You're one in a googol: optimizing genes for protein expression (original) (raw)
Abstract
A vast number of different nucleic acid sequences can all be translated by the genetic code into the same amino acid sequence. These sequences are not all equally useful however; the exact sequence chosen can have profound effects on the expression of the encoded protein. Despite the importance of protein-coding sequences, there has been little systematic study to identify parameters that affect expression. This is probably because protein expression has largely been tackled on an ad hoc basis in many independent projects: once a sequence has been obtained that yields adequate expression for that project, there is little incentive to continue work on the problem. Synthetic biology may now provide the impetus to transform protein expression folklore into design principles, so that DNA sequences may easily be designed to express any protein in any system. In this review, we offer a brief survey of the literature, outline the major challenges in interpreting existing data and constructing robust design algorithms, and propose a way to proceed towards the goal of rational sequence engineering.
Keywords: heterologous expression, synthetic biology, codon bias, gene optimization, gene design algorithms
1. Introduction
At the heart of biotechnology is our ability to cause a cell to produce a protein it would not normally make. These proteins may be useful in themselves, for example as therapeutics or industrial catalysts. They may enable a cell to produce new compounds or to interact with other cells in a novel way. Whether a protein is a modified version of proteins that are naturally produced by the intended expression host or comes from another kingdom, its sequence must be encoded in a gene that the host cell recognizes as instructions to produce appropriate amounts of the specified amino acid sequence.
Synthetic biologists envision a future in which combinations of well-characterized sequence elements lead to predictable outcomes, enabling the rational design of biological circuits and novel metabolic pathways (Andrianantoandro et al. 2006; Heinemann & Panke 2006; Drubin et al. 2007; Sayut et al. 2007; Tyo et al. 2007). Attempts to characterize and employ sequences that control transcription, mRNA stability and the initiation of translation are underway in many synthetic biology laboratories (Yokobayashi et al. 2002; Sprinzak & Elowitz 2005; Heinemann & Panke 2006; Dasika & Maranas 2008; Michalodimitrakis & Isalan 2009). One aspect of the design process that needs more attention, however, is the treatment of coding sequences. As in biotechnology, these are at the centre of synthetic biology: it is proteins that will catalyse the reactions in a novel metabolic pathway or be the signal transducers or the new biomaterials. There is an implicit assumption that, because we know the genetic code, it will be straightforward to choose a DNA sequence to encode any protein. But we need to think about more than the sequences that will ensure enough mRNA and an adequate rate of translational initiation: the codon choices themselves must not limit expression under the anticipated conditions of use.
Today, researchers can obtain genes by cloning from cDNA libraries or polymerase chain reaction (PCR) amplification from the source organism. Increasingly, they are also turning to direct synthesis of genes whose sequences appear in rapidly expanding sequence databases, but whose physical location is frequently obscure (Venter et al. 2004). When a gene is synthesized, it is generally modified from the natural version. These modifications are made to simplify subsequent manipulations (adding or eliminating restriction sites, for example), but also for a much more significant reason: natural genes are often poorly expressed in heterologous hosts, even when the expression system is related to the organism from which the gene originated.
Several different prokaryotic and eukaryotic systems are now available for heterologous expression, offering flexibility for a variety of protein types and applications. For improved results, these systems can be further manipulated by changing environmental conditions such as temperature or media components, by changing the intracellular environment by altering tRNA levels, and by changing the context and copy number of the gene itself (Hannig & Makrides 1998; Baneyx 1999; Aricescu et al. 2006). Despite so many options, natural genes are still frequently recalcitrant to expression in any host system useful for an intended application. Synthetic biology applications may also be particularly inflexible in their choice of host, especially if the characterization of synthetic parts turns out to be very host specific. Successful expression of target proteins, particularly high-yield expression, may therefore only be achieved by adaptation of the gene sequence to the desired expression system.
Unfortunately, robust rules for designing a gene for heterologous expression are not available, despite much study. There are two main reasons for this: (i) synthetic genes have only been widely and cheaply available for a few years, so systematic, well-controlled studies of the relationship between gene design parameters and expression have not been practical and (ii) protein synthesis is a complex process and probably depends on multiple properties of the gene sequence in addition to host-specific variables and environmental conditions.
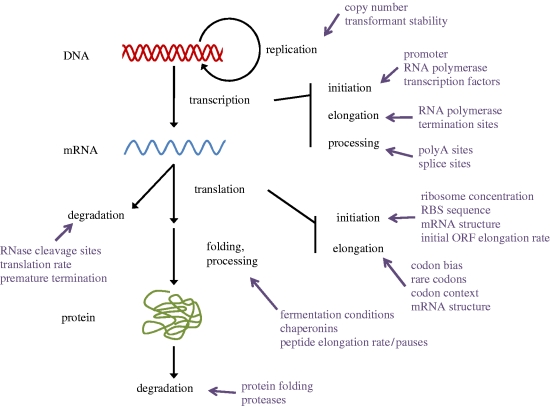
Design of an ‘optimal’ gene (which we define in this review as one in which the codon choices do not limit expression) requires a thorough understanding of the interaction of the gene sequence with the expression environment and specification of the desired goal (expression level, solubility, localization of expressed protein, etc.). One does not need to dig deeply into the scientific literature to realize that the relationship between sequence, host and expression properties is complex (figure 1). It is also clear that there are big gaps in the data available to map this relationship. Previous studies have generally focused on one or a few rules applied to design a single modified version of a gene (Gustafsson et al. 2004; Wu et al. 2007_a_,b). If expression is improved, there is a good chance that the result is described in the published literature; if not, it is probably not reported. As none of these rules has reliably improved expression, the tendency has been to layer more and more rules on top of each other, greatly complicating the gene design task and creating problems of prioritization when applying several conflicting principles. The problem is that the design rules are often only weakly supported by anecdotal data and little is known about their general applicability and relative significance. In many cases, they also have no obvious grounding in biochemical explanations of protein expression. A robust and generally applicable gene design method must instead be based on well-established relationships that are validated by thorough experimentation. The ability to create synthetic gene sets with variations in synonymous gene coding will be essential to elucidate these relationships.
