Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila (original) (raw)
Abstract
In Drosophila, three types of endogenous small RNAs—microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and endogenous small-interfering RNAs (endo-siRNAs or esiRNAs)—function as triggers in RNA silencing. Although piRNAs are produced independently of Dicer, miRNA and esiRNA biogenesis pathways require Dicer1 and Dicer2, respectively. Recent studies have shown that among the four isoforms of Loquacious (Loqs), Loqs-PB and Loqs-PD are involved in miRNA and esiRNA processing pathways, respectively. However, how these Loqs isoforms function in their respective small RNA biogenesis pathways remains elusive. Here, we show that Loqs-PD associates specifically with Dicer2 through its C-terminal domain. The Dicer2–Loqs-PD complex contains R2D2, another known Dicer2 partner, and excises both exogenous siRNAs and esiRNAs from their corresponding precursors in vitro. However, Loqs-PD, but not R2D2, enhanced Dicer2 activity. The Dicer2–Loqs-PD complex processes esiRNA precursor hairpins with long stems, which results in the production of AGO2-associated small RNAs. Interestingly, however, small RNAs derived from terminal hairpins of esiRNA precursors are loaded onto AGO1; thus, they are classified as a new subset of miRNAs. These results suggest that the precursor RNA structure determines the biogenesis mechanism of esiRNAs and miRNAs, thereby implicating hairpin structures with long stems as intermediates in the evolution of Drosophila miRNA.
Keywords: esiRNA, Dicer2, Loquacious, miRNA, Drosophila
INTRODUCTION
In RNA silencing, small RNAs of 20–30 nucleotides (nt) guide Argonaute proteins, the main catalytic factors involved in RNA silencing, to the target RNAs to be silenced (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Siomi 2009). Endogenous small RNAs in animals can be divided into three subgroups: microRNAs (miRNAs) (21–23 nt), PIWI-interacting RNAs (piRNAs) (24–30 nt), and endogenous small interfering RNAs (endo-siRNAs or esiRNAs) (∼21 nt) (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Kuramochi-Miyagawa 2009; Siomi and Siomi 2009). miRNAs are encoded by their own genes and are expressed ubiquitously (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Siomi 2009). piRNAs are mainly derived from intergenic repetitive regions of the genome, including transposons and even non-transposon loci, which are generically called piRNA clusters (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Kuramochi-Miyagawa 2009; Siomi and Siomi 2009). Unlike miRNAs, piRNAs are primarily expressed in germline cells. esiRNAs also mainly originate from transposons and repetitive genomic elements but are not germline-specific. Thus, piRNAs and esiRNAs are distinct from each other. They are also loaded onto different Argonaute partners: piRNAs are associated with germline-specific Argonaute members, the PIWI proteins, whereas esiRNAs interact with ubiquitously expressed Argonaute (AGO) proteins. miRNAs are also loaded onto AGO proteins (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Kuramochi-Miyagawa 2009; Siomi and Siomi 2009).
The processing pathway of miRNAs has been extensively studied (Bartel 2009; Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Siomi 2009). In Drosophila, the primary transcripts of miRNA genes, pri-miRNAs, are first processed in the nucleus into miRNA precursors, pre-miRNAs, by a complex of Drosha, an RNase III enzyme, and Pasha, a double-stranded RNA-binding domain (dsRBD)-containing protein (Denli et al. 2004). After export to the cytoplasm by Exportin5 (Shibata et al. 2006; Okamura et al. 2007), pre-miRNAs are secondarily processed into mature miRNAs by the Dicer1–Loqs complex (an RNase III enzyme and a dsRBD-containing protein, respectively; Loqs is also known as R3D1) (Lee et al. 2004; Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). Following this two-step maturation, miRNAs are predominantly loaded onto AGO1 and exert their function in regulating gene silencing (Okamura et al. 2004; Saito et al. 2005).
The esiRNAs in Drosophila are generated by Dicer2 and are loaded onto AGO2. In dicer2 mutants, the accumulation of esiRNAs is severely impaired, and some transposons are abnormally overexpressed (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). This strongly supports the idea that Dicer2 is involved in the esiRNA processing pathway (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). Dicer2, the other RNase III enzyme in Drosophila, was originally shown to be required for the production of exogenous siRNAs (exo-siRNAs) from long, completely annealing dsRNAs (Bernstein et al. 2001; Lee et al. 2004; Pham et al. 2004). During exo-siRNA processing, Dicer2 is associated with R2D2 (Liu et al. 2003), one of the dsRBD-containing proteins. Without R2D2, Dicer2 is destabilized in vivo (Liu et al. 2003). However, using in vitro assay systems, the exo-siRNA excising activity of Dicer2 was not affected by the addition or depletion of R2D2 (Liu et al. 2003). Thus, R2D2 might only be necessary for stabilizing Dicer2 in the exo-siRNA-producing pathway.
Like dicer2 mutants, loqs mutants fail to accumulate esiRNAs in vivo, especially those originating from non-transposon-type precursors: long stem–loop structures containing many mismatch pairs in their stems (Ghildiyal et al. 2008; Okamura et al. 2008). This result suggests that Loqs is needed for the esiRNA processing pathway, in addition to its requirement in the miRNA processing pathway. The loqs gene in Drosophila is known to give rise to four isoforms, Loqs-PA to Loqs-PD (Drysdale et al. 2005; Förstemann et al. 2005; Hartig et al. 2009; Zhou et al. 2009). Loqs-PD is very similar in size to Loqs-PC, and the sequences of Loqs-PC and Loqs-PD differ only at the very C-terminal end. Recent studies have indicated that Loqs-PB and Loq-PD are involved in the miRNA and esiRNA processing pathways, respectively (Hartig et al. 2009; Zhou et al. 2009). However, how these two isoforms function in small RNA biogenesis pathways remains elusive.
