogenous mRNA targeted by dsRNA accumulates to some level in the nucleus before being destroyed.
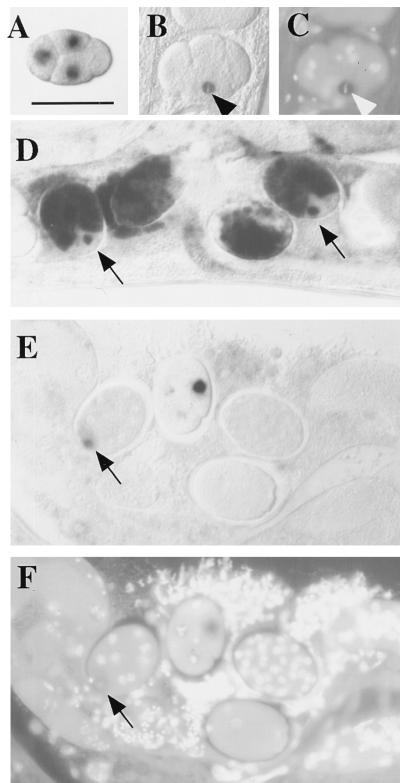
Figure 2.
Analysis of target RNA distribution in RNAi-treated embryos. Embryos shown are from an integrated (homozygous) transgenic line carrying a pes-10_∷_gfp fusion. The pattern of RNA products was analyzed by whole-mount in situ hybridization, with and without prior injection of dsRNA covering the 5′ half of the gfp coding region. (A) Control (uninjected) 4-cell embryo showing transcripts of a pes-10_∷_gfp translational fusion as they first accumulate in the nuclei of the 3 somatic blastomeres of the 4-cell embryo (anterior at left; dorsal at top). This corresponds to the pattern of pes-10 expression described by Seydoux et al. (15). [Bar = 50 μm (for all panels).] (B) An equivalent embryo after injection of dsRNA targeting gfp. Transcripts are still present in the nucleus of cells that have recently commenced transgene expression. As shown in this figure, a relatively strong signal was often detected in the EMS blastomere (arrowhead) of the 4- to 6-cell stage embryo, even while undergoing early stages of mitosis. These transcripts generally do not accumulate to the same levels observed in untreated embryos. (C) Image of embryo in B stained with the DNA-binding dye 4′,6-diamidino-2-phenylindole (DAPI), showing EMS in metaphase (arrowhead). (D) Untreated embryos. Transcripts are eventually transported out of the nucleus and accumulate in the cytoplasm. Nascent transcripts continue to be detected in later emerging somatic blastomeres (arrows). (E) RNAi-treated embryos of stages similar to those depicted in D. Whereas nascent transcripts from a recently emerging somatic blastomere are detected (arrow), transcripts do not accumulate to high levels and are never detected in the cytoplasm. (F) Image of DAPI-stained embryos shown in E. Images of in situ results were obtained with Nomarski differential interference contrast (DIC) microscopy; images of DAPI-stained embryos were obtained with epifluorescence microscopy.
Is Translational Surveillance Involved in RNAi?
The smg (_s_uppressor affecting _m_essa_g_e stability) gene products have been identified as constituting a system that degrades translationally aberrant mRNAs and may play a role in normal mRNA degradation (30–32). Given the suggestions above that RNA decay had some role in the RNAi mechanism, we asked whether the SMG system was required for RNAi to operate. We used a smg-3 mutant host, which has been shown in genetic studies to lack SMG activity and hence to stabilize a variety of prematurely terminated or aberrant mRNAs (30). Despite the inability of Smg-3 animals to degrade these aberrant mRNAs, the strain was evidently still capable of RNAi. As in wild-type animals (3), when we injected smg-3 mutant animals with dsRNA targeted against mex-3 (a highly abundant maternal mRNA) we observed disappearance of the targeted RNA (Fig. 3 A and B) and universal embryonic arrest consistent with loss of MEX-3 function.