Figure 1.
Factors influencing protein expression. Several factors that act along the path of expression from DNA to mRNA to protein are shown, any of which could be altered by or could affect the impact of gene design. RBS, ribosome-binding site.
In this review, we discuss the difficulties with current gene design methods, focusing on the interdependence and conflicts of common design principles. The principles that are currently applied are evaluated and challenges in incorporating them into algorithms for automated design are described. Finally, we propose practical ways to resolve current uncertainties that limit the potential of synthetic genes.
2. The gene design challenge
The standard genetic code encodes the 20 ubiquitous amino acids by 61 nucleotide triplets (codons). An amino acid may be encoded by as few as one or as many as six codons. This redundancy means that a protein can be encoded by many alternative nucleic acid sequences; a 300 amino acid protein of average amino acid composition could be encoded by more than 10100 different gene sequences. If the codon choice at each position is considered an independent variable, the possibilities would be distributed over an intractable, high-dimensional sequence space. Methodical gene optimization is thus only practical if the governing variables can be dramatically reduced and/or general rules exist to limit the considered possibilities.
A reasonable body of published work exists in which significant changes in expression are found in genes that have been resynthesized according to simple design rules (Gustafsson et al. 2004; Wu et al. 2007_a_,b). This is encouraging because it suggests that the optimization problem can be reduced to a manageable number of variables to describe a gene sequence. What has not yet emerged is the identity of the most important sequence variables or their contributions to protein expression. Part of the problem is one of variable definition—we know categorically what is important but not exactly what, how and/or when. For example, codon bias is widely thought to affect expression, but which codons are important, how best they should be biased, whether the same biases are important for all proteins and how this relates to the expression host is much less clear. Compounding this lack of clarity, we do not know how to prioritize or compromise with interdependent variables. For example, consider a protein containing many tyrosine residues that is encoded by following the rule ‘maximize GC content’. The resulting expression levels may then be ascribed to the overall GC content of the gene, since that was the design rule used. However, suppose that the expression levels were primarily influenced by the choice of UAC instead of UAU to encode every tyrosine. A second gene containing no tyrosines encoded by ‘maximizing GC content’ would be completely unaffected by the choice of tyrosine codons, so that the rule may result in quite different expression properties in the second case. In §3, the case for and limitations of various gene design variables previously associated with expression level are discussed.
3. Gene design principles
3.1. Origins of codon usage bias
Grantham et al. (1981) identified biases in the codons that were used to encode amino acids in the 161 full or partial mRNA sequences present in the nucleic acid sequence database at that time. These biases differed depending on which organism the gene came from. The authors speculated that these differences in codon bias may play a role in protein expression levels, and proposed a biophysical basis for this. We now have a much more detailed understanding of how gene expression is regulated including synthesis, processing and degradation of mRNA and initiation of translation. The role of natural codon biases, however, is still very poorly understood.
Why some organisms show marked bias and why organisms often differ dramatically has been the subject of much speculation (Holm 1986; Eyre-Walker & Bulmer 1993, 1995; Eyre-Walker 1996; Akashi 2001; Knight et al. 2001; Akashi & Gojobori 2002; Rocha 2004; Marquez et al. 2005; Suzuki et al. 2008; Yang & Nielsen 2008). Codon bias may serve to make the translational process more efficient. Biases can reduce the diversity of isoacceptor tRNAs required, perhaps reducing the metabolic load (Rocha 2004). This may particularly be beneficial to organisms that spend much of their life cycle in rapid growth. Several other constraints, not directly related to expression yield, are also likely to influence codon bias. These include altering the likelihood and directionality of amino acid substitutions (mutational bias) and selection for GC content (Eyre-Walker & Bulmer 1995; Eyre-Walker 1996; Knight et al. 2001; Marquez et al. 2005; Antezana & Jordan 2008; Yang & Nielsen 2008).
Whatever the evolutionary factors contributing to codon usage biases, the relevance of natural biases to designing genes for heterologous expression is not clear. Weak correlations between codon bias and expression of individual intragenomic genes have been observed in yeast and bacteria, but these genes are very rarely expressed at the high levels that are often desirable in biotechnological applications (more than 10 or 20% of cell protein). Furthermore, the cellular protein expression machinery has several means to control expression other than by control of translational elongation step rates. Indeed, in thorough studies where protein expression has been normalized relative to mRNA levels, correlations between expression and codon bias have all but disappeared (Gouy & Gautier 1982; Sharp & Li 1987; dos Reis et al. 2003; Jansen et al. 2003; Friberg et al. 2004; Lu et al. 2007; Wu et al. 2007_a_,b). Nevertheless, genes that are designed using different codon biases often do have significantly different expression properties, suggesting that bias or covariant gene variables are important (Gustafsson et al. 2004).
