Management of High-Throughput DNA Sequencing Projects: Alpheus (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 10.
Published in final edited form as: J Comput Sci Syst Biol. 2008 Dec 26;1:132. doi: 10.4172/jcsb.1000013
Abstract
High-throughput DNA sequencing has enabled systems biology to begin to address areas in health, agricultural and basic biological research. Concomitant with the opportunities is an absolute necessity to manage significant volumes of high-dimensional and inter-related data and analysis. Alpheus is an analysis pipeline, database and visualization software for use with massively parallel DNA sequencing technologies that feature multi-gigabase throughput characterized by relatively short reads, such as Illumina-Solexa (sequencing-by-synthesis), Roche-454 (pyrosequencing) and Applied Biosystem’s SOLiD (sequencing-by-ligation). Alpheus enables alignment to reference sequence(s), detection of variants and enumeration of sequence abundance, including expression levels in transcriptome sequence. Alpheus is able to detect several types of variants, including non-synonymous and synonymous single nucleotide polymorphisms (SNPs), insertions/deletions (indels), premature stop codons, and splice isoforms. Variant detection is aided by the ability to filter variant calls based on consistency, expected allele frequency, sequence quality, coverage, and variant type in order to minimize false positives while maximizing the identification of true positives. Alpheus also enables comparisons of genes with variants between cases and controls or bulk segregant pools. Sequence-based differential expression comparisons can be developed, with data export to SAS JMP Genomics for statistical analysis.
Keywords: Alpheus, sequencing-by-synthesis, pyrosequencing, GMAP, GSNAP, resequencing, transcriptome sequencing
Introduction
High-throughput DNA sequencing using the Illumina GA series, the Roche 454 and the ABI SOLiD platforms have enabled a plethora of methods previously either prohibitively expensive or technologically impractical to be developed. Among these are whole genome shotgun sequencing [WGSS] and whole transcriptome shotgun sequencing [WTSS]. The output from these technologies currently ranges from 1–20 gigabases of raw sequence information per experiment, with a relatively high error rate compared to Sanger sequencing. The sheer quantity of output, the relative shortness of reads and the frequency of errors have created problematic areas for data management in terms of organization, analysis and information extraction. While several genome browsers currently exist (e.g., Ensembl (Flicek, et al., 2008), the UCSC Genome Browser (Mangan, et al., 2008), and the Broad Institute Integrative Genomics Viewer [http://www.broad.mit.edu/igv/], these tools presently include a significant structural overhead that make their application to so-called generation-2 sequencing efforts generally intractable. We have developed a streamlined application focusing on the acquisition, storage, analysis, organization, exploration and export of high-throughput sequencing data.
In this paper, we will describe the infrastructure and application necessary to perform these tasks, and focus on three of many applications in resequencing: genome variant detection, transcript expression analysis and protocol evaluation and analysis.
Genome Variant Detection
In DNA analysis, non-synonymous genetic variations (nsVariants; nsV) that cause an amino acid change are critical to understanding various diseases and traits (i.e., phenotypes) in all organisms. Single nucleotide polymorphisms (SNPs) represent the most frequent type of DNA variation, often having a neutral effect on phenotype (a synonymous SNP); nsVariants result in an amino acid change in the protein products of genes, and thus are believed to have the most significant impact on phenotype (Ramensky, et al., 2002). Aberrations resulting in point mutations, genome rearrangements, and insertions/deletions (indels) have been linked to tumorigenesis (Morozova and Marra, 2008).
SNPs have traditionally been found using Sanger sequencing methods at considerable cost. Microarray-based studies have also been used to detect known SNPs (Ropers, 2007), and the International HapMap Project was developed to determine patterns of human heritability to improve the success of genetic heritability studies (Manolio, et al., 2008). However, while array-based analysis has improved and has led to the discovery of almost 100 loci for nearly 40 common diseases and traits (Manolio, et al., 2008), the large number of unknown SNPs as well as poor hybridization of probes has caused some frustration on the part of researchers (Ropers, 2007). Sequencers such as Illumina Genome Analyzer (sequencing-by-synthesis; Illumina, Inc., San Di-ego, CA]), Roche-454 GS20 (pyrosequencing; Roche Applied Science, Inc., Indianapolis, IN) and Applied Biosystems SOLiD (sequencing-by-ligation; Applied Biosystems, Foster City, CA) have recently been used to perform massive sequencing of human and plant genomes and transcriptomes at low cost as compared to Sanger sequencing methods. Direct sequencing is likely to replace indirect approaches (SNP-HapMap), making it possible to screen entire genomes to examine introns, UTRs, and promoter regions as well as exons for likely pathogenic variation (Mardis, 2008). The generation-2 sequencing technologies enable comprehensive resequencing of common, complex disorders and feature relatively deep coverage. However, the nature of the technology also leads to relatively high sequence error rates that can cause false discovery of SNPs that can be expensive and time-consuming to validate.
Expression Analysis
Evolving from the WTSS methodologies, a variety of transcriptome sequencing methodologies are developing collectively referred to as RNA-seq (Lister, et al., 2008; Marioni, et al., 2008; Mortazavi, et al., 2008; Nagalakshmi, et al., 2008; Pan, et al., 2008; Wang, et al., 2009; Wilhelm, et al., 2008). Transcriptome sequencing has evolved from the low-coverage EST and cDNA projects which provided early gene discovery projects, and resulted in such databases as the NCBI dbEST and the Unigene databases (Wheeler, et al., 2008). With a variety of human, animal and plant genomes now completely sequenced, and vast assemblies of cDNAs and ESTs, WTSS projects have evolved into gene discovery, novel exon determination (Shin, et al., 2008; Wang, et al., 2008), whole transcriptome differential expression analysis (Mudge, et al., 2008), and quantitative tag-based methodologies, the latter with sensitivities in the 1–10 molecule per cell range (t Hoen, et al., 2008). Moreover, miRNA and other small transcribed non-coding RNAs can be captured using modified RNA isolation protocols (Chellappan and Jin, 2009; Hafner, et al., 2008; Lu, et al., 2007), and sequenced in parallel with transcriptomes. This has resulted in the discovery of a significant number of new miRNAs in animals (Burnside, et al., 2008) and plants (Lu, et al., 2008; Lu, et al., 2006; Lu, et al., 2007), and the technology has developed to the point that parallel sequencing of mRNAs and miRNAs on the same libraries have been used to develop the concept of an RNA degradome (Addo-Quaye, et al., 2008; Addo-Quaye, et al., 2008; German, et al., 2008).
Our focus has been the development of a hardware and software infrastructure sufficiently robust as to support both variant detection and RNA expression analysis. This hardware and software infrastructure serves multiple purposes. First, it provides a data management system for the data acquired in multiple internal and contract sequencing projects, as well as a gateway to statistical analysis tools. Second, it provides us with a querying mechanism for information derived from these projects necessary for publication of large-scale sequencing results (Mudge, et al., 2008; Sugarbaker, et al., 2008). Third, because publications frequently focus on the narrow subset of the information pertinent to that paper, while WTSS generally provides much more information than might necessarily be published, it provides a mechanism for both validation of results presented by external reviewers and users, and a resource that can be queried by the community for additional information that may not have been captured in or the focus of the publication. Finally, the primary national archive for data of this type is the NCBI Short Read Archive [SRA; (Wheeler, et al., 2008) and http://www.ncbi.nlm.nih.gov/Traces/sra], designed in large part to serve the needs of the 1000 Genomes Project (Siva, 2008) and http://www.1000genomes.org. While the logic of this design is indisputable, it remains that data deposited in the SRA will require a significant computational effort to realign to reference sequence data.
