Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase (original) (raw)
WEE1 and CHK1 jointly regulate Cdk activity to prevent DNA damage during replication.
Abstract
Maintenance of genome integrity is of critical importance to cells. To identify key regulators of genomic integrity, we screened a human cell line with a kinome small interfering RNA library. WEE1, a major regulator of mitotic entry, and CHK1 were among the genes identified. Both kinases are important negative regulators of CDK1 and -2. Strikingly, WEE1 depletion rapidly induced DNA damage in S phase in newly replicated DNA, which was accompanied by a marked increase in single-stranded DNA. This DNA damage is dependent on CDK1 and -2 as well as the replication proteins MCM2 and CDT1 but not CDC25A. Conversely, DNA damage after CHK1 inhibition is highly dependent on CDC25A. Furthermore, the inferior proliferation of CHK1-depleted cells is improved substantially by codepletion of CDC25A. We conclude that the mitotic kinase WEE1 and CHK1 jointly maintain balanced cellular control of Cdk activity during normal DNA replication, which is crucial to prevent the generation of harmful DNA lesions during replication.
Introduction
The genome is inherently unstable as the result of spontaneous chemical reactions as well as exposure to a wide variety of genotoxic agents. To deal with environmental and endogenously arising DNA lesions, cells have evolved responses that coordinate cell cycle progression and DNA repair pathways to ensure the integrity of the genome. Failure to maintain genomic integrity is a threatening condition; as examples, chromosomal aberrations and rearrangements are associated with cancer and contribute to carcinogenesis (Halazonetis et al., 2008).
When cells replicate their DNA in S phase, they generate structures that are sensitive to both endogenous and exogenous insults. Furthermore, oncogenes can induce lesions at replication forks, and subsequent induction of the DNA damage response occurs early during tumorigenesis and has been proposed to act as a barrier to tumor progression (Bartkova et al., 2006; Di Micco et al., 2006). This barrier can be impaired by several mechanisms such as p53 mutations, allowing cancer development. The continuous formation of DNA double-strand breaks (DSBs) during replication may also contribute to the genomic instability that characterizes human cancers (Halazonetis et al., 2008).
To deal with a variety of DNA lesions, cells harbor pathways composed of large networks of damage sensors, signal transducers, and effectors (Kastan and Bartek, 2004). The initial response to replicative stress is activated mainly by the ATR (ataxia telangiectasia and Rad3-related protein) kinase, which targets proteins such as p53, H2AX, and the CHK1 kinase. ATR and CHK1 are critical for the response to exogenous DNA-damaging agents. In addition, they are indispensable for regulating several processes during the unperturbed cell cycle where they control cell cycle progression by regulating replication and mitotic events (Ben-Yehoyada et al., 2007; Chen and Poon, 2008).
A key target of CHK1 is the CDC25A phosphatase, which is an activator of Cdks. CHK1 phosphorylation of CDC25A accelerates its degradation, leading to a slowing down of DNA replication and preventing entry into mitosis until the damage has been repaired (Bartek and Lukas, 2007). CHK1-mediated phosphorylation and inhibition of the CDC25–Cdk pathway are implicated in the cell cycle checkpoint control of G1/S, S, and G2/M phases. CHK1 suppresses replicative damage through regulation of DNA replication (Syljuåsen et al., 2005). Additionally, CHK1 has been shown to induce repair of DNA lesions by stimulating homologous recombination (HR) repair (Sørensen et al., 2005) and DNA cross-link repair (Wang et al., 2007).
To identify critical regulators of genome integrity, we screened a human cell line with a kinome siRNA library for genes for which depletion leads to DNA damage in the absence of exogenous insults. The results show that the mitotic kinase WEE1 is crucial for genome integrity during S phase in a manner dependent on Cdk activity. WEE1 controls a branch parallel to CHK1–CDC25A. These two pathways converge to control Cdk activity during normal S-phase progression to avoid the generation of harmful DNA lesions.
