Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine (original) (raw)
Abstract
In herpesviruses and many bacterial viruses, genome-packaging is a precisely mediated process fulfilled by a virally encoded molecular machine called terminase that consists of two protein components: A DNA-recognition component that defines the specificity for packaged DNA, and a catalytic component that provides energy for the packaging reaction by hydrolyzing ATP. The terminase docks onto the portal protein complex embedded in a single vertex of a preformed viral protein shell called procapsid, and pumps the viral DNA into the procapsid through a conduit formed by the portal. Here we report the 1.65 Å resolution structure of the DNA-recognition component gp1 of the Shigella bacteriophage Sf6 genome-packaging machine. The structure reveals a ring-like octamer formed by interweaved protein monomers with a highly extended fold, embracing a tunnel through which DNA may be translocated. The N-terminal DNA-binding domains form the peripheral appendages surrounding the octamer. The central domain contributes to oligomerization through interactions of bundled helices. The C-terminal domain forms a barrel with parallel beta-strands. The structure reveals a common scheme for oligomerization of terminase DNA-recognition components, and provides insights into the role of gp1 in formation of the packaging-competent terminase complex and assembly of the terminase with the portal, in which ring-like protein oligomers stack together to form a continuous channel for viral DNA translocation.
Keywords: terminase, DNA-packaging, oligomer, bacteriophage, protein:DNA interaction
In many tailed double-stranded DNA (dsDNA) bacteriophages and herpesvirus, the newly synthesized viral DNA in the infected host cell is present as a concatemer that is comprised of multiple copies of unit-length genome. The concatemeric DNA is packaged into a preformed capsid precursor termed procapsid through a channel formed by a portal protein complex embedded in a unique vertex of the procapsid (1–5). The DNA-packaging process involves a precisely coordinated sequence of molecular events, driven by a molecular machine called terminase consisting of two proteins: A DNA-recognition component and a catalytic component, also known as the “small” and “large” subunit, respectively. The terminase specifically binds to concatemeric viral DNA through the DNA-recognition component and cleaves it using the catalytic component, generating a new terminus for the DNA. This terminus is threaded through the portal protein channel embedded in the procapsid, and the catalytic component pumps the DNA into the procapsid in an ATP-dependent manner. When an appropriate amount of DNA is inserted, the terminase cuts the DNA again, dissociates from the procapsid, and can start the next cycle of DNA-packaging. Many of these features in DNA-packaging are also shared by adenoviruses (6). The x-ray structures of the catalytic component from bacteriophage T4 and the portals from various phages were previously described (7–10). However, it is not known how the terminases are assembled and how they interact with the portals and procapsids.
The terminases of many tailed dsDNA bacteriophages and herpesviruses contain protein components that are responsible for selection of viral DNA for packaging by recognizing sequence-specific motifs on viral DNA called packaging signals. These DNA-recognition components are small proteins of ∼140–210 amino acid residues in tailed bacteriophages, whereas they are large proteins with ∼700–900 amino acid residues in herpesviruses, which presumably encode additional biological functions such as interactions with other viral or host factors (11). The terminase DNA-recognition components of bacteriophages T4, T7, SPP1 and P22 have been reported to form oligomers (12–16). In bacteriophage lambda, the two components of terminase, gpNu1 and gpA, formed heterooligomers (17). Formation of oligomers has been shown to be critical for the function of the DNA-recognition components (15, 18).
Bacteriophage Sf6 is a member of the Podoviridae family of tailed dsDNA bacteriophage and an evolutionarily close relative of bacteriophage P22 (19, 20). Sf6 infects Shigella flexneri, an important human pathogen that can cause acute diarrhoea and bacillary dysentery. Sf6 is of particular interest because of its ability to alter the host’s serotype and virulence by changing host lipopolysaccharide structure via horizontal gene transfer (21). The Sf6 terminase consists of the DNA-recognition component gp1 and the catalytic component gp2 (19). Here we present the atomic structure of the terminase DNA-recognition component gp1 of bacteriophage Sf6 at 1.65 Å resolution. To our knowledge, this is the first atomic structure for any full-length terminase DNA-recognition component. The structure shows a ring-like molecular assembly suited to mediating protein:protein and protein:DNA interactions in formation of the packaging-competent terminase complex and terminase:portal assembly, indicating a central role of gp1 in initiation and progression steps of serial viral DNA-packaging.
