Gata6 is an important regulator of mouse pancreas development (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 18.
Abstract
Gata4, Gata5, and Gata6 represent a subfamily of zinc-finger transcriptional regulators that are important in the development and differentiation of numerous tissues, including many endodermally-derived organs. We demonstrate that Gata4 and Gata6 have overlapping expression patterns in the early pancreatic epithelium. Later, Gata4 becomes restricted to exocrine tissue and Gata6 becomes restricted to a subset of endocrine cells. In addition, we show Gata6, but not Gata4, physically interacts with Nkx2.2, an essential islet transcription factor. To begin determining the roles that Gata4 and Gata6 play during pancreatic development, we expressed Gata4-Engrailed and Gata6-Engrailed dominant repressor fusion proteins in the pancreatic epithelium and in the islet. At e17.5, transgenic Gata6-Engrailed embryos exhibit two distinct phenotypes: a complete absence of pancreas or a reduction in pancreatic tissue. In the embryos that do form pancreas, there is a significant reduction of all pancreatic cell types, with the few differentiated endocrine cells clustered within, or in close proximity to, enlarged ductal structures. Conversely, the majority of transgenic Gata4-Engrailed embryos do not have a pancreatic phenotype. This study suggests that Gata6 is an important regulator of pancreas specification.
Keywords: Gata4, Gata6, Pancreas development, Islet
Introduction
The pancreas regulates glucose homeostasis in two important ways: the endocrine pancreas produces and secretes hormones, while the exocrine pancreas produces and secretes digestive enzymes. The endocrine cells cluster into islets, comprised of α, β, δ, PP, and ε cells that produce glucagon, insulin, somatostatin, pancreatic polypeptide, and ghrelin, respectively. In the mouse, development of the pancreas begins with patterning of the foregut endoderm around embryonic day (e) 8.5. Cells are committed to a pancreatic fate by virtue of their unique position within the endoderm and their proximity to adjacent tissues (Bort et al., 2004; Kim et al., 1997; Kim and Melton, 1998). In response to signals from surrounding tissues, evagination of the endoderm leads to formation of the dorsal and ventral pancreatic buds. From e9.5 to birth, the cells of the pancreatic epithelium execute a complex program of proliferation, branching, and differentiation that results in the formation of three primary structures: the endocrine pancreas, the exocrine pancreas, and the ductal network (Pictet and Rutter, 1972; Slack, 1995).
Numerous transcription factors have been implicated in the sophisticated regulatory cascade that occurs during pancreas development. Before specification of the pancreatic epithelium, Pdx1 and Ptf1a can be detected in endodermal regions that will form the dorsal and ventral buds. Pdx1 and Ptf1a null mice do not develop a mature pancreas, demonstrating that both transcription factors are required for pancreas development (Jonsson et al., 1994; Krapp et al., 1998; Offield et al., 1996). Early Pdx1 expression is necessary for regionalization of the primitive gut endoderm and specification of the early pancreatic epithelium, while a second wave of expression is necessary for maturation of β cells and insulin regulation (Guz et al., 1995; Wilding and Gannon, 2004) as well as formation of the exocrine pancreas (Hale et al., 2005). The basic helix–loop–helix (bHLH) factor Ptf1a is initially required for specification of all pancreatic cell types, and has been shown to regulate exocrine differentiation and enzyme expression during later developmental stages (Kawaguchi et al., 2002; Rose et al., 2001).
Other transcription factors are also required for differentiation of the endocrine pancreas. The bHLH transcription factor Ngn3 is transiently expressed in all cells of the endocrine lineage and is recognized as a marker of endocrine progenitor cells (Gu et al., 2002). Nkx6.1 is required for proper differentiation of β cells in the developing pancreas (Sander et al., 2000). Pax6 is required for normal development of the islet cell types and has been shown to function as an activator of endocrine hormone gene transcription (Sander et al., 1997). The homeodomain-containing transcription factor Nkx2.2 is also required for proper differentiation of endocrine cell types (Sussel et al., 1998). Recent studies in our lab have centered on identifying transcription factors that work in concert with Nkx2.2 to regulate gene expression during pancreas development. Potential candidates include members of the Gata transcription factor family because they are known to interact with Nkx2 family members in other organs. For example, Gata6 and Nkx2.1 interact during lung epithelial development (Bruno et al., 2000; Liu et al., 2002a; Weidenfeld et al., 2002) and Nkx2.5 interacts with Gata4 and Gata5 during heart development (Stennard et al., 2003).
In the mouse, the Gata transcription factor family has six members and these zinc finger DNA binding proteins recognize the consensus sequence A/T-GATA-A/G. Gata1, Gata2, and Gata3 are known to be important for the differentiation of various cell types in the hematopoietic system while Gata4, Gata5, and Gata6 have been implicated in the development of organs derived from endoderm and mesoderm (Molkentin, 2000; Patient and McGhee, 2002). Gata4 is expressed in the primitive endoderm, heart, liver, small intestine, and gonads of the embryonic mouse (Arceci et al., 1993). Characterized as an important activator of many cardiac genes, Gata4 is required for cardiomyocyte maturation and formation of the heart tube. In addition, Gata4 is essential for ventral morphogenesis as Gata4 null embryos die between e7.5 and e9.5 due to defects in development of the visceral endoderm (Kuo et al., 1997; Molkentin et al., 1997; Narita et al., 1997). Expression of Gata6 during embryonic development is observed in the primitive streak, allantois, muscle, heart, lung, and gut (Morrisey et al., 1996). Gata6 plays an important role in lung development, specifically in branching morphogenesis and later stage differentiation of the lung epithelium (Keijzer et al., 2001; Koutsourakis et al., 2001; Liu et al., 2002b; Yang et al., 2002). Mice deficient for Gata6 die between e5.5 and e7.5, due to a defect in the formation of extraembryonic tissues in the developing blastocyst (Koutsourakis et al., 1999; Morrisey et al., 1998).
Recently, there have been two conflicting reports on Gata expression in the pancreas. Ritz-Lazer et al. (Ritz-Laser et al., 2005) used RT-PCR to demonstrate that Gata4 and Gata6 are both present in the islet and in endocrine cell lines, and suggest that Gata4 activates glucagon expression in vitro. However, Ketola et al. (Ketola et al., 2004) reported that Gata6 is expressed in the endocrine pancreas, while Gata4 expression is restricted to the exocrine pancreas. Further, they suggest that Gata4 and Gata6 are not expressed in the early pancreatic epithelium. The experiments described in our report attempt to resolve this incongruence and extend the analysis to attempt to determine the in vivo function of these Gata factors during embryonic pancreas development.
In this study, we demonstrate that both Gata4 and Gata6 are expressed at the onset of embryonic mouse pancreas development. Further, comprehensive expression analysis confirms that Gata4 and Gata6 are expressed in mutually exclusive pancreatic domains at later developmental stages. Since both Gata4 and Gata6 null mice die prior to formation of the pancreatic buds, we utilized dominant repressor Gata transgenic mice as an alternative in vivo approach to investigate the role of Gata factors in pancreas development. This strategy has been used successfully to study Gata6 during various stages of embryonic lung development (Koutsourakis et al., 2001; Liu et al., 2002b; Yang et al., 2002) and the role of ETS domain transcription factors in mouse development (Liu et al., 2003). We generated transient transgenic mice that express the repression domain of the Drosophila Engrailed protein fused to either full-length Gata4 or Gata6 or the Gata6 DNA binding domain. Expression of each fusion protein is driven by the pancreas-specific Pdx1 promoter element (Apelqvist et al., 1997; Li and Edlund, 2001; Stoffers et al., 1999). From this work, we conclude that Gata6 plays an important role in pancreas specification as well as the development of endocrine cell types.
Materials and methods
Construction of the Gata transgenes and generation of transgenic embryos
Three Gata transgenic constructs were created as follows: The repression domain of the Drosophila Engrailed (EnR) gene (encoding amino acids 1–298) was PCR-amplified from the pCS2:EnR plasmid (K. Artinger, UCHSC). Primers included 5′ _Bam_HI and 3′ _Xho_I sites for cloning. A FLAG tag was added, in frame, by cloning the EnR sequence into the _Bam_HI and _Xho_I sites of the pCMV-Tag4B vector (Stratagene) to produce pCMV-Tag4B-EnR. Full-length Gata6 cDNA, full-length Gata4 cDNA, and the Gata6 DNA binding domain (encoding amino acids 232–354) were PCR-amplified from cDNA and cloned into the pCR®-Blunt II-TOPO® vector (Invitrogen). The Gata cDNA inserts were released by digestion and cloned into the _Bam_HI site of pCMV-Tag4B-EnR to produce pCMV-Tag4B-G6FLEnR, pCMV-Tag4B-G4FLEnR, and pCMV-Tag4B-G6DBDEnR. Finally, the fusion constructs were PCR-amplified from their respective pCMV-Tag4B vectors and blunt-end cloned into the _Eco_RI site of the pPDX1_-Eco_RI vector (J. Jensen, UCHSC) to produce Pdx1:G6FLEnR, Pdx1:G4FLEnR, and Pdx1:G6DBDEnR. A control transgenic construct was created as follows: The repression domain of the Drosophila Engrailed gene (EnR) was PCR-amplified from the pCS2:EnR plasmid using primers that added a 5′ _Eco_RI site and nuclear localization sequence (encoding PKKKRKV) as well as a 3′ _Xho_I site. The PCR product was cloned into the pCR®-Blunt II-TOPO® vector to produce TOPO:NLSEnR. A FLAG tag was added, in frame, by cloning the NLSEnR sequence into the _Eco_RI and _Xho_I sites of pCMV-Tag4B to produce pCMV-Tag4b-NLSEnR. The NLSEnR construct was PCR-amplified from pCMV-Tag4b-NLSEnR and blunt-end cloned into the _Eco_RI site of the pPDX1_-Eco_RI vector to produce Pdx1: NLSEnR. Pdx1:G6FLEnR, Pdx1:G6DBDEnR, and Pdx1:NLSEnR were linearized with _Dra_III, while Pdx1:G4FLEnR was linearized with _Sap_1. The transgenic constructs were subsequently purified using the QIAquick Gel Extraction Kit (Qiagen). Each construct was injected into FVB fertilized oocytes, which were subsequently transferred into pseudopregnant females (UCCC Transgenic/Knockout Core Facility).