Results and discussion
High-throughput siRNA screen reveals a key role for Cdk regulators in the maintenance of genomic integrity
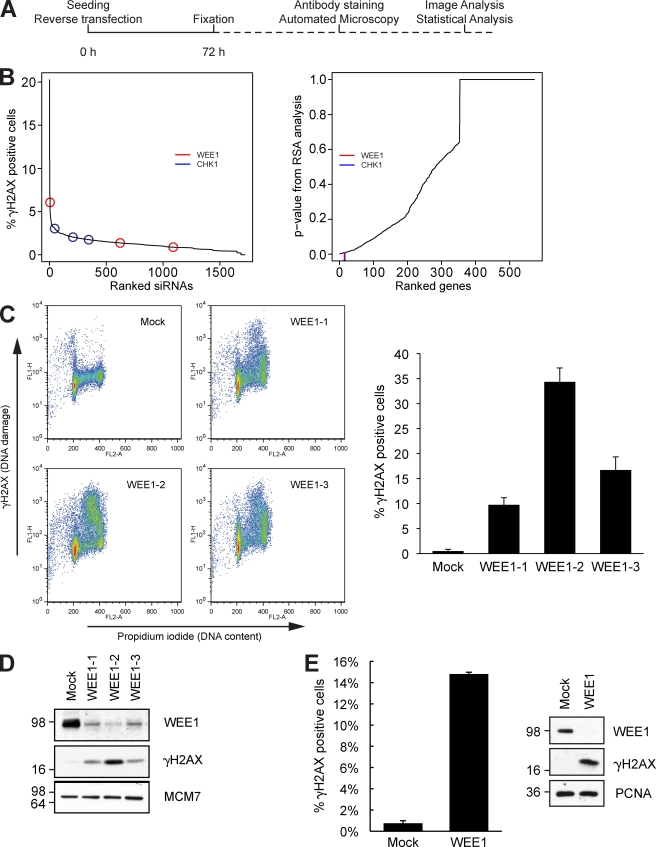
To uncover important factors involved in maintaining genomic integrity in human cells, we performed a robot-automated high-throughput screen with a human kinome siRNA library. The human osteosarcoma cell line U2OS was chosen as the model system because it has low levels of spontaneous DNA damage and is easy to transfect and handle. U2OS cells were reverse transfected with siRNA and placed in a standard cell incubator. After 72 h of depletion, cells were fixed, and the DNA damage marker phosphorylated H2AX (γ-H2AX) was visualized by immunofluorescent staining (Fig. 1 A). We then performed automated microscope image acquisition (Fig. S1 A), and a statistical analysis was performed to estimate candidate genes (König et al., 2007). The analysis revealed that the majority of siRNAs did not lead to marked DNA damage, suggesting that most kinases are dispensable for genomic integrity in this cell line (Fig. 1 B). The entire statistically analyzed results of the screen are included in a supplemental Excel file, which includes nonkinase genes that were included as controls (Table S1). 15 kinases had a p-value below 0.01 (Fig. S1 B), and further analysis using the PANTHER classification system revealed that these kinases were mostly in functionally separate pathways (Fig. S1 C). Additional analysis by text mining revealed that several of these kinases have not been previously associated with a phenotype similar to genome integrity and DNA damage (Table S1, graphs 1–15). However, the networks around each of these kinases contain many proteins that have been associated with such a phenotype. Also, analysis by text mining revealed that several of these kinases are indirectly connected to each other via one shared interaction partner (Table S1, Targets Subnetwork tab). While this manuscript was under review, a recent study with a quite similar approach was published (Paulsen et al., 2009). Kinases such as PLK1 and CHK1 scored in both screens; interestingly, several kinases were identified in only one of the screens, suggesting that siRNA screens are complementary. This is likely because of the large number of variables in these large-scale screening approaches, including differing siRNA sequences, choice of cell lines, growth conditions, and duration of siRNA treatment.
Figure 1.
High-throughput siRNA screen identifies WEE1 as a regulator of genomic integrity. (A) Schematic overview of the robot-automated high-throughput siRNA screen. U2OS cells were reverse transfected, and after incubation for 72 h, cells were stained with anti–γ-H2AX antibody and Hoechst. (B) Individual siRNAs were ranked according to the percentage of cells that were γ-H2AX positive (left), and genes were ranked according to their RSA-derived p-value (right). The positions of WEE1 and CHK1 are indicated. (C) U2OS cells were transfected with three different siRNAs targeting WEE1 and harvested for flow cytometric analysis after 48 h. The bar plot is the mean of the percentage of γ-H2AX–positive cells of three experiments. (D) Immunoblot of cells treated as in C. (E) TIG-3-tert cells were transfected with WEE1 siRNA for 48 h and analyzed by flow cytometry and immunoblotting. (C and E) Error bars indicate SD from three experiments. (D and E) Molecular mass is indicated in kilodaltons. PCNA, proliferating cell nuclear antigen.
We noticed that WEE1 and CHK1, both negative regulators of Cdk activity, were among our top-ranked candidates (Fig. 1 B and Fig. S1 B). We then focused our analysis on WEE1 and its downstream targets CDK1 and -2. To validate the findings of the screen and exclude possible off-target effects of the siRNA, we transfected cells with the three siRNAs targeting WEE1 from the library. Cells were analyzed by flow cytometry 48 h after transfection, which showed that the depleted cells had high levels of phosphorylated H2AX in mid to late S phase, thereby confirming our screen results and indicating that the DNA damage is linked to replication (Fig. 1 C). Immunoblotting confirmed that WEE1 was effectively depleted by the three siRNAs and that they all caused increased γ-H2AX levels (Fig. 1 D). WEE1 depletion inhibited the phosphorylation of CDK1 on Tyr15, which is consistent with the published function of WEE1 (Fig. S1 D; Coleman and Dunphy, 1994; Kellogg, 2003). In line with this, the kinase activities of immunoprecipitated cyclin A and cyclin B1 were rapidly increased upon WEE1 depletion (Fig. S1 E; Chow et al., 2003). To understand whether the effect of WEE1 siRNA was specific to transformed cells, we treated TIG-3-tert cells (nontransformed telomerized fibroblasts) with WEE1 siRNA. Depletion of WEE1 also induced marked DNA damage in these cells (Fig. 1 E) as well as other cell lines tested, including BJ-tert fibroblasts and HeLa cells (not depicted).
To be certain that the γ-H2AX staining reflected actual DNA damage, we cast live cells in agarose plugs and ran pulsed field gel electrophoreses (PFGEs). After 24-h transfection with WEE1 siRNA, the cells had accumulated substantial damage in the form of DSBs as well as smaller fragments (Fig. S1 F).