Results
Overall Structure of gp1.
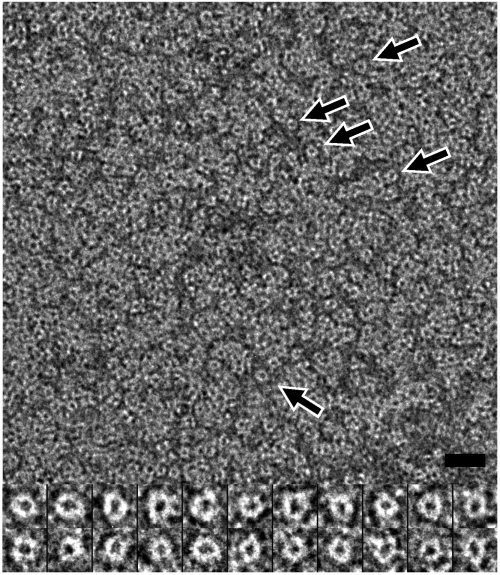
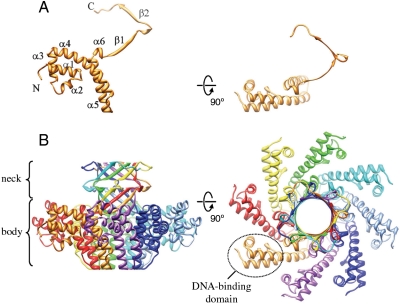
The 140-residue gp1 overexpressed in E. coli shows a major peak in size-exclusion chromatography, corresponding to the molecular mass of an octamer (Fig. S1). This oligomer sometimes remains stable in SDS-PAGE (Fig. S1). Negative staining electron microscopy of purified gp1 showed ring-like particles with an approximate outer diameter of 80 Å (Fig. 1). The purified gp1 was crystallized into the space group _P_4212 with unit cell dimensions of a = 88.9 and c = 72.6 Å. The structure was determined at 1.65 Å resolution using multiwavelength anomalous dispersion phasing (Table S1). The structure shows a ring-like octameric assembly (Fig. 2). The eightfold rotational axis of the octamer is coincidental with the crystallographic c axis, and the crystallographic asymmetric unit contains two gp1 molecules. Residues 10–139 and 10–132 are modeled for the two molecules respectively. The gp1 monomer displays a highly extended conformation, beginning with a compact DNA-binding domain (see below in Section gp1:DNA Interaction) formed by three N-terminal α-helices, followed by two long antiparallel α-helices in the central domain that pack against each other, and a lengthy C-terminal region that reaches as far as 40 Å away (Fig. 2, Fig. S2). This C-terminal region consists of two tandem β-strands separated by a 310 helix. While all the other helices are nearly ideally straight, the longest helix α4 (residues 54–76) is bent significantly by ∼45°.
Fig. 1.
Electron micrograph of purified gp1 negatively stained with 1% uranyl acetate. Ring-like particles distribute throughout the micrograph, and some are indicated by arrows. Closeups of representative ring-like particles were shown in the lower panel. Bar, 200 and 97 Å in the upper and lower, respectively.
Fig. 2.
Overall structure of gp1. Ribbon diagrams of the gp1 monomer (A) and the octameric assembly (B). The diagrams in the right column are rotated by 90° about the horizontal axis from those in the left column. (A) Secondary structure elements are labeled sequentially, and the N and C termini are indicated. (B) The eight monomers are ramp-colored, and one of the DNA-binding domains is indicated by a dashed ellipsoid. The gp1 monomer colored in gold on the left (B) is in identical orientation and scale with the one on the left in (A).