Embryo fixation and tissue preparation
15.5 to 17.5 days following injection of the transgenic construct, embryos were harvested and fixed overnight in 4% paraformaldehyde. Following cryoprotection in 30% sucrose overnight at 4°C, the tissue was embedded and frozen in OCT. Embryos were sectioned at 8–10 µm.
PCR genotyping
DNA was extracted from tail tissue removed from the embryos upon dissection. Primers and conditions used for genotyping transgenic mice are documented in Table 1.
Table 1.
PCR primers and conditions
| Transgene | Primers | Conditions |
|---|---|---|
| Pdx1:G6FLEnR | (KD71) 5′-GTGCCTCGACCACTTGCTATGAAA-3′ | 96° 5 m; 35 cycles: 96° 1 m, 55° 1 m, 72° 1 m; 72° 5 m |
| (KD73) 5′-TGGTGTGCGTCTGATTGTGGAAAC-3′ | ||
| Pdx1:G6DBDEnR | (KD76) 5′-AAATGCAGCATCTTCACCACCAGC-3′ | 96° 5 m; 35 cycles: 99° 1 m, 55° 1 m, 72° 1 m; 72° 5 m |
| (KD79) 5′-TAGGCAGCCGGATTGAAGCAGTTA-3′ | ||
| Pdx1:G4FLEnR | (KD25) 5′-GGCGTGGAAATATTCTTATTGG-3′ | 96° 5 m; 40 cycles: 96° 1 m, 48° 1 m, 72° 1 m; 72° 5 m |
| (KD31) 5′-TCAAACATATCGAGATTGGGG-3′ | ||
| Pdx1:NLSEnR | (TM9) 5′-TCCTGCCTTTCTCTTTATGGTTACAA-3′ | 95° 5 m; 35 cycles: 95° 30 s, 54° 30 s, 72° 1 m; 72° 5 m |
| (KD22) 5′-GCTAAACTCCAGCAGATCCAC-3′ |
Immunohistochemistry and in situ hybridization
All immunohistochemistry was performed on frozen sections as previously described (Norgaard et al., 2003). Primary antibodies were used as indicated: guinea pig α-insulin 1:1000 (Linco), rabbit α-glucagon 1:1000 (Phoenix Pharmaceuticals), rabbit α-amylase 1:1000 (Sigma), rabbit α-somatostatin 1:200 (Phoenix Pharmaceuticals), rabbit α-ghrelin 1:1000 (Phoenix Pharmaceuticals), Fluorescein-Dolichos biflorus Agglutinin 1:100 (DBA-Lectin, Vector Laboratories), rabbit α-Pdx1 1:1000 (Chemicon), rabbit α-Pax6 1:300 (Chemicon), rabbit α-Nkx6.1 1:800 (Novo Nordisk A/S), goat α-Gata4 1:100 (Santa Cruz Biotechnology), mouse α-Vimentin 1:25 (DakoCytomation). Secondary antibodies were used as indicated: Alexa Fluor 488 and 594 1:400 (Molecular Probes), and HRP-conjugated antibodies according to manufacturer’s instructions (Vectastain ABC Kit).
RNA in situ hybridization was performed as previously described (Prado et al., 2004) using antisense riboprobes transcribed from linearized plasmids. Riboprobes were generated for Gata4, Gata6, Ngn3, and Nkx2.2 from full-length cDNA. A riboprobe targeting mRNA encoding the Drosophila EnR repression domain was generated from the pCS2:EnR plasmid.
Microscopy and imaging
Images were obtained with a Leica DM5000 microscope and an Evolution MP Color camera, and were processed using ImagePro software from Media Cybernetics.
Cell culture
AR42J cells were cultured and maintained according to protocols from American Type Culture Collection (Manassas, VA).
Protein–protein interactions
The yeast two-hybrid assay was performed using the Clontech Matchmaker Two-hybrid system. The Nkx2.2 bait clone was introduced into the pAS2-1 (GAL4 DNA binding domain) vector and the Gata prey plasmids were cloned into the pACT2 (GAL4 activation domain) vector. All assays were performed in the yeast Y190 strain according to manufacturer’s directions. To generate an Nkx2.2 two-hybrid bait construct without autonomous activation activity, we identified and deleted two distinct activation domains of the Nkx2.2 gene: amino acids 20–97 and amino acids 244–273 (Grasch and Sussel, unpublished data). Specifically, full-length Nkx2.2 cDNA was cloned into pBluescript and an _Nco_I site was introduced at the Nkx2.2 start codon by PCR-mediated site-directed mutagenesis. A _Xho_I site was then introduced by PCR-mediated site-directed mutagenesis at base pair 400 and the resulting plasmid was digested with _Xho_I and religated to delete the sequence between the engineered _Xho_I site and the endogenous _Xho_I site at base pair 637 (amino acids 20–97 of the Nkx2.2 protein). The resulting Nkx2.2–_NcoI_Δ20–97 vector was digested with NcoI and PstI to truncate the Nkx2.2 protein at amino acid 244 and delete the 3′ activation domain. (A _Pst_1 site is located at base pair 1067 in the Nkx2.2 cDNA.) The resulting fragment was cloned into the _Nco_I and _Pst_I sites of the pAS2-1 vector (Clontech). Full-length Gata4 and Gata6 coding sequences were cloned in frame into the pACT2 vector (Clontech). pACT:Gata6-N-terminus was created by digesting the pACT:Gata6 full-length with _Sac_I and religating to delete C-terminal Gata6 sequences between the endogenous _Sac_I site and the pACT polylinker site, thereby removing the C-terminal portion of Gata6 containing the zinc fingers. Colony lift filter β-galactosidase assays were performed as recommended by the manufacturer (Clontech).
In vitro pull-down assays were performed using bacterially-generated Maltose-binding protein (MBP)-Nkx2.2 fusion protein and 35S-labeled in vitro translated Gata6. Full-length Nkx2.2 was cloned in frame into the pMAL-c2X vector (New England Biolabs) and transformed into the BL21 E. coli strain (Stratagene). Extracts containing the MBP-Nkx2.2 fusion protein were prepared as described (Sepulveda et al., 1998), and MBP-fusion proteins were immobilized on amylose beads according to the manufacturer’s instructions. Full-length Gata6 was cloned into a pBluescript vector downstream from the T7 promoter. Binding assays were performed with labeled Gata6 protein synthesized in vitro using the TNT® Coupled Reticulocyte Lysate System (Promega) in the presence of 35S-labeled cysteine and 35S-labeled methionine (Amersham) according to the manufacturer’s instructions. Immobilized MBP-Nkx2.2 fusion protein and MBP control protein were incubated with 35S-labeled proteins for 1–2 h at 4°C while rotating in protein binding buffer containing 20 mM HEPES (pH 7.9), 150 mM KCl, 25 mM MgCl2, 1 mM DTT, 10% glycerol, 0.1% Triton X-100, 0.1% NP-40 (IGEPAL CA-630, Sigma). Beads were washed three times in protein binding buffer. Bound proteins were eluted from the beads into SDS-sample buffer by boiling and resolved by SDS-PAGE. Gels were dried onto Whatman paper and analyzed for autoradiography using a Storm phosphoimager (Molecular Devices).
For co-immunoprecipitation assays, COS-7 cells were transfected with pcDNA3:Gata6 and pcDNA3:Myc-Nkx2.2 using FuGene 6 (Roche) following the manufacturer’s instructions. Cells were harvested 2–3 days post-transfection by incubating in PBS lysis buffer containing either 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100 and protease inhibitors (Complete mini, Roche) in PBS or EBC200 buffer (50 mM Tris–HCl (pH 8), 0.2 M NaCl, 0.5% NP-40, 0.1 M NaF, 0.2 mM Na3VO4, and protease inhibitors) on ice for 30 min. The lysates were collected and cleared by centrifugation at 14,000 × g for 10 min at 4°C. Total protein concentration in the lysates was determined using the BioRad DC protein assay. 500 µg of lysate total protein was immunoprecipitated with protein G-sepharose beads (Sigma) overnight with 1 µg α-Gata6 antibody (rabbit polyclonal, Santa Cruz Biotechnology) or 1 µg α-myc antibody (mouse monoclonal, Sigma). The beads were pelleted and washed three times in either cold PBS (if PBS lysis buffer used) or cold NETN (if EBC200 used; 20 mM Tris (pH 8), 0.1 M NaCl, 1 mM EDTA, 0.5% NP40) and then resuspended in SDS-loading buffer. Bound proteins were eluted by boiling, resolved by SDS-PAGE, and Western blotted. The blots were blocked and incubated in PBS containing 3% milk and α-Nkx2.2 antibody 1:100 (goat polyclonal, Santa Cruz) for 1–4 h. After two 10-min washes in PBS, the blots were incubated in alkaline phosphatase-conjugated secondary antibody 1:2500 (Sigma) in PBS/3% milk for 1–2 h. For detection of bound proteins, the blots were washed twice in PBS for 10 min, rinsed in water, and then incubated in BM Purple (Roche).
Luciferase assays
Luciferase assays were performed as described (Kathiriya et al., 2004).