WEE1 depletion leads to DNA damage during DNA replication in U2OS cells
Ablation of WEE1 has previously been shown to induce cell death (Yuan et al., 2003; Tominaga et al., 2006). It was a possibility that DNA damage after WEE1 knockdown was a secondary event occurring after induction of cell death pathways. To investigate this, we performed more detailed analysis of the early events after WEE1 knockdown. Immunoblotting revealed that WEE1 depletion lead to an activated DNA damage response (Fig. 2 A), as measured by increased phosphorylation of the ATR targets H2AX, replication protein A (RPA), and CHK1 (Ser317). Flow cytometry analysis revealed that γ-H2AX staining was visible already 12 h after transfection and was strong 24 h after (Fig. 2 B). Next, we asked whether the DNA damage arises in actively replicating cells. We depleted WEE1 for 12 h and then added the nucleotide analogue EdU for 4 and 8 h before fixation. We costained against γ-H2AX and EdU, followed by analysis of cells with flow cytometry. The results of this experiment establish that the DNA damage exclusively occurs in EdU-positive cells (Fig. S2, A and B). In a second set of experiments, we depleted WEE1 for 18 and 24 h and then added the nucleotide analogue EdU for 10 min before fixation. Our flow cytometry analysis of these samples revealed that most of the cells with DNA damage were actively replicating at 18 h after WEE1 depletion (Fig. S2 C); however, at later stages of depletion, these cells ceased replication and arrested in S phase.
Figure 2.
WEE1 depletion leads to DNA damage in newly replicated DNA. (A) U2OS cells were transfected with WEE1 siRNA and processed for immunoblotting. Molecular mass is indicated in kilodaltons. PCNA, proliferating cell nuclear antigen. (B) Flow cytometry analysis for γ-H2AX and propidium iodide (PI) of cells 12, 24, 36, and 48 h after transfection with WEE1 siRNA. The boxed areas indicate γ-H2AX–positive cells.The bar plot is the mean of the percentage of γ-H2AX–positive cells of three experiments. (C) Immunofluorescence confocal microscopy of cells 12 and 18 h after transfection with WEE1 siRNA. To detect ssDNA, cells were cultured in medium with 10 µM BrdU throughout the experiment; DNA was not denatured before staining against BrdU, γ-H2AX, and DAPI. (D) For the detection of replication-associated DSBs, cells were labeled with methyl-[14C]thymidine (14C-TdR) for 30 min. Control and WEE1 samples were labeled 12 h after siRNA transfection. Cells were harvested 24 h after transfection, and DNA fragments were separated by PFGE. IR, ionizing radiation. (E) U2OS cells were depleted for the DNA replication proteins MCM2 and CDT1 for 48 h. WEE1 was depleted the last 20 h. Cells were processed for flow cytometry analysis with staining of γ-H2AX and DNA (PI). (B and E) Error bars indicate SD from three experiments. Bar, 10 µm.
We hypothesized that DNA damage from WEE1 depletion was characterized by single-stranded DNA (ssDNA) formation, a common intermediate at replication-associated lesions. To test this, we incubated cells with BrdU and treated them with WEE1 siRNA. Cells were then fixed and immunostained for γ-H2AX and BrdU in the absence of HCl and DNase treatment. Confocal microscopy revealed strong pannuclear γ-H2AX staining, and the anti-BrdU staining revealed ssDNA at very early time points (Fig. 2 C). This could indicate processing of DNA lesions in areas of DNA replication, where ssDNA is a key intermediate in DNA damage signaling and DNA repair processes. A key remaining question was whether the lesions were arising in areas of replicating DNA. To investigate this, we pulsed cells for 30 min with [14C]thymidine and ran PFGE to analyze the DNA fragments. Importantly, cells depleted for WEE1 experienced marked DNA damage in newly replicated areas. This was in contrast to cells treated with ionizing radiation, which barely induced damage in replicating areas even though the degree of damage as measured by PFGE was markedly higher (Fig. 2 D). To further analyze the connection between the damage phenotype and DNA replication, we codepleted WEE1 and the important replication proteins CDT1 and MCM2 (Tada, 2007). In both cases, the depletion of CDT1 and MCM2 significantly reduced damage after WEE1 knockdown (Fig. 2 E). Short-term depletion of CDT1 or MCM2 did not lead to major perturbations of cell cycle progression. Together, these data substantiate the novel concept that WEE1 plays a major role in supporting correct DNA replication, thereby preventing loss of genomic integrity. Cessation of DNA replication at later stages after WEE1 knockdown correlated with premature entry into mitosis, as measured by increased histone H3 Ser10 phosphorylation and DNA condensation (unpublished data). Premature mitotic entry is commonly observed after loss of WEE1 (Russell and Nurse, 1987; Harvey and Kellogg, 2003; Stumpff et al., 2004; Tominaga et al., 2006). However, because DNA damage after WEE1 knockdown initiates rapidly in newly replicated areas, our data suggest that this damage is independent of premature mitotic entry.
