Self vs. non-self discrimination during CRISPR RNA-directed immunity (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 28.
Published in final edited form as: Nature. 2010 Jan 13;463(7280):568–571. doi: 10.1038/nature08703
Abstract
All immune systems must distinguish self from non-self to repel invaders without inducing autoimmunity. Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci protect bacteria and archaea from invasion by phage and plasmid DNA through a genetic interference pathway1–9. CRISPR loci are present in ~ 40% and ~90% of sequenced bacterial and archaeal genomes respectively10 and evolve rapidly, acquiring new spacer sequences to adapt to highly dynamic viral populations1, 11–13. Immunity requires a sequence match between the invasive DNA and the spacers that lie between CRISPR repeats1–9. Each cluster is genetically linked to a subset of the cas (CRISPR-associated) genes14–16 that collectively encode >40 families of proteins involved in adaptation and interference. CRISPR loci encode small CRISPR RNAs (crRNAs) that contain a full spacer flanked by partial repeat sequences2, 17–19. CrRNA spacers are thought to identify targets by direct Watson-Crick pairing with invasive “protospacer” DNA2, 3, but how they avoid targeting the spacer DNA within the encoding CRISPR locus itself is unknown. Here we have defined the mechanism of CRISPR self/non-self discrimination. In Staphylococcus epidermidis, target/crRNA mismatches at specific positions outside of the spacer sequence license foreign DNA for interference, whereas extended pairing between crRNA and CRISPR DNA repeats prevents autoimmunity. Hence, this CRISPR system uses the base-pairing potential of crRNAs not only to specify a target but also to spare the bacterial chromosome from interference. Differential complementarity outside of the spacer sequence is a built-in feature of all CRISPR systems, suggesting that this mechanism is a broadly applicable solution to the self/non-self dilemma that confronts all immune pathways.
S. epidermidis strain RP62a (ref. 20) contains a CRISPR locus that includes a spacer (spc1) that is identical to a region of the nickase (nes) gene found in nearly all sequenced staphylococcal conjugative plasmids (Fig. 1a and Supplementary Fig. 1a), including those that confer antibiotic resistance in methicillin- and vancomycin-resistant Staphylococcus aureus strains21–23. The S. epidermidis CRISPR system limits conjugation between staphylococci by an _spc1_-directed interference pathway3. As in other species2, 7, 17–19, 24, 25 the S. epidermidis CRISPR locus is transcribed and processed into crRNAs3, and northern analysis indicates that most _spc1_-containing crRNAs are ~49 nucleotides long (Supplementary Fig. 1b). During CRISPR interference in S. epidermidis, target specificity appears to be achieved by direct pairing of the spc1 crRNA with the plasmid DNA3. DNA targeting most likely underlies CRISPR interference in many other bacterial and archaeal species as well2, 26. Since a sequence match also exists between the crRNA and the CRISPR locus DNA that encodes it, a central issue in CRISPR interference is therefore how the crRNA identifies bona fide targets without attacking the CRISPR locus in the host chromosome.
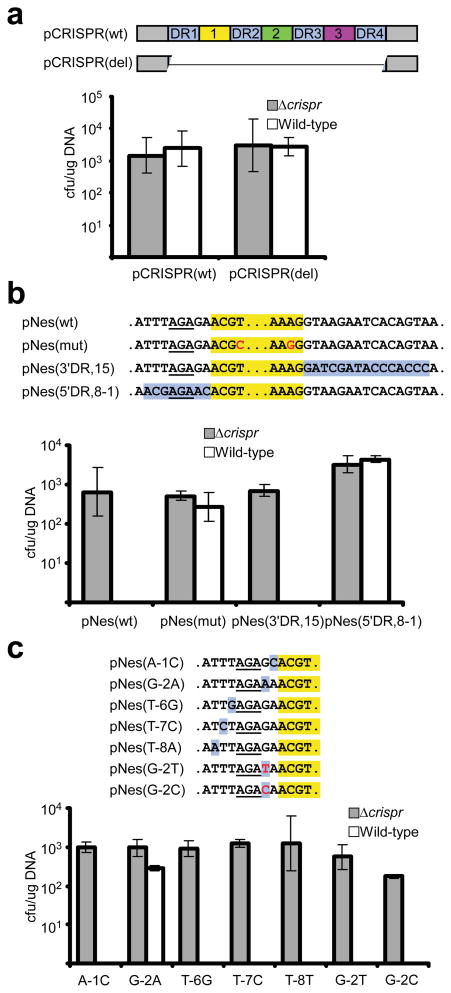
Figure 1. Protection of nes target by spacer flanking sequences.