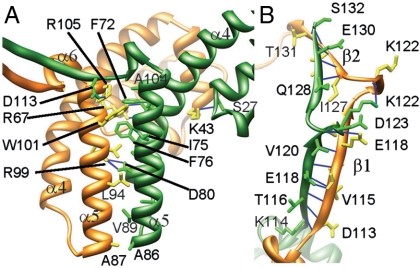
The overall gp1 octameric assembly resembles a vase with a wider body formed by the two long α-helices, a narrower neck formed by interweaved C-terminal β-strands, and eight peripheral appendages each formed by the N-terminal DNA-binding domain (Fig. 2). The assembly shows a diameter of 99 Å and a height of 66 Å. The outer diameters of the bottom end of the body and the top end of the neck are 40 and 48 Å, respectively. The eight monomers are closely packed, and as much as 26% of the total solvent accessible area, or 1,928 _Å_2, is buried for each monomer. Extensive intermolecular interactions contribute to the formation of the assembly, through bundled helices α4 and α5 in the central domains and a β-barrel formed by the C-terminal domains. The helix α5 from one molecule packs against a crevice formed by helices α4 and α5 from the neighboring molecule (Fig. 3), forming a three-helix bundle, and this helix bundling is mediated by intermolecular interactions of an array of hydrophobic residues F72, I75, F76, A79, A82, A86, V89, V96, L103 and A104 from the first molecule and residues A87, L94, W101 and M106 from the second. Three pairs of electrostatic interactions between acidic residues E73, D80, and D113 from the first molecule and arginine residues 67, 99, and 105 from the second, respectively, also contribute to the intermolecular interactions. The C-proximal β-strands form a second nexus of interactions that hold up the assembly (Fig. 3). These β-strands form a substructure reminiscent of β-barrels in many integral membrane proteins such as bacterial porin (22). Nevertheless, unlike the antiparallel β-strands in typical β-barrel structures, the β-strands in the gp1 assembly are parallel. The N-terminal DNA-binding domain participates minimal interactions with neighboring molecules, with only a hydrogen-bond between the carboxyl oxygen atom of S27 from one molecule and the side chain amino group of residue K43 from the second molecule. Thus, the DNA-binding domains appear to be hanging at the periphery of the whole assembly, supported by the long curved helix α4.
Fig. 3.
Intermolecular interactions in the gp1 assembly. Ribbon diagrams of the gp1 body region (A) and neck region (B) show side chains of residues involved in intermolecular interactions. The intermolecular hydrogen-bonds are shown as blue lines. The two adjacent gp1 monomers are in gold and green, respectively. Numbers of some residue as well as secondary structural elements are labeled to facilitate identification.
Architecture of the Channel in the gp1 Assembly.
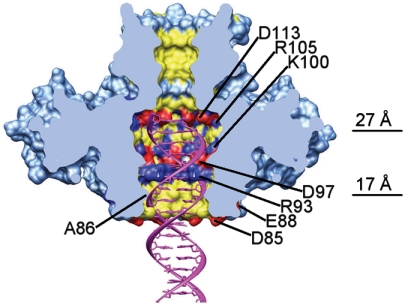
The eight copies of gp1 molecules form a ring-like architecture that embraces a continuous channel throughout the whole assembly (Fig. 4). The inner surface of the channel is dominated by side chains of positively and negatively charged residues including D97, D113, R93, K100, and R105, as well as hydrophobic residues A86, A90, L94, and W101 in the body region and V115, L119, I127, and I129 in the neck region. The channel in the gp1 assembly can be apparently divided into two segments: A wider part in the body region and a narrower part in the neck region. The diameter of the channel in the body region ranges from 17 Å near the bottom opening to 27 Å at the widest position (Fig. 4). In a related bacteriophage SPP1, the modeled dodecameric portal protein complex showed a channel with a narrowest inner diameter of 18 Å (10). The overall diameter of the channel in the body region nicely accommodates a B-form dsDNA (Fig. 4), suggesting that the channel is involved in DNA translocation. The two constrictions, one near the bottom opening formed by side chains of residues A86 and the other formed by side chains of residues R93, must closely interact with translocating DNA (Fig. 4).