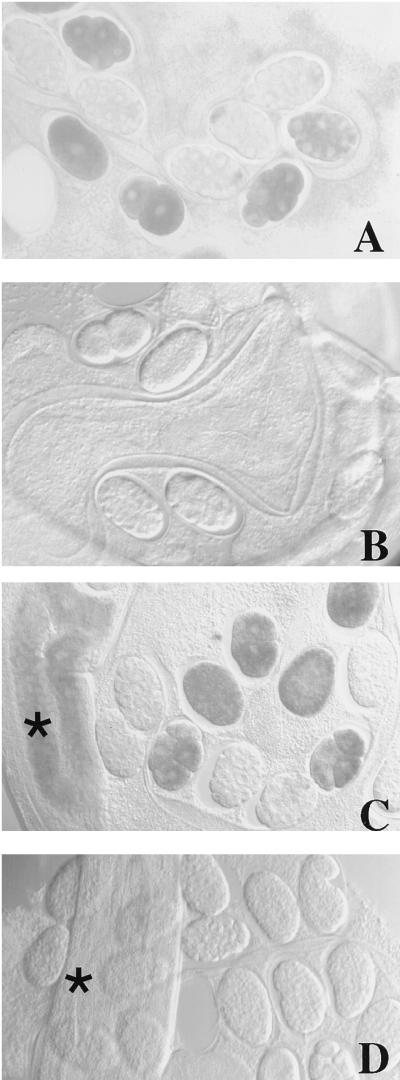
Figure 3.
Loss of maternal mex-3 RNA in RNAi-treated smg-3 and wild-type hermaphrodites and their progeny as revealed by whole-mount in situ hybridization. mex-3 message is abundant and correctly localized in the embryos of a smg-3 mutant (A) but disappears from the progeny of smg-3 mutant hermaphrodites injected with mex-3 dsRNA (B). (C) In wild-type hermaphrodites mex-3 message is detected throughout the gonad (indicated by the asterisk at far left) as well as in embryos. (D) Injection of mex-3 dsRNA results in complete loss of detection of endogenous mex-3 in the maternal germ line (here splayed out) as well as in all F1 progeny. Images were obtained with Nomarski differential interference contrast microscopy. As a control for the in situ hybridization experiments, we determined that loss of hybridization signal was specific to the targeted mRNA: levels of another germ-line mRNA, cey-2, were unaffected when adult hermaphrodites were injected with mex-3 dsRNA (data not shown). (×240)
The lack of SMG requirement for effective RNAi suggests the involvement of other RNA-degradation mechanisms. The SMG system is not essential for viability (30, 32). It will be interesting to determine whether the degradative processes responsible for RNAi are essential for the organism.
Experiments with mex-3 also revealed another aspect of RNA-mediated interference in C. elegans. mex-3 RNA is a maternal product that is “masked” in the distal portion of the maternal germ line and is not translated until late oogenesis (10). We have observed that mex-3 endogenous message is lost from the whole germ line in both wild-type (Fig. 3 C and D) and smg-3 (data not shown) mutant hermaphrodites injected with mex-3 dsRNA. Both this result and the effect of dsRNA on nuclear transcripts indicate that translation is not required for mRNAs to serve as targets of RNAi.
The preceding data strongly suggest that RNAi targets a post-transcriptional process. Our working model has three components.
(i) Exogenous dsRNA has no direct effect on the targeted gene or on the initial biosynthesis of corresponding endogenous transcripts. Evidence for this includes the lack of observed changes in the endogenous gene and the transient appearance of primary transcripts.
(ii) Exogenous dsRNA causes early degradation of homologous mRNA and/or mRNA precursor molecules. This hypothesis is based on the observed decrease in steady-state nuclear RNA levels for the targeted gene, combined with the complete loss of cytoplasmic RNA.
(iii) This degradation process can occur after splicing but prior to transport from the nucleus. Evidence for this includes the behavior of operons, the requirement for exon sequences in the interfering dsRNA (3), and the timing of observed RNA decay. None of the experiments described here address the important question of whether preexisting mRNAs in the cytoplasm can be targeted for degradation by RNAi.