Methods
Sequencing
While Alpheus is capable of handling Sanger, Roche 454, Illumina GA2 and SOLiD data, much of our focus has been on Illumina GA2 output. The output sequence data from the GA2 is intermediate in size (36–106 bp) compared to the ABI instrument (26 bp) and Roche 454 (200–450bp).
Base-calling
Base-calling is generally performed using instrument-specific software.
Hardware: Clustering and Alignments
Alignments are performed on a cluster of 50 SunBlade X6220s with 2 dual core processors with 16 GB of RAM and 146 GB local disk per blade. Cluster management is provided by Platform Rocks (Platform Computing, Markham, Ontario, Canada); resource management is provided by Platform LSF/HPC. At present, we are experimenting with the deployment of the alignment resources at the New Mexico Computing Applications Center (NMCAC) on an SGI/Intel Altix cluster (14,336 cores).
The distributed WX2 database cluster (see below) consists of 4 Sun X4240 servers each with two quad core processors, 64 GB RAM, 16 × 146 GB disks, and additional Sun x4140 server with 2 quad core processors, 64 GB RAM and 8 × 146 GB disks. The X4240 servers host the relational database, and the X4140 acts as the application server.
Database Management Systems
Alpheus was designed with an underlying relational database management system. The current installation is on Sybase 12.5.4. We are, however, presently experimenting with an implementation on the Kognitio’s (Berkshire, UK & Chicago, IL, USA) WX2 analytical database.
Alignments
We have tested a variety of alignment tools for resequencing. These include GMAP (Wu and Watanabe, 2005), Blast (Altschul, et al., 1997), MegaBlast (Zhang, et al., 2000) and Eland [Illumina, Inc.]. Though the pipeline is insensitive to the source of the alignments, our workhorse alignment software remains GMAP, originally developed to by Tom Wu at Genentech, Inc., to align EST and cDNA data to reference genomes and transcriptomes, but which can be parameterized to handle short-read data. GMAP implements a collection of sophisticated algorithms producing gene models associated with cDNA sequences through comparison with a genomic reference. Following an initial mapping step, the cDNA sequence is aligned to its mapping target, establishing an approximate gene structure. This structure is refined through the use of a novel splice site inference algorithm, ultimately producing its gene model. It is notable that GMAP gene models accommodate the presence of microexons.
Genomic mapping in GMAP makes use of exact searches based upon 24 bp oligomers. Beginning from the ends of the query cDNA sequence, oligomers are mapped onto the genomic reference. The resulting maps are examined for reasonable levels of proximity. Expecting that the cDNA is “expanded” in the genome sequence, this process is continued from the ends inward along the cDNA in order to accumulate additional supporting evidence of proximal genomic coordinates. The mapping is complete if this process produces a small number of significant candidate regions on the genome; otherwise a different strategy is employed. In this case, oligomers are sampled from the interior of the cDNA instead. Segments of the genomic reference with sufficient density and colinearity of these 24-mers define the genomic map.
GMAP’s algorithm for approximate alignment of the cDNA to these mapped regions allows for local mismatches, cDNA insertions, and genomic insertions. Each position of the cDNA is associated with an 8-mer (e.g., an eight-nucleotide oligomer beginning at that position), and that 8-mer associated with several coordinates on the genomic reference. From this perspective, various alignments can be created by running along the cDNA, selecting monotonically increasing (or decreasing) genomic coordinates. GMAP produces its “approximate alignment” using dynamic programming, identifying the highest scoring path through this set of feasible alignments.
Refinement of this approximate alignment between the cDNA and the genomic segment, to localize introns, for example, is performed by a technique Wu & Watanabe refer to as “sandwich dynamic programming.” This method involves the computation of two global alignment matrices (Needleman-Wunsch) in the vicinity of significant gaps in the approximate alignment, and the selection of an optimal transition from one to the other. Scoring includes rewards for transitions occurring across standard splicing donor-acceptor pairs.
Our experience with GMAP has shown it to be robust and reliable, and well-suited for a role in high-volume processing pipelines such as Alpheus.
Wu has also made his recently-developed GSNAP software available to us (T. Wu, personal communication). GSNAP was specifically designed to handle short-read data (26–100 bp), including paired-end sequence data. Paired-end data includes both 5′- and 3′- ends of the clonally amplified fragments, and is particularly useful in resequencing of genomic samples (which include duplicate genes, introns, and intergenic regions). Paired ends are also useful in de novo sequencing applications (i.e., those sequences for which no reference genome exists).
GSNAP is a program for aligning short reads to a reference sequence, typically a genome, but possibly a set of transcripts. The program is designed to be both fast and flexible. GSNAP is designed to be fast through its use of multiple specialized algorithms, each one handling a different mapping type. The program is flexible in that it can handle arbitrarily long read lengths, which is becoming important as sequencing technologies are currently generating reads of 100 bp and longer. The program can also find and report alignments containing multiple mismatches, a single insertion or a single deletion, or a combination of these. The program is also able to map transcriptional reads that span an exon-exon boundary in a reference genome, including distant translocation events.
GSNAP also has flexibility in the types of input data it can process, including single-end, paired-end and circular-end reads. Paired-end reads are obtained when the sample DNA or RNA is fragmented into uniform lengths, typically 200—500~bp, and sequences obtained from both ends. Circular-end reads are obtained when long fragments of 10,000 bases or more are circularized with a 200–500 bp linker and then cut at the appropriate places to provide reads at the ends of the original long fragment.
The ability of GSNAP to handle paired-end reads, circular-end reads, and arbitrarily distant translocation events depends on having random access to the entire genome. Most other short-read mapping programs do not have this capability, because they are designed to have only sequential access. The prevailing architecture is to index a given dataset of short reads, and then to use that data-based index to scan the reference sequence. In contrast, GSNAP is one of only a few programs that depend on pre-indexing the reference genome or transcriptome.
The reference indices used by GSNAP are in the same format as those used by GMAP, and a reference index per genome or transcriptome serves both programs. A reference index is built by scanning the reference sequence for 12-mers, at a sampling interval of 3 bp. The positions for each sampled 12-mer are sorted and stored in a positions file. An offsets file contains pointers to the positions for each sampled 12-mer. Therefore, a reference index allows a program to find a uniform sampling of genomic or transcriptional positions for any given 12-mer. In addition to the offsets and positions files, the pre-indexing process also stores a compressed version of the reference sequence that essentially stores each 32 nucleotides in three 4-byte words. This compression allows storage of non-ACGT characters in the reference genome.