Fig. 4.
Structure of the gp1 channel. The molecular surface of the gp1 assembly is shown in light blue. The positive, negative, and hydrophobic residues in the channel are colored in blue, red, and yellow, respectively. Charged residues forming rings in the channel are labeled. The widest position of the channel in the gp1 body and the position of the constriction formed by residues A86 are indicated by lines and values of diameters. The front half of gp1 was computationally removed to show the internal surface of the channel. Also shown is a B-form double-stranded DNA (Magenta) fitted into the channel.
The channel in the neck region formed by the gp1 C-terminal domain is significantly narrower, with an overall inner diameter of ∼10 Å (Fig. 4). The sharp transition between the wider body channel and the narrower neck channel is made by residues 107–112, which form a short α-helix (Fig. 4; helix α6 in Fig. 2). It was reported that the C-terminal region of phage lambda terminase DNA-recognition component gpNu1 interacted with the terminase large subunit (23). This implies that the neck region may undergo conformational change upon molecular association with the terminase large subunit such that the narrow channel in this region is widened for DNA translocation, which may serves as an additional regulatory mechanism.
Gp1:DNA Interaction.
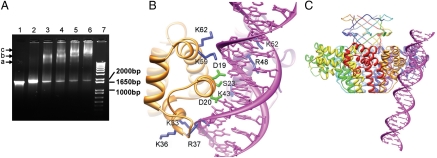
A large region of ∼1,800 bp in Sf6 genomic DNA that encompasses gp1- and a part of gp2-coding sequences was implicated to harbor the packaging signal (19). To characterize the binding of gp1 to the viral genomic DNA, we performed electrophoretic mobility shifting assay using the DNA corresponding to the region in the Sf6 genome that encodes gp1 and gp2 (the gp1–gp2 coding DNA). The assay shows a hierarchical mode of interactions between gp1 and the DNA (Fig. 5A). Three bands are visible, which may correspond to three species of nucleoprotein complexes formed by gp1 and DNA. When increasing the gp1 concentration, species a and b become less abundant, whereas species c becomes more abundant (Fig. 5A, lanes 3 to 6) and is dominant at higher concentration of gp1 (Fig. 5A, lane 6). Gp1 also binds to nonspecific DNA (Fig. S3), as was observed in other phage terminase small subunits including lambda gpNu1, T4 gp16, and P22 gp3 (12, 24, 25). However, gp1 binding with nonspecific DNA showed a relatively smeared band (Fig. S3) and didn’t show the characteristic three bands as in binding with the gp1–gp2 coding DNA.
Fig. 5.
DNA-binding of gp1. (A) Electrophoretic mobility shift assay of DNA-binding of gp1. Lane 7, 1 kb Plus DNA ladder (Invitrogen). The positions of three DNA species of 1,000, 1,650, and 2,000 bp are indicated. Lane 1, the DNA encompassing the gp1- and gp2-coding regions (1,836 bp) alone. Lane 2–6, the same amount of DNA as in Lane 1 but incubated with 25 μM, 50 μM, 75 μM, 100 μM, and 125 μM gp1, respectively prior to being loaded onto the gel. The bands corresponding to three species of nucleoprotein complexes are indicated with arrows and labeled with a, b, and c, respectively. (B) Modeled molecular interactions of two adjacent gp1 DNA-binding domains (Gold and Purple Ribbon Diagrams) with DNA (Magenta). Side chains of residues putatively involved in contact with DNA backbones and the major groove are shown in blue and green, respectively. Notice that three residues, K43, R48, and K52, belong to the gp1 monomer behind the DNA. (C) A panoramic view of the gp1 octameric assembly with the putative bound DNA. The view is nearly the same as in (B).
This pattern of retardation behavior, that is, presence of multiple bands in electrophoretic mobility shift assay, is reminiscent of the terminase DNA-recognition subunit gpNu1 of bacteriophage lambda (24), for which three gpNu1-binding sites were identified nearby the gpNu1-coding region in the phage DNA (26). Thus, it’s likely that there are multiple gp1-binding sites on the gp1–gp2 coding DNA, which results in multiple species of nucleoprotein complexes, and gp1 binds to these sites hierarchically in a way similar to lambda gpNu1.