DNA damage after WEE1 depletion is dependent on CDK1 and -2
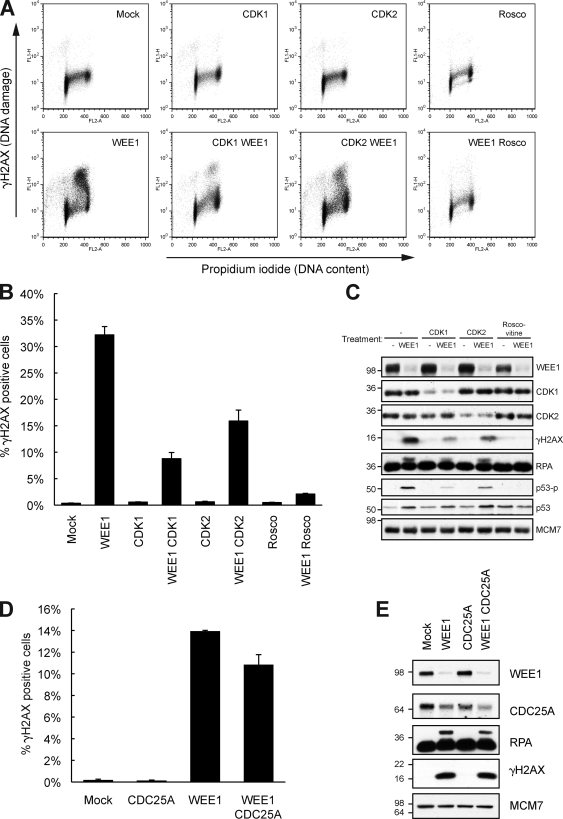
WEE1 is a kinase that phosphorylates and thereby inhibits CDK1 and -2, which normally promote S phase and mitotic entry (Fig. S1, D and E; Coleman and Dunphy, 1994; Kellogg, 2003). This prompted us to investigate whether CDK1 and -2 played a role in the S-phase damage observed after WEE1 depletion. We codepleted WEE1 with either CDK1 or -2 for 36 h and analyzed the cells by flow cytometry. Knockdown of CDK1 rescued the WEE1 depletion phenotype (Fig. 3, A and B), with no visible effects on the cell cycle (Fig. S2 D). We further depleted CDK2 with WEE1, and this also had an inhibitory effect on the induction of DNA damage. Furthermore, we observed similar findings in TIG-3-tert cells depleted for WEE1 and CDK1 and -2 (Fig. S2 E). To substantiate these data, we depleted WEE1 in the presence of the Cdk inhibitor roscovitine, and as expected, roscovitine completely repressed the phenotype. Immunoblotting confirmed that depletion of CDK1 and -2 suppressed WEE1-mediated DNA damage as well as phosphorylated RPA (Fig. 3 C). Thus, the excess DNA damage after WEE1 depletion is mainly through deregulated CDK1 and -2 activity. These data highlight the crucial role played by enzymes that restrain Cdk activity in maintaining the integrity of the genome in S phase. The inhibitory Cdk phosphorylation mediated by WEE1 is antagonized by CDC25A, the phosphatase which activates Cdks in S and G2 phases of the cell cycle (Chen and Poon, 2008). To investigate the role of CDC25A in DNA damage after WEE1 siRNA transfection, we cotransfected WEE1 and CDC25A for 24 h. Flow cytometry revealed almost similar amounts of γ-H2AX–positive cells after CDC25A and WEE1 codepletion compared with WEE1 depletion alone (Fig. 3 D). Consistently, WEE1-depleted cells and WEE1- and CDC25A-codepleted cells had equal amounts of γ-H2AX and phosphorylated RPA, as measured by immunoblotting (Fig. 3 E), although CDC25A was partly down-regulated after WEE1 depletion alone because of an activated DNA damage response (Fig. 2 A). Thus, the DNA damage induced by WEE1 depletion appears largely independent of CDC25A.
Figure 3.
DNA damage accumulating after WEE1 depletion is dependent on CDK1 and -2 but not CDC25A. (A) U2OS cells were transfected with WEE1 siRNA with and without 100 nM CDK1 or -2 siRNA for 36 h or with 25 µM of the Cdk inhibitor roscovitine (Rosco) added 12 h after transfection. Cells were processed for flow cytometry analysis with staining of γ-H2AX and DNA (PI). (B) Mean of three experiments treated as in A. (C) Immunoblot of cells treated as in A. (D) U2OS cells were transfected with a WEE1 and CDC25A siRNA pool (50 nM of each siRNA), harvested after 24 h and stained for γ-H2AX and with PI and analyzed by flow cytometry. (B and D) Error bars indicate SD from three experiments. (E) Immunoblot of cells treated as in D. (C and E) Molecular mass is indicated in kilodaltons.