3.2. Biochemistry of codon usage bias
Synonymous codon choice may influence heterologous expression yield by limiting the translational elongation rate. For each codon along a message, the translational elongation step rate is probably primarily determined by the concentrations of cognate and competing EF-Tu.aa-tRNA ternary complexes in the cell and rate constants for complex selection at the ribosomal A-site (Varenne et al. 1984; Curran & Yarus 1989; Gromadski & Rodnina 2004; Wintermeyer et al. 2004; Rodnina et al. 2005). There is also evidence that the rate of tRNA selection at the A-site may be significantly influenced by the tRNA and codon occupying the ribosomal P-site, causing local context effects (Yarus & Folley 1985; Gouy 1987; Folley & Yarus 1989; Gutman & Hatfield 1989; Irwin et al. 1995; Boycheva et al. 2003; Moura et al. 2005; Buchan et al. 2006). Finally, and of particular relevance when high levels of heterologous protein are expressed, the concentrations of free amino acids and charged tRNA in the cell could change significantly, altering the relative translation step rates for different codons (Dong et al. 1995; Elf et al. 2003; Dittmar et al. 2005; Elf & Ehrenberg 2005_a_,b).
A detailed mechanistic model of expression incorporating accurate rates for all steps in the synthesis pathway and context dependence is not imminent even for Escherichia coli and even further off for many other potentially useful hosts. Even without complete understanding, however, the biochemical principles of expression can inspire some reasonable guesses about design criteria. In E. coli, there is some correlation between codon usage frequency and observed cognate tRNA level, which is more pronounced at higher growth rates (Ikemura 1981; Bulmer 1987; Dong et al. 1996). Translation of a gene containing many codons that are rarely used in the host organism will therefore generally use cognate tRNAs that are present at low levels in the cell in a large number of steps in translational elongation. This would be expected to impair expression, an effect that is indeed observed (Chen & Inouye 1990; Kane 1995; Cruz-Vera et al. 2004). Also consistent with this mechanistic explanation, E. coli strains expressing boosted levels of such tRNAs from plasmid-borne genes can in some cases support increased expression levels of genes containing rare codons (Kane 1995; Burgess-Brown et al. 2008).
Although rare codons may often be translated at lower rates, the relationship between their use and expression yield is not a simple one. In fact, in some cases, the inclusion of rare codons may even improve yield perhaps by controlling the ribosomal traffic along the translated message or by introducing translational pauses at strategic positions, such as domain boundaries, to help promote proper protein folding (Angov et al. 2008; Tsai et al. 2008). Also, position and sequence context of the rare codons can significantly affect their impact. In certain contexts, rare codons have been shown to increase translational errors (Del Tito et al. 1995; Kane 1995; Kurland & Gallant 1996; You et al. 1999; Kerrigan et al. 2008). In particular, consecutive rare codons within the first codons of a message may be especially deleterious, whereas in some cases rare codons may be distributed downstream of the initial coding sequence with little effect (Varenne & Lazdunski 1986; Chen & Inouye 1990, 1994).
How to abstract gene design principles from non-rare codon biases in an expression host's genome is even less clear. One idea that is commonly cited, despite the gradual evaporation of experimental support, is that higher levels of expression can be obtained by maximizing high-frequency codons within a gene. This idea is an extension of Grantham's work (Grantham et al. 1981), coupled with the observation that some highly expressed genes in E. coli and Saccharomyces cerevisiae (predominantly ribosomal proteins) are more biased in codon usage than the average for the genome (Sharp & Li 1987). In this line of reasoning, the codon that is used most frequently in these highly expressed genes is considered an ‘ideal’ codon. How closely a gene conforms to this ideal can then be quantified as the codon adaptation index (CAI; Sharp & Li 1987); a gene of maximal CAI equal to 1 is one that uses only the most frequent codon in the high expressor subset to encode each amino acid. Although the CAI of a gene has often been cited as a predictor of the expression level of a protein, there is no demonstrated causality. In E. coli and S. cerevisiae, where protein and mRNA levels have both been measured, there is no meaningful correlation between CAI and protein yield per mRNA transcript, suggesting that CAI is not a measure of translational efficiency (Friberg et al. 2004; Lu et al. 2007).
From a purely biochemical perspective, simply maximizing the CAI of a gene might be problematic, especially for applications where the target protein is to be expressed at much higher levels than any single natural protein. While it would favour the use of tRNAs present at higher levels in a non-expressing cell, limiting the used tRNA pool to just one or two isoacceptors per amino acid could limit the maximal synthesis flux and increase translational errors (Kane 1995; Kurland & Gallant 1996). A balance between tRNAs used that maximizes the availability of aa-tRNA for production while maintaining a non-limiting elongation rate is probably preferable.
An additional complication is that different proteins with different amino acid compositions will stress the translational process differently (Kane 1995). For example, the optimal balance of serine codons in a gene encoding a protein containing only a few serine residues may be quite different from the optimal balance of serine codons where the protein is 20 per cent serine. In the latter case, the rates at which serine tRNAs are recharged may have a significant impact on translation rate. Likewise, the effect of codon bias may depend on the expression level itself. The prevalence of serine in a protein, for example, may not matter if the protein is produced at a low level owing to other expression limiting factors, such as low mRNA level or slow translational initiation. In such a case, the stress on serine tRNAs would be low, independent of the protein serine content.