The structure of the isolated N-terminal 68-residue DNA-binding domain of the bacteriophage lambda terminase DNA-recognition component gpNu1 was determined by NMR, showing that the α helix (residues 17–27) and the wing (residues 31–39) in the winged helix-turn-helix motif of gpNu1 were the structural determinants for phage lambda DNA-recognition (27). To understand the DNA-recognition mechanism of gp1, we performed three-dimensional structure alignment between the DNA-binding domains of gp1 and gpNu1. The overall folds of the structures are readily superimposable (Fig. S4). The α-helix (residues 17–27) and the wing (residues 31–39) in the winged helix-turn-helix motif of gpNu1 that were implicated in phage lambda DNA-recognition was aligned with α1 (residues 16–26) and the loop composed of residues 37–42 in gp1, respectively (Fig. S4). This suggests that helix α1 and the loop of residues 37–42 are directly involved in gp1:DNA interaction.
We manually superimposed a dsDNA onto the gp1 assembly by maintaining the typical scheme of interactions between DNA and a winged helix-turn-helix motif (Fig. 5B, C). The DNA fits nicely into an intermolecular crevice formed by two adjacent DNA-binding domains. The helix α1 of gp1 fits into the DNA major groove, with side chains of residues D19, D20, and S23 pointing into the DNA major groove, which may directly contact the bases of the DNA and be responsible for specificity of gp1:DNA-recognition. Positively charged residues K59, K62, K33, and R37 from two flanking regions make electrostatic interactions with the DNA backbone phosphate (Fig. 5B). A K59E mutant of gp1 failed to bind to DNA (Fig. S3), supporting this model and confirming that the N-terminal domain of gp1 is directly involved in DNA-binding. Positively charged residues K43, R48, and K52 from the adjacent gp1 molecule appear to make additional electrostatic interactions with the DNA backbone, helping hold the DNA in place (Fig. 5B).
Discussion
Oligomerization of the Terminase DNA-Recognition Components.
The Sf6 gp1 structure shows a ring-like octameric assembly. The central domain contributes to formation of the oligomer through bundling of helices from adjacent monomers. The C-terminal domain of gp1 forms a β-barrel, also contributing to assembly of the octamer. DNA-recognition components of terminase from phage T4 (12), T7 (16), SPP1 and a related Bacillus phage (28), and P22 (13, 25) all formed oligomers as deduced from biochemical or electron microscopic analysis, although oligomerization states ranged from octamer to decamer. In addition, a phage lambda in vitro packaging-competent complex formed by the small subunit gpNu1 and the large subunit gpA displayed a 8∶4 stoichiometry, suggestive of a gpNu1 octamer (29). Analysis of primary sequences and structure prediction identified coiled-coil motifs in several terminase DNA-recognition components (15), and it was demonstrated that oligomerization through coiled-coil motifs was required for function of the DNA-recognition component of phage T4 (15). In particular, gpNu1 of phage lambda was predicted to contain two consecutive coiled-coil motifs with high probability in approximate ranges of residues 50–75 and 85–110 (15), which match well with the two long helices α4 and α5 in the present structure of Sf6 gp1 in terms of their lengths and their relative positions in the polypeptides (Fig. S2). Part of the first coiled-coil motif in gpNu1 (residues 50–75) showed partially mobile helical structure in the NMR studies (27), which is close to the long helix α4 in gp1 in our superimposition of the two DNA-binding domains (Fig. S4) and could extend into a longer helix that is involved in oligomerization. Thus, the ring-like oligomerization through helix bundling as observed in the present Sf6 gp1 structure may represent a common structural scheme in terminase DNA-recognition components. It is worth pointing out that this similarity in three-dimensional structures for terminase DNA-recognition components may not be apparent at the primary sequence level, as analysis of the Sf6 gp1 primary sequence only showed a weakly probable coiled-coil region in gp1 at residues 48–68 (Fig. S5), and inspection of the gp1 sequence didn’t identify apparent heptad repeats. Indeed, these terminase DNA-recognition components show very low sequence identity. For example, the sequence identity of Sf6 gp1 with lambda gpNu1, T4 gp16, and P22 gp3 are 16%, 11%, and 15%, respectively. These indicate that the structural and functional relationships among these proteins are better understood at the three-dimensional level. The Sf6 gp1 structure shows an octamer. Nevertheless, no direct structural data are available for any terminase holoenzyme that includes both small and large subunits, or in complex with the portal or procapsid. Thus, further studies are required to reveal the stoichiometry of in vivo oligomers of terminase DNA-recognition components, which could vary among different phages as shown in heterologously expressed and purified proteins (see above).