The GSNAP program then processes short reads to find alignments of the following types:
- Exact matches
- Single mismatches
- Multiple mismatches, possibly with a single insertion or deletion
- Exon-exon matches, local (if the splicing flag is selected)
- Exon-exon matches, distant (if the splicing flag is selected)
The combination of mismatches, insertions, and deletions is handled by assigning a penalty value to an insertion or deletion. For example, if the user specifies a maximum of four mismatches, and an indel penalty of 3, then the program will find alignments with a single indel and up to 1 mismatch.
The user may specify a minimum and maximum search level. The minimum search level specifies that the user wants all mapping results up to the chosen level. After the minimum level of search is performed, the program reports all results accumulated through that level. If there are no results found so far, the program then proceeds to subsequent levels through the maximum and reports the results at the first successful level.
The program has a specialized algorithm to solve each alignment type. The algorithm for finding exact matches is essentially an intersection operation. The program identifies a set of 12-mers that span the given short read, and takes the intersection of their corresponding reference positions. Because our reference sequence is sampled every third nucleotide, the ends of the short read may be represented by a 10-mer or 11-mer. These cases are handled by substituting all possible nucleotides in the overhanging positions and treating these cases as the union of the corresponding reference positions.
The algorithm for finding single mismatches is an intersection operation that allows one 12-mer to be left out. The program must then compare the short read against the reference sequence. This comparison operation makes use of the compressed version of the reference sequence that was built during the pre-indexing process.
The algorithm for finding multiple mismatches is essentially a union operation. The program looks up the reference positions for every 12-mer in the short read, and then uses a heap-based priority queue to merge these lists of positions into segments. Based on this information, the program can compute a minimum bound, or floor, for each segment, which is the number of mismatches possible for that mapping. If this floor is less than the maximum number of mismatches specified by the user, the program performs a comparison against the compressed reference sequence. The resulting segments are stored for later use in finding insertions and deletions, and for finding exon-exon alignments.
Insertions and deletions, or indels, are identified in two separate algorithms. The first algorithm finds indels that occur in the middle of the short read, between the first and last 12-mer. It finds pairs of segments within a user-specified distance of each other (default 30 bp for deletions and 9 bp for insertions). The algorithm then tests each pair against the compressed reference sequence to see if an alignment is possible within the allowed number of mismatches. The second algorithm finds indels that occur in the ends, within the first or last 12-mer. Each segment can be scored for a floor as if the first or last 12-mer were excluded, and segments that have a sufficiently small floor are compared against the compressed reference sequence for a possible alignment having the allowed number of mismatches or less.
To find exon-exon alignments, the program identifies segments that contain a likely donor or acceptor splice site. These are sites containing the canonical GT or AG and having adjacent nucleotides that score sufficiently high when compared against a probabilistic splice site model. For local exon-exon alignments, the program identifies pairs of segments, one with a donor and one with an acceptor, that are within a certain distance on the genome corresponding to a maximal intron length (default 100,000 bp). If a local exon-exon alignment cannot be found, the program attempts to find distant exon-exon alignments by pairing segments with donor and acceptor splice sites, regardless of their genomic distance.
Alpheus Pipeline
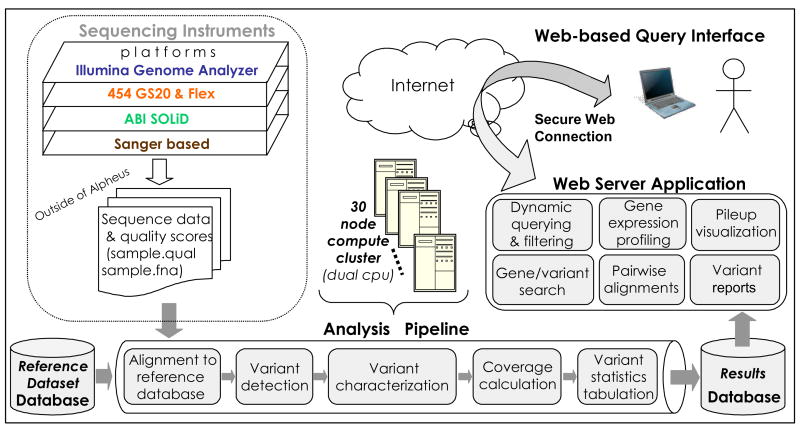
Alpheus is a multi-component system that includes processing and analytical pipelines, information storage and retrieval services, and web-based applications (see Figure 1). The pipeline begins with quality assessment of new read data, stores the data, maps the reads onto genome and transcriptome references and creates alignments, computes coverage statistics for these references, performs variant prediction, and stores the computed results for reuse. With respect to mapping and alignment, the system is significantly flexible, typically customized to the kind of read data being processed. This is necessary since by design Alpheus is intended to accommodate high volume data produced by different technologies including Illumina GA2, Roche 454, SOLiD, and Sanger. The system itself, developed in Java, makes extensive use of clear data abstraction, tiered architecture principles, adapters, etc., in order to support this fundamental flexibility and to facilitate its continuing improvement. The Alpheus implementation is based upon industrial grade technologies (Java, Sybase), and makes use of community standards (e.g., XML, GFF3), software (e.g., BioJava), and current best practices.
Figure 1.
Key components of Alpheus and data flow through the system.
Available mapping and alignment methods include GMAP and GSNAP, as described earlier, as well as MegaBLAST and Eland. Potential variants are identified using an Enumeration/Characterization module, which makes use of the computed alignments. This module reports synonymous and nonsynonymous SNPs, indels, premature stop codons, and candidate alternative splicing. The variant module can accommodate different read and library types. Read coverage is reported by gene, transcript, and chromosome. Other modules address sequencing-based gene expression and small/micro RNA studies.
Alpheus provides researchers who lack programming or bioinformatic sophistication the ability to explore and analyze tens of gigabases of sequence results and hundreds of samples through the Alpheus web tools. In addition to a project summary, users can view read data, coverage statistics, variant data, and perform sophisticated differential analysis. Data is accessible to clients by download, and as discussed later, by export for use in other analytical tools such as SAS JMP Genomics.
Alpheus Inputs: Read and Reference Data
There are a number of formats in current use for sequence data, both reads and reference. Sanger reads typically are presented in FASTA format. Roche 454 quality scores are supplied in a similar fashion: FASTA format with tagged to match their associated reads. Illumina reads are provided in FASTQ format, similar to FASTA but with read and quality data residing in the same file. Reference data includes not only genomic or transcript reference sequences, but also annotations (e.g., gene, CDS) on the references. These are most often available as GFF3 or Genbank feature table formats. Parsers for all of these are used in Alpheus.
Alignment to reference library and variant enumeration/characterization. GMAP and GSNAP provide essential mapping and alignment for short read data, and these results are used for identification of variants. Alignments to the references are made, typically require 95 percent identity and an identity count of 34 bp for a 36 bp read. Best-match alignments for the reads are stored in the database; all alignments equivalent to the best-match are stored which is important in the case of hits to shared exons in alternate splicing. All positions at which a read differs from the aligned reference sequence are enumerated. Contiguous indel events are treated as single polymorphisms. All occurrences of potential polymorphisms in reads with respect to a given position are unified as a “single polymorphism,” with associated statistics on frequency, alignment quality, base quality, and other attributes that may be used to assess the likelihood that the polymorphism is a true variant. Candidate variants are further characterized by type (SNP, indel, alternate splicing or stop codon) and as synonymous variant (sV) or non-synonymous variant (nsV).