Many examples exist where changes in synonymous codon usage have a dramatic effect on the yield of heterologously expressed protein, but drawing conclusions about optimal codon biases from these data is very difficult (Gustafsson et al. 2004). Published examples differ widely in many respects including the expression host, regulatory elements associated with the gene and, most importantly, the protein being expressed. Furthermore, the sample sets in such examples are generally small, usually describing only two genes, one natural and one whose codon bias has been altered. Now that genes can be synthesized quickly and cheaply, it should be possible to construct sets that are diversified systematically to experimentally test the effects of changes in bias of codons for each amino acid, the occurrence of codon pairs, GC%, the use of rare codons and other potentially important determinants of expression level. The effects of other factors on expression can also be tested, including those described in the following sections.
3.3. Codon bias at the start of the open reading frame
Numerous lines of evidence suggest that the initial 15–25 codons of the open reading frame deserve special consideration in gene optimization (Eyre-Walker & Bulmer 1993; Chen & Inouye 1994; Stenström et al. 2001_a_,b; Stenström & Isaksson 2002; Gonzalez de Valdivia & Isaksson 2004, 2005). Natural E. coli genes show a distinct bias in codon usage for the initial 25 codons compared with the overall genomic bias (Eyre-Walker & Bulmer 1993; Chen & Inouye 1994; Stenström et al. 2001_b_). In fact, rare codons are enriched in this initial leader for reasons that are not clear. Studies have shown that the impact of rare codons on translation rate is particularly strong in these first codons, especially within the first six triplets (Chen & Inouye 1990, 1994).
Ribosomes in the initial phase of elongation appear to be particularly prone to abortive termination, perhaps owing to an increased rate of peptidyl-tRNA drop-off (Gonzalez de Valdivia & Isaksson 2004, 2005). Early rare and NGG codons may accelerate premature termination by stalling elongation (Gonzalez de Valdivia & Isaksson 2005). These codon effects appear to be independent of alterations in mRNA secondary structure that might also stall early elongation or prevent initiation. As translational initiation depends on the rates of both ribosome binding and clearing of the ribosome-binding site (RBS) after initial elongation (approx. 13–20 codons), slow translation through the initial leader may reduce or eliminate any benefits of a strong RBS sequence.
3.4. mRNA structure
Gene design strategies often seek to minimize mRNA structure. Structures that involve or otherwise occlude the RBS and/or start codon in genes expressed in prokaryotes can impair expression, presumably by interfering with ribosomal binding and translational initiation (Kozak 1986; de Smit & van Duin 1990, 1994; Griswold et al. 2003; Studer & Joseph 2006). For this reason, gene design strategies often consider such structure in coding of the first several amino acids. Voigt and co-workers have recently developed an algorithm for designing prokaryotic RBSs to achieve desired rates for initiation of translation considering the structure of the mRNA and the affinity of the RBS for the ribosome (http://www.voigtlab.ucsf.edu/software/).
As with codon bias, considerations of the effects of mRNA structure within the open reading frame are not straightforward. While some RNA structures, particularly pseudoknots, have been shown to cause translational pauses (Kontos et al. 2001; Hansen et al. 2007; Wen et al. 2008), a clear relationship of RNA structure strength, type and distribution to translation rate is lacking. Ribosomes possess an intrinsic helicase activity that allows translation through even very strong hairpins and may preclude many structures from limiting the translation rate (Takyar et al. 2005). An actively translated message can be densely packed with ribosomes, unwinding structure as they move along. For this reason and others, structures predicted by RNA folding algorithms may not reliably represent actual mRNA structures in vivo (Meyer & Miklos 2004, 2007). Relevant structures may be those restricted to windows along the mRNA where structure could form between ribosomes. The lengths and lifetimes of such windows would be dependent on translational kinetics and would probably vary significantly along the message. These many layers of uncertainty greatly obscure a rational approach to general mRNA structure optimization. As structure minimization strategies can greatly influence other gene parameters, such as codon bias, it is critical that systematic analysis of the benefits of various mRNA structure treatments be performed.
3.5. Gene design and protein structure
Although much of the forgoing discussion has implicitly assumed that maximizing the rate of translational elongation is unequivocally desirable, this is not entirely accurate. Often the expressed protein must be properly folded to be useful. There have been several recent reports describing the effects of synonymous codon changes on protein folding (Thanaraj & Argos 1996; Angov et al. 2008; Tsai et al. 2008). It has been suggested that too rapid translation may not allow for efficient ‘self’ or chaperone-aided folding and that strategically placed slower codons or codon runs, perhaps at protein domain boundaries, could maximize folding efficiency while maintaining a high overall translation rate (Angov et al. 2008). Unfortunately, there are even less data from which to derive rules for such designs than there are for understanding codon bias. Developing rules for designing genes to express soluble active protein should be facilitated by synthesis and testing of varied sets of genes, as described above for penetrating the mysteries of codon bias and other gene variables.
3.6. Potentially deleterious motifs
Depending on the host expression system, there are a number of sequence motifs to be avoided in gene design. These comprise an expanding list of sequence element classes that could have negative effects on expression of a target protein. For example, in an E. coli system expressing a gene under control of a T7 promoter, one would wish to avoid both class I and II transcriptional termination sites. Shine–Dalgarno-like sequences within the coding sequence may cause incorrect downstream initiation or translational pauses in prokaryotic hosts. In eukaryotic hosts, potential splice signals, polyadenylation signals and other motifs affecting mRNA processing and stability are generally to be avoided. Other classes of deleterious motifs include sequences that promote ribosomal frameshifts and pauses (Kurland & Gallant 1996; Kontos et al. 2001; Hansen et al. 2007). For many of these motifs, polyadenylation sites in particular, the relationship of sequence and impact on expression is not yet well understood. With further work, we expect that the list of toxic and regulatory motifs will grow, but also that rules for avoiding them in gene design will be better defined.