In this study, we found that the four Loqs isoforms, Loqs-PA to Loqs-PD, associate differently with endogenous Dicer1 and Dicer2 and that the C-terminal region of Loqs-PD determines the specific association with Dicer2. The Dicer2–Loqs-PD complex is able to process esiRNAs, but not miRNAs, from their corresponding precursors in in vitro assay systems, while the Dicer1–Loqs-PA and Dicer1–Loqs-PB complexes were able to process miRNA precursors, but not esiRNA precursors. The Dicer2–Loqs-PD complex also contained R2D2 and thus showed an ability to process long dsRNAs into siRNAs in vitro. However, Loqs-PD, but not R2D2, enhanced the esiRNA excision activity of Dicer2. These results indicate that Dicer2 is able to form a ternary complex with Loqs-PD and R2D2 in vivo but that only Loqs-PD, associated with Dicer2, is required to produce esiRNAs from their own precursors. Mutational analyses of esiRNA long stem–loop-type precursors revealed that the Dicer1–Loqs-PB complex prefers to process hairpins with stems of ∼22 nt, similar to miRNA precursors, whereas the Dicer2–Loqs-PD complex prefers to process precursors with longer stems. Indeed, small RNAs derived from shorter esiRNA precursors, similar to the size of miRNA precursors, were found to be loaded onto AGO1; thus, they are classified as an alternative source of miRNA-type regulatory RNAs, and we have termed them semiRNAs (siblings of esiRNA miRNAs). Together, these results suggest that the RNA precursor structure serves as the determinant for the esiRNA and miRNA processing pathways.
RESULTS
Interaction of Loqs isoforms with Dicer1 and Dicer2
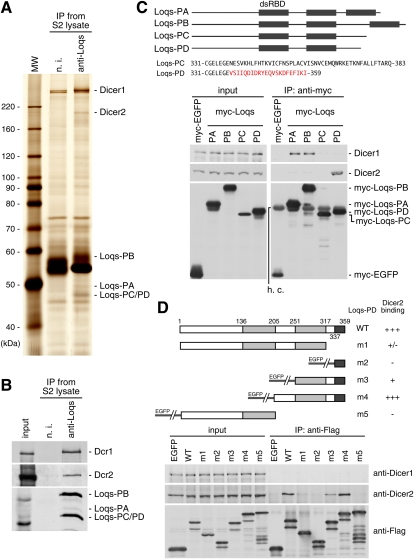
Loqs complexes were immunopurified from S2 cells using anti-Loqs antibodies (Miyoshi et al. 2009). Silver staining of the immunoprecipitates revealed that two prominent proteins coimmunoprecipitated with Loqs isoforms Loqs-PA, Loqs-PB, and Loqs-PC/PD (Fig. 1A; Förstemann et al. 2005; Hartig et al. 2009; Zhou et al. 2009). (Loqs-PB is also known as R3D1-L, but hereafter we use Loqs-PB for uniformity [Jiang et al. 2005].) Mass spectrometry analysis revealed that the two Loqs-interacting proteins were Dicer1 (∼300 kDa) and Dicer2 (∼200 kDa). Western blot analysis of the immunoprecipitates (Fig. 1A) confirmed their associations (Fig. 1B). While this work was in progress, two groups reported that all Loqs isoforms interacted with exogenously expressed Dicer2 in S2 cells (Hartig et al. 2009; Zhou et al. 2009). It was also found that Loqs-PB strongly interacted with Dicer1 whereas Loqs-PC and Loqs-PD did not show such activity (Hartig et al. 2009; Zhou et al. 2009). Since we have monoclonal antibodies for Dicer1 and Dicer2 (Miyoshi et al. 2009), we examined the binding specificity of each Loqs isoform with endogenous Dicer1 and Dicer2. myc-tagged Loqs-PA and Loqs-PB specifically interacted with endogenous Dicer1 (Fig. 1C). In contrast, myc-tagged Loqs-PD showed a capacity to interact with endogenous Dicer2, but not with Dicer1 (Fig. 1C). Although there were some discrepancies between our results and the previous results of Hartig et al. (2009) and Zhou et al. (2009), our findings are at least consistent with the finding that only Loqs-PD is involved in the esiRNA processing pathway (see below). No Loqs-PC interaction with Dicer1 or Dicer2 was detected, although Loqs-PC and Loqs-PD differ only at their C-terminal ends (Fig. 1C). Through immunofluorescence analyses using an anti-myc antibody, we found that Loqs-PC expressed in S2 cells localized predominantly in large foci in the cytoplasm (Supplemental Fig. 1), whereas other Loqs isoforms, such as Loqs-PB, localized evenly in the cytosol (Förstemann et al. 2005; Saito et al. 2005). This unique localization of Loqs-PC might be related to the observation that little or no Loqs-PC interacts with either Dicer1 or Dicer2.
FIGURE 1.