Data Model
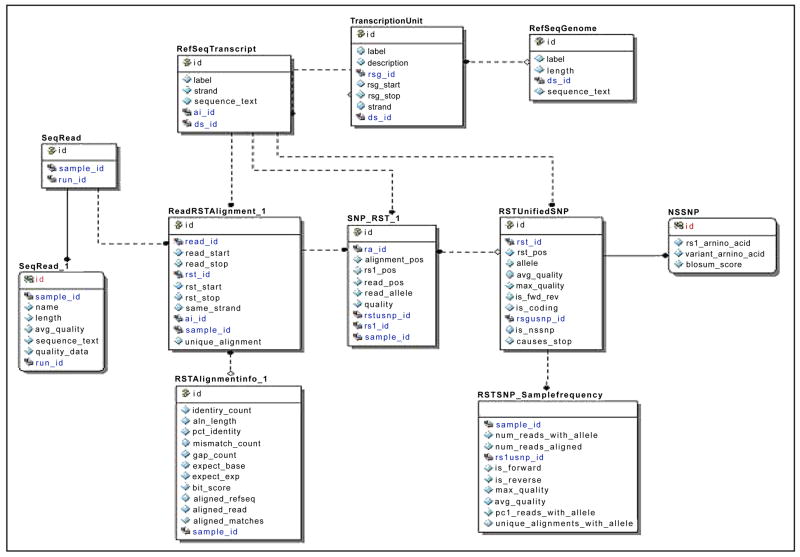
Data is stored in sample-specific tables, which are created dynamically by the Alpheus pipeline as each sample is processed. Sample-specific tables are differentiated from each other through the use of a sample-specific suffix that identifies each table as belonging to the sample with that primary key identifier. Transcriptome reference sequences are stored in the RefSeqTranscript table; genome reference data (e.g. chromosomes) is stored in the RefSeqGenome table. Gene data, including the genomic position of the gene, is stored in the Transcription Unit table. Figure 2 shows a sub-section of the Alpheus database that stores transcriptome alignments and substitution sequence variants. Data stored in sample-specific tables include a record of all sequences and their accompanying quality scores, (SeqRead_1), read alignments to the transcriptomic reference (RSTAlignmentInfo_1, ReadRSTAlignment_1), and all substitution variants detected in each read (SNP_RST_1). Sequence variants are initially recorded for each read, then “unified” into the RSTUnifiedSNP table where each unique combination of reference position and allele is recorded once. If it can be determined that a substitution results in an amino acid change, a row is stored for the SNP in the NSSNP table, which records the reference and variant amino acid, as well BLOSUM62 (Henikoff and Henikoff, 1993) matrix score for the amino acid shift. Sample-specific statistics such as the number of reads showing the variant allele, the total number of reads covering the variant position and quality metrics for reads showing the allele are stored in the RSTSNP_SampleFrequency table. Transcriptomic indels are stored in tables parallel to the substitution tables. All positions are recorded with start and stop coordinates rather than a single position. Similarly, genomic alignment, substitution, insertion/deletion and frequency tables are stored in another parallel set of tables, with the difference that reference coordinates are recorded on a genomic rather than a transcriptomic reference entity.
Figure 2.
Partial schema of Alpheus. Transcriptome alignments and substitution sequence variants are stored in this core schema, as described in detail in Materials and Methods.
Alpheus Queries
The Alpheus system features a web-interface in which researchers select between two principal types of queries by selecting boxes or items from pull-down menus. First, a collection of sequences from an individual sample or set of samples can be queried for particular events (principally, nucleotide variants or loci expressed at a certain level). Second, collections of sequences from case-control cohorts can be queried for particular events that differ in frequency or magnitude between groups. The query interface provides a considerable degree of flexibility in inclusion or exclusion of particular samples in group comparisons and in cutoffs for magnitude of change, event type, coverage, quality score or event frequency. This allows, for example, a query to be performed with or without the inclusion of an outlier sample. Queries can be performed in sequences aligned to more than one reference (for example, against alignments to RefSeq transcript or a genome reference, which can return quite different datasets [Wang et al., 2008]). Typically, an investigator will perform a query repetitively, modifying filters, cutoffs and samples based on results returned. A researcher is able provide a set of candidate genes or known features when an instance of Alpheus is developed for a particular project. Such a reference gene or event set can provide the investigator with guidance regarding optimal design of a filter set (by, for example, optimizing a query so that it will return a gene set that the investigator knows to be altered in that experiment). Alpheus offers extensive link-outs to accessory data that can greatly assist in annotating results of queries or assessing putative biological significance on a gene-by-gene basis. These include gene- or variant-specific link-outs to Entrez gene, Genbank, OMIM and dbSNP. Alpheus also enables drill down from gene lists to individual gene-associated events to sequences and alignments associated with specific variants. With these tools, an investigator can undertake exploratory, iterative analyses of large or complex datasets that provide an understanding of data complexity, stratification, limitations and confounding effects. Having undertaken such exploratory analyses, an investigator can perform a final query with optimized cutoffs and filters, returning a final data set for downstream analyses.
Statistics and Analysis: SAS JMP Genomics
After querying and adjustment of filters to provide appropriate data screening for sequencing errors, coverage, and differentially expression, data returned by queries can be exported to Excel or SAS JMP Genomics SAS, Inc., Cary, NC) format. Because of the quantitative and reproducible nature of data derived by direct sequencing (Marioni, et al., 2008), we have collaborated with Russ Wolfinger of SAS to develop a suite of statistical analyses comparable to those used for microarrays (Mudge, et al., 2008). Utilizing experimental and instrument metadata associated with the sequencing run in Alpheus and stored in a locally-developed laboratory information management system (LIMS), general assessment approaches such as distribution analyses, correlation analysis at the sample and gene level, and principal components analysis (PCA) can be applied to summarized sequence counts and proportions of variant and reference alleles. JMP Genomics quality assessment tools are particularly useful for partitioning variance due to experimental and technical factors, and for guiding the decision for which factors should be included in downstream modeling. Additionally, JMP Genomics tools can be applied in identifying potential outlier samples which should be excluded prior to pursuing more detailed analyses.
Once outliers have been detected and removed, JMP Genomics provides a variety of modeling methods, including analysis of variation (ANOVA) and association testing approaches, that can be used to detect differences between groups of sequenced samples. Examples include comparing transcript counts or variant proportions between cases and controls, or examining changes in treatment effects for different sample groups over time. More complex analyses can include combining different data types derived from sequence data to relate variant sequences to gene expression changes, for example to identify cis-acting eQTLs (Kingsmore et al., submitted).