4. Integrating principles into design algorithms
Several algorithms have been developed which allow researchers to manipulate various gene design parameters (Grote et al. 2005; Jayaraj et al. 2005; Villalobos et al. 2006; Ferro et al. 2007; Wu et al. 2007_a_,b). Ideally, an algorithm should be based on an accurate predictive model of the relationship between design parameters and expression yield. To develop such a model, it is critical to first identify the sufficient subset of predictive design variables for explaining expression. There is good reason to hope that careful experimentation will allow reasonable quantification of the effects of codon bias, mRNA structure and other factors on heterologous expression in various expression systems.
Prioritization of the expression-determining variables is also necessary to create a robust design algorithm. It may not be sufficient or practical to simply apply standard criteria independently to a number of design parameters, as the parameters themselves may not be fully independent of each other. Avoidance of possible deleterious motifs, particularly those that are ambiguously defined or otherwise common, can conflict with codon usage and other design parameters. Common design requirements such as the removal of restriction sites, avoidance of dam or dcm methylation sites overlapping with restriction sites or elimination of extended coding sequences in other reading frames also constrain codon choices.
Optimization of multiple constraints based on anecdotal information and accepted but often unsubstantiated lore is particularly problematic. This ‘system voodoo’ can so significantly limit the available DNA sequences as to actually preclude adequate expression! It is impossible to overstate the value of experimental support for assigning importance to the impact of sequence variables on expression.
4.1. Managing constraints
Irrespective of the specific variables, gene design will always involve several, often conflicting, types of sequence constraints. Meeting these various constraints simultaneously necessitates development of sophisticated algorithms. The most useful algorithms would allow flexibility in prioritization of constraints, as appropriate for different applications and design goals. In some cases, compromises may be acceptable for some of the parameters, for example minimizing repetitive sequences instead of eliminating them completely. In other cases, the algorithm might have the choice of meeting at least one of a set of constraints. For example, the gene design may require that either EcoRI or HindIII sites not be present in the resulting DNA sequence.
Another important criterion is algorithm efficiency or run time. For a complex set of design constraints, optimization can be computationally intensive, especially for large genes. In some cases, running the algorithm for a few days or weeks might be acceptable, as long as all the goals are met. In other cases, the algorithm might need to be executed for a large library of genes in a quick manner, so that post-optimization analysis may be performed and new goals defined. Finally, there will often be cases where the optimization problem is so difficult, owing to particulars of the amino acid sequence to be encoded and/or the combination of constraints applied, that design goals cannot be reached within a reasonable time and an exhaustive search of all parameter space is unrealistic. Thus, a practical algorithm must employ some kind of heuristic, perhaps Monte Carlo random walks, simulated annealing or a genetic algorithm, to efficiently search parameter space.
Typical gene design parameters vary in nature and present different problems in optimization. Particularly challenging are parameters of a distributed nature such as codon bias or repeats in the DNA sequence, which are necessarily interdependent with other parameters. Codon bias for at least one amino acid will change whenever a new codon is chosen anywhere in the sequence to modify any other design parameter. Changing a codon to eliminate repetition of one sequence element within the gene may introduce other, different repeated elements. Design algorithms must use optimization methods that are suited to the nature of the parameters involved.
4.2. Choosing optimization methods
For any optimization method, the starting point can be very important. Ideally, it should be chosen as close as possible to the expected optimum and in a way that is not deterministic. That way, if the starting point proves unacceptable, the algorithm can be restarted with a new starting point. One way to select a starting point that is non-deterministic and focuses on search space near a typical optimization goal is to select a codon for each amino acid, based on the probability of that codon occurring as given by a target codon bias table.
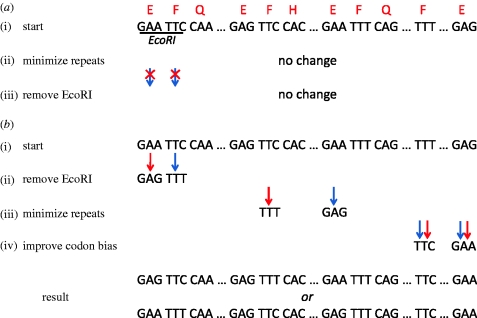
As is common with multidimensional optimization problems, there are many ways of navigating the search space and each has its benefits depending on the optimization requirements (figure 2). The search hierarchy must accommodate the interdependencies and priorities of the constraints. Otherwise, conflicts might cause the algorithm to get cornered in a local optimum and not reach the design goals.
Figure 2.
Choosing an appropriate design algorithm. A simple example is shown of how two different algorithms for the same optimization problem are affected by sequence constraints. The coding sequence encodes five peptide segments of a protein, which may or may not be contiguous. The initial starting sequence is one possibility, chosen to match the target codon bias of the gene. The optimization constraints for both algorithms are that (i) no EcoRI is allowed, (ii) codon usage ratios for E (GAG/GAA) and F (TTC/TTT) must be equal to 1, and (iii) direct sequence repeats greater than seven nucleotides should be minimized. Iterations involve single codon replacements and a greedy search is followed. Thus, replacements are allowed only if improvement is achieved. At each step, no worsening of previously applied constraints is allowed. The algorithm in (a) begins by minimizing repeat elements and then tries to remove EcoRI sites without increasing the number of repeats. Since either possible substitution to remove the EcoRI site will add new repeats, no change is allowed and the algorithm fails to reach its goals. In (b), because the hard constraint of restriction site removal is applied first, the algorithm has two routes (red versus blue arrows) to successfully reach the goals.