Endogenously arising DNA damage after CHK1 depletion is dependent on CDC25A
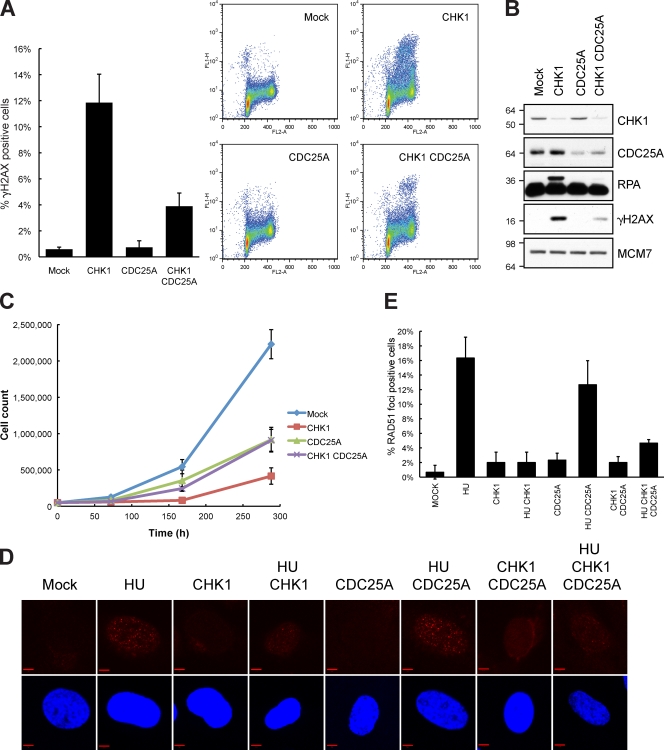
Inhibition and depletion of CHK1 led to DNA damage in S phase via deregulation of Cdk activity (Fig. S3 A). CHK1 inhibits CDC25A directly; thus, DNA damage occurring upon CHK1 inhibition has been suggested to be mediated through deregulation of the CDC25A–Cdk pathway, leading to elevated Cdk activity (Syljuåsen et al., 2005). We tested this hypothesis by codepleting CHK1 and CDC25A with siRNA. To obtain efficient down-regulation of CDC25A, we used a pool of two siRNAs targeting CDC25A (Fig. S3 B). CHK1-depleted cells accumulated substantial DNA damage, whereas cells treated with both CHK1 and CDC25A siRNA developed significantly less DNA damage when analyzed by flow cytometry (Fig. 4 A). Immunoblotting revealed that CDC25A knockdown not only suppressed γ-H2AX but also phosphorylation of RPA (Fig. 4 B). Similar results were obtained in TIG-3-tert cells (Fig. S3 D). CHK1 inhibition with inhibitors leads to DNA damage shortly after drug addition (Syljuåsen et al., 2005). To investigate whether this rapid response was dependent on CDC25A, we treated U2OS cells for 2 h with the CHK1 inhibitor Gö6976 48 h after transfection with CDC25A siRNA. In line with our results for CHK1 depletion by siRNA (Fig. 4, A and B), the CDC25A depletion significantly repressed DNA damage in the S phase after Gö6976 treatment (Fig. S3 C).
Figure 4.
Endogenously arising DNA damage after CHK1 depletion is dependent on CDC25A. (A) U2OS cells were transfected with CHK1 siRNA and a CDC25A siRNA pool (50 nM of each siRNA). After 48 h, cells were fixed, stained for γ-H2AX and with PI, and analyzed by flow cytometry. The bar plot is the mean of the percentage of γ-H2AX–positive cells of three experiments. (B) Cells were treated as in A and prepared for immunoblotting. Molecular mass is indicated in kilodaltons. (C) TIG-3-tert cells were transfected with the indicated siRNA and, 24 h later, trypsinized and seeded. Cells were counted at the indicated time. Data are the mean of three experiments. (D) TIG-3-tert cells were transfected with CHK1 siRNA and a CDC25A siRNA pool and, after 24 h, 2 mM HU was added. Confocal microscopy was performed on cells immunostained with an antibody against RAD51 and DAPI for DNA. (E) Quantification of D. 100 cells were counted in triplicates. (A, C, and E) Error bars indicate SD from three experiments. Bars, 5 µm.
CDC25A plays a role in the unperturbed cell cycle (Ben-Yehoyada et al., 2007); to address the concern that the CDC25A siRNA was slowing down replication, which would affect the result, we pulsed cells with BrdU after 48 h of CDC25A siRNA transfection and chased the cells for 8 h. Flow cytometric analysis revealed that the progression through S phase at these time points was largely unaffected by the lower cellular concentration of CDC25A (Fig. S3 E). Thus, repression of DNA damage after CDC25A depletion is not caused by markedly slowed progression through S phase or G1-phase arrest.
Finally, we asked whether CDC25A accumulation could be an important determinant of the proliferative capability of cells depleted for CHK1 because coknockdown of CDC25A with CHK1 abolished the majority of DNA damage. Importantly, TIG-3-tert cells depleted for CHK1 proliferated very poorly, whereas this phenotype was partly rescued by codepletion of CDC25A. In fact, cells depleted for both CDC25A and CHK1 proliferated with similar kinetics as CDC25A-depleted cells (Fig. 4 C). Inspired by this result, we wondered whether the role of CHK1 in the cellular response to replication fork stalling induced by hydroxyurea (HU) could be mediated partly via the CHK1–CDC25A pathway, perhaps operating in a cooperative fashion with the CHK1–RAD51 pathway. To address this issue, we treated cells with siRNA to CDC25A and CHK1 in the presence and absence of HU. CHK1 depletion largely prevented the formation of RAD51 foci after HU. As CHK1 and CDC25A codepletion also led to a marked reduction in RAD51 foci formation after HU, we found little evidence for a major role of the CHK1–CDC25A pathway in controlling RAD51 foci formation after HU (Fig. 4, D and E). In line with this result, WEE1 depletion could not block RAD51 foci formation after HU (Fig. S3 G). We noticed that cells depleted for CHK1 could form RAD51 foci, although at much reduced numbers (Fig. 4 E). Because HR is required to support proliferation, the ability to form some RAD51 foci may provide some HR repair to help cells cope with the DNA damage after CHK1 depletion. In support of this, our proliferation assays revealed that RAD51-depleted cells were more affected than CHK1- and CDC25A-codepleted cells (Fig. S3 F).