Interaction of Loqs isoforms with Dicer1 and Dicer2. (A) The Loqs-containing complexes were immunoprecipitated from S2 cells using anti-Loqs antibodies and the proteins contained in the immunoprecipitates were visualized by silver staining. The protein bands corresponding to Loqs-PA, Loqs-PB, and Loqs-PC/Loqs-PD are indicated. Two proteins coimmunopurified with Loqs were identified as Dicer1 and Dicer2 by mass spectrometry analyses. (n.i.) Nonimmune IgG used as a negative control. (B) Western blot analyses using anti-Dicer1 and anti-Dicer2 antibodies confirmed that the immunoprecipitates obtained using anti-Loqs antibodies (panel A) contain both Dicer proteins, along with all Loqs isoforms. (C) Loqs-PA and Loqs-PB interact with Dicer1, whereas Loqs-PD interacts with Dicer2. Each Loqs isoform was expressed in S2 cells, and the anti-myc immunoprecipitated complexes were probed with anti-Dicer1 and anti-Dicer2 antibodies. (h.c.) Heavy chain of anti-c-myc antibody. (D) Deletion mutant analysis of Loqs-PD to determine the domain necessary for interacting with Dicer2. Various deletion mutants of Loqs-PD (m1–m5) were expressed in S2 cells, and their interactions with endogenous Dicer2 were examined. The Loqs-PD-m4 mutant interacted with Dicer2 as efficiently as wt Loqs-PD. The dsRNA-binding domains (light gray boxes) and C-terminal region (dark gray boxes) found exclusively in Loqs-PD but not in other Loqs isoforms, respectively, are indicated (upper diagram). The lower panel shows immunoblotting results of the protein–protein interaction assays. EGFP was used as a negative control.
We then determined the domain of Loqs-PD that binds with Dicer2. A series of deletion mutants of Loqs-PD, m1 to m5, was produced and expressed in S2 cells as Flag-tagged peptides. Loqs-PD-m2 to Loqs-PD-m5 were fused with EGFP to attain a size similar to that of wild-type (wt) Loqs-PD. Western blot analysis using anti-Dicer2 antibodies on anti-Flag immunoprecipitates showed that the Loqs-PD-m4 mutant, which contains the second dsRBD and both flanking regions, was able to interact with Dicer2 as efficiently as wt Loqs-PD (Fig. 1D). Loqs-PD-m3, containing the second dsRBD and the C-terminal flanking region, associated weakly with Dicer2. The C-terminus-lacking mutant (Loqs-PD-m1) and the C-terminal end of Loqs-PD per se showed little or no binding with Dicer2. These results demonstrate that the peptide covering amino acid 206 to the end of Loqs-PD (Gly206–Ile359) is the domain that interacts with Dicer2 (Fig. 1D). Thus, the Loqs-PD-specific C-terminal sequence may help the folding of the second dsRBD so that Dicer2 can only bind Loqs-PD. A previous study showed that Dicer1 interacts strongly with the third dsRNA-binding domain of Loqs-PB (Ye et al. 2007). Together, these data clarify the selectivity of Loqs isoforms for Dicer proteins.
The Dicer2–Loqs-PD complex excises esiRNAs from precursors
It was recently shown in S2 cells that depletion of Loqs-PB interfered with miRNA processing, whereas Loqs-PD depletion affected esiRNA processing (Hartig et al. 2009; Zhou et al. 2009). By performing RNAi in S2 cells, followed by Northern blot analyses for multiple miRNAs (bantam, miR-2b, and miR-8) and non-transposon-derived esiRNAs (esiRNA-sl-1 and CG18854A), we confirmed that the requirement of Loqs isoforms in the miRNA and esiRNA processing pathways indeed differ from one another (Supplemental Fig. 2). The level of an esiRNA derived from transposon DM1731, esiRNA-1731, was also significantly lowered by Loqs-PD depletion (Supplemental Fig. 2). Thus, Loqs-PD is also necessary for the production of transposon-derived esiRNAs.