If there is only one constraint (codon bias, for example) or multiple constraints with non-conflicting goals, then a ‘greedy’ algorithm may suffice to rapidly find an optimum. At each iteration of such an algorithm, the sequence is scored based on the optimization parameters. If all the goals have not been met, a random codon position is changed and the resulting sequence is scored. If the new sequence is improved, it becomes the starting point for the next iteration. Otherwise, the previous sequence is kept and changed randomly again in the next iteration. This continues until all goals are met or a minimum is reached.
If the constraints are interdependent, as the most proposed gene optimization parameters are, a greedy algorithm may be prone to getting trapped in undesired local optima as it will always work towards the optimum closest to the starting point. One way of getting around this problem is to apply simulated annealing (Kirkpatrick et al. 1983; Rodrigo et al. 2007; Rocha et al. 2008). In this method, worse scoring sequences can also be selected as the next current state, but at a given probability (‘temperature’). As the iterations step forward, the temperature is dropped progressively, decreasing the probability of accepting a sequence that scores lower than its predecessor. This ‘cooling’ results in an algorithm that initially samples a broad region of search space and then slowly becomes greedier in its heuristic, eventually becoming a simple greedy algorithm once it reaches zero. Generally, the ‘best state’ observed during the iterations is taken as the final result.
Another method for avoiding local optima, particularly as parameter space becomes large and interdependency is high, is to simultaneously follow multiple search paths and choose those that perform best. In a ‘genetic algorithm’, for example, a population of different current states is maintained (Mitchell 1998; Patil et al. 2005; Rocha et al. 2008). With each generation, the best individual sequences are selected as parents for the next generation. These are randomly mutated and recombined. The best of the resulting progeny is then selected and iterations continue until convergence in performance of the population is reached. The combination of multiple starting points and diversification through mutation and recombination efficiently searches a large expanse of sequence space, avoids single suboptimal solutions and is more likely to find a true optimum for complex multivariate problems.
4.3. Algorithms for exploring parameter space
Creating an optimization algorithm to find sequences that meet multiple interdependent constraints is only half the battle. The functionality of sequences designed by these algorithms will generally be limited by how well the constraints that the algorithm imposes match parameters that actually affect expression. Robust optimization algorithms will therefore require data and development of valid models describing the design–expression relationship. One way of approaching this problem is to coevolve the design algorithms together with these models. Initial algorithms can be used to independently vary parameters thought to be important, with experimental measurements and data modelling allowing these hypotheses to actually be tested. As more data are gathered, unimportant parameters will be discarded, new parameters may be added and remaining parameters will be reprioritized in the model. Thus, the optimization algorithm and our understanding of how to design genes for protein expression will be refined together.
5. Future prospects
Rapid expansion of sequence databases and development of gene synthesis technologies have greatly increased the repertoire of protein sequences to which biological researchers have access. Natural, derivative or novel sequences of interest may be directly obtained by researchers with minimal expertise in molecular biology. Although the rules for deciphering a DNA sequence to determine the amino acid sequence of the encoded protein were established over 40 years ago, the rules for designing DNA sequences to express an encoded protein are still not well understood. Fortunately, the methods for determining such rules are very familiar to both scientific and engineering traditions, merged in the field of synthetic biology. Reliable criteria for designing expressible genes will help to enable synthetic systems, where a gene encoding any protein may be slotted between reusable control elements, combined into new biosynthetic pathways or biological circuits without having to suffer through extensive trial and error just to get the gene to express.
Footnotes
One contribution to a Theme Supplement ‘Synthetic biology: history, challenges and prospects’.