a, Direct repeats (DR1–4, purple boxes) and spacers (1–3, colored boxes) of the S. epidermidis RP62a CRISPR locus were cloned into pC194 generating pCRISPR(wt) and its deletion variant, pCRISPR(del). b, pNes(wt) and pNes(mut) contain wild-type and mutated nes target sequence of pG0400 (highlighted in yellow, with mutations in red). In pNes(5′DR,8-1) and pNes(3′DR,15) nes target flanks were replaced by repeat sequences present upstream and downstream of spc1 (highlighted in purple), respectively. c, Individual nucleotides upstream of nes target were replaced by those present upstream of spc1 (highlighted in purple). The G at position −2 was also changed to C and T (in red). The AGA sequence (underlined) is shared by both the nes target and spacer 5′ flanking sequences. All plasmids were transformed into S. epidermidis RP62a and its isogenic Δ_crispr_ mutant. The average of at least three independent measures of the transformation efficiency (determined as cfu/μg DNA) is reported and error bars indicate 1 s.d.
We previously demonstrated that CRISPR interference can prevent transformation of pC194-based plasmids that contain a target sequence3. To test whether CRISPR spacers have an intrinsic ability to evade interference, we cloned the repeat/spacer sequences of the RP62a CRISPR locus, along with ~200 base pairs (bp) from either side of the repeats and spacers, into pC194 (Fig. 1a). The resulting plasmid, pCRISPR(wt), which contains three possible interference targets (spc1, spc2 and spc3), was transformed into wild-type and Δ_crispr_ cells. Unlike the nes protospacer-containing plasmid, pCRISPR(wt) transformation efficiency was similar in both strains [Fig. 1a; Supplementary Table 1 shows the transformation efficiency values for all plasmids here described]. These results indicate that the potential targets present in the CRISPR locus are specifically exempted from CRISPR interference.
We hypothesized that differences between flanking regions of spacers and targets – i.e., the presence or absence of repeats – could provide the basis for self/non-self discrimination. To test this, we replaced 15 bp from either side of the nes target with the corresponding spc1_-flanking repeat sequences. The resulting plasmids [pNes(5′DR,15) and pNes(3′DR,15)] were tested for CRISPR interference by transformation into wild-type and Δ_crispr cells (Fig. 1b and Supplementary Fig. 2a,b). Only pNes(5′DR,15) escaped interference, indicating that repeat sequences upstream of a target (i.e., adjacent to the 5′ end of the crRNA spacer sequence) can protect that target. Similar experiments further narrowed the protective region to the eight bp closest to the target (Fig. 1b and Supplementary Fig. 2b). This region contains five mutations in the nes upstream sequence, since the 5′-AGA-3′ sequence from position −5 (i.e., 5 bps upstream of the start of the nes protospacer) to −3 are shared with spc1 5′ flank (Supplementary Fig. 2a). Each of these five mutations was individually tested for its effect on CRISPR interference (Fig. 1c). Only the guanosine-to-adenosine change at position −2 (G-2A) conferred protection. These results demonstrate that repeat sequences upstream of spacers prevent CRISPR interference, and point to position −2 as an important determinant of this effect.
Short (2–4 bp), conserved sequences called “CRISPR motifs”12, 24, 25 or “protospacer adjacent motifs” (PAMs)27 have been found near protospacers in other CRISPR systems, and mutations in these motifs can compromise interference12, 25. A CRISPR motif has not been defined in S. epidermidis protospacers, of which only two have been specified3. To test whether G-2 is part of a conserved motif important for interference, we tested plasmids carrying the mutations G-2C and G-2T. Surprisingly, unlike the G-2A mutation, C and T transversions had no effect on transformation efficiency (Fig. 1c). This result was corroborated in a conjugation assay3 using pG0400 G-2A or G-2C mutants (Supplementary Fig. 3) and excludes the possibility that a G at position -2 is simply a crucial CRISPR motif residue. Instead, this observation suggests that only an A at position −2 – i.e., the nucleotide present in the repeats – allows protection, and that any deviation from this nucleotide enables interference.