The wealth of pattern discovery tools available to visualize patterns in high-dimensional genomics data is also a particular strength of JMP Genomics. In addition to hierarchical clustering, other methods such as K-Means clustering, principal components analysis, and distance matrix creation offer visual representations of patterns which connect gene sets. After identifying interesting gene or variant subsets through pattern discovery and modeling tools in JMP Genomics, predictive modeling tools can also be used to discover well-supported subsets of these which best predict classes of samples. Extensive cross-validation options are available to ensure the selection of high-quality predictor profiles. The application of this software is demonstrated in our examples below.
Results and Examples
As examples of the efficacy of Alpheus in practical large-scale data analysis, we enumerate three examples of work made possible using the software system. Specifically, 1.) A variant discovery project in which mesothelioma tissues were explored for SNP discovery; 2. A schizophrenia project which utilized the gene expression capabilities; and 3.) A protocol evaluation project, in which the effects of modifying protocols were evaluated.
Variant Discovery: Mesothelioma
One of Alpheus primary functions is for discovery of single nucleotide polymorphisms (SNPs). Alpheus was successfully used to determine candidate causal single nucleotide variants (SNVs) in malignant pleural mesothelioma (MPM) tissues, an asbestos-related, rapidly fatal cancer (Sugarbaker, et al., 2008). It is known that cancers arise due to multiple mutations; however the causative mutations remain largely unknown. To determine possible mechanisms that lead to the development of MPM transcriptome sequencing was performed to determine SNVs. Whole-transcriptome 454 pyrosequencing was performed on cDNA from tumors of four MPM patients as well as an adenocarcinoma lung tumor and normal lung tissue from an MPM patient. For the six samples > 260 Mb of the transcriptome sequence were obtained by shotgun 454 pyrosequencing with the 454 Life Sciences GS20. More than 98% of the 15 million, approximately 105 bp sequence reads aligned to human mRNA and DNA databases. Transcript sequences mapped to 19,306 human reference mRNAs present in the RefSeq mRNAs database (Pruitt, et al., 2005). In each sample, approximately 15,000 known RefSeq genes were detected by one or more reads, with approximately 10,000 genes with at least 20 or more reads per gene, corresponding to 1X coverage. To facilitate analysis and visualization, the data was imported into Alpheus. Filter parameters include patient sample, gene name, read coverage, variant frequency, variant type, variant location and links to NCBI sequence and gene function databases.
Due to the high number of false positive SNVs characteristic of early high-throughput shotgun sequencing, the data was filtered to determine true mutations. Inclusion criteria were that the variant must be present in at least four reads covering the base position, present in at least 30% of the total number of reads covering the variant, have a GS20 quality score = 20, be observed in reads from both orientations and be located within a read that is >90% identical along the entire length of the target RefSeq mRNA sequence. Under these constraints there was 96% sensitivity in identification of 2,465 annotated SNPs found in dbSNP (www.ncbi.nlm.nih.gov/projects/SNP). The authors found 100% agreement of 94 SNVs that were independently confirmed using conventional Sanger sequencing. The six tissues sequenced contained 659 to 1,155 known RefSeq genes with at least one coding SNV (cSNV). Within the MPM tissues 153–220 genes contained previously uncharacterized cSNV – which represent candidate causal mutations. The four tumors contained a total of 619 nonredundant, previously uncharacterized cSNVs and 2,369 known SNVs. nsV not present in dbSNP were further explored due to the possible functional relevance. Twelve nsV were common to all five tumors but absent in the normal lung, four were common only to MPM tissues; sequencing of the genomic DNA determined that they were germ-line variants and not mutations. 54 of 69 nsV tested within the mesothelioma tissue were present in the genomic DNA, indicating polymorphisms and not mutations. The remaining 15 nsV were tumor-specific relating to somatic mutations (n = 7), RNA editing (n = 1), loss of heterozygosity due to chromosomal deletions (n = 3), and epigenetic silencing (n = 3). The frequency of the seven nsV somatic mutations were evaluated in 49 additional MPM tumors by genotyping cDNA and gDNA in the specific exons affected by the individual mutations. Three of the mutations were present in 4–6% of a larger cohort of MPM tissues; COL5A2 mutation in 3 of 53 (c2773t, NM_000393.3); UQCRC1 mutation in 3 of 53 (g851a, NM_003365.2) and MXRA5 mutation in 2 of 53 (c7862a, NM_015419.1).
Differential Gene Expression: Schizophrenia
The schizophrenia data described below is based on 20 transcriptome samples, >475 million reads, >16 gigabases of high-quality base-called data, and comprises more than 825 gigabytes of data inserted in to the database.
Schizophrenia genome convergence analysis using Alpheus
One of Alpheus primary functions is to assist in the analysis of large genome-wide association studies with the use of JMP Genomics statistical software. Mudge, et al. (Mudge, et al., 2008) used mRNA transcripts isolated from 20 postmortem cerebellums to find candidate genes possibly involved in schizophrenia (SCZ). While several studies have identified candidate SCZ risk genes, few have been replicated or translated into causal alleles, diagnostics or therapeutics. Deep sequencing was performed on mRNA transcripts of 14 SCZ patients and 6 matched controls. 12.5–38.7 million high quality sequences of 32–36 bp were generated per sample. The sequences were aligned to the human genome and RefSeq transcript databases using the GMAP algorithm allowing for < 2 mismatches. Interestingly 43.5 ± 6.7% of sequences aligned to a transcript while 69.4 ± 9.6% to the genome. There was little difference in the total number of transcripts (33,200 ± 1,000) between samples, corresponding to 85 ± 3% of RefSeq transcript entries. 12.5 million sequences per sample were sufficient to reach a plateau in the number of transcripts detected, with deeper sequencing resulting in a linear increase in average depth of coverage. Alpheus was used to normalize the reads per million. Results were imported into JMP Genomics and read frequencies were Log10 transformed which improved overlaid kernel density estimates, univariate distribution and Mahalanobis distances. Using unsupervised PCA (with Pearson product-momentum correlation) SCZ patients were easily distinguished between controls. Principal components of variance (with Pearson correlations) were used to survey diagnosis against other sources of variability such as patient, sample and experimental parameters. The diagnosis attributed to 45.3% of variance with cause of death (9.6%), instrument (12.5%), year sequenced (19%) and post-mortem interval (0.1%) as the other major components of variance with a low unknown residual variance (13.5%). Analysis-of-variance was performed using the diagnosis as the discriminatory effect and the major non-diagnosis components of variance as fixed effects. Following FDR-correction 88 genes exhibited differences in expression in genome-aligned read frequencies and 152 genes differed significantly in transcript-aligned reads, 25 genes were common to both genome- and transcript-aligned sequences. GO annotation determined that 23 genes significantly affected were related to Golgi function or presynaptic vesicular transport and GABAergic neurotransmission which may define a unifying molecular hypothesis for dysfunction in cerebellar cortex in SCZ.
Protocol Evaluation using Transcriptome Sequencing
Whole Blood RNA Isolation
Human whole blood samples were collected in PAX gene tubes (Qiagen Inc., Valencia, CA) from one of the co-authors at three random time points over a two-month period. The samples were frozen at −80°C for at least 24h. RNA isolation was performed per manufacture’s instructions.