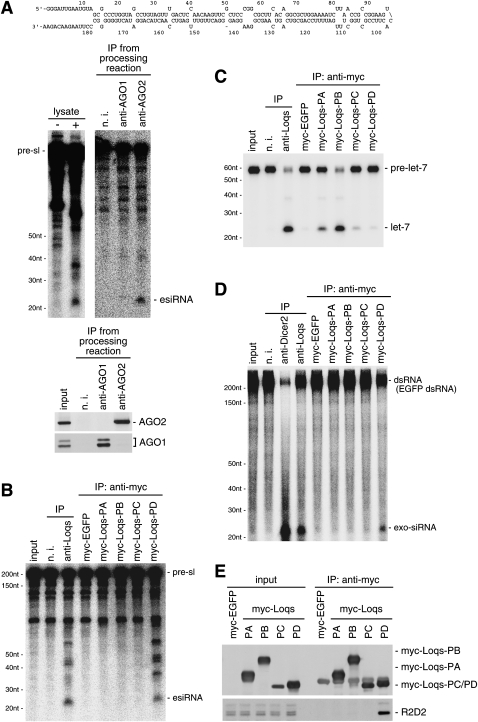
We then assessed if the Dicer2–Loqs-PD complex was able to process esiRNA precursors into mature esiRNAs in vitro. A long stem–loop-type esiRNA precursor (pre-esiRNA), which contains the sequence of esiRNA-sl-1, was designed (pre-sl) (Fig. 2A) and utilized in the assays. We confirmed that small RNAs of ∼21 nt derived from the precursor were loaded onto AGO2, as expected, but not onto AGO1 in S2 lysates (Fig. 2A). All Loqs isoforms tagged with a myc peptide were expressed in S2 cells following transfection. Immunoprecipitation was performed 24 h post-transfection using an anti-myc antibody to isolate each complex. The Loqs-PA and Loqs-PB complexes that contain Dicer1 (shown in Fig. 1C) showed no activity to produce esiRNAs from the precursor (Fig. 2B). Loqs-PC, whose interaction with Dicer1 and Dicer2 was under the limit of detection by Western blot analysis (Fig. 1C), also showed no such activity. However, the Loqs-PD complex containing Dicer2 was able to process the esiRNA precursor to the same extent as the complex immunoprecipitated from S2 cells using anti-Loqs antibodies (Fig. 2C). When an miRNA precursor, pre-let-7, was used as an alternative in the assays, the Loqs-PD–Dicer2 complex produced only a background signal (Fig. 2C). Pre-let-7 was efficiently processed to mature let-7 by both of the Loqs-PA and Loqs-PB complexes, as expected. The Loqs-PB complex showed a relatively higher activity than the Loqs-PA complex, although both complexes contain Dicer1 at a similar level (Fig. 1C), suggesting that the Dicer1–Loqs-PB complex is more suited for miRNA processing. Loqs-PC showed a slightly stronger signal than myc-EGFP, indicating that although the interaction with Dicer1 was not detected by Western blot analysis (Fig. 1C), Loqs-PC might be associated with Dicer1 at a very low level. It should be noted that the expression levels of Loqs isoforms seemed to vary in tissues (Supplemental Fig. 3), suggesting a model in which changes in the ratio of these isoforms can affect processing of pre-miRNAs and pre-esiRNAs in vivo.
FIGURE 2.
Small RNA excision activities of the Dicer2–Loqs-PD complex immunopurified from S2 cells. (A) esiRNAs excised from the precursor (pre-sl; the nucleotide sequence and a possible structural formation are shown at the top; the structural formation was predicted by RNA & DNA folding applications; http://mfold.bioinfo.rpi.edu/) were found to be loaded onto AGO2, but not onto AGO1 in S2 cell lysates. (Lysate −) esiRNA precursors incubated without S2 lysate, (lysate +) esiRNA precursors incubated with S2 lysate. (Lower panels) Western blot results showing the specificity of immunoprecipitation. (B) The Dicer2–Loqs-PD complex was capable of excising esiRNAs from the precursor. Loqs-containing complexes immunopurified from S2 cells using anti-Loqs antibodies (anti-Loqs) were employed as positive controls. (C) The Dicer2–Loqs-PD complex failed to excise let-7 from the precursor (pre-let-7), whereas the Loqs-PA and Loqs-PB complexes specifically containing Dicer1 were able to process the precursor. (D) The Dicer2–Loqs-PD complex was also able to excise exo-siRNAs from the precursors (EGFP dsRNAs). (E) Western blot analysis using anti-R2D2 antibodies showed that the Dicer2–Loqs-PD complex also contained the R2D2 protein.
When the Loqs-PD complex was incubated with a long, completely complementary dsRNA (EGFP-dsRNA), which serves as an exo-siRNA precursor, the long dsRNA was processed to mature siRNAs of ∼22 nt (Fig. 2D). Therefore, we examined if Dicer2, when associated with Loqs-PD, also contained R2D2. Western blot analysis using anti-R2D2 antibodies revealed that R2D2 was detectable in the Loqs-PD complex, albeit at low levels (Fig. 2E). The existence of R2D2 was unique to the Loqs-PD complex, suggesting that Dicer2 might be able to associate with Loqs-PD and R2D2 simultaneously in a dsRNA-independent manner.
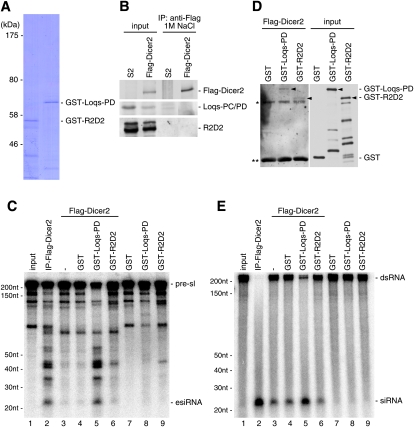
Loqs-PD, but not R2D2, enhances the esiRNA processing activity of Dicer2 in vitro
Loqs-PB specifies miRNA precursors as a substrate of Dicer1 by physically associating with Dicer1 (Saito et al. 2005). In in vitro assay systems, Loqs-PB was also able to promote the miRNA processing activity of Dicer1 (Jiang et al. 2005; Saito et al. 2005). To examine whether Loqs-PD in the esiRNA processing pathway showed similar effects to Loqs-PB in the miRNA processing pathway, esiRNA processing assays were performed using a glutathione S-transferase (GST)-tagged, recombinant Loqs-PD protein produced in and purified from Escherichia coli (Fig. 3A). As a control, GST-tagged R2D2 was also produced (Fig. 3A). Flag-Dicer2 protein was expressed in S2 cells following transfection and purified using an anti-Flag antibody under harsh conditions containing 1 M NaCl, which should disrupt most protein–protein interactions occurring in vivo. Western blot analysis using anti-Loqs and anti-R2D2 antibodies revealed that Loqs-PC/PD and R2D2 were not detected in the purified Dicer2 complex (Fig. 3B). The purified Dicer2 alone did not show a significant activity to excise esiRNAs from the precursors (Fig. 3C, lane 3). When GST or GST-tagged R2D2 was added to the reaction mixture, the production of esiRNA was unchanged. However, when GST-tagged Loqs-PD was added instead, we found that the esiRNA production activity increased to a level similar to that of the Flag-Dicer2 complex immunoprecipitated from S2 cells using anti-Flag antibodies (Fig. 3C, lane 2). GST-Loqs-PD alone did not show this esiRNA processing activity (Fig. 3C, lane 8). Both GST-Loqs-PD and GST-R2D2 interacted to an approximately equal degree with Flag-Dicer2 immunoprecipitated in a buffer containing 1 M NaCl (Fig. 3D). We also examined whether the addition of GST-Loqs-PD affects the exo-siRNA processing ability of Dicer2 and revealed that GST-Loqs-PD but not GST-R2D2 enhances this activity (Fig. 3E). These results correlate well with the earlier observation that R2D2 is not necessary for Dicer2 to process long dsRNAs into exo-siRNAs (Liu et al. 2003). Taken together, these results indicate that Loqs-PD enhances Dicer2 activity to process esiRNAs from their precursors by associating with Dicer2.