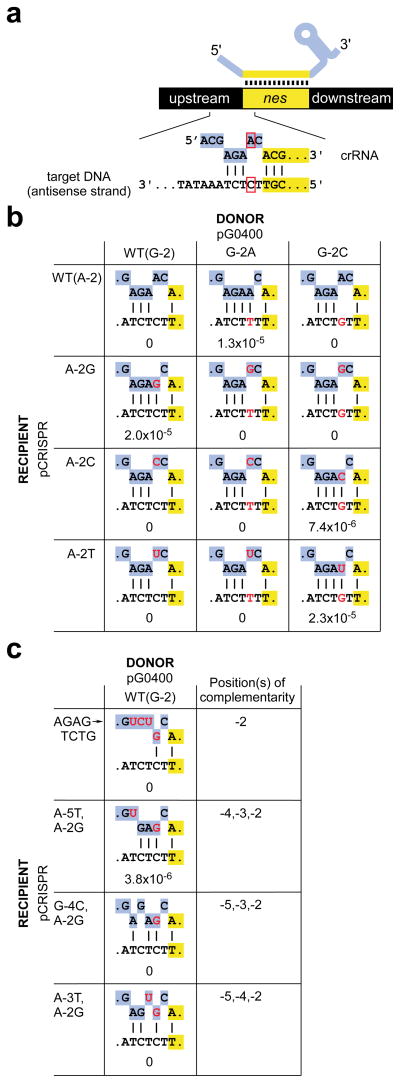
In light of this observation, we considered the complementarity of crRNAs with target protospacers and with CRISPR sequences: although the spacer region of a crRNA can pair with target DNA (Fig. 2a) and CRISPR DNA alike, only the CRISPR DNA will be fully complementary with the CRISPR repeat sequences at the crRNA termini. We therefore hypothesized that specific base pairs in the crRNA/DNA heteroduplex outside of the spacer enable protection, thereby providing a mechanism to avoid autoimmunity. To explore this possibility we introduced compensatory mutations in the crRNA (Fig. 2a, b) by changing sequences upstream of spc1 in pCRISPR(wt), which can complement the interference deficiency of the Δ_crispr_ strain. The wild-type spc1 crRNA contains an adenosine at position −2, so we generated the A-2G, A-2C and A-2T mutants. We then tested each for interference with conjugation of wild-type pG0400 as well as its G-2A and G-2C mutant derivatives. In agreement with our hypothesis, the nes target was protected from interference only when pairing was possible at position -2: rA-dT, rG-dC, and rC-dG Watson-Crick appositions each resulted in evasion of the CRISPR system by the conjugative plasmid (Fig. 2b). All crRNA mutants were functional and therefore correctly processed2, since all pCRISPR plasmids were able to restore interference in Δ_crispr_ cells with pG0400 derivatives that were mismatched at position −2 (Fig. 2b). Interestingly, a rU-dG wobble apposition also protected the conjugative plasmid from interference while a rG-dT wobble apposition did not, despite the greater stability of rG-dT pairs in an otherwise Watson-Crick-paired heteroduplex28. These results provide strong evidence that crRNA/target noncomplementarity at position −2 is important for interference. Further mutagenesis of the −5 to −2 sequence of spc1 crRNA indicated that the minimal complementarity required for protection involves positions −4, −3 and −2 (Fig. 2c and Supplementary Fig. 4). Altogether these results indicate that protection of the nes target during conjugation in S. epidermidis requires complementarity between crRNA and target upstream flanking sequences at these positions, and strongly suggest that the mechanism of protection requires base-pair formation in this region (Fig. 4a). Furthermore, the base pairing implied by our compensatory analyses reinforces our earlier conclusion3 that the target of the S. epidermidis spc1 crRNA is the antisense strand of the nes DNA locus, not the sense-oriented mRNA.
Figure 2. Complementarity between crRNA and target DNA flanking sequences is required for protection.
a, Schematic of the complementarity between the flanking sequences of crRNA (top, highlighted in purple) and target DNA (bottom). The red box indicates the nucleotides mutated in the experiments shown in b. b, c, Conjugation assays of pG0400 and its mutant variants, using as a recipient the Δ_crispr_ strain harboring different pCRISPR plasmids. Mutations are shown in red. Conjugation efficiency was determined as transconjugant cfu/recipient cfu; the average of at least 3 independent experiments is reported.