Both WEE1 and CHK1 are crucial for negatively regulating Cdk. Whereas WEE1 inhibits Cdks by direct phosphorylation, CHK1 negatively controls Cdk activity indirectly through its control over CDC25A stability (Ben-Yehoyada et al., 2007). Cdk activity is also negatively regulated by MYT1 (Chen and Poon, 2008), and siRNA depletion of this kinase also leads to DNA damage, although not as notable as WEE1 knockdown (unpublished data). In conclusion, our data support a model in which genomic integrity during DNA replication is maintained by the precise regulation of Cdk activity mediated through two parallel pathways by WEE1 and CHK1–CDC25A. Future directions of our research deal with the elucidation of the key targets of Cdk activity that control genomic integrity.
Materials and methods
Cell culture and chemicals
The human U2OS osteosarcoma cell line and telomerized human normal fetal lung fibroblast cell line TIG-3 was grown in Dulbecco’s modified Eagle’s medium (Invitrogen) with 10% FBS. For siRNA-mediated knockdown, U2OS cells were transfected using Oligofectamine (Invitrogen) according to the manufacturer’s protocol. TIG-3 cells were reverse transfected using Lipofectamine RNAiMAX (Invitrogen). The following oligonucleotide sequences were ordered from Sigma-Aldrich: WEE1-1, 5′-GGAAAAAGGGAAUUUGAUG[dT][dT]-3′; WEE1-2, 5′-GGGAAUUUGAUGUGCGACA[dT][dT]-3′; WEE1-3, 5′-GGUAUAUUCAUUCAAUGUC[dT][dT]-3′; CDK1, 5′-GAUGUAGCUUUCUGACAAAAA[dT][dT]-3′; CDK2, 5′-GUUUCAGUAUUAGAUGCAC[dT][dT]-3′; CDC25A-1, 5′-GGCGCUAUUUGGCGCUUCA[dT][dT]-3′; CDC25A-2, 5′-CCUGACCGUCACUAUGGAC[dT][dT]-3′; CHK1, 5′-GGGAUAUUAAACCAGAAAA[dT][dT]-3′; RAD51, 5′-UUGAGACUGGAUCUAUCAC[dT][dT]-3′; MCM2, 5′-UCAUCGGAAUCCUUCACCA[dT][dT]-3′; and CDT1, 5′-GCGCAAUGUUGGCCAGAUC[dT][dT]-3′.
Experiments with siRNA-transfected cells were performed as indicated in the figure legends. Inhibition of CHK1 kinase activity was achieved by the addition of 100 nM Gö6976 (EMD). Cdk activity was inhibited by the addition of 25 µM roscovitine (EMD).
Robot-automated screen
The automated screen was performed using a liquid handling station (STAR; Hamilton Robotics). U2OS cells were reverse transfected with an siRNA library targeting the human kinome (Applied Biosystems). The library is comprised of three independent siRNA constructs per target, with each construct being individually spotted. In brief, 4 µl siRNA was added to 2.5 µl OptiMEM (Invitrogen) to each well of a 384-well plate (Corning). To this, a 6.4 µl OptiMEM/0.1 µl Oligofectamine mix was added and left to stand for 15 min, after which 27 µl of cells was added to give a total cell density of 2,800 cells per well. The final concentration of siRNA was 100 nM. Cells were incubated for 72 h, followed by Hoechst staining (bisBenzimide H 33342; Sigma-Aldrich), 4% paraformaldehyde fixation, permeabilization, and phosphorylated H2AX antibody staining, in conjunction with an Alexa Fluor 594 secondary antibody. Cells were left at 4°C in PBS before imaging. The cells were imaged on an IN Cell Analyzer 1000 (GE Healthcare), and 10 images per well were acquired using a 20× objective to count ∼2,000 cells per well. The acquired images were then analyzed using the IN Cell Analyzer Workstation 3.5 software (GE Healthcare).
Statistical analysis
To minimize the risk of misinterpretation of gene activity because of off-target effects of the siRNAs, we computed a statistical score that modeled the probability of a gene “hit” based on the collective activities of multiple siRNAs per gene using the statistical method redundant siRNA activity (RSA) analysis (König et al., 2007). Using the RSA method, we first ranked the siRNAs according to their score. Afterward, a p-value was assigned to each gene, and this value indicated how likely it was to observe this distribution of siRNAs for one gene at a high position in the list by chance. The calculation of the p-values was based on the iterative hypergeometric distribution (König et al., 2007). For this study, the RSA analysis was performed based on the percentage of γ-H2AX–positive cells.
Text mining protein interaction networks and phenotype associations
The protein interaction networks were derived from text mining interactions partners from a resource that integrates protein interaction phrases retrieved from the Biomolecular Interaction Network Database (BIND; Bader et al., 2003) with the cocitations of protein pairs extracted from the same sentence as these interaction terms in the >19 million abstracts in the Medline database. In this resource, the protein citations and their symbols, names, and synonyms are extracted from Medline using the PubGene gene and biological association database (Jenssen et al., 2001). Each of the target genes are connected to putative interaction partners in the larger network we generated if the interaction partner shares connections to at least two of the top-ranked target genes. Protein interaction networks were also generated from this resource around the individual top-ranked target genes to illustrate their highest-scoring article gene cocitations from the PubGene database.