References
- Akashi H. 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11, 660–666. ( 10.1016/S0959-437X(00)00250-1) [DOI] [PubMed] [Google Scholar]
- Akashi H., Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA. 99, 3695–3700. ( 10.1073/pnas.062526999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Basu S., Karig D. K., Weiss R. 2006. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2, 2006.0028 ( 10.1038/msb4100073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angov E., Hillier C. J., Kincaid R. L., Lyon J. A. 2008. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS ONE. 3, e2189 ( 10.1371/journal.pone.0002189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antezana M. A., Jordan I. K. 2008. Highly conserved regimes of neighbor-base-dependent mutation generated the background primary-structural heterogeneities along vertebrate chromosomes. PLoS ONE. 3, e2145 ( 10.1371/journal.pone.0002145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu A. R., et al. 2006. Eukaryotic expression: developments for structural proteomics. Acta Crystallogr. D: Biol. Crystallogr. 62, 1114–1124. ( 10.1107/S0907444906029805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10, 411–421. ( 10.1016/S0958-1669(99)00003-8) [DOI] [PubMed] [Google Scholar]
- Boycheva S., Chkodrov G., Ivanov I. 2003. Codon pairs in the genome of Escherichia coli. Bioinformatics. 19, 987–998. ( 10.1093/bioinformatics/btg082) [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Aucott L. S., Stansfield I. 2006. tRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Res. 34, 1015–1027. ( 10.1093/nar/gkj488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. 1987. Coevolution of codon usage and transfer RNA abundance. Nature. 325, 728–730. ( 10.1038/325728a0) [DOI] [PubMed] [Google Scholar]
- Burgess-Brown N. A., Sharma S., Sobott F., Loenarz C., Oppermann U., Gileadi O. 2008. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr. Purif. 59, 94–102. ( 10.1016/j.pep.2008.01.008) [DOI] [PubMed] [Google Scholar]
- Chen G., Inouye M. 1990. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 18, 1465–1473. ( 10.1093/nar/18.6.1465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. T., Inouye M. 1994. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 8, 2641–2652. ( 10.1101/gad.8.21.2641) [DOI] [PubMed] [Google Scholar]
- Cruz-Vera L. R., Magos-Castro M. A., Zamora-Romo E., Guarneros G. 2004. Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. Nucleic Acids Res. 32, 4462–4468. ( 10.1093/nar/gkh784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. 1989. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 209, 65–77. ( 10.1016/0022-2836(89)90170-8) [DOI] [PubMed] [Google Scholar]
- Dasika M. S., Maranas C. D. 2008. OptCircuit: an optimization based method for computational design of genetic circuits. BMC Syst. Biol. 2, 24 ( 10.1186/1752-0509-2-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Tito B. J., Jr, Ward J. M., Hodgson J., Gershater C. J., Edwards H., Wysocki L. A., Watson F. A., Sathe G., Kane J. F. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 177, 7086–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl Acad. Sci. USA. 87, 7668–7672. ( 10.1073/pnas.87.19.7668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. 1994. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 244, 144–150. ( 10.1006/jmbi.1994.1714) [DOI] [PubMed] [Google Scholar]
- Dittmar K. A., Sørensen M. A., Elf J., Ehrenberg M., Pan T. 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6, 151–157. ( 10.1038/sj.embor.7400341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Nilsson L., Kurland C. G. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 177, 1497–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Nilsson L., Kurland C. G. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663. ( 10.1006/jmbi.1996.0428) [DOI] [PubMed] [Google Scholar]
- dos Reis M., Wernisch L., Savva R. 2003. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 31, 6976–6985. ( 10.1093/nar/gkg897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. A., Way J. C., Silver P. A. 2007. Designing biological systems. Genes Dev. 21, 242–254. ( 10.1101/gad.1507207) [DOI] [PubMed] [Google Scholar]
- Elf J., Ehrenberg M. 2005a. Near-critical behavior of aminoacyl-tRNA pools in E. coli at rate-limiting supply of amino acids. Biophys. J. 88, 132–146. ( 10.1529/biophysj.104.051383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J., Ehrenberg M. 2005b. What makes ribosome-mediated transcriptional attenuation sensitive to amino acid limitation?. PLoS Comput. Biol. 1, e2 ( 10.1371/journal.pcbi.0010002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J., Nilsson D., Tenson T., Ehrenberg M. 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 300, 1718–1722. ( 10.1126/science.1083811) [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. 1996. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy?. Mol. Biol. Evol. 13, 864–872. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Bulmer M. 1993. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 21, 4599–4603. ( 10.1093/nar/21.19.4599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Bulmer M. 1995. Synonymous substitution rates in enterobacteria. Genetics. 140, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro A., Giugno R., Pigola G., Pulvirenti A., Di Pietro C., Purrello M., Ragusa M. 2007. Sequence similarity is more relevant than species specificity in probabilistic backtranslation. BMC Bioinform. 8, 58 ( 10.1186/1471-2105-8-58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folley L. S., Yarus M. 1989. Codon contexts from weakly expressed genes reduce expression in vivo. J. Mol. Biol. 209, 359–378. ( 10.1016/0022-2836(89)90003-X) [DOI] [PubMed] [Google Scholar]
- Friberg M., von Rohr P., Gonnet G. 2004. Limitations of codon adaptation index and other coding DNA-based features for prediction of protein expression in Saccharomyces cerevisiae. Yeast. 21, 1083–1093. ( 10.1002/yea.1150) [DOI] [PubMed] [Google Scholar]
- Gonzalez de Valdivia E. I., Isaksson L. A. 2004. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 32, 5198–5205. ( 10.1093/nar/gkh857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez de Valdivia E. I., Isaksson L. A. 2005. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. FEBS J. 272, 5306–5316. ( 10.1111/j.1742-4658.2005.04926.x) [DOI] [PubMed] [Google Scholar]