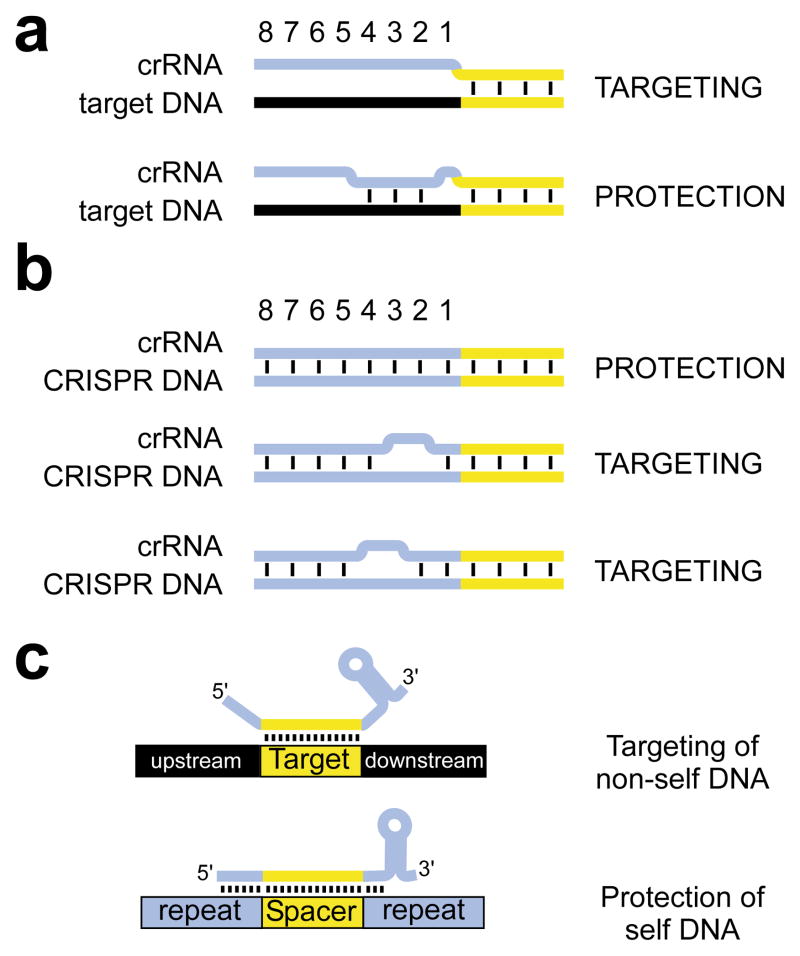
Figure 4. Requirements for targeting and protection during CRISPR immunity.
a, In S. epidermidis, CRISPR interference is enabled by mismatches between target DNA and crRNA sequences upstream of the spacer. Formation of at least 3 base pairs at positions −4, −3 and −2 eliminates targeting. b, Complementarity between the S. epidermidis CRISPR locus and the crRNA 5′ terminus protects it from interference. Disruption of base pairing at positions −4,−3 or −3,−2 eliminates protection. c, General model for the prevention of autoimmunity in CRISPR systems. The ability of crRNA termini (5′, 3′, or both) to base pair with potential targets enables discrimination between self and non-self DNA during CRISPR immunity.