To assign a phenotype association to the target genes and the their interaction partners in the networks, we manually compiled a list of 66 relevant terms from the Gene Ontology and Medical subject headings that are associated to the phenotype of genome integrity and DNA damage. The terms are listed in Table S1. The number of times a target gene or an interaction partner is cocited in an article with the phenotype terms according to the PubGene database was recorded. We then used the Cytoscape network visualization software (Shannon et al., 2003) to visualize the networks and color code the genes in the network according to their strength of association to the phenotype in the literature. In this color-coding scheme, the intensity of green color of a gene in a protein interaction network is governed by the number of times that gene is cocited with the combined phenotype terms in Medline. This intensity was illustrated as an attribute of the gene in the Cytoscape software using the logarithm of the number of articles as the score of color intensity for each gene. Genes that were not associated to the phenotype in Medline articles were colored gray. Furthermore, in the visualization of the protein interaction networks, the target genes were shaped as diamonds with red outer edges, and connections between the targets were colored red.
Immunoblotting and antibodies
Cells were lysed on ice in radioimmunoprecipitation assay buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 1% IgePal630, 1% deoxycholic acid [Na salt], 0.1% SDS, 1 mM PMSF, 5 µg/ml leupeptin, 1% aprotinin, 50 mM NaF, and 1 mM DTT). Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were incubated in primary antibody diluted in 5% milk, followed by incubation with secondary antibody (peroxidase-labeled anti–mouse or –rabbit IgG; 1:10,000; Vector Laboratories). Films were developed using an x-ray machine (Valsoe; Ferrania Technologies). Phospho-CHK1 antibody (CHK1-pSer317) and phospho-p53 (pSer15) antibody were purchased from Cell Signaling Technology. Antibodies against MCM7 (DCS-141) and CHK1 (DCS-310) have been described previously (Sørensen et al., 2005). Anti-Wee1 (C-20), anti-CDC25A (F-6), anti–cyclin A1 (H-432), anti–p-Cdc2 p34 (Thr14/Tyr15), anti-Cdk2 (D-12), and anti-RAD51 (H-92) were purchased from Santa Cruz Biotechnology, Inc. Anti–phosphorylated H2AX (JBW103) and anti–phosphorylated histone H3 Ser10 were purchased from Millipore. Anti–proliferating cell nuclear antigen (PC10) was purchased from Abcam, and anti-RPA (Ab-3) and anti-Cdc2/Cdk1 (Ab-1) were purchased from EMD. Antiactin (clone C4) was purchased from Millipore. p53 (DO-1) was a gift from K. Helin (Biotech Research and Innovation Centre, University of Copenhagen, Copenhagen, Denmark). Monoclonal anti-BrdU (clone BU-1) antibody was purchased from GE Healthcare.
Flow cytometry
Cells were prepared for flow cytometry as previously described (Jørgensen et al., 2007). For BrdU pulse-chase proliferation assays, cells were pulse labeled with 10 µM BrdU (Roche) for 5 min, fixed in 70% ethanol, and incubated in 2 M HCl for 30 min. Cells were stained with mouse antibody to BrdU (1:100) for 1 h. For EdU analysis, cells were either pulsed for 10 min with 10 mM EdU at 18 or 24 h after transfection or labeled continuously with 0.5 mM EdU from 12 h after transfection. Cells were fixed in 70% ethanol and stained with mouse anti–γ-H2AX (1:500; Millipore) for 1 h, followed by 30-min incubation with Alexa Fluor 405 anti–mouse IgG (1:500; Invitrogen). EdU was detected with a Click-iT EdU Cell Proliferation Assay kit (Invitrogen), and DNA was stained with CellCycle 633 (Invitrogen) according to the manufacturer’s instruction. Flow cytometry analysis was performed on a BD LSRII using BD FACSDiva software. The EdU-positive and -negative populations were gated, and the percentage of γ-H2AX–positive cells within each population was determined.
Microscopy and immunofluorescence
Microscopy and immunofluorescence were performed as previously described (Jørgensen et al., 2007). For detection of RAD51 foci, cells were fixated in 3% newly made paraformaldehyde for 20 min at room temperature. The cells were rinsed twice in PBS 0.05% Tween 20, permeabilized in PBS with 0.3% Triton X-100, and rinsed once in PBS 0.05% Tween 20 before incubation of primary antibodies overnight, incubation with Alexa Fluor 488 or 594 anti–mouse or –rabbit secondary antibodies for 1 h, and mounting using Vectashield mounting medium (Vector Laboratories) containing DAPI. Pictures were acquired on a microscope (Axiovert 200M LSM 520; Carl Zeiss, Inc.) using a 63× c-Apochromat objective with NA 1.2 in H2O. Pictures were taken at room temperature using LSM 510 META software and LSM image examiner software (Carl Zeiss, Inc.). The pictures were exported in preparation for printing using Photoshop (Adobe) and Illustrator (Adobe).