Sequencing-by-synthesis
Transcriptomic libraries were prepared using Illumina’s standard protocols as previously described (Mudge, et al., 2008) except with either poly-A selection, ribominus [Ribo (−)] selection, or zinc fragmentation after poly-A tail selection. Briefly, following RNA quality assessment using Bioanalyzer 2100 (Agilent Inc., Santa Clara, CA), poly A+ RNA was isolated from 1 ì g total RNA by two rounds of oligo-dT selection (Invitrogen Inc., Santa Clara, CA.) or rRNA depletion using a RiboMinus kit (Invitrogen) per manufacturer’s instructions. For one sample, zinc fragmentation was performed for 10m at 70°C using Ambion RNA Fragmentation Reagents (Applied Biosystems Inc., Austin TX) and ethanol/glycogen precipitated. All mRNA samples were then annealed to high concentration of random hexamers and reverse transcribed. Following second strand synthesis, end repair and A-tailing, adapters complementary to sequencing primers were ligated to cDNA fragment ends. Libraries were size fractionated on agarose gels, 200 bp fragments excised and amplified by 15 cycles of polymerase chain reaction. Following quality assessment, single-stranded cDNA fragments were randomly annealed to the surface of a flow cell. Annealed fragments were extended with DNA polymerase and unlabeled dNTPs in a solid phase “bridge amplification.” The resultant double strand fragments were denatured and bridge amplification repeated for 35 cycles, generating approximately 30–40 million clusters. Subsequently, 36 cycles of sequencing-by-synthesis chemistry were performed in Illumina Genome Analyzer II instruments (Illumina Inc.) with dNTPs featuring cleavable dyes and reversible terminators. Following base extensions, four images are taken upon laser excitation. Incorporation of the next base occurred after removal of the blocked 3′-terminus and fluorescent tag of the previously incorporated nucleotide. High quality sequence reads, as defined by default filtering parameters used in the Illumina GA Pipeline GERALD stage, were retained.
Read Alignment-based Gene Expression Profiling
High quality reads were aligned to the human genome, Build 36.2, RefSeq Transcript database, Release 22 (Pruitt, et al., 2005), Unigene Hs, build 215 using the algorithm GMAP (Wu and Watanabe, 2005) and Alpheus. Adjustments for SBS reads were oligomer overlap interval = 3 nt, identities = 34/36. A read was denoted aligned to a locus if its sequence alignment to the genomic reference sequence (NCBI build 36.2) fell within the boundaries of the locus coordinates on the chromosome. Locus boundaries on the genome were defined by NCBI annotations, as reported through the Nucleotide database. Reads with a single best alignment or with equally good alignments to alternative transcripts of the same locus were considered uniquely aligned. Aligned read frequencies (per million reads) were calculated for each sample and locus expression and locus using Alpheus.
Statistical Analysis
Read frequencies were log2 transformed prior to evaluation of inter-sample differences. Overlaid kernel density estimates, correlation coefficients of pairwise sample comparisons and unsupervised PCA (by Pearson product-momentum) of read frequencies were performed with JMP Genomics. Heat maps were compared by selecting genes with a least a two-fold difference of expression within one of the comparison groups and then hierarchal clustering of the data set was performed.
Results
Library Prep Techniques Leads to Highly Variably Transcriptomic Expression Profiles
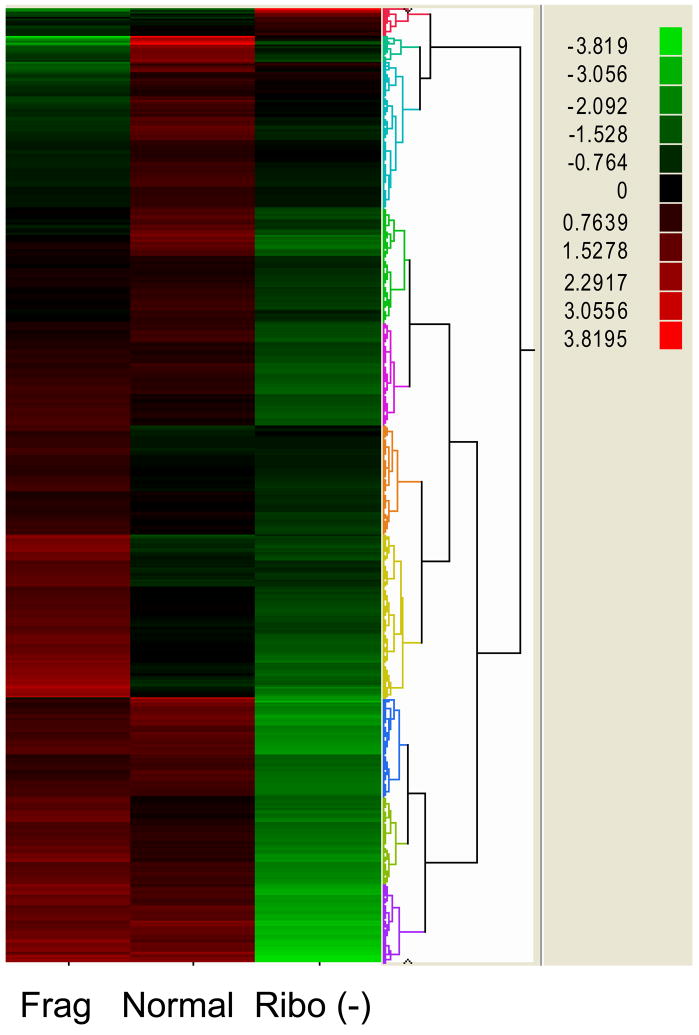
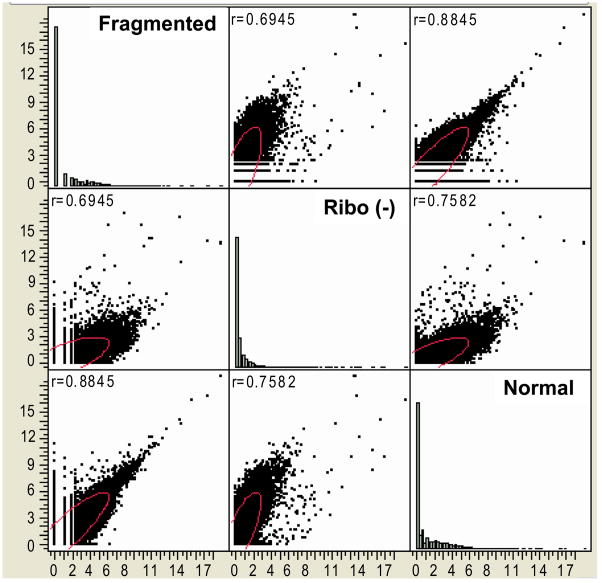
To compare the differences in transcriptomic expression variance in the Ilumina library prep protocols we compared expression data of the normal Illumina library preparation protocol, a zinc mRNA fragmentation protocol after normal library prep, or after performing ribosomal exclusion. Whole blood was collected in PAX gene tubes from one of the coauthors at three separate time points over a three-month time period. It is believed that zinc fragmentation would improve 3′-bias, while ribosomal exclusion would allow for screening of non-gene related transcribed RNA. After SBS and pipelining into Alpheus, uniquely aligned expression data was imported into JMP Genomics for statistical analysis. After import, the data was log2 transformed. While the all of the samples were from the same individual we found vastly different expression results from the three library preparation techniques as demonstrated by parallel plots (fig. 3) and the unsupervised PCA (fig. 4). To determine changes in expression, we subtracted log transformed values to find genes that had at least a two-fold difference in expression. Under these constraints, 13,791 genes out of 33,887 total genes were at least two-fold different in expression. As visualized by the hierarchal cluster analysis, overall expression was much lower in the ribo(−) selection as compared to the normal technique and the fragmented technique (fig. 5). The pairwise comparisons of fragmented versus non-fragmented protocols (r2= 0.8845) were fairly regular as compared to fragmented versus ribo(−) (r2 = 0.6945) and non-fragmented versus ribo(−) (r2 = 0.7582) (fig. 6). We were not able to determine any major advantage using the fragmented technique over the non-fragmented technique in terms of 3′ bias or overall expression data (data not shown). However, the data did highlight expression differences of the two protocols (fig. 5). Both the non-fragmented and fragmented techniques were preferred over ribo(−) selection as it appeared robust changes in transcriptomic expression was muted using the ribo(−) technique as well as a very poor pairwise correlation results as previously mentioned.