Gp1:DNA Interaction and Formation of a Ternary Prepackaging Complex.
The Sf6 gp1 octamer presents eight DNA-binding domains at the periphery, indicating a capability of simultaneously binding to multiple sites on DNA. The electrophoretic mobility shifting assay of Sf6 gp1 with gp1-gp2 coding DNA showed multiple species of nucleoprotein complexes, which was not observed in gp1 binding with nonspecific DNA. The observed DNA-binding can largely be nonspecific DNA-protein interactions in view of high concentrations of protein used in the assays, and further analysis is required to define the nature of these complexes at the molecular level. Nevertheless, resemblance of these observations with those of lambda gpNu1 (24) favors a model in which gp1 can simultaneously binds to multiple sites on the DNA. In phage lambda, three segments of sequence-specific DNA (R-elements) in the viral genome contribute to DNA-binding of gpNu1, and the segment in the middle is arranged in the opposite direction with respect to the other two (26). The NMR structure of the dimeric DNA-binding domain of gpNu1 suggested that simultaneous binding of gpNu1 with multiple sites on viral DNA lead to DNA bending or looping (27). It was reported that binding of the terminase DNA-recognition component G1P of phage SPP1 induced DNA looping, and interestingly, multiple sites for G1P binding are present in the SPP1 genome-packaging initiation region (28). Taken together, these data suggest that, like other terminase DNA-recognition components, the oligomeric Sf6 gp1 may bind to multiple sites of the viral DNA, which may induce bending or looping of the viral DNA. This change in DNA conformation may be important for preparing a mature form of DNA for cleavage by the terminase catalytic subunit (large subunit) gp2 to create the new DNA terminus, which is to be inserted into the procapsid. It is clear that formation of a ternary prepackaging complex among the two terminase subunits and the viral DNA is a critical step in the initiation phase of viral DNA-packaging, and Sf6 gp1 appears to play a central role in that it directs the assembly of the ternary prepackaging complex through interactions with DNA and gp2.
Implication for Terminase:Portal Assembly.