Results
Gata6 and Gata4 are expressed in the embryonic mouse pancreas
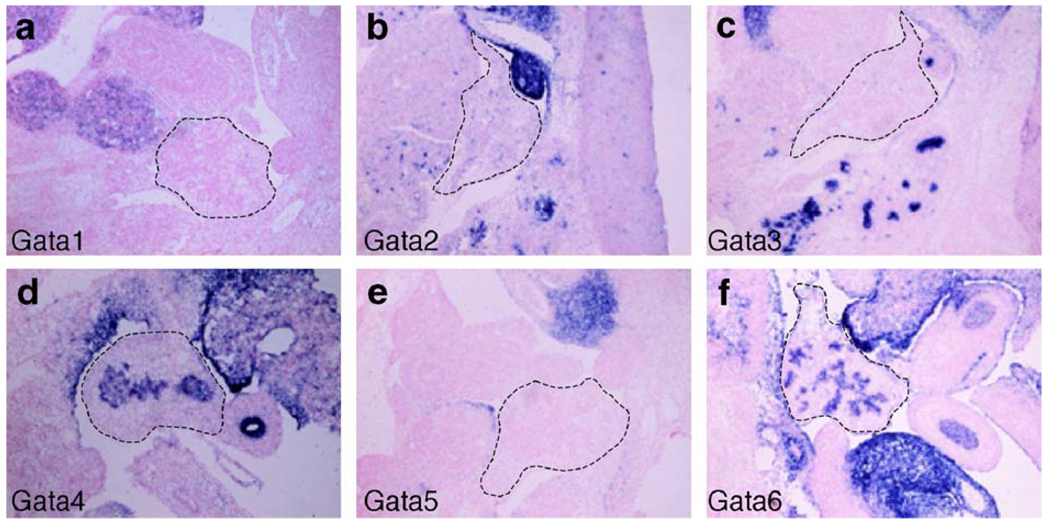
To begin to ascertain whether Gata factors are involved in the development of the embryonic mouse pancreas, we first determined which of the known mouse Gata family members are expressed during pancreas development. RNA in situ hybridization using RNA probes specific for each of the six Gata transcription factors was performed on frozen sections of e12.5 embryos. Our analysis shows that mRNA for Gata4 and Gata6 is expressed in the developing pancreas (Fig. 1). mRNA could not be detected for Gata1, Gata2, Gata3, or Gata5 in the pancreas, although we detected strong expression of these genes in other organs including the kidney, liver, and brain (Fig. 1 and data not shown). We performed RNA in situ hybridization at additional developmental stages between e9.5 and e16.5. This time period encompasses both initial pancreas specification (~ e9.5) and the secondary transition (~ e12.5–e15.5), a stage of extensive expansion and differentiation of the pancreatic epithelium. Gata4 and Gata6 expression is maintained throughout these stages of pancreas development (Fig. 1–Fig. 3 and data not shown). To confirm the RNA in situ hybridization results and rule out the possibility that Gata1, Gata2, Gata3, and Gata5 are expressed at levels below detection by non-radioactive RNA in situ hybridization, we performed RT-PCR on an e12.5 pancreas cDNA library using gene-specific primers for each of the six Gata factor family members. In addition, we assessed expression data previously generated from Affymetrix microarray analyses of cDNA generated from e12.5 pancreas RNA (Prado et al., 2004). Consistent with the RNA in situ hybridization expression data, Gata4 and Gata6 mRNA was strongly detected in the e12.5 pancreas, but Gata1, Gata2, Gata3 or Gata5 mRNA was not present (data not shown).
Fig. 1.
In situ hybridization of e12.5 tissue using antisense RNA probes for each Gata transcription factor family member reveals that only Gata4 and Gata6 are expressed in the embryonic mouse pancreas (d, f). Gata1, Gata2, Gata3, and Gata5 are not expressed in the developing mouse pancreas (a–c, e). Pancreas outlined with dashed line.
Fig. 3.
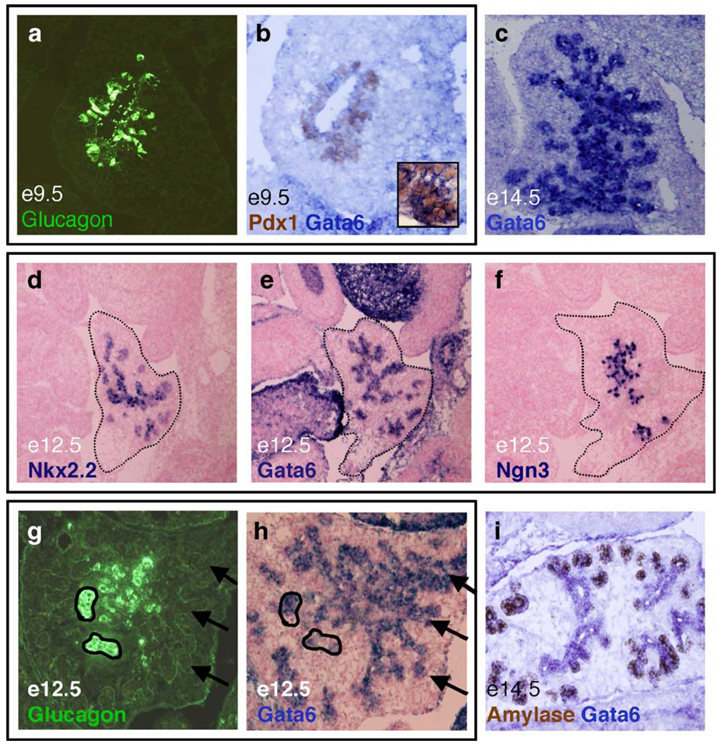
Gata6 mRNA expression analysis. At e9.5, Gata6 mRNA (blue) is expressed globally throughout the pancreatic epithelium, along with Pdx1 (brown, b) and glucagon (green, a); a and b are adjacent sections. By e14.5, Gata6 mRNA is highly expressed in the pancreatic epithelium (c). At e12.5, Gata6 mRNA expression overlaps with the endocrine transcription factors Nkx2.2 and Ngn3 (d–f). Images d–f represent near adjacent sections. At e12.5, Gata6 mRNA (blue, h) is expressed in some, but not all cells that express glucagon (green, g); g and h represent the same section; Gata6 in situ analysis was performed first, followed by immunofluorescence with the α-glucagon antibody. Gata6 mRNA (blue) is localized to the ducts at e14.5 and becomes extinguished from amylase-expressing (brown) exocrine cells as they bud from the duct (i).
Gata4 and Gata6 expression becomes restricted to distinct pancreatic lineages at later developmental stages
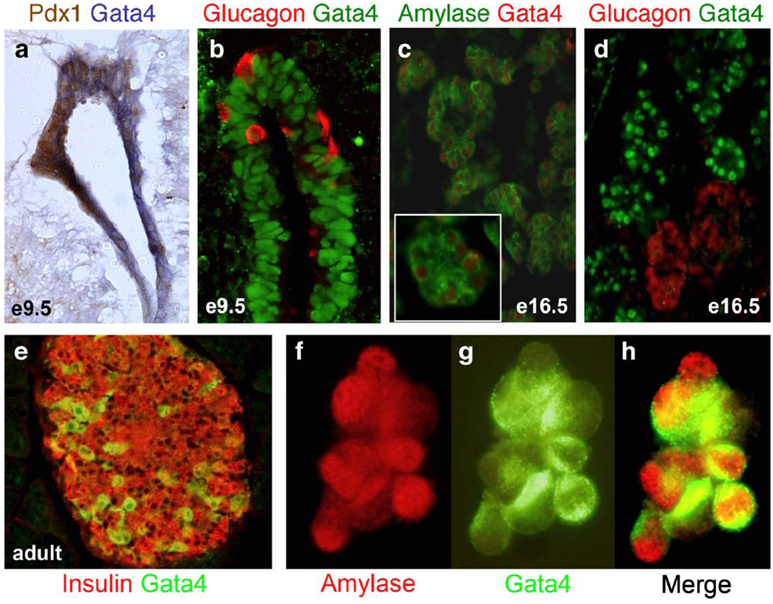
To further characterize the spatial and temporal expression of Gata4 and Gata6 within the developing pancreas, we compared expression of Gata4 mRNA, Gata4 protein, and Gata6 mRNA with the expression of pancreatic cell type-specific markers at different embryonic stages. We were unable to analyze Gata6 protein expression because a reliable Gata6 antibody is not available. In contrast to previous studies (Ketola et al., 2004), our analysis shows that both Gata4 and Gata6 are expressed throughout the pancreatic epithelium at e9.5, when the pancreas first begins to form. Gata4 mRNA is expressed from the onset of dorsal bud evagination and overlaps extensively with Pdx1 (Fig. 2a). In addition, immunofluorescence studies demonstrate that Gata4 protein is detected throughout the pancreatic epithelium at e9.5 but is absent from glucagon-expressing cells (Fig. 2b), suggesting that Gata4 is excluded from endocrine cells at the earliest stage of pancreas development. Gata4 mRNA is observed in most pancreatic epithelial cells by e11.5 (data not shown), but expression progressively moves toward the tips of the branching ducts (i.e., the presumptive acini) as development proceeds (Fig. 2c, inset). By e16.5, Gata4 protein expression is restricted to the exocrine pancreas where it is observed in the nucleus of cells expressing amylase and carboxypeptidase A (Fig. 2c and data not shown). However, Gata4 is excluded from both the ducts and developing islets, and is not found in cells expressing glucagon (Fig. 2d and data not shown). At all stages of embryonic pancreas development, the pattern of Gata4 protein expression is consistent with the pattern of Gata4 mRNA expression and Gata4 protein is localized to the nucleus. By e18.5, a subset of acini continues to express Gata4 (data not shown). Our in vivo results are supported by immunofluorescence analysis in the AR42J exocrine pancreas cell line, confirming that Gata4 is present in cells also expressing the exocrine enzyme amylase (Figs. 2f–h). Interestingly, in the adult pancreas, Gata4 is expressed in a subset of α and β cells of the adult islet, but is no longer detectable in the exocrine tissue (Fig. 2e and data not shown). Nuclear and non-nuclear expression of Gata4 can be detected in the adult islet.