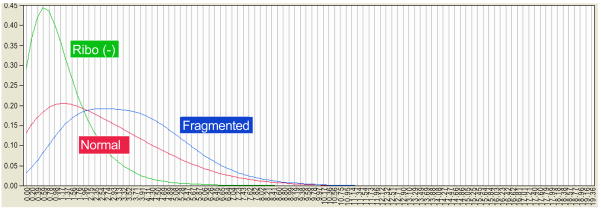
Figure 3.
Overlaid kernel density estimates of gene expression by sequence read frequencies. Gene expression of whole blood mRNA for normal Illumina library prep (red), fragmented after poly-A selection, and with Ribo(−) exclusion. The X-axis show log2 transformed gene expression values, while the Y-axis shows kernel densities. Without log transformation, samples showed greater variability in kernel densities and sequence read frequencies showed near exponential decay.
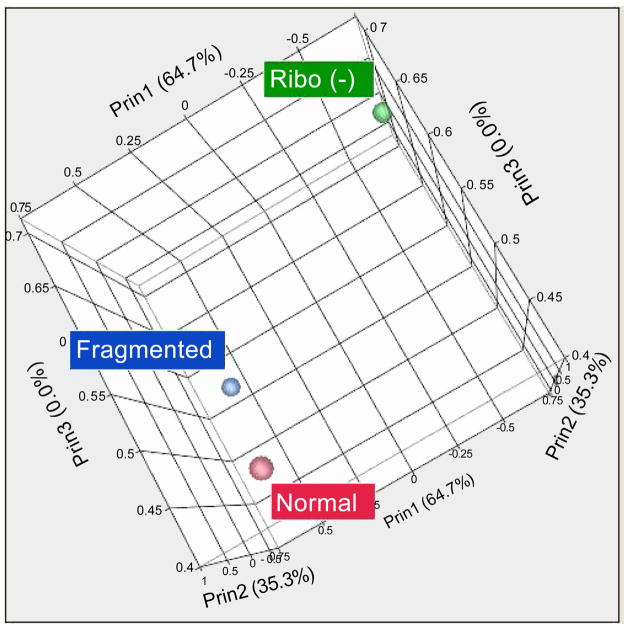
Figure 4.
Unsupervised PCA of expression data. Three dimensional plot of unsupervised PCA by Pearson product-moment correlation of log sequence expression. Normal (Red) and fragmented (Blue) libraries are more similar than the Ribo(−) prepped libraries (blue).
Figure 5.
Hierarchal clustering of log transformed expression data. 13,791 genes out of 33,887 total genes were at least two-fold different. Most genes had much higher expression in both normal and fragmented library preps than Ribo(−). Normal and fragmented prep had 6,577 genes that were at least two fold different. 10,404 genes were different between normal and ribo(−), while 11,104 genes were two fold different between fragmented and ribo(−) library preps
Figure 6.
Pairwise sample correlations of Log2 transformed read frequencies, showing pairwise correlation coefficients. Pairwise comparisons suggest fairly linear distribution of gene expression of the normal library technique versus the fragmented technique, while there is much great frequency distribution between ribo (−) and the normal and fragmented techniques.
Discussion
Alpheus is a modular, robust data management and exploratory tool. It has been utilized for multiple high-throughput genome and transcriptome sequencing projects with differing objectives (total data presently stored in the production databases is approximately seven terabytes). Because the environment of second-generation sequencing is evolving extremely rapidly, we have scoped this project not to encompass all aspects of annotation, but rather to leverage existing, supported resources. For example, while performing essential alignment services, we rely on RefSeq, OMIM, pathway databases and other functional annotation resources for primary information. Where necessary and appropriate (e.g., in differential gene expression and exon discovery projects), we extract information from our database for further analysis. For example, in quests for new or un-annotated genes and exons in specific tissues, treated or pathologic tissues, we query the database for alignment information represented in the genome sequence, but not present in the transcriptome-based unigene assemblies. Occasionally, these are represented as gene models; however, frequently, even in heavily annotated reference sequences (e.g., human and the model plant, Arabidopsis thaliana), reads identify un-identified areas. An example of this is demonstrated above for mesothelioma; however, the value of this query type has been shown in a variety of animal and plant projects (manuscripts in preparation).
The design of the resource is amenable to expansion to accommodate new sequencing technologies. While genome and transcriptome resequencing has been the focus of many projects, particularly variant discovery and differential gene expression, a module to accommodate miRNA sequencing is being added with relative ease. Developments in sequencing technologies have thus far driven our development efforts. In recent months, additions to the repertoire of production-level technologies are helping us prioritize the evolution of Alpheus. These technologies include epigenetic mapping (e.g., methylome analysis, (Butcher and Beck, 2008; Lister, et al., 2008; Pomraning, et al., 2008) and combined RNA-seq techniques, such as those which define the RNA degradome (Addo-Quaye, et al., 2008) and quantification of alternative splice isoform and alternative polyadenylation (Wang et al., 2008). As per-run data outputs increase, methods for bar-coding or indexing of individual libraries so that samples can be combined in sequencing runs and later deconvoluted become more important, and difficult to decipher on a production scale (Craig, et al., 2008; Goossens, et al., 2008; Hillier, et al., 2008). Analysis of structural variation in shotgun sequences from sets of individual eukaryotic genomes (such as the 1000 genomes project) is also driving the evolution of Alpheus. While much of our work is in higher eukaryotes with genomes ranging from 1–3 gigabases, bacterial and lower eukaryotes have genomes that range from 3–50 megabases, and survey level sequencing (5–10x coverage) can be accommodated in a mixed samples. Finally, metagenomes will present a particular challenge. Metagenomes are mixtures of microorganisms from environmental, plant and animal samples, most of which are unculturable and unidentified, which represent generally stable populations. The aggregate representation of the DNA sequence of the population is thus referred to as the metagenome (review, (Medini, et al., 2008). At present, the sequencing issues are approachable on a production scale, but the informatics issues remain subtle (review, (Kunin, et al., 2008). Automating and organizing the subtlety of metagenomic analysis will be challenging, to say the least.