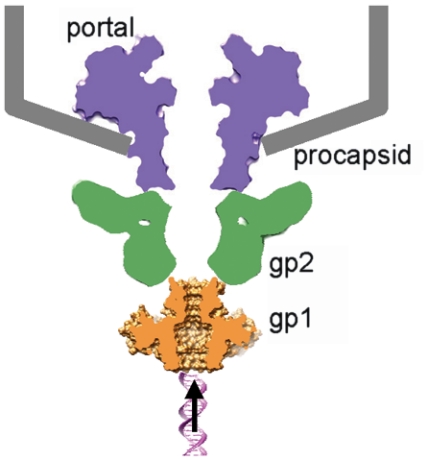
The terminase must dock onto the procapsid through association with the portal to start DNA translocation. Unlike sequence diversity in terminase small subunits, most phage terminase large subunits show conserved ATPase activity and nuclease activity associated with respective domains of the proteins, which is also true in herpesvirus (1, 7). Previous data suggested molecular interactions between terminase large subunits and portals in bacteriophages lambda (30), T3 (31), and T4 (32), and conserved polypeptide segments containing acidic and hydrophobic residues near the C termini of terminase large subunits of these three phages were identified to be potential portal-interacting sites. Direct contact between terminase large subunits and portal was also evident in an in vitro DNA-packaging system for phages T4 consisting of the terminase large subunit and the procapsid (7, 33) and an intermediate resolution electron cryomicroscopic analysis of phi29 procapsid captured in the process of DNA-packaging (9). On the other hand, it was reported that the C-terminal region of phage lambda terminase small subunit gpNu1 (DNA-recognition component) was responsible for interaction with the terminase large subunit (23). The present Sf6 gp1 octamer structure shows a channel suited for DNA passage. These data converge to a model for Sf6 terminase:portal assembly, in which coaxial stacking of ring-like oligomers of the two terminase subunits and the portal forms a continuous channel for viral DNA translocation (Fig. 6). In this model, the terminase large subunit is sandwiched by the portal and the terminase small subunit. Nevertheless, current data does not rule out an alternative model, in which the terminase small subunit acts as an adapter between the portal and the terminase large subunit (Fig. S6). This alternative model may not necessarily be incompatible with previous data suggesting interactions between portals and terminase large subunits, because: (i) the terminase large subunit gp17 in T4 showed a multidomain architecture with extended linker regions among domains (7) which can potentially allow conformational change needed to reach the portal, and (ii) terminase small subunits stimulate the ATPase activity of large subunits, an activity required for DNA-packaging, as shown in phages T4 (34, 35), SPP1 (36) and lambda (37), and this stimulation may result from conformational change of large subunits induced by binding of small subunits (38). A feature of this alternative model is that both terminase subunits participate in molecular interactions with the portal. Further studies are required to unveil the molecular mechanisms for terminase assembly and terminase:portal interactions, and the present structure of gp1 makes Sf6 a convenient model system in this regard.
Fig. 6.
A hypothetical model for coaxial assembly of terminase with portal and procapsid. The Sf6 terminase small subunit gp1 (Gold), the large subunit gp2 (Green), the portal (Purple), and the procapsid (Gray). A fragment of dsDNA is shown underneath to indicate the direction of DNA translocation. The shapes of the portal and the terminase large subunit are schematically shown based on x-ray structures of the phage SPP1 portal (RCSB PDB code 2jes) and the phage T4 gp17 (RCSB PDB code 3cpe).
Materials and Methods
Protein Purification, Crystallization and Data Collection.
The Sf6 gp1 gene was cloned from the Sf6 genomic DNA, and the protein was overexpressed in E. coli and purified as described in the SI Text. Crystals of gp1 were grown at 20 ºC by vapor diffusion in hanging drops containing equal volumes of protein solution (10 mg/mL) and reservoir solution with 100 mM CAPS pH 10.5, 0.2 M Li2SO4, 1.2 M NaH2PO4/0.8 M K2HPO4 and 5% glycerol. Crystals belonged to the space group _P_4212 with unit cell dimensions of a = 88.9 Å and c = 72.6 Å. For cryogenic experiments, crystals were soaked in the mother liquor plus 1.0 M Li2SO4 for 30 s, transferred to 2.0 M Li2SO4 and 5% glycerol for 5 min, and frozen in liquid nitrogen. The native x-ray dataset was collected at 100 K at the SSRL Beamline 11-1 on a Mar325 detector and the Se-Met datasets were collected at the SSRL Beamline 7-1 on a CCD detector. The diffraction data were processed with HKL2000 (39). X-ray data processing statistics are summarized in Table S1.
Structure Determination.
The structure of gp1 was determined with the 1.86 Å selenium multiwavelength anomalous dispersion data using the program SOLVE/RESOLVE (40). Automated model building and refinement was performed with the program PHENIX (41), and the resolution was extended to 1.65 Å for the native dataset. The final model building was done with the program O (42) and the final refinement of the coordinates was carried out using the program CNS (43). Refinement statistics are summarized in Table S1. The final model consists of two molecules of gp1 in the crystallographic asymmetric unit.
Supplementary Material
Supporting Information
Acknowledgments.