FIGURE 3.
Loqs-PD, but not R2D2, enhanced the Dicer2 esiRNA processing activity in vitro. (A) Coomassie brilliant blue staining of GST-tagged, recombinant Loqs-PD and R2D2 proteins produced in and purified from E. coli. (B) Flag-tagged Dicer2 protein expressed in S2 cells following transfection was affinity-purified with an anti-Flag antibody in a buffer containing 1 M NaCl. Western blot analysis revealed that the Dicer2 fraction isolated under such harsh conditions contained only traces of R2D2 and Loqs proteins. (C) In vitro esiRNA processing assays using the purified Dicer2 obtained in B. While Dicer2 alone did not excise esiRNAs from the precursor (pre-esiRNA; lane 3, Fig. 2A), addition of GST-Loqs-PD (lane 5), but not of GST (lane 4) and GST-R2D2 (lane 6), increased the esiRNA production activity of Dicer2 to the same extent as that of Flag-Dicer2 immunoprecipitated from S2 cells with an anti-Flag antibody (lane 2). GST-Loqs-PD alone did not show any esiRNA processing activity (lane 8). These results indicate that Loqs-PD appears to enhance the ability of Dicer2 to process esiRNAs from their precursors by associating with Dicer2. (D) Both GST-tagged Loqs-PD and R2D2, but not GST itself, were able to bind with Flag-Dicer2 purified from S2 cells under a high-salt condition as in Figure 4B. Note that only the full-length proteins (black arrowheads) seemed to be bound with Dicer2. (*) Heavy chains of anti-Flag antibody, (**) light chains of anti-Flag antibody. (E) GST-Loqs-PD (lane 5) but not GST-R2D2 (lane 6) enhances the ability of Dicer2 to process long dsRNAs into exo-siRNAs.
Substrate preference of the Dicer1–Loqs-PB and Dicer2–Loqs-PD complexes
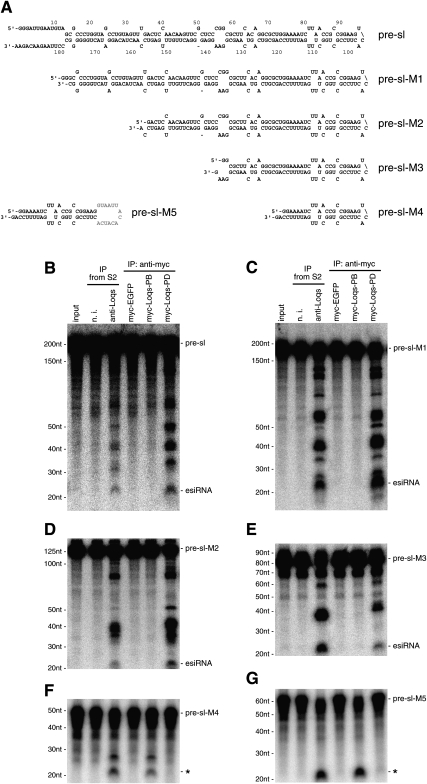
How do the Dicer1–Loqs-PB and Dicer2–Loqs-PD complexes determine their substrates? To address the question, we produced a series of deletion mutants of pre-sl (Fig. 2A; mutants are summarized in Fig. 4A) and performed esiRNA processing assays using both Dicer–Loqs complexes immunoisolated from S2 cells. The anti-Loqs immunoprecipitates and the immunoisolated myc-tagged EGFP complex were also employed as controls in these assays. Pre-sl, pre-sl-M1, pre-sl-M2, and even the pre-sl-M3 mutant, which contains a 37-nt-long stem, were specifically processed by the Dicer2–Loqs-PD complex (Fig. 4B–E). On the other hand, the shorter pre-sl mutant, pre-sl-M4, which contains a 22-nt-long stem, as in most typical miRNA precursors, was processed by the Dicer1–Loqs-PB complex but not by the Dicer2–Loqs-PD complex (Fig. 4F). Another mutant, pre-sl-M5, which contains the same stem as pre-sl-M4 but with the loop swapped with that of pre-let-7, was also processed by the Dicer1–Loqs-PB complex (Fig. 4G). These data suggest that the length of the stem, but not the loop, of the stem–loop-structured, non-transposon-derived esiRNA precursors largely determines recognition by different Dicer–Loqs complexes.