The observed potency of the interfering RNA requires that our models go beyond a traditional antisense mechanism that would require stoichiometric interaction between injected RNA sequences and the native transcript. One conceivable source for the potency of RNAi would be a replication-based mechanism whereby dsRNA could be amplified after injection into the animal. Replication of interfering RNA has been suggested for certain cases of cosuppression in plants (25). We have yet to uncover any evidence that the introduced dsRNA replicates in C. elegans. In particular, if introduced dsRNA were replicating, then in situ hybridization with an antisense probe overlapping or covering the region of interference might be expected to show a strong signal from newly synthesized sense strands. This idea was tested with mex-3, modifying the procedure used in Fig. 3 so that the probe for in situ hybridization corresponded precisely to the segment used for interference. We detected no sense copies of the dsRNA in these experiments; we similarly failed to detect antisense copies of the dsRNA in experiments using a mex-3 sense probe corresponding to the dsRNA segment (data not shown).
Our failure to detect replication of the injected dsRNA has led us to models in which dsRNA induces a catalytic mechanism to destroy homologous cellular RNAs (Fig. 4). We envision a three-step model. First, dsRNA would form part of a specialized ribonucleoprotein complex, with partial unwinding of the duplex allowing homology-based target recognition. Second, recognized segments of the cellular RNA would be “marked” as a consequence of interaction with the dsRNA complex. Possible marking mechanisms include cleavage, covalent modification (e.g., by dsRNA-dependent adenosine deaminase), or changes in the spectrum of associated proteins. Third, target RNAs would be rendered undetectable. This third step could conceivably involve any covalent change that renders the RNA fully inaccessible to in situ hybridization. Although cleavage, deamination of adenosines, or a change in localized protein coating for the targeted region might play a role in the initial marking, the eventual result of RNA interference is a failure to detect any region of the targeted mRNA. In interpreting our data, it should be noted that in most cases we have used RNA probes for in situ hybridization that examine a different region of the message from that directly targeted by dsRNA. Given the observed failure to detect any part of the affected RNA, we have been led in our working model to propose that RNAs targeted by RNAi are subject to rapid degradation. The final degradation could be closely coupled to the initial interaction with the interfering RNA, or alternatively could involve activation of a secondary process using more general components of the RNA degradation mechanisms within the cell. We have ruled out the obligate involvement of one such mechanism (the SMG system). Since the SMG system is nonessential for survival of the organism, there must be other systems for physiological RNA degradation. These intrinsic mechanisms have yet to be characterized; thus it remains to be seen if they might play a role in dsRNA-mediated degradation.
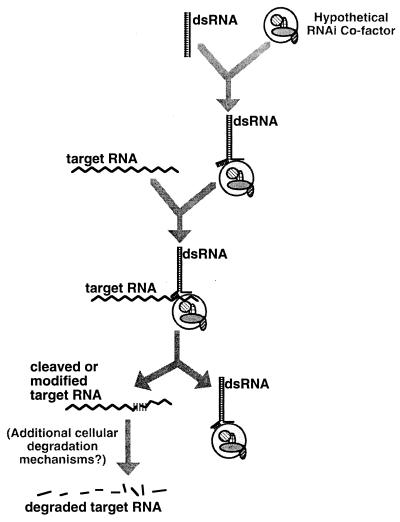
Figure 4.
Possible model for dsRNA-mediated genetic interference in C. elegans. Upon introduction into the cell, dsRNA is proposed to complex with a (hypothetical) protein or ribonucleoprotein complex that allows unwinding of an arbitrary segment of the duplex. The complex would then search by homology for corresponding segments of cellular RNA. Recognition of cellular RNAs would be followed by a process marking the target RNA for degradation. Possible “marking” mechanisms include direct cleavage of the target RNA, covalent modification (e.g., by adenosine deaminase), or the recruitment or removal of specific RNA-binding proteins. Whatever the mechanism of this initial interaction, our data suggest a rapid subsequent degradation of the entire targeted transcript. The secondary degradation machinery could involve a combination of components specific to RNA interference and/or more general catabolic mechanisms.