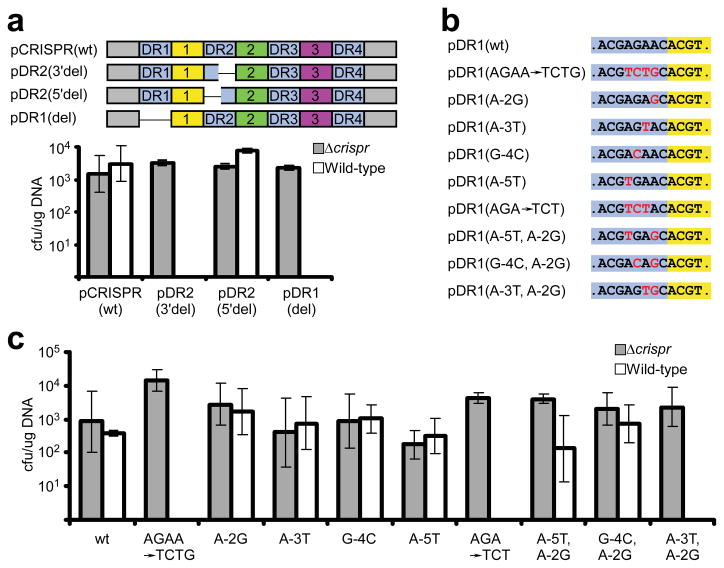
If base pairs at positions −4, −3 and −2 confer protection on an otherwise susceptible target, then abolition of base pairing in the same region should confer susceptibility on an otherwise protected CRISPR locus. Deletion analyses in pCRISPR (Fig. 3a and Supplementary Fig. 5a) demonstrated that sequences immediately upstream of spacers prevent autoimmunity. We then tested the effect of substitutions on the direct repeat upstream of spc1 (DR1) on CRISPR protection (Fig. 3b, c). To avoid the expression of mutant crRNAs that would complicate our analyses, we generated pDR1(wt), a deletion variant of pCRISPR(wt) that lacks the 200 bp preceding the CRISPR locus. This plasmid is unable to produce functional crRNAs (data not shown) but is protected from interference in wild-type cells (Fig. 3b,c). In contrast, plasmid pDR1(AGAA→TCTG), which contains mutations at positions −5 to −2, was subject to CRISPR interference. This indicates that this region is critical for protection of the CRISPR locus. Mutagenesis of this and other regions of spc1 upstream flanking sequences (Fig. 3b, c and Supplementary Fig. 5b) indicated that at least two consecutive mismatches from positions −4 to −2 are required to eliminate protection of the CRISPR locus (Fig. 4b).
Figure 3. Mutations in upstream flanking sequences of CRISPR spacers elicit autoimmunity.
a, Deletions were performed in the flanking repeats of spc1: the 3′ half of DR2, the 5′ half of DR2, and all of DR1 were deleted from pCRISPR(wt) in pDR2(3′del), pDR2(5′del) and pDR1(del), respectively. b, c, Substitutions (red) were introduced in the 5′ flanking sequence (highlighted in purple) of spc1 (highlighted in yellow), generating different pDR1 variants that were tested by transformation. All plasmids were transformed into S. epidermidis RP62a and Δ_crispr_ strains. The average of at least three independent measures of the transformation efficiency (determined as cfu/μg DNA) is reported and error bars indicate 1 s.d.
CRISPR systems show a high degree of diversity in cas gene content4, 9, 15, 16, and therefore mechanistic differences between different CRISPR/cas subtypes are likely to exist. However, differential crRNA pairing potential with CRISPR loci and invasive targets outside of the spacer region (Fig. 4c) is intrinsic to all CRISPR systems, and therefore the mechanism of self/non-self discrimination that we have defined in S. epidermidis could apply broadly. The specific bps that are monitored could vary between CRISPR/cas systems, and we speculate that protein components of the system may “proofread” paired vs. unpaired structures at these sites in a manner that either aborts or enables later steps in interference, respectively. Our findings also highlight the importance of the previously noted 5′-terminal homogeneity of crRNAs2,3,17, which consistently contain ~8 nt of upstream repeat sequences. Finally, our results are inconsistent with a critical role for the CRISPR motif12, 25, 27 during the interference phase of CRISPR immunity. No significant similarity exists between the two known targets of the S. epidermidis CRISPR locus (Supplementary Fig. 5c) and therefore we cannot determine if the nucleotides that are important for discrimination are also part of a CRISPR motif. However, our mutagenesis experiments indicate that no specific flanking nucleotides are required for interference: the decisive characteristic is noncomplementarity with the crRNA rather than nucleotide identity per se. By extension, it is conceivable that CRISPR motif mutants previously shown to evade interference12, 25 could reflect a failure of self/non-self discrimination via crRNA/target base pairing, rather than a requirement for specific nucleotide identity within the CRISPR motif. We therefore suspect that CRISPR motifs are more important during the acquisition of new spacers27. In summary, our results reveal the mechanism of self/non-self discrimination in this recently discovered immune system and will facilitate efforts to exploit CRISPR interference for biotechnological applications.