We thank the staff at the Stanford Synchrotron Radiation Laboratory for assistance in x-ray data collection. Electron micrographs were recorded on a Tecnai F20 G2 transmission electron microscope at the microscopy and analytical imaging laboratory at the University of Kansas. Research was supported by the University of Kansas, the National Institutes of Health Grant R21AI076249 and a subproject from the National Institutes of Health Center of Biomedical Research Excellence program P20RR17708 (to L.T.), and by the National Institutes of Health Grant R01AI074825 (to S.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.K. is a guest editor invited by the Editorial Board.
Data deposition: The coordinates of the Sf6 gp1 structure have been deposited in the RCSB Protein Data Bank with the accession code 3hef.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 3.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol 1. New York City: Plenum Press; 1988. pp. 15–91. [Google Scholar]
- 4.Trus BL, et al. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78(22):12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalano CE. Viral genome packaging machines: Genetics, structure, and mechanism. Georgetown, Tex., New York: Landes Bioscience/Eurekah.com; Kluwer Academic/Plenum Publishers; 2005. p. 153. [Google Scholar]
- 6.Ostapchuk P, Hearing P. Control of adenovirus packaging. J Cell Biochem. 2005;96(1):25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- 7.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135(7):1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Guasch A, et al. Detailed architecture of a DNA translocating machine: The high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315(4):663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 9.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26(7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7(2):107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272(6):3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 13.Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383(3):494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278(26):23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 15.Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: Insights into DNA packaging initiation and assembly of packaging motor. J Mol Biol. 2006;358(1):67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 16.White JH, Richardson CC. Gene 18 protein of bacteriophage T7. Overproduction, purification, and characterization. J Biol Chem. 1987;262(18):8845–8850. [PubMed] [Google Scholar]
- 17.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J Mol Biol. 2005;347(3):523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Berton N, Manning MC, Catalano CE. Domain structure of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999;38(43):14238–14247. doi: 10.1021/bi991408f. [DOI] [PubMed] [Google Scholar]
- 19.Casjens S, et al. The chromosome of Shigella flexneri bacteriophage Sf6: Complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol. 2004;339(2):379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 20.Gemski P, Jr, Koeltzow DE, Formal SB. Phage conversion of Shigella flexneri group antigens. Infect Immun. 11(4):685–691. doi: 10.1128/iai.11.4.685-691.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg AA, Wollin R, Gemski P, Wohlhieter JA. Interaction between bacteriophage Sf6 and Shigella flexner. J Virol. 1978;27(1):38–44. doi: 10.1128/jvi.27.1.38-44.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol R. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frackman S, Siegele DA, Feiss M. The terminase of bacteriophage lambda. Functional domains for cosB binding and multimer assembly. J Mol Biol. 1985;183(2):225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- 24.Yang Q, Hanagan A, Catalano CE. Assembly of a nucleoprotein complex required for DNA packaging by bacteriophage lambda. Biochemistry. 1997;36(10):2744–2752. doi: 10.1021/bi9622682. [DOI] [PubMed] [Google Scholar]
- 25.Nemecek D, et al. Subunit conformations and assembly states of a DNA-translocating motor: The terminase of bacteriophage P22. J Mol Biol. 2007;374(3):817–836. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker A, Murialdo H. Bacteriophage lambda DNA: The beginning of the end. J Bacteriol. 1990;172(6):2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Beer T, et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9(5):981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 28.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J Mol Biol. 1995;252(4):386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 29.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45(51):15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 30.Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J Mol Biol. 1995;245(2):141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 31.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245(5):635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 32.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: Interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289(2):249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 33.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363(4):786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 34.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. Biol Chem. 2000;275(47):37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 35.Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278(7):4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 36.Gual A, Camacho AG, Alonso JC. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J Biol Chem. 2000;275(45):35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, et al. Cloning, expression, and characterization of a DNA binding domain of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999;38(1):465–477. doi: 10.1021/bi981271d. [DOI] [PubMed] [Google Scholar]
- 38.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279(39):40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Terwillinger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information