FIGURE 4.
esiRNA processing using pre-sl deletion mutants. (A) The nucleotide sequences and possible structural formation of various pre-sl deletion mutants (pre-sl-M1 to pre-sl-M5) are shown. Pre-sl-M5 contains the loop of pre-let-7 (gray) instead of the pre-sl loop. (B) esiRNA processing was performed with pre-sl as in Figure 2B. The anti-Loqs immunoprecipitates and both Dicer1–Loqs-PB and Dicer2–Loqs-PD complexes immunoprecipitated with an anti-myc antibody, as in Figure 2B, were utilized as the enzymatic sources. The myc-EGFP complex was employed as a negative control. (C) esiRNA processing with pre-sl-M1, as in B. (D) esiRNA processing with pre-sl-M2, as in B. (E) esiRNA processing with pre-sl-M3, as in B. (F) pre-sl-M4 processing, as in B. Note that the precursor was now processed by myc-Loqs-PB but not by the myc-Loqs-PD complex. (G) pre-sl-M5 processing, as in B.
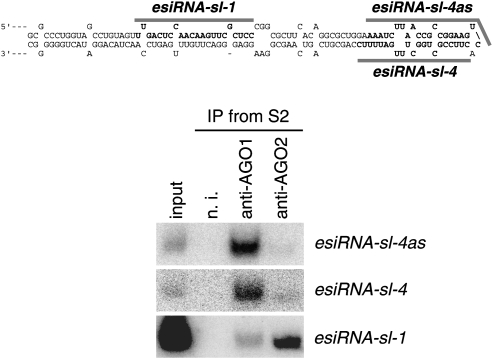
miRNAs are processed from corresponding precursors by the Dicer1–Loqs-PB (or Dicer1–Loqs-PA) complex and loaded onto AGO1 in vivo. We then examined if small RNAs derived from processed intermediates of esiRNA precursors, similar to miRNA precursors (like pre-sl-M4 in Figure 4), are loaded onto AGO2 in vivo. AGO1 and AGO2 were immunoprecipitated from S2 cells, and RNA fractions associated with both proteins were probed with DNA oligos recognizing esiRNA-sl-1, esiRNA-sl-4, and esiRNA-sl-4as (Fig. 5). esiRNA-sl-1 was detected as expected mostly in the RNA pool immunoisolated with AGO2; in contrast, both esiRNA-sl-4 and esiRNA-sl-4as were found predominantly with AGO1. β-elimination experiments confirmed that esiRNA-sl-4 and esiRNA-sl-4as can be categorized as AGO1 binders (data not shown). These results suggest that esiRNA precursors are processed by the Dicer2–Loqs-PD complex until the length of the stem is long enough to be recognized by the particular complex. However, once the stem becomes too short for Dicer2–Loqs-PD-dependent processing, as in miRNA precursors, the Dicer1–Loqs-PB (or PA) complex comes into contact with it and processes it further into small RNAs. We consider these final small RNA products as a unique subset of miRNAs, and suggest naming them semiRNAs, or siblings of esiRNA miRNAs.
FIGURE 5.
Small RNAs (esiRNA-sl-4 and esiRNA-sl-4as) arising from the region most proximal to the loop in the hairpin esiRNA precursor are loaded predominantly onto AGO1 in S2 cells. esiRNA-sl-1 was loaded onto AGO2 as expected. RNA pools immunoisolated with AGO1 and AGO2 from S2 cells were probed with DNA oligos for esiRNA-sl-1, esiRNA-sl-4, and esiRNA-sl-4as.
DISCUSSION
siRNAs in Drosophila are produced from completely matched dsRNA precursors by the Dicer2–R2D2 complex (Liu et al. 2003), whereas miRNAs are excised from pre-miRNAs by Dicer1–Loqs (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). Therefore, it was believed that siRNAs and miRNAs could be easily distinguished from one another based on their origin and processing factors. However, the discovery of esiRNAs made it difficult to distinguish these two classes of small RNAs (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). This is because esiRNA production requires Dicer2 and Loqs, the processing factors required in siRNA and miRNA pathways, respectively (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). From these findings, it was speculated that small RNA biogenesis systems in Drosophila cells could intersect with each other and that they are even more complex than previously conceived. However, two groups recently showed that the requirement for Loqs in the esiRNA and miRNA processing pathways differs in regard to the isoforms: Loqs-PD is required for esiRNA production, whereas Loqs-PB functions in miRNA production (Hartig et al. 2009; Zhou et al. 2009). In this study, we not only confirmed the functional differences of Loqs-PB and Loqs-PD, but we also showed that Loqs-PA, which is the most abundant Loqs isoform in testes, is involved in miRNA, but not in esiRNA, production. We also showed that Dicer2 was able to simultaneously associate with Loqs-PD and R2D2 (Fig. 6), and that Loqs-PD, but not R2D2, was able to stimulate esiRNA production from their precursors. This result correlated well with a previous observation that depletion of R2D2 did not significantly affect esiRNA production in Drosophila cells (Czech et al. 2008; Okamura et al. 2008). The association of Dicer2 with two distinct factors, Loqs-PD and R2D2, might be quite reasonable for its function because in this situation Dicer2 does not have to select or change its partner (either Loqs-PD or R2D2) for the processing of different precursors (either esiRNA or exo-siRNA precursors).