Fig. 2.
Gata4 mRNA and protein expression analysis. At e9.5, Gata4 RNA (blue, a) and protein (green, b) are expressed globally in the pancreatic epithelium along with Pdx1 (brown; a) and glucagon (red; b). By e16.5, Gata4 expression becomes restricted to the exocrine pancreas (red, c; green, d), where it is coexpressed with amylase (green, c) but not with glucagon (red, d). Gata4 is expressed (red) in the nucleus of cells that express amylase (green) in the cytoplasm (c, inset). Gata4 (green) is co-expressed with a subset of insulin cells (red) in the adult islet (e). Gata4 (green) is coexpressed with amylase (red) in the AR42J exocrine cell line (f, g, h).
Similar to Gata4, Gata6 mRNA is expressed globally in the e9.5 dorsal pancreatic bud and is co-expressed with Pdx1 (Fig. 3b). In contrast to Gata4, Gata6 mRNA is expressed in cells also expressing glucagon at this stage (Figs. 3a and b). As development proceeds, Gata6 expression becomes restricted to cells of the endocrine pancreas and the ductal epithelium (Fig. 3c). By e12.5, Gata6 is expressed throughout the differentiating pancreatic epithelium in regions similar to endocrine transcription factor Nkx2.2 (Figs. 3d and e) and a marker of endocrine progenitor cells, Ngn3 (Figs. 3e and f). During the secondary transition, glucagon is coexpressed in some, but not all cells that express Gata6 (Figs. 3g and h). Between e14.5 and e15.5, Gata6 mRNA is expressed strongly in the pancreatic epithelial ducts and becomes down-regulated in cells differentiating into acini (Fig. 3i).
Nkx2.2 physically interacts with Gata6, but not Gata4
Gata4 and Gata6 are known to interact with tissue-specific transcription factors to regulate genes expressed during the differentiation of numerous embryonic tissues. We originally pursued our analysis of the Gata factors in the pancreas due to the close link between Gata factors and Nkx2 factors in the development of several different organs (Stennard et al., 2003). Early in pancreas development, expression of Gata4 and Gata6 overlaps with Nkx2.2, a transcription factor known to be essential for normal pancreas development (Prado et al., 2004; Sussel et al., 1998). Later in development, only Gata6 is coexpressed with Nkx2.2 in endocrine cells of the islet. To determine whether Gata4 and/or Gata6 are able to interact with Nkx2.2, we performed a series of yeast two-hybrid assays in which a derivative of Nkx2.2 that lacks its two activation domains (see Materials and methods) was fused to the GAL4 DNA binding domain and used as bait (BD-Nkx2.2). Chimeric proteins containing full-length Gata4 and Gata6 fused to the GAL4 activation domain (AD) were tested for their ability to co-activate GAL4 UAS-lacZ with BD-Nkx2.2. As shown in Supplemental Fig. 1a, BD-Nkx2.2 alone or in combination with AD-Gata4 is incapable of activating GAL4 UAS-lacZ, whereas coexpression of BD-Nkx2.2 with AD-Gata6 results in the activation of lacZ expression. Furthermore, coexpression of BD-Nkx2.2 with a C-terminal truncation of AD-Gata6 (pACT:Gata6-N-terminal), which deletes the zinc finger domain and the C-terminal extension required for interaction between Gata4 and Nkx2.5 (Durocher et al., 1997), prevented the interaction and failed to activate lacZ expression. These results suggest that Gata6, but not Gata4, interacts with Nkx2.2.
We confirmed the physical association of Gata6 and Nkx2.2 using in vitro pull down assays and co-immunoprecipitation assays of mammalian cell extracts. Full-length Nkx2.2 was fused with maltose binding protein (MBP) to generate MBP-Nkx2.2. Immobilized MBP-Nkx2.2 retained 35S-labeled Gata6, whereas immobilized MBP alone did not (Supplemental Fig. 1b). Co-immunoprecipitation was performed on whole cell extracts from COS-7 cells transfected with Gata6 and myc-tagged Nkx2.2, individually or in combination. Immunoprecipitation of extracts using α-Gata6 antibody or α-myc antibody followed by Western blotting for Nkx2.2 confirmed that Nkx2.2 interacts with Gata6 in vivo (Supplementary Fig. 1c). These results suggest that Nkx2.2 and Gata6 may cooperate in the regulation of early pancreas or islet specific gene expression, similar to Nkx2.5 and Gata4 cooperativity in heart-specific gene expression (Durocher et al., 1997; Sepulveda et al., 1998). We are currently attempting to identify direct targets of Nkx2.2 and Gata6 in the islet.
Production of Gata dominant repressor transient transgenic mice
The dynamic expression patterns of Gata4 and Gata6 in the pancreas, combined with the physical interaction between Nkx2.2 and Gata6, suggest that Gata4 and Gata6 may have overlapping functions during the initial specification of the embryonic mouse pancreas, but may be uniquely involved in the differentiation of endocrine cell types (Gata6) or exocrine cell types (Gata4). To investigate whether Gata factors play a functional role in the developing mouse pancreas, we utilized a dominant repressor transient transgenic mouse system modeled after experiments assessing the role of Gata6 in lung development (Koutsourakis et al., 2001; Liu et al., 2002b; Yang et al., 2002). As illustrated in Supplemental Fig. 2 and described in Materials and methods, we designed two Gata6 dominant repressor transgenes (Pdx1:G6FLEnR and Pdx1:G6DBDEnR), one Gata4 dominant repressor transgene (Pdx1:G4FLEnR), and one control transgene (Pdx1:NLSEnR). Based on the previous studies, we predicted that fusion of the Engrailed repression domain to full-length Gata4 or Gata6 would repress genes normally activated by Gata4 and Gata6, respectively. Further, since the DNA binding domains of Gata4 and Gata6 are highly conserved at the DNA and protein levels, we hypothesized that fusion of the Engrailed repressor domain to the Gata6 DNA binding domain would result in repression of all Gata4 and Gata6 target genes. The control transgene, Pdx1:NLSEnR, was not expected to affect pancreas development, similar to the findings of Yang et al. (Yang et al., 2002) who expressed an EnR-only control transgene in the lung epithelium without phenotypic consequences. Expression of each fusion protein is driven by the 4.5 kb Pdx1 promoter element, which is active starting at e8 in the developing endoderm and becomes restricted to the developing pancreas around e9 (Gannon et al., 2000; Norgaard et al., 2003). The Pdx1 promoter is well characterized, recapitulates endogenous Pdx1 expression which is largely pancreas-specific, and mirrors the early pancreas expression pattern of Gata4 and Gata6 (Apelqvist et al., 1997; Li and Edlund, 2001; Stoffers et al., 1999). While later stage expression of Pdx1 is predominantly restricted to the endocrine pancreas (similar to Gata6), Pdx1 is also expressed at lower levels in exocrine cells and is required for proper differentiation of these cell types (Hale et al., 2005).
Transient transgenic mice carrying each of the Gata derivatives or the control were generated after several attempts to establish stable mouse lines proved unsuccessful. We chose to analyze transient transgenic embryos due to the possibility that the Gata transgenes were significantly disrupting pancreas development, resulting in neonatal lethality. Pregnant mice were sacrificed and embryos were harvested between 15.5 and 17.5 days following pronuclear injection. For each of the four transgenes, phenotypic analysis was performed on transgenic and wildtype littermates. For this study, we generated twenty-five embryos which carried a transgene and assessed transgene expression by RNA in situ analysis on sectioned pancreatic tissue. Of these, two Pdx1:G6FLEnR, one Pdx1:G6DBDEnR, one Pdx1:NLSEnR (control), and eight Pdx1:G4FLEnR embryos expressed the transgene. Three additional Pdx1: G6FLEnR embryos exhibited a dramatic “absence of pancreas” phenotype (see below); assessment of transgene expression in the absent tissue was not possible. Injected embryos that did not contain the transgene were used as littermate controls.