Although the dsRNA-triggered RNA-degradation model described in this paper accounts for many of the observations regarding RNA-mediated interference in C. elegans, it is still conceivable (and perhaps likely) that additional mechanisms will be involved. In plants, viral RNAs that have been engineered to contain precise homology to cellular genes can trigger two apparently distinct processes: degradation of the homologous cellular mRNA (33) and methylation of the corresponding cellular gene (34). The first mechanism is thought to be responsible for many transient cosuppression effects, whereas the RNA-dependent methylation has been proposed to play a role in producing slower (but potentially more stable) changes in gene expression. For all of the genes we tested in C. elegans, RNAi effects were not heritable; nonetheless, Mello and colleagues (35) have observed a multigenerational effect for a small number of genes. It will be interesting to determine whether dsRNA-triggered RNA decay plays a role in these processes, and if so how the interference state might become heritable for a subset of genes.
Acknowledgments
We thank G. Seydoux, S. Dymecki, S. Strome, T. Blumenthal, B. Meyer, C. Mello, B. Kelly, S. Kostas, K. Liu, L. Timmons, J. Hsieh, B. Harfe, M. Hsu, J. Fleenor, and S. Parrish for helpful discussions. We also thank M. Wallenfang, G. Seydoux, and J. Priess for providing some of the clones used in this study. This research was supported in part by the National Institute of General Medical Sciences (Grants R01-GM37706 to A.F. and GM17164 to M.K.M.).
ABBREVIATIONS
dsRNA
double-stranded RNA
RNAi
RNA interference
References
- 1.Proud C. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs B L, Langland J O. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Montgomery M K, Fire A. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 5.Matzke M A, Matzke A J. Cell Mol Life Sci. 1998;54:94–103. doi: 10.1007/s000180050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nellen W, Lichtenstein C. Trends Biochem Sci. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-c. [DOI] [PubMed] [Google Scholar]
- 7.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 10.Draper B W, Mello C C, Bowerman B, Hardin J, Priess J R. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- 11.Jantsch-Plunger V. Ph.D. thesis. Vienna, Austria: Univ. of Vienna; 1993. [Google Scholar]
- 12.Benian G, Kiff J, Neckelmann N, Moerman D, Waterston R. Nature (London) 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- 13.Benian G, L’Hernault S, Morris M. Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner R W, Sun L. Nature (London) 1998;391:744–745. doi: 10.1038/35750. [DOI] [PubMed] [Google Scholar]
- 15.Seydoux G, Fire A. Development (Cambridge, UK) 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- 16.Seydoux G, Fire A. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism, Methods in Cell Biology. Epstein H F, Shakes D C, editors. Vol. 48. San Diego: Academic; 1995. pp. 323–337. [DOI] [PubMed] [Google Scholar]
- 17.Harfe B, Fire A. Development (Cambridge, UK) 1998;125:421–429. doi: 10.1242/dev.125.3.421. [DOI] [PubMed] [Google Scholar]
- 18.Patel N H, Goodman C S. In: Nonradioactive Labeling and Detection of Biomolecules. Kessler C, editor. Berlin: Springer; 1992. pp. 377–381. [Google Scholar]
- 19.Selker E U. Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 20.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O’Connell M A, Samuel C E, Herbert A. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 21.Fire A, Albertson D, Harrison S W, Moerman D G. Development (Cambridge, UK) 1991;113:503–514. doi: 10.1242/dev.113.2.503. [DOI] [PubMed] [Google Scholar]
- 22.Gaudet J, VanderElst I, Spence A M. Mol Biol Cell. 1996;7:1107–1121. doi: 10.1091/mbc.7.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doniach T, Hodgkin J A. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- 24.Kimble J, Edgar L, Hirsh D. Dev Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- 25.Wassenegger M, Pelissier T. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 26.Zorio D A, Cheng N N, Blumenthal T, Spieth J. Nature (London) 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]
- 27.Clark S G, Lu X, Horvitz H R. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–412. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korf I, Fan Y, Strome S. Development (Cambridge, UK) 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- 30.Pulak R, Anderson P. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 31.Morrison M, Harris K S, Roth M B. Proc Natl Acad Sci USA. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angell S M, Baulcombe D C. EMBO J. 1997;16:3675–3684. doi: 10.1093/emboj/16.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassenegger M, Heimes S, Riedel L, Sanger H L. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 35.Tabara H, Grishok A, Mello C. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]