As discussed in the introduction, we consider the Alpheus infrastructure as an adjunct to publication of the results of high-throughput sequencing efforts. The NCBI Trace Archive evolved from a need to present the raw data from genome and transcriptome sequencing projects to allow the validation of experimental results, and to make the data available to a larger community than the sequencing teams involved in the project. These resources continue to be mined using new algorithmic methods, and frequently re-assembled by other groups. The NCBI Short Read Archive is designed to serve a similar purpose. However, the computational problem of utilizing this data, either resequencing alignments or particularly de novo sequencing efforts, remains challenges. A single deep transcriptome alignment can take up to 1600 CPU hours to completely analyze. Alpheus can be used to present the underlying assumptions used in complex experiments, and particularly to make the entire analyzed data set available to the community in a fashion that it not only can be examined, but can be further explored by others in the research community. As implemented, Alpheus can present virtually any level of access, from the complete sequencing data set and underlying query tools to the narrower data set necessary to confirm published experiments. The mesothelioma and schizophrenia results are examples of this, with varying levels of accessibility to data at the web sites.
The scale of contemporary high-throughput sequencing has migrated DNA data acquisition from a simple tool to an essential platform for systems biology. We have developed Alpheus as a data management and exploration tool to complement experimentation and provide leads in a plethora of human, agricultural and basic biological research. As discussed, we intend to continue to develop the system to extend into new technologies and research arenas.
Acknowledgments
Alpheus is trademarked to the National Center for Genome Resources. We appreciate the collaboration and trust of Dr. Thomas Wu of Genentech, Inc., for allowing us access to unreleased versions of GMAP, as well as for pre-publication access to source code for GSNAP. We are grateful for the collaboration of Drs. Russell Wolfinger, Wendy Czika, Tzu-Ming Chu and Shannon Conners of SAS JMP Genomics for their active collaboration with NCGR in developing robust statistical methods for sequence-based analysis using the SAS framework. The New Mexico Computing Applications Center (NMCAC) has provided access to and support for the SGI/Intel supercomputer, Encanto, on which future development work is being carried out We particularly acknowledge Drs. Thomas Bowles and William Feiereisen of the Governors Office of the State of New Mexico in this respect. Dr. Tom Brettin of Los Alamos National Laboratories has collaborated on the development of NMCAC projects. Dr. Damian Gessler has provided input to the development of this paper. This work was supported by and established cross-disciplinary, collaborative partnership of universities and national laboratories. The work on Characterization of candidate SZ nsSNPs and CD indels (nsV) was performed by research staff at the National Center for Genome Resources (NCGR), Santa Fe, NM, UNM and MIND Institute (both Albuquerque, NM), and, notably, by three faculty and five minority undergraduate students at Northern New Mexico College (NNMC), Espanola, NM, and two volunteers (a family physician in Los Alamos, NM, and a physician-software engineer in Santa Fe, NM). Alpheus development was funded internally at NCGR and by the International Mesopthelioma program (led by Dr. David Sugarbaker and Dr. Raphael Bueno at the Brigham and Women’s Hospital, Boston, MA), by grant support from NCRR (5P20RR016480-08, led by Dr. Jeff Arterburn at New Mexico State University), from NIAID (under Contract number HHSN266200400064C, led by Dr. Jeff Gulcher of deCODE Genetics, Reykjavik, Iceland), from the Nancy Davis Foundation for Multiple Sclerosis, Los Angeles, CA, and by Macrogen Inc, Seoul, Korea. A deo lumen, ab amicis auxilium.
Abbreviations
ANOVA
Analysis of variation
cSNV
Coding SNV
indels
Insertions/deletions
MPM
Malignant pleural mesothelioma
nsV
Non-synonymous genetic variations
SRA
NCBI Short Read Archive
PCA
Principal components analysis
ribo(−)
Ribominus
SNPs
Single nucleotide polymorphisms
SNVs
Single nucleotide variants
sV
Synonymous variant
WGSS
Whole genome shotgun sequencing
WTSS
Whole transcriptome shotgun sequencing
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Addo QC, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addo QC, Miller W, Axtell MJ. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics (Oxford, England) 2008 doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, et al. Deep sequencing of chicken microRNAs. BMC genomics. 2008;9:185. doi: 10.1186/1471-2164-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher LM, Beck S. Future impact of integrated high-throughput methylome analyses on human health and disease. Journal of genetics and genomics = Yi chuan xue bao. 2008;35:391–401. doi: 10.1016/S1673-8527(08)60057-0. [DOI] [PubMed] [Google Scholar]
- 6.Chellappan P, Jin H. Discovery of Plant MicroRNAs and Short-Interfering RNAs by Deep Parallel Sequencing. Methods in molecular biology (Clifton, NJ) 2009;495:1–12. doi: 10.1007/978-1-59745-477-3_11. [DOI] [PubMed] [Google Scholar]
- 7.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nature methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, et al. Ensembl 2008. Nucleic acids research. 2008;36:D707–714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature biotechnology. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 10.Goossens D, Moens LN, Nelis E, Lenaerts AS, Glassee W, et al. Simultaneous mutation and copy number variation (CNV) detection by multiplex PCR-based GS-FLX sequencing. Human mutation. 2008 doi: 10.1002/humu.20873. [DOI] [PubMed] [Google Scholar]
- 11.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods (San Diego Calif) 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- 13.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, et al. Whole-genome sequencing and variant discovery in C. elegans. Nature methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 14.Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, O’Malley RC, Tonti FJ, Gregory BD, Berry CC, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA poly-merase-2 mutant. Genome research. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods (San Diego, Calif) 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Mangan ME, Williams JM, Lathe SM, Karolchik D, Lathe WC., 3rd UCSC genome browser: deep support for molecular biomedical research. Biotechnology annual review. 2008;14:63–108. doi: 10.1016/S1387-2656(08)00003-3. [DOI] [PubMed] [Google Scholar]
- 20.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. The Journal of clinical investigation. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 24.Morozova O, Marra MA. From cytogenetics to next-generation sequencing technologies: advances in the detection of genome rearrangements in tumors. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2008;86:81–91. doi: 10.1139/O08-003. [DOI] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS ONE. 2008;3:e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science (New York, N.Y) 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 29.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods (San Diego, Calif) 2008 doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research. 2005;33:D501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic acids research. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ropers HH. New perspectives for the elucidation of genetic disorders. American journal of human genetics. 2007;81:199–207. doi: 10.1086/520679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin H, Hirst M, Bainbridge MN, Magrini V, Mardis E, et al. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC biology. 2008;6:30. doi: 10.1186/1741-7007-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siva N. 1000 Genomes project. Nature biotechnology. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 35.Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic acids research. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 41.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics (Oxford, England) 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]