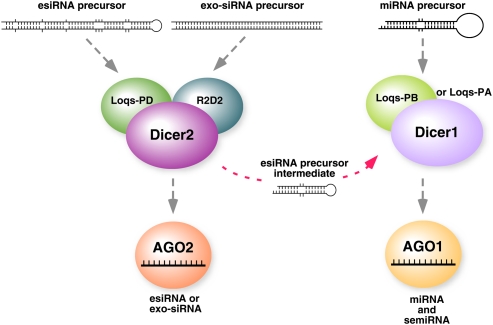
FIGURE 6.
A model for esiRNA, exo-siRNA, and miRNA biogenesis pathways in Drosophila. The Dicer1–Loqs-PB (and the Dicer1–Loqs-PA) complex processes miRNA precursors, whereas the Dicer2–Loqs-PD complex processes esiRNA precursors. The Dicer2–Loqs-PD complex also contains R2D2 and thus processes long dsRNAs into siRNAs. The shorter esiRNA precursor intermediates, which are structurally similar to miRNA precursors, are processed by the Dicer1–Loqs-PB complex; this process gives rise to semiRNAs, a new subset of miRNAs.
How do Dicer proteins distinguish Loqs-PA/Loqs-PB over Loqs-PD, or vice versa? A previous study clearly indicated that Dicer1 binds strongly with the third dsRNA-binding domain of Loqs-PB but not with other truncated versions containing the first and second dsRBD (Ye et al. 2007). Loqs-PD contains the first and second dsRBD but lacks the third. Thus, these previous results support our findings that Loqs-PD did not bind to Dicer1. Deletion analyses of Loqs-PD showed that truncation of the C-terminal end (42 amino acids) caused a severe decrease in binding with Dicer2. However, the C-terminal region itself was obviously insufficient for the association. In addition to the C-terminal region, the second dsRBD and its upstream region of ∼50 amino acids were also needed for tight binding with Dicer2 (Fig. 1D). Loqs-PB shares with Loqs-PD the second dsRBD and its two flanking regions. Considering also that even Loqs-PC showed no binding to Dicer2, the C-terminal end of Loqs-PD, which is absent from Loqs-PC, is likely to determine the specific binding with Dicer2.
Accumulated evidence suggests that several miRNAs in animals and plants originate from genomic repetitive elements and transposons (Chapman and Carrington 2007). A recent study also showed that unlike Saccharomyces cerevisiae, other budding-yeast species, Saccharomyces castellii and Candida albicans, have systems to generate small interfering RNAs, which mostly correspond to transposable elements and to Y′ subtelomeric repeats residing in their genomes; however, they do not have miRNAs (Drinnenberg et al. 2009). These data together suggest that miRNAs may have arisen much later in evolution than esiRNAs. This assumption further implies that in Drosophila, dicer1 may have arisen as a result of duplication of a proto _dicer2_-like gene and that such gene duplication might have helped the production of miRNAs originating from genomic repetitive elements. Drosophila has maintained miRNA and esiRNA pathways independently from each other, probably because both pathways are essential; the miRNA system is used for regulating gene expression and the esiRNA system functions as an innate immune system. However, in higher animals, like mammals, only one Dicer, which is likely to be dedicated to the miRNA pathway, is maintained. This is because through evolution, mammals have acquired a more elaborate immune system, namely, ”adaptive immunity” to protect themselves from pathogenic invaders, such as viruses.
In addition, it can be speculated that in parallel with miRNA evolution and dicer gene duplication in Drosophila, the loqs gene has evolved to give rise to various isoforms that differentially associate with Dicer1 and Dicer2 and function independently in processing miRNAs and esiRNAs because both pathways are indispensable for Drosophila.
MATERIALS AND METHODS
Plasmid constructs
The myc-tagged constructs for Loqs-PB, Loqs-PA and EGFP have been described previously (Förstemann et al. 2005). The construct for myc-tagged Loqs-PD (Hartig et al. 2009) was produced using the strategy described in Förstemann et al. 2005 with the primers 5′-AGCGGATCCATGGAACAAAAACTTATTTCTGAAGAAGACTTGGAACAAAAACTTATTTCTGAAGAAGACTTGGCCGTGAGTATCATTCAAGACATC-3′ and 5′-CAAAGCGGCCGCTTAGATCTTGATGAACTC-3′ and the construct for myc-tagged Loqs-PC was produced with the primers 5′-CAAGGATCCAACGAATCTGTAAAGCACCT-3′ and 5′-CAAAGCGGCCGCCTACTGCGGGGCTGTAAATAAG-3′. All constructs for Loqs-PD deletion mutants were generated by PCR using full-length myc-tagged Loqs-PD plasmid as a template. PCR products were subcloned into pRH3×Flag (Saito et al. 2005) or pRH3×Flag-EGFP (Saito et al. 2005). To obtain a cDNA encoding Dicer2, poly(A)+ RNAs were purified from S2 cells and reverse transcription polymerase chain reaction (RT-PCR) was carried out using a ProSTAR Ultra-HF RT-PCR system (Stratagene). To produce a 3×Flag-tagged Dicer2 protein, full-length Dicer2 cDNA was subcloned into pRH3×Flag (Saito et al. 2005). To produce GST-tagged proteins, Loqs-PD and R2D2 cDNAs were subcloned into the expression vector, pGEX-6p-2 (GE Healthcare). The fusion proteins were induced and purified from E. coli, as described by the manufacturer.