Analysis and classification of Gata dominant repressor transient transgenics
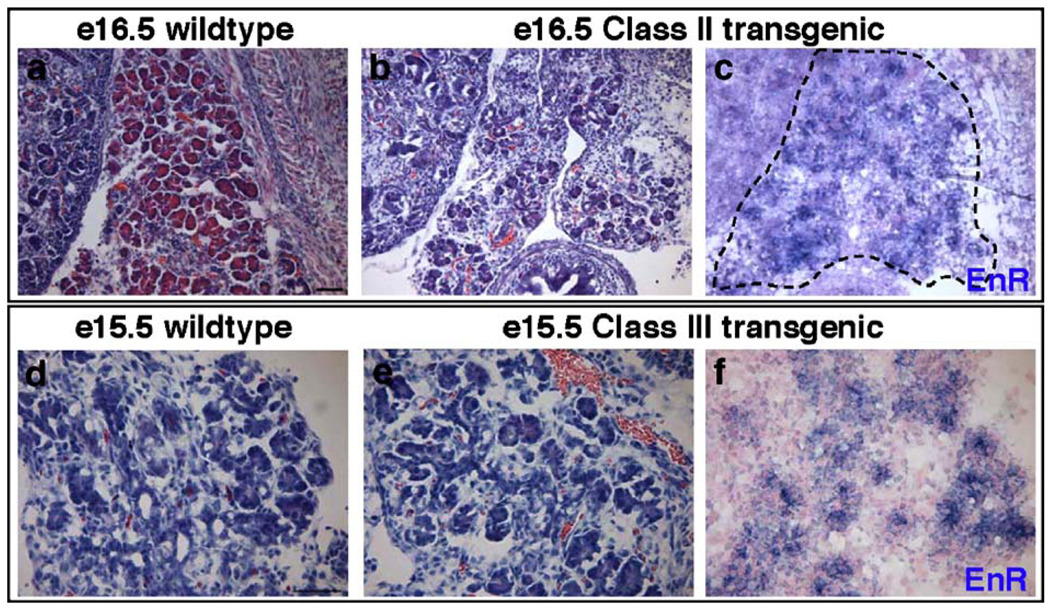
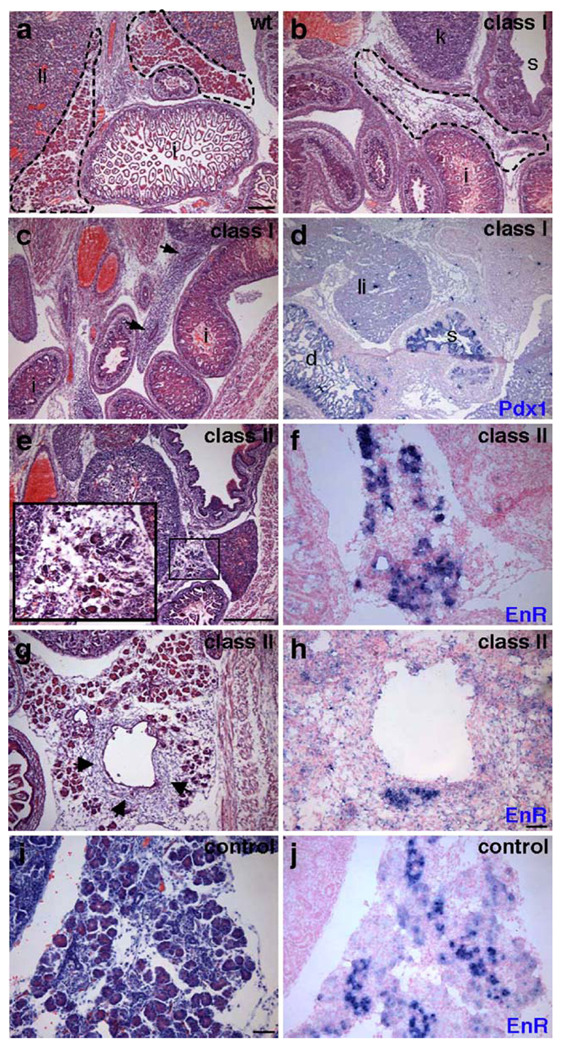
To assess whether the Gata dominant repressor transgenes affect development of the embryonic mouse pancreas, we first performed histological analysis of the late stage embryos. Frozen sections from transgenic and wildtype embryos were stained with hematoxylin and eosin (H&E). As summarized in Table 2, we observed three distinct pancreatic phenotypes in the transgenic embryos: absence of pancreas (hereafter referred to as Class I); severely disrupted pancreas morphology (Class II); and little to no observable phenotype (Class III). All transgenic embryos exhibited overall normal morphology of non-pancreatic tissue. Three Pdx1:G6FLEnR embryos exhibited a Class I defect, completely lacking a pancreas or any recognizable pancreatic rudiment. Two Pdx1:G6FLEnR embryos and the Pdx1: G6DBDEnR embryo had Class II defects. A Class III phenotype was not observed for any Gata6 transgenic embryo. Interestingly, only one Pdx1:G4FLEnR transgenic embryo displayed disrupted islet morphology, while seven Pdx1:G4FLEnR transgenic embryos had no observable phenotype despite strong transgene expression (Fig. 7). The Pdx1:NLSEnR embryo had normal pancreas morphology and no apparent phenotype (Fig. 4i and data not shown).
Table 2.
Summary of transgenic phenotypes
| Phenotype | Class I | Class II | Class III |
|---|---|---|---|
| Pdx1:G6FLEnR | 3 | 2 | 0 |
| Pdx1:G6DBDEnR | 0 | 1 | 0 |
| Pdx1:G4FLEnR | 0 | 1 | 7 |
Fig. 7.
Analysis of Gata4 transgenic embryos. H&E staining (a, b, d, e). RNA in situ analysis of transgene expression (c, f). One Pdx1:Gata4FLEnR transgenic embryo exhibited a Class II phenotype (b) in comparison to its e16.5 wildtype littermate (a). The Class II transgenic embryo expressed the Gata4-repressor transgene at significant levels throughout the pancreas (c). Seven Pdx1:Gata4FLEnR transgenic embryos were morphologically normal (e) compared to their wildtype littermates (d), despite expression of the Gata4-repressor transgene (f). Scale bar 100 µm.
Fig. 4.
Histological analysis of Gata6 transgenic mice. H&E stained sections of an e17.5 Pdx1:G6FLEnR wildtype littermate (a) and two e17.5 Pdx1:G6FLEnR transgenic littermates with a Class I (no pancreas) phenotype (b, c). One embryo exhibited morphological evidence of endodermal-like epithelium (c, arrows); however, this tissue did not express pancreatic markers. In situ hybridization using a Pdx1 RNA probe shows that duodenum formation is not disrupted in Class I transgenic embryos (d). Endocrine, exocrine, and ductal pancreas morphology is severely disrupted in Class II Pdx1:G6FLEnR and Pdx1: G6DBDEnR transgenic embryos (e, g). Excess mesenchymal tissue surrounds the enlarged duct of one Class II transgenic embryo (g, arrows). Morphology of the pancreas is normal in Pdx1:NLSEnR control embryos (i). In situ hybridization using an RNA probe for Engrailed reveals Gata6-repressor transgene expression in embryos with a Class II phenotype (f and h) and expression of the transgene in Pdx1:NLSEnR controls (j). Scale bar is 500 µm for panels a–d and i–j. Scale bar is 100 µm for panels e–h. 1, liver; i, intestine; k, kidney; s, stomach; d, duodenum. Pancreas outlined with dashed line; dashed line in b indicates expected location of pancreas in Class I transgenic.
The pancreas is absent in Class I Gata6 repressor transgenics
Following initial analysis and classification of the transgenic embryos, a more thorough investigation of the Pdx1:G6FLEnR and Pdx1:G6DBDEnR phenotypes was pursued. Strikingly, in three independent e17.5 Pdx1:G6FLEnR transgenic embryos, pancreas formation appeared to be completely disrupted (Class I phenotype; Figs. 4b–d). Although there was no morphological evidence of the pancreas, it was important to determine whether any pancreatic rudiments or cell types had formed, as seen in Pdx1 null mice (Offield et al., 1996), or whether there were ectopic islet cells, as observed in Ptf1a null mice (Krapp et al., 1998). Tissue sections throughout each of the three Class I Gata6 transgenics were analyzed with a panel of diagnostic pancreatic markers to ascertain whether pancreatic rudiments were present anywhere within these Pdx1:G6FLEnR embryos. Using immunohistochemical analysis, we were unable to detect expression of amylase, insulin, glucagon, somatostatin, PP, or ghrelin (data not shown). Furthermore, we were unable to detect pancreatic expression of the markers Pdx1 and Ptf1a (data not shown), although Pdx1 was still expressed in the duodenum and stomach (Fig. 4d). In addition, we did not detect pancreatic expression of Ngn3, Nkx2.2, Nkx6.1, or Pax6, although these transcription factors were still expressed in non-pancreatic tissues (data not shown). Although we cannot rule out the possibility that the pancreas initially formed and subsequently degenerated, our analyses suggest that no pancreatic rudiments or cell types have formed in the Class I Pdx1:G6FLEnR embryos.
Pancreas development is disrupted in the second class of Gata6 repressor transgenics
In another subset of Pdx1:G6FLEnR and Pdx1:G6DBDEnR embryos we observed a second phenotypic class in which there was a severe loss of differentiated pancreas cell types although a small amount of pancreatic tissue remained (Class II phenotype; Figs. 4e and g). Class II transgenic embryos form both the dorsal and ventral pancreatic domains; however, there is less overall pancreatic tissue and islets are not properly formed (Figs. 5b, d, f and h). In addition, fewer acini are present and some embryos display a disruption of acinar cell morphology that correlates with a reduction in enzyme expression (Fig. 5d, arrow). Interestingly, a phenotypic difference was not observed between Class II Pdx1:G6FLEnR embryos and the Pdx1:G6DBDEnR embryo. In addition, the phenotype was similar in each of the Class II embryos and we did not observe a gradient of phenotypes between the Class I and Class II phenotypes, suggesting that Gata6 may function at two discrete stages in pancreatic development (see Discussion).
Fig. 5.