Methods summary
Bacterial strains and growth conditions
S. epidermidis wildtype (RP62a, ref. 20) and Δ_crispr_ (LAM104, ref. 3) and S. aureus (RN4220, ref. 29) strains were grown in brain-heart infusion (BHI) and tryptic soy broth media, respectively. When required, the medium was supplemented with antibiotics as follows: neomycin (15 μg/ml) for selection of S. epidermidis; chloramphenicol (10 μg/ml) for selection of pC194-based plasmids; and mupirocin (5 μg/ml) for selection of pG0400-based plasmids. E. coli DH5α cells were grown in LB medium, supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) when necessary.
Conjugation and transformation
Conjugation and transformation were performed as described previously3 with the following modification: transformations of S. epidermidis were recovered at 30 °C in 150 μl of BHI for 6 hs. Corroboration of the presence of the desired plasmid in transconjugants or transformants was achieved by extracting DNA of at least two colonies or agar of empty plates, performing PCR with suitable primers and sequencing the resulting PCR product.
Online Methods
DNA cloning
All plasmids used in this study were constructed by cloning CRISPR or nes sequences into the _Hin_dIII site of pC194 (ref. 30). Inserts were generated by PCR using primers and templates described in Supplementary Table 2. Supplementary Table 3 contains primer sequences. To introduce mutations, internal primers carrying the desired substitution were used with external primers, thus generating two PCR products that were then joined by nested PCR using the external primers. Amplified DNA was cloned into pCR2.1 vector (Invitrogen), the resulting plasmid purified and the insert sequenced. Inserts were cut from pCR2.1 with _Hin_dIII, purified and ligated into pC194. Ligation products were transformed into S. aureus OS2 cells31 and colonies containing the desired insert were identified by PCR using primers P86/P87.
pG0400 mutagenesis
Mutations were introduced by allelic exchange as previously described3. P15 and P18 primers were used with reverse and forward primers containing the desired mutation, respectively, to amplify 1 kb of nes sequence. PCR products were then joined by nested PCR using P15/P18 primers and recombined into pKOR1 (ref. 32). Recombination products were transformed into E. coli DH5α, and the resulting plasmids were purified, sequenced and transformed into S. aureus RN4220 (pG0400) (ref. 33) for allelic exchange. pG0400(G-2A), pG0400(G-2C) and pG0400(G-2T) mutants were generated using the knock-out constructs pLM397, pLM406 and pLM405, respectively. Inserts contained in these plasmids were generated by joining the PCR products obtained with the following primer pairs: pLM397, P15/P190 and P191/P18; pLM406, P15/P208 and P209/P18; pLM405, P15/P206 and P207/P18. See Supplementary Table 3 for primer sequences.
Northern blot analysis
RNA extraction of S. epidermidis and oligonucleotide labelling was performed as indicated in ref. 3. Northern blot analysis was carried out according to Pall et al. 34 using UV crosslinking and a hybridization temperature of 37°C.
Supplementary Material
1
2
3
4
Acknowledgments
We are indebted to Nancy Fang for cloning assistance and members of our laboratory for critical reading of the manuscript. We thank Vince Gerbasi and Joao Marques for experimental advice. L.A.M. is a Fellow of The Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by a grant from the National Institutes of Health, USA, to E.J.S.
Footnotes
Author contributions. L.A.M. designed experiments with input from E.J.S. L.A.M. conducted experiments. L.A.M. and E.J.S. analyzed data, interpreted experiments, and wrote the paper.
Author information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 6.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 7.Lillestøl RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 9.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 12.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 14.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 15.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang TH, et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang TH, et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–81. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 20.Gill SR, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Climo MW, Sharma VK, Archer GL. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 23.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 24.Lillestøl RK, et al. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 25.Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova II, Severinov K. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Lett. 2009;296:110–116. doi: 10.1111/j.1574-6968.2009.01626.x. [DOI] [PubMed] [Google Scholar]
- 26.Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 27.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 28.Sugimoto N, Nakano M, Nakano S. Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry. 2000;39:11270–11281. doi: 10.1021/bi000819p. [DOI] [PubMed] [Google Scholar]
- 29.Kreiswirth BN, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 30.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 32.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Morton TM, Johnston JL, Patterson J, Archer GL. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother. 1995;39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4