Immunoprecipitation, silver staining, and Western blot analysis
Immunoprecipitation from S2 cell lysates was performed using a modified protocol of a previously reported method (Miyoshi et al. 2005). Antibodies were cross-linked to Gammabind sepharose beads (GE Healthcare) with 20 mM dimethyl pimelimide. To obtain myc-tagged Loqs proteins, immunoprecipitation was performed using a monoclonal anti-c-myc antibody (9E10, catalog number M4439, Sigma-Aldrich). To purify Flag-Dicer2, immunoprecipitation was performed using anti-Flag M2 agarose beads (Sigma-Aldrich). After immunoprecipitation, the beads were washed extensively with a buffer containing 30 mM HEPES (pH 7.4), 150 mM potassium acetate, 5 mM magnesium acetate, 5 mM DTT, 0.1% NP40, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 0.5% aprotinin. Silver staining and Western blotting were performed as described (Miyoshi et al. 2005). Anti-Loqs, anti-Dicer1, and anti-Dicer2 antibodies were used as described (Miyoshi et al. 2009).
Northern blot analysis
Northern blot analysis was carried out essentially as described previously (Saito et al. 2005). Oligo DNAs used as probes were: esiR-sl-4, 5′-GAAAATCAAAGCCAGCGGAAG-3′; esiRNA-sl-4as, 5′-GACTTCCGGCGGTTAAGATTT-3′.
In vitro processing assays
DNA fragments coding pre-esiRNA, pre-sl, and the pre-sl mutants, pre-sl-M1 to pre-sl-M5, were obtained from PCR reactions. The primers used were: 5′-TAATACGACTCACTATAGGGATTGAATGTAGCGCCCTGG-3′ and 5′-TTCTGTTCTTAAGGGCCCCCCAGTAT-3′ for pre-sl; 5′-TAATACGACTCACTATAGGGCGCCCTGGTAGCCTGTAGTT-3′ and 5′-GCCCCCCAGTATCCTGTAGTTGG-3′ for pre-sl-M1; 5′-TAATACGACTCACTATAGACTCCAACAAGTTCGCTCCCGG-3′ and 5′-TGGACTCAAACAAGTCCCTC-3′ for pre-sl-M2; 5′-TAATACGACTCACTATAGGCGCTTCACAGGCGCTGGAAAA-3′ and 5′-CTTCCGCTCGACAGACGCTG-3′ for pre-sl-M3. The template DNAs were produced by annealing sets of oligo DNAs for pre-sl-M4 and pre-sl-M5. The oligo DNAs used were: 5′-TAATACGACTCACTATAGGAAAATCTTAACCGCCGGAAGTCACTTCCGCTGGCTTTGATTTTCCAG-3′ and 5′-CTGGAAAATCAAAGCCAGCGGAAGTGACTTCCGGCGGTTAAGATTTTCCTATAGTGAGTCGTATTA-3′ for pre-sl-M4; 5′-TAATACGACTCACTATAGGAAAATCTTAACCGCCGGAAGGTAATTACACATCACTTCCGCTGGCTTTGATTTTCCAG-3′ and 5′-CTGGAAAATCAAAGCCAGCGGAAGTGATGTGTAATTACCTTCCGGCGGTTAAGATTTTCCTATAGTGAGTCGTATTA-3′ for pre-sl-M5. A DNA fragment coding for dsRNA (EGFP dsRNA) was amplified from epEGFP-C1 (Clontech) by PCR using the primers: 5′-TAATACGACTCACTATAGGGATGGTGAGCAAGGGCGAGGA-3′ and 5′-TAATACGACTCACTATAGGGTAGGTCAGGGTGGTCACGAG-3′. The PCR products and annealed oligo DNAs were used as templates for an in vitro transcription reaction using MEGAscript T7 kits (Ambion) in the presence of 32P-UTP. These transcripts and 5′-32P end-labeled pre-let-7 (Miyoshi et al. 2009) were self-annealed. In vitro processing reactions of pre-miRNA (pre-let-7), pre-esiRNAs, and long dsRNA (EGFP dsRNA) with purified myc-Loqs proteins or Flag-Dicer2 were performed as described (Saito et al. 2005). For processing using Flag-Dicer2, high-salt (1 M NaCl) purified Flag-Dicer-2 was added in the presence or absence of bacterially produced GST-Loqs-PD or GST-R2D2. After 90 min incubation at 26°C, RNAs were purified with ISOGEN LS (Nippon Gene) and separated on 8% acrylamide denaturing gels containing 1× TBE buffer, 8 M urea, and 25% formamide.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank K. Förstemann for sharing unpublished data and for helpful discussions. We also thank Q. Liu and K. Saito for antibodies, technical assistance, advice, and discussion; T.N. Okada, T. Sunohara, A. Ishizuka, and H. Tsukumo for technical assistance and reagents; and other members of the Siomi laboratory for discussions and comments on the manuscript. This work was supported by a Boehringer Ingelheim Fonds fellowship to J.V.H., MEXT grants to H.S., and NEDO (New Energy and Industrial Technology Development Organization) grants to M.C.S. M.C.S. is supported by CREST from JST.
Footnotes
REFERENCES
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA FlyBase Consortium. FlyBase: Genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Böttcher R, Saito K, Förstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Kuramochi-Miyagawa S. RNA silencing in germlines—exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol. 2009;21:426–434. doi: 10.1016/j.ceb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA- generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]