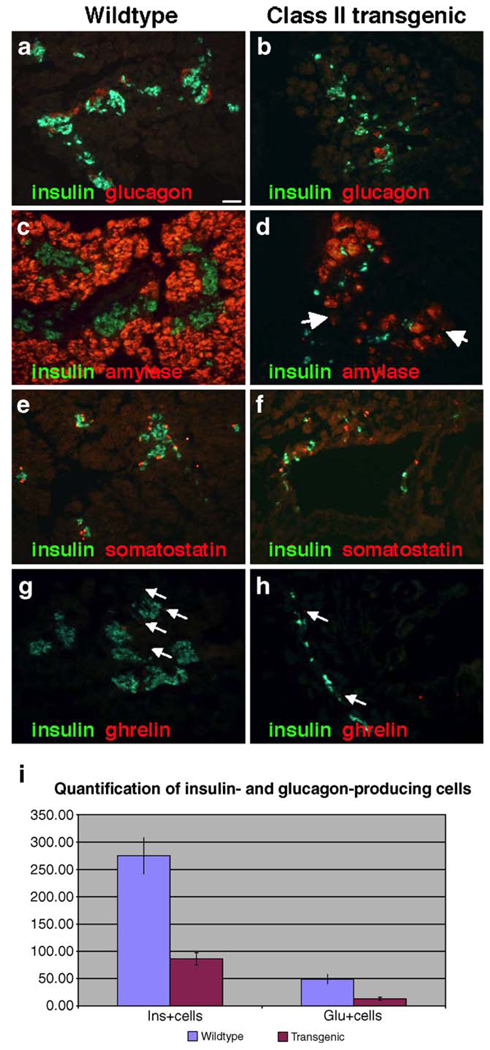
Immunohistochemical staining for pancreatic hormones and enzymes. In comparison to the pancreas of the wildtype littermate (a, c, e, and g), Class II transgenic embryos have reduced numbers of cells expressing insulin (b, d, f, and h), glucagon (b), amylase (d, arrows) somatostatin (f) and ghrelin (h, arrows). Scale bar 100 µm. There was a > 3-fold reduction in the number of insulin-expressing cells (p<0.001) and an approximately 4-fold reduction in the number of glucagon-expressing cells (p<0.001) when comparing wildtype and Class II littermates (i). Insulin-positive and glucagon-positive cells were counted on ten sections each from two wildtype and two Class II transgenic embryos.
To explore the possibility that the less severe phenotype associated with the Class II Gata6 transgenic mice is due to weaker or mosaic expression of the transgene, we performed in situ hybridization using an RNA probe for the Engrailed portion of the transgene in these embryos. Strong expression of the transgene was detected in differentiated regions of the pancreas in all Pdx1:G6FLEnR and Pdx1:G6DBDEnR embryos with a Class II phenotype (Figs. 4f and h, and data not shown). Similar to the Class I embryos, transgene expression could no longer be detected in undifferentiated regions of the pancreas, regions that had differentiated into exocrine cells, or tissue that appeared more mesenchymal in structure (Figs. 4g and h). It remains possible that timing or initiation of transgene expression may be affecting the phenotype; however, this cannot be explored in transient transgenic embryos.
Class II Gata6 repressor embryos have fewer differentiated cells
Histological analysis of the Class II Gata6 embryos suggested that few differentiated pancreatic structures had formed. To assess the degree to which cell differentiation was disrupted in these mice, we performed immunofluorescence to detect the presence and localization of exocrine enzymes and endocrine hormones. Exocrine tissue is clearly present; however, the relative numbers of acini clusters are reduced concomitant with a reduction in amylase expression (Fig. 5d). Furthermore, there appear to be acini clusters that express reduced levels of amylase (Fig. 5d, arrows). In addition, these embryos lack the well-formed islets normally seen at this stage of development, and have a significant reduction in the number of cells expressing insulin (Figs. 5b, d, f, h, and i), glucagon (Figs. 5b and i), somatostatin (Fig. 5f), and ghrelin (Fig. 5h). The hormone-producing cells that are present tend to be scattered throughout the pancreas and around the ductal tissue. To quantify the loss of insulin and glucagon cells in the Class II embryos, we quantified insulin-positive and glucagon-positive cells from two independent Class II transgenic embryos as well as their wildtype littermates. This analysis revealed a 3- to 4-fold reduction in the number of insulin-expressing and glucagon-expressing cells in the Gata6 transgenic embryos (Fig. 5i). Traditional morphometric analysis of the islet was not possible due to the severe disruption of general pancreas and islet morphology.
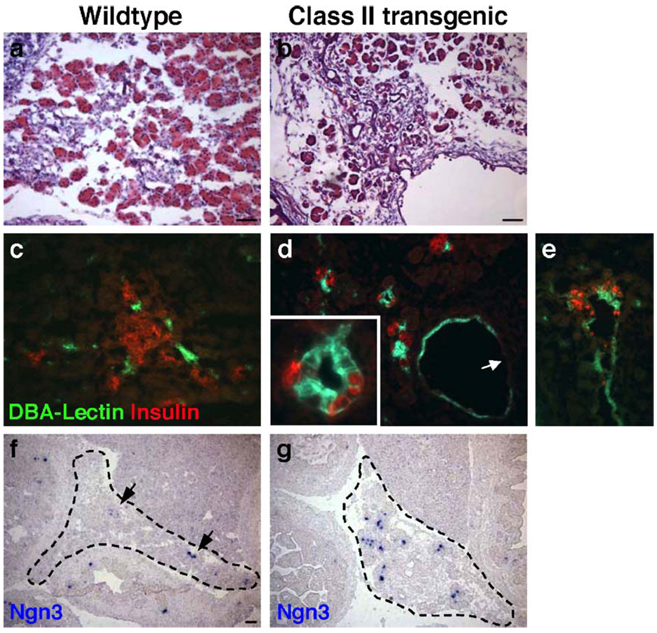
In comparison to wildtype littermates, Class II Gata6 transgenic embryos also display aberrant development of the ductal epithelium (Figs. 6b, d and e; Figs. 4g and h). Ducts in a Class II embryonic pancreas are generally larger, greater in number, and morphologically abnormal compared to ducts in a typical e17.5 pancreas. To determine whether the ducts of Class II embryos were epithelial in nature, we performed immunofluorescence using a fluorescein-conjugated antibody against DBA-Lectin, a marker of pancreatic ductal epithelium. In the wildtype littermate, DBA-Lectin marks discrete areas of ductal epithelia that permeate throughout the pancreas (Fig. 6c). By contrast, in the Class II embryos, DBA-Lectin delineates greatly enlarged ducts that are epithelial in nature (Figs. 6d and e). Interestingly, DBA-Lectin is not expressed in all duct cells in these embryos (Fig. 6d, arrow). In some cases, the epithelia of these oversized ducts are surrounded by a stratified cell type (Fig. 4g, arrows) that express the mesenchymal marker, Vimentin (data not shown). In wildtype embryos, pancreatic mesenchyme is significantly reduced by e17.5.
Fig. 6.
Analysis of aberrant duct development. H&E staining (a, b); immunofluorescence staining of insulin (red) and the ductal marker DBA-Lectin (green, c–e). In comparison to the wildtype littermate (a, c), Class II transgenic embryos display aberrant and enlarged duct development (b, d, e). Some cells of the enlarged ducts do not stain for the ductal marker DBA-Lectin (d, arrow). Of the few endocrine cells that differentiate in the Class II embryos, many are localized to the ducts (d and e; inset in d). RNA in situ analysis of Ngn3 (f, g). Class II embryos display increased numbers of Ngn3+ cells (g) in comparison to wildtype littermates (f). Ngn3 staining was performed on two sections each from two wildtype and two Class II transgenic embryos. Scale bar 100 µm.
In Class II embryos, we also observe a significant reduction in the overall numbers of endocrine cells and a large proportion of the cells that remain are localized to the ducts (Figs. 6d and e; Fig. 5f). Occasionally, cells that express the ductal marker DBA-Lectin also express hormones, such as insulin (Fig. 6d inset). Potentially, these cells represent an endocrine progenitor cell population. To determine whether Class II embryos maintain a larger pool of endocrine progenitors than wildtype littermates, we performed RNA in situ hybridization to analyze expression of Ngn3, a transient marker of all endocrine progenitor cells. By e17.5 in a wildtype embryo, the endocrine progenitor population has decreased to a low level and few cells express Ngn3 (Fig. 6f, arrows). In comparison, Class II embryos show an apparent increase in the number of Ngn3-expressing endocrine progenitor cells, despite a clear reduction in pancreatic mass (Fig. 6g). However, due to loss of overall pancreatic tissue and significant disruption to pancreas morphology in the Class II embryos, it is unclear whether this increase is statistically significant.
To further investigate the differentiation state of the pancreatic islet cell types in Class II Gata6 embryos, we analyzed expression of the transcription factors Pdx1, Nkx6.1, and Pax6. Pdx1 is normally expressed in endocrine progenitors along with mature β and δ cells (Guz et al., 1995; Ohlsson et al., 1993). Nkx6.1 is expressed in duct cells as well as terminally-differentiated β cells, while Pax6 is expressed in all mature endocrine cell types (Sander et al., 1997; Sander et al., 2000; St-Onge et al., 1997). Consistent with the reduction in total pancreatic islet tissue, there is a reduction in the numbers of cells expressing these transcription factors in the transgenic embryos (Supplementary Figs. 3b, d, and f). In addition, cells expressing these transcription factors in the Class II embryos are often localized to the periphery of the ducts (Supplementary Figs. 3d and f). For example, the majority of Nkx6.1-expressing cells are within the ductal epithelium in Class II embryos (Supplementary Fig. 3d, inset,). Since Gata factors are known to co-regulate one another, we also analyzed Gata4 expression in Class II Gata6 embryos by performing immunofluorescence using an α-Gata4 antibody. Although the total number of exocrine cells is reduced, Gata4 expression is maintained in the nucleus of these cells (data not shown).
The loss of pancreatic mass in the Class II Gata6 transgenics could be due to a block in cell differentiation, a reduction in cell proliferation, and/or an increase in cell death. To determine if there are any abnormalities in cell death or proliferation, we compared levels of apoptosis and cellular proliferation between transgenic and wildtype embryos. We observed no change in the level of cell death in Class II or Class I embryos, as determined by TUNEL assay at e17.5 (data not shown); however, these embryos have such an extreme loss of pancreatic tissue by e17.5 that it is possible there is substantial cell death at an earlier period, which is difficult to assess in the transient transgenic system. We also performed immunohistochemical analysis using an antibody against phospho-Histone H3, which marks cells in M phase, and determined that there is no significant difference in the proliferative state of pancreatic cell types between the wildtype and Class II transgenic littermates (data not shown). The absence of obvious proliferative defects or increased cell death at e17.5 supports our previous findings that suggest Pdx1:G6FLEnR and Pdx1:G6DBDEnR may cause a disruption of pancreatic cell differentiation, however, we cannot rule out that increased cell death or decreased proliferation occurred at time points prior to our analysis at e17.5.