PFGE
25-cm2 flasks were inoculated with 106 cells 24 h before siRNA transfection. Pulse labeling of the replication forks with [14C]thymidine (4.8 µM; 9.25 kBq/ml) for 30 min was performed 12 h after transfection. Cells were harvested 24 h after transfection. Irradiated samples were labeled immediately before irradiation and harvested immediately. Harvested cells were melted into 1% agarose plugs (InCert Agarose; Lonza; 5 × 105 cells/plug) and incubated for 48 h in 0.5% EDTA, 1% _N_-laurylsarcosyl, and 1 mg/ml proteinase K at 20°C. Plugs were washed four times in Tris-EDTA buffer before they were loaded onto a 1% agarose (chromosomal grade) gel and separated by PFGE for 20 h (CHEF-DR II system; Bio-Rad Laboratories; 120° angle, 60–240-s switch time, and 4 V/cm). DNA was visualized on the gel by ethidium bromide staining. For detection of radioactivity, the gel was dried and exposed to a phosphoimager plate for 96 h. Quantification was performed in Image Gauge software (FLA-3000; Fujifilm).
Online supplemental material
Fig. S1 shows the top-ranking genes from the screen as well as additional data on WEE1 depletion. Fig. S2 shows that DNA damage after WEE1 depletion occurs in S phase and that it is dependent on CDK1 and -2. Fig. S3 shows that DNA damage after CHK1 depletion is dependent on CDK1 and -2 and CDC25A and also contains data on the role of RAD51 after CHK1 and WEE1 depletion. Table S1, included as an Excel file, shows the results from the siRNA screen as well as bioinformatical analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905059/DC1.
Acknowledgments
We thank the Danish Cancer Society, the Novo Nordisk Foundation, the Lundbeck Foundation, the Danish Medical Research Council, the Norwegian Cancer Society, Harald Andersens Legat, and the South-Eastern Norway Regional Health Authority for their support of this work.
Footnotes
Abbreviations used in this paper:
DSB
double-strand break
HR
homologous recombination
HU
hydroxyurea
PFGE
pulsed field gel electrophoresis
PI
propidium iodide
RPA
replication protein A
RSA
redundant siRNA activity
ssDNA
single-stranded DNA
References
- Bader G.D., Betel D., Hogue C.W. 2003. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31:248–250 10.1093/nar/gkg056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Lukas J. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238–245 10.1016/j.ceb.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M., Gautier J., Dupré A. 2007. The DNA damage response during an unperturbed S-phase. DNA Repair (Amst.). 6:914–922 10.1016/j.dnarep.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Chen Y., Poon R.Y. 2008. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front. Biosci. 13:5016–5029 [DOI] [PubMed] [Google Scholar]
- Chow J.P., Siu W.Y., Ho H.T., Ma K.H., Ho C.C., Poon R.Y. 2003. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J. Biol. Chem. 278:40815–40828 10.1074/jbc.M306683200 [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Dunphy W.G. 1994. Cdc2 regulatory factors. Curr. Opin. Cell Biol. 6:877–882 10.1016/0955-0674(94)90060-4 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 444:638–642 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Harvey S.L., Kellogg D.R. 2003. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 13:264–275 10.1016/S0960-9822(03)00049-6 [DOI] [PubMed] [Google Scholar]
- Jenssen T.K., Laegreid A., Komorowski J., Hovig E. 2001. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 28:21–28 10.1038/88213 [DOI] [PubMed] [Google Scholar]
- Jørgensen S., Elvers I., Trelle M.B., Menzel T., Eskildsen M., Jensen O.N., Helleday T., Helin K., Sørensen C.S. 2007. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179:1337–1345 10.1083/jcb.200706150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature. 432:316–323 10.1038/nature03097 [DOI] [PubMed] [Google Scholar]
- Kellogg D.R. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 116:4883–4890 10.1242/jcs.00908 [DOI] [PubMed] [Google Scholar]
- König R., Chiang C.Y., Tu B.P., Yan S.F., DeJesus P.D., Romero A., Bergauer T., Orth A., Krueger U., Zhou Y., Chanda S.K. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 4:847–849 10.1038/nmeth1089 [DOI] [PubMed] [Google Scholar]
- Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 35:228–239 10.1016/j.molcel.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1987. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 49:559–567 10.1016/0092-8674(87)90458-2 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen C.S., Hansen L.T., Dziegielewski J., Syljuåsen R.G., Lundin C., Bartek J., Helleday T. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7:195–201 10.1038/ncb1212 [DOI] [PubMed] [Google Scholar]
- Stumpff J., Duncan T., Homola E., Campbell S.D., Su T.T. 2004. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr. Biol. 14:2143–2148 10.1016/j.cub.2004.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuåsen R.G., Sørensen C.S., Hansen L.T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25:3553–3562 10.1128/MCB.25.9.3553-3562.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S. 2007. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 12:1629–1641 10.2741/2175 [DOI] [PubMed] [Google Scholar]
- Tominaga Y., Li C., Wang R.H., Deng C.X. 2006. Murine Wee1 plays a critical role in cell cycle regulation and pre-implantation stages of embryonic development. Int. J. Biol. Sci. 2:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kennedy R.D., Ray K., Stuckert P., Ellenberger T., D’Andrea A.D. 2007. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 27:3098–3108 10.1128/MCB.02357-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Xie Y.M., Chen I.S. 2003. Depletion of Wee-1 kinase is necessary for both human immunodeficiency virus type 1 Vpr- and gamma irradiation-induced apoptosis. J. Virol. 77:2063–2070 10.1128/JVI.77.3.2063-2070.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]