The majority of Pdx1:G4FLEnR transgenic embryos do not display disrupted pancreas morphology or development
Both Gata4 and Gata6 are expressed globally throughout the pancreatic epithelium at the earliest stages of pancreas development, but their expression becomes restricted to mutually exclusive domains at later stages of development. Consequently, it was possible the Pdx1:G4FLEnR transgene would produce phenotypes that affect early pancreas development as well as differentiation of exocrine cell types. We analyzed eight Gata4 transgenic embryos along with their wildtype littermates. RNA in situ hybridization to detect the Engrailed portion of the transgene demonstrated that all eight Gata4 transgenic embryos strongly express the Gata4 dominant repressor transgene (Figs. 7c, f, and data not shown). Surprisingly, in contrast to the Gata6 transgenics, most of the Gata4 transgenic embryos appear to be phenotypically normal (Fig. 7e) with no obvious defects in endocrine hormone, exocrine enzyme, and pancreatic transcription factor expression (data not shown). To ensure that the G4FLEnR construct was functional, we assessed its ability to interfere with Gata4 transcriptional activation. Since Gata4 has been shown to be essential for the activation of the a myosin heavy chain gene (αMyHC) (Molkentin et al., 1994; Kathiriya et al., 2004) we determined that the G4FLEnR fusion protein is able to effectively block Gata4-dependent activation of an αMyHC-luciferase reporter in HeLa cells (Supplementary Fig. 4). This suggests the G4FLEnR fusion protein is functional in an in vitro reporter assay, however we cannot rule out that the Gata4 dominant repressor transgene is less effective in the pancreas in an in vivo context. One Gata4 transgenic embryo exhibited a phenotype similar to the Class II Gata6 transgenic embryos: there is a reduction in the mass of exocrine and endocrine tissue and a corresponding loss of hormone and enzyme expression (Fig. 7e and data not shown). This phenotype may be due to off-target effects of the G4FLEnR transgene. Conditional knockout analysis of Gata4 in the pancreas will help resolve this issue.
Discussion
Gata factor family members are required for the proper development of many essential cell types, tissues, and organs in vertebrates (Patient and McGhee, 2002). The objectives of this study were to determine which Gata factors are expressed in the embryonic mouse pancreas and whether Gata factors play a functional role during pancreas development. Our analyses demonstrate that Gata4 and Gata6 are expressed at the onset of pancreas development throughout the pancreatic epithelium. As development proceeds, Gata6 becomes restricted to the endocrine pancreas and ducts, while Gata4 becomes restricted to the exocrine pancreas. Other Gata factor family members could not be detected in the developing pancreas. Using a dominant repressor transgenic approach that has proven successful in the analysis of Gata function in lung development, we determined that Gata6 plays a predominant role in the initial specification of the pancreas and in pancreatic cell type differentiation. Consistent with this finding, we demonstrated that Gata6 physically interacts with the essential pancreatic transcription factor, Nkx2.2. These studies are the first to demonstrate the importance of Gata6 function in the vertebrate pancreas.
Gata4 and Gata6 are dynamically expressed in the developing pancreas
The results of the Gata4 and Gata6 expression analysis performed in this study are largely consistent with a previous study that reported Gata6 expression in the endocrine pancreas and Gata4 expression in the exocrine pancreas during mouse embryogenesis (Ketola et al., 2004). However, we also revealed overlapping expression domains for Gata4 and Gata6 in the early pancreatic epithelium. In support of our findings, Chiang and Melton (Chiang and Melton, 2003) reported Gata4 and Gata6 expression in RNA isolated from total e12.5 pancreas and in almost every individually isolated e10.5 pancreatic cell assayed. Gata5 was detected in a small number of the individually isolated e10.5 pancreatic cells; however, we were unable to detect significant Gata5 pancreatic expression in any of our assays. Both our results and the Ketola study conflict with the study of Ritz-Laser et al. (Ritz-Laser et al., 2005) that suggests Gata4 is expressed in the developing endocrine pancreas and activates glucagon gene expression. We were unable to detect coexpression of Gata4 and glucagon at any time during embryonic development. We do observe Gata4 expression in the islet of adult mice, where it is coexpressed with a subset of glucagon-expressing cells. This may indicate that Gata4 regulates glucagon in the adult islet. Consistent with our expression data, we do not observe an effect on glucagon expression in the embryonic islet for the majority of the embryos expressing a Gata4-repressor transgene. We are currently following up these studies using a conditional allele of Gata4 to specifically delete Gata4 from the embryonic a cells and assess how this affects glucagon gene expression.
Disruption of Gata6, but not Gata4, function causes abnormal pancreatic development
The strong and dynamic expression patterns of Gata4 and Gata6 suggested that they may both function in pancreatic development. We predicted that Gata4 and Gata6 might function redundantly at early stages of pancreas formation when their expression overlaps, but that Gata4 would function specifically in the differentiation of exocrine cells and that Gata6 would be involved in the differentiation of endocrine cells. Surprisingly, several full-length Gata6-EnR transgenics completely blocked pancreas formation, indicating that Gata6 may function non-redundantly in early pancreatic specification. In addition, two Gata6 transgenic embryos and the Gata6DBD transgenic embryo severely disrupted pancreatic cell development.
An unexpected result from these studies was the finding that all but one of the Gata4 transgenics did not have an overt pancreatic phenotype, suggesting that Gata4 is not required for early pancreatic specification. In the majority of the Gata4 transgenic embryos, we also did not observe a defect in the development and differentiation of the exocrine acinar cells. However, a caveat of these studies is the use of the Pdx1 promoter to drive expression of the Gata4 transgene. Although Pdx1 is required for exocrine cell differentiation, it is only expressed at low levels in the acinar cells. Therefore, it remains possible that expression levels of the Gata4 transgene are not sufficient to interfere with endogenous Gata4 activity in the later stage pancreas. Recent functional analysis of Gata4 in zebrafish has demonstrated that Gata4 is required for the formation of many organs, including the liver, intestine, and pancreas. Interestingly, Gata4 morphant embryos are able to initiate endocrine pancreas budding, but have specific defects in exocrine pancreas development (Holtzinger and Evans, 2005). In light of the zebrafish studies, we cannot eliminate the possibility that Gata4 functions at some point during exocrine development.
Of note, one Gata4 transgenic embryo did display a disrupted pancreatic phenotype similar to the Class II Gata6 transgenics. It is possible that this single embryo reveals a role for Gata4 in pancreas development that is not observed in the other Gata4 transgenics due to differences in timing or levels of transgene expression caused by position effect. However, a disproportionate number of Gata4 transgenic embryos do not have an obvious phenotype, although they express the transgene in comparatively similar expression domains and at equal or higher levels than the Class II Gata4 transgenic embryo. We are unable to assess the onset of transgene expression in our transient transgenic system, so this issue remains unresolved. An alternative explanation is that the Gata4 transgene interferes with Gata6 function in the single Class II Gata4 embryo. Several studies in cardiomyocytes have demonstrated that Gata4 and Gata6 bind to similar DNA consensus sequences (Charron et al., 1999). In addition, due to Gata4 and Gata6 overlapping expression patterns and variable or absent phenotypes in mouse knockout studies, there has been ongoing speculation that these proteins can partially compensate for each other during heart development. Therefore, it is possible that the Gata4 transgene could interfere with Gata6 activity. However, it is unclear why this phenotype is observed infrequently.
Gata6 repressor disrupts two discrete stages of pancreatic development
Expression of the Gata6 dominant repressor transgenes results in two distinct phenotypes, allowing us to evaluate how Gata6 functions during early and late stages of pancreas development. Examples of phenotypic diversity are well-documented in experimental transgenic systems and may be related to the specific site of integration as well as number of transgene copies. Phenotypic variation has also been observed in Gata mutational, knockdown, and knockout analyses. In zebrafish, there is considerable variation in the phenotypes of the faust (Gata5) and Gata6 morphant embryos, which either completely lack myocardial tissues or have minor myocardiac phenotypes (Heicklen-Klein et al., 2005; Peterkin et al., 2003; Reiter et al., 2001). While the majority of Gata4 null mice die at gastrulation, 60–70% of the Gata4 null mice that survive gastrulation are able to form normal differentiated myocardium and the remainder have defects in myocardial differentiation (Kuo et al., 1997; Molkentin et al., 1997). The observed phenotypic differences may be due to partial compensation by other Gata factors. Alternatively, evidence is accumulating that members of the Gata transcription factor family may establish and regulate chromatin domains to control cell type differentiation (Bresnick et al., 2005; Martowicz et al., 2005). It is possible that a threshold of Gata activity is required to regulate chromatin, and in some embryos, this threshold can be reached even in the absence of a particular Gata factor. In these embryos, development will proceed normally until the next Gata-dependent regulatory time point is encountered. Once transcriptional targets of Gata6 have been identified in the pancreas, we will be able to more precisely assess the mechanism of Gata function.
Three of the Pdx1:G6FLEnR transgenic embryos analyzed did not form a pancreas, suggesting that Gata6 is required for the initial specification of the embryonic mouse pancreas. The block in pancreas development does not appear to be caused by an early foregut endoderm defect because the duodenum, which arises from foregut endoderm adjacent to the pancreas and also expresses Pdx1, appears to develop normally in these mice (Fig. 4d). Furthermore, recent studies in mouse using a tetraploid complementation approach to rescue Gata6 function in extra-embryonic tissues demonstrate that general specification of ventral endoderm does not require Gata6 (Zhao et al., 2005). In zebrafish, Gata6 morphant studies result in a complete block of liver development; however, the endodermal markers FoxA2 and Sox17 are expressed normally which indicates that Gata6 is not involved in general endoderm specification (Holtzinger and Evans, 2005). Pancreas formation was not assessed in the zebrafish study. By e17.5 in mouse, the dorsal pancreas is normally found between the stomach and the intestine while the ventral pancreas is found adjacent to the liver. Analysis of the Pdx1:G6FLEnR embryos that lacked pancreatic tissue demonstrated that these regions were compacted and largely devoid of recognizable tissues or structures. However, in two of the embryos there appear to be small patches of undifferentiated epithelium that resembled e10.5 pancreatic epithelium (Fig. 4c). This tissue did not express Pdx1, glucagon, or insulin, suggesting that it may represent undifferentiated endoderm that was blocked prior to pancreas specification. These results indicate that Gata6 may lie upstream of the earliest essential pancreatic transcription factors Pdx1 and Ptf1a. Deletion of either Pdx1 or Ptf1a in mice severely disrupts pancreas development and differentiation, but early pancreatic rudiments are able to form (Krapp et al., 1998; Offield et al., 1996).
The second class of Gata6 transgenics has a less severe phenotype. A particularly striking feature of these embryos is the presence of significantly enlarged ducts. This phenotype may result from a failure of the ductal epithelium to differentiate, causing an accumulation of precursor cells. Consistent with this possibility, we observe a modest increase in the number of Ngn3-expressing cells and a higher proportion of ductal cells expressing Nkx6.1 and Pdx1. It also appears that the Gata6 transgene is interfering with normal cell type maturation; many of the differentiated islet cells that form are aberrantly localized within the duct cells or are located immediately adjacent to the ductal epithelium. Only a small percentage of differentiated cell types remain, which may be evidence of a migration defect due to the absence of the correct local environmental signals. Finally, the enlarged duct structures do not exhibit normal branching. Gata6 has been shown to be an important regulator of branching and morphogenesis in the lung (Koutsourakis et al., 2001; Liu et al., 2002b) and therefore may play a similar role in the developing pancreas.
Gata6 and Nkx2.2 may co-regulate islet-specific transcription factors
Although disruption of Gata6 function blocks the earliest stages of pancreas development, it remains possible that Gata6 also functions at later stages in the islet where it is expressed at high levels in a subset of endocrine cells. In support of this, we determined that Gata6 interacts with the essential islet transcription factor, Nkx2.2. It is believed that specific functions of Gata4, Gata5, and Gata6 in cardiogenesis are determined by specific protein–protein interactions. Interestingly, Nkx2.5 can interact with Gata4 and Gata5 in the heart, but does not interact with Gata6 (Durocher et al., 1997). This indicates that not all Gata factors are able to interact with every Nkx2 family member, and the specific interactions that can form may be critical in the regulation of cell-type specific targets. Additionally, Gata6 appears to have more extensive functions in the pancreas than Nkx2.2, since the Gata6 transgenic phenotype is more severe than the phenotype caused by a deletion of Nkx2.2. Identification of Nkx2.2 and Gata6 co-regulated genes in the islet is a current focus of our research.
Supplementary Material
Supplemental figures 1-4
Acknowledgments
We thank J. Jensen (Barbara Davis Center, UCHSC) and K. Artinger (UCHSC) for the generous gifts of the Pdx1 promoter vector and pCS2-EnR vector, respectively. We thank D. Srivastava (UCSF) and C. Glembotski (UCSD) for Gata-reporter vectors. Special thanks to Keith Anderson for assistance with the luciferase assays. We thank Lee Niswander, Kristin Artinger and members of the Sussel lab for critical reading of the manuscript. This research was supported by the NIH NIDDK (L.S.) and a Juvenile Diabetes Foundation Postdoctoral fellowship (D.G.). Additional support was provided by the University of Colorado Cancer Center Transgenic/Knockout Core and the Diabetes and Endocrinology Research Center (NIH P30 DK57516).
Footnotes
References
- Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr. Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort R, Martinez-Barbera JP, Beddington RS, Zaret KS. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development. 2004;131:797–806. doi: 10.1242/dev.00965. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Martowicz ML, Pal S, Johnson KD. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 2005;205:1–9. doi: 10.1002/jcp.20393. [DOI] [PubMed] [Google Scholar]
- Bruno MD, Korfhagen TR, Liu C, Morrisey EE, Whitsett JA. GATA-6 activates transcription of surfactant protein A. J. Biol. Chem. 2000;275:1043–1049. doi: 10.1074/jbc.275.2.1043. [DOI] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev. Cell. 2003;4:383–393. doi: 10.1016/s1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Herrera PL, Wright CV. Mosaic Cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis. 2000;26:143–144. doi: 10.1002/(sici)1526-968x(200002)26:2<143::aid-gene13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hale MA, Kagami H, Shi L, Holland AM, Elsasser HP, Hammer RE, Macdonald RJ. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev. Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Heicklen-Klein A, McReynolds LJ, Evans T. Using the zebrafish model to study GATA transcription factors. Semin. Cell Dev. Biol. 2005;16:95–106. doi: 10.1016/j.semcdb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kathiriya IS, King IN, Murakami M, Nakagawa M, Astle JM, Gardner KA, Gerard RD, Olson EN, Srivastava D, Nakagawa O. Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 2004;279:54937–54943. doi: 10.1074/jbc.M409879200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Keijzer R, van Tuyl M, Meijers C, Post M, Tibboel D, Grosveld F, Koutsourakis M. The transcription factor GATA6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development. 2001;128:503–511. doi: 10.1242/dev.128.4.503. [DOI] [PubMed] [Google Scholar]
- Ketola I, Otonkoski T, Pulkkinen MA, Niemi H, Palgi J, Jacobsen CM, Wilson DB, Heikinheimo M. Transcription factor GATA-6 is expressed in the endocrine and GATA-4 in the exocrine pancreas. Mol. Cell. Endocrinol. 2004;226:51–57. doi: 10.1016/j.mce.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Pancreas development in the chick embryo. Cold Spring Harbor Symp. Quant. Biol. 1997;62:377–383. [PubMed] [Google Scholar]
- Kim SK, Melton DA. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M, Keijzer R, Visser P, Post M, Tibboel D, Grosveld F. Branching and differentiation defects in pulmonary epithelium with elevated Gata6 expression. Mech. Dev. 2001;105:105–114. doi: 10.1016/s0925-4773(01)00386-0. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development [corrected and republished with original paging, article originally printed in Development 1999 Feb;126(4):723–732] Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Li H, Edlund H. Persistent expression of Hlxb9 in the pancreatic epithelium impairs pancreatic development. Dev. Biol. 2001;240:247–253. doi: 10.1006/dbio.2001.0440. [DOI] [PubMed] [Google Scholar]
- Liu C, Glasser SW, Wan H, Whitsett JA. GATA-6 and thyroid transcription factor-1 directly interact and regulate surfactant protein-C gene expression. J. Biol. Chem. 2002a;277:4519–4525. doi: 10.1074/jbc.M107585200. [DOI] [PubMed] [Google Scholar]
- Liu C, Morrisey EE, Whitsett JA. GATA-6 is required for maturation of the lung in late gestation. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2002b;283:L468–L475. doi: 10.1152/ajplung.00044.2002. [DOI] [PubMed] [Google Scholar]
- Liu C, Ikegami M, Stahlman MT, Dey CR, Whitsett JA. Inhibition of alveolarization and altered pulmonary mechanics in mice expressing GATA-6. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2003;285:L1246–L1254. doi: 10.1152/ajplung.00443.2002. [DOI] [PubMed] [Google Scholar]
- Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J. Biol. Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Kalvakolanu DV, Markham BE. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Mol. Cell. Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 1997;189:270–274. doi: 10.1006/dbio.1997.8684. [DOI] [PubMed] [Google Scholar]
- Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev. Biol. 2003;264:323–338. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr. Opin. Genet. Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. EMBO J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pictet R, Rutter WJ. Development of the Embryonic Endocrine Pancreas. Washington, DC: Williams and Wilkins; 1972. [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Ritz-Laser B, Mamin A, Brun T, Avril I, Schwitzgebel VM, Philippe J. The zinc finger-containing transcription factor Gata-4 is expressed in the developing endocrine pancreas and activates glucagon gene expression. Mol. Endocrinol. 2005;19:759–770. doi: 10.1210/me.2004-0051. [DOI] [PubMed] [Google Scholar]
- Rose SD, Swift GH, Peyton MJ, Hammer RE, MacDonald RJ. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 2001;276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol. Cell. Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, Zorn AM, Harvey RP. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev. Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Heller RS, Miller CP, Habener JF. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology. 1999;140:5374–5381. doi: 10.1210/endo.140.11.7122. [DOI] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Weidenfeld J, Shu W, Zhang L, Millar SE, Morrisey EE. The WNT7b promoter is regulated by TTF-1, GATA6, and Foxa2 in lung epithelium. J. Biol. Chem. 2002;277:21061–21070. doi: 10.1074/jbc.M111702200. [DOI] [PubMed] [Google Scholar]
- Wilding L, Gannon M. The role of pdx1 and HNF6 in proliferation and differentiation of endocrine precursors. Diabetes/Metab. Res. Rev. 2004;20:114–123. doi: 10.1002/dmrr.429. [DOI] [PubMed] [Google Scholar]
- Yang H, Lu MM, Zhang L, Whitsett JA, Morrisey EE. GATA6 regulates differentiation of distal lung epithelium. Development. 2002;129:2233–2246. doi: 10.1242/dev.129.9.2233. [DOI] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures 1-4