- Gouy M. 1987. Codon contexts in enterobacterial and coliphage genes. Mol. Biol. Evol. 4, 426–444. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10, 7055–7074. ( 10.1093/nar/10.22.7055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. 1981. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 9, r43–r74. ( 10.1093/nar/9.1.213-b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold K. E., Mahmood N. A., Iverson B. L., Georgiou G. 2003. Effects of codon usage versus putative 5′-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expr. Purif. 27, 134–142. ( 10.1016/S1046-5928(02)00578-8) [DOI] [PubMed] [Google Scholar]
- Gromadski K. B., Rodnina M. V. 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 13, 191–200. ( 10.1016/S1097-2765(04)00005-X) [DOI] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Munch R., Nortemann B., Hempel D. C., Dieter J. 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 1, W526–W531. ( 10.1093/nar/gki376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353. ( 10.1016/j.tibtech.2004.04.006) [DOI] [PubMed] [Google Scholar]
- Gutman G. A., Hatfield G. W. 1989. Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl Acad. Sci. USA. 86, 3699–3703. ( 10.1073/pnas.86.10.3699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig G., Makrides S. C. 1998. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol. 16, 54–60. ( 10.1016/S0167-7799(97)01155-4) [DOI] [PubMed] [Google Scholar]
- Hansen T. M., Reihani S. N., Oddershede L. B., Sorensen M. A. 2007. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 104, 5830–5835. ( 10.1073/pnas.0608668104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M., Panke S. 2006. Synthetic biology-putting engineering into biology. Bioinformatics. 22, 2790–2799. ( 10.1093/bioinformatics/btl469) [DOI] [PubMed] [Google Scholar]
- Holm L. 1986. Codon usage and gene expression. Nucleic Acids Res. 14, 3075–3087. ( 10.1093/nar/14.7.3075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151, 389–409. ( 10.1016/0022-2836(81)90003-6) [DOI] [PubMed] [Google Scholar]
- Irwin B., Heck J. D., Hatfield G. W. 1995. Codon pair utilization biases influence translational elongation step times. J. Biol. Chem. 270, 22 801–22 806. ( 10.1074/jbc.270.39.22801) [DOI] [PubMed] [Google Scholar]
- Jansen R., Bussemaker H. J., Gerstein M. 2003. Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res. 31, 2242–2251. ( 10.1093/nar/gkg306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraj S., Reid R., Santi D. V. 2005. GeMS: an advanced software package for designing synthetic genes. Nucleic Acids Res. 33, 3011–3016. ( 10.1093/nar/gki614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane J. F. 1995. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6, 494–500. ( 10.1016/0958-1669(95)80082-4) [DOI] [PubMed] [Google Scholar]
- Kerrigan J. J., McNulty D. E., Burns M., Allen K. E., Tang X., Lu Q., Trulli J. M., Johanson K. O., Kane J. F. 2008. Frameshift events associated with the lysyl-tRNA and the rare arginine codon, AGA, in Escherichia coli: a case study involving the human relaxin 2 protein. Protein Expr. Purif. 60, 110–116. ( 10.1016/j.pep.2008.02.016) [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. 1983. Optimization by simulated annealing. Science. 220, 671–680. ( 10.1126/science.220.4598.671) [DOI] [PubMed] [Google Scholar]
- Knight R. D., Freeland S. J., Landweber L. F. 2001. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2, 0010.1–0010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H., Napthine S., Brierley I. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21, 8657–8670. ( 10.1128/MCB.21.24.8657-8670.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 83, 2850–2854. ( 10.1073/pnas.83.9.2850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C., Gallant J. 1996. Errors of heterologous protein expression. Curr. Opin. Biotechnol. 7, 489–493. ( 10.1016/S0958-1669(96)80050-4) [DOI] [PubMed] [Google Scholar]
- Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124. ( 10.1038/nbt1270) [DOI] [PubMed] [Google Scholar]
- Marquez R., Smit S., Knight R. 2005. Do universal codon-usage patterns minimize the effects of mutation and translation error?. Genome Biol. 6, R91 ( 10.1186/gb-2005-6-11-r91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer I. M., Miklos I. 2004. Co-transcriptional folding is encoded within RNA genes. BMC Mol. Biol. 5, 10 ( 10.1186/1471-2199-5-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer I. M., Miklos I. 2007. SimulFold: simultaneously inferring RNA structures including pseudoknots, alignments, and trees using a Bayesian MCMC framework. PLoS Comput. Biol. 3, e149 ( 10.1371/journal.pcbi.0030149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalodimitrakis K., Isalan M. 2009. Engineering prokaryotic gene circuits. FEMS Microbiol. Rev. 33, 27–37. ( 10.1111/j.1574-6976.2008.00139.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. 1998. An introduction to genetic algorithms. Cambridge, MA: MIT Press [Google Scholar]
- Moura G., Pinheiro M., Silva R., Miranda I., Afreixo V., Dias G., Freitas A., Oliveira J. L., Santos M. A. 2005. Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol. 6, R28 ( 10.1186/gb-2005-6-3-r28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K. R., Rocha I., Forster J., Nielsen J. 2005. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform. 6, 308 ( 10.1186/1471-2105-6-308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P. 2004. Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 14, 2279–2286. ( 10.1101/gr.2896904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M., Maia P., Mendes R., Pinto J. P., Ferreira E. C., Patil K., Nielsen J., Rocha I. 2008. Natural computation meta-heuristics for the in silico optimization of microbial strains. BMC Bioinform. 9, 499 ( 10.1186/1471-2105-9-499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M. V., Gromadski K. B., Kothe U., Wieden H. J. 2005. Recognition and selection of tRNA in translation. FEBS Lett. 579, 938–942. ( 10.1016/j.febslet.2004.11.048) [DOI] [PubMed] [Google Scholar]
- Rodrigo G., Carrera J., Jaramillo A. 2007. Genetdes: automatic design of transcriptional networks. Bioinformatics. 23, 1857–1858. ( 10.1093/bioinformatics/btm237) [DOI] [PubMed] [Google Scholar]
- Sayut D. J., Kambam P. K., Sun L. 2007. Engineering and applications of genetic circuits. Mol. Biosyst. 3, 835–840. ( 10.1039/b700547d) [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. ( 10.1093/nar/15.3.1281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D., Elowitz M. B. 2005. Reconstruction of genetic circuits. Nature. 438, 443–448. ( 10.1038/nature04335) [DOI] [PubMed] [Google Scholar]
- Stenström C. M., Isaksson L. A. 2002. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 288, 1–8. ( 10.1016/S0378-1119(02)00501-2) [DOI] [PubMed] [Google Scholar]
- Stenström C. M., Holmgren E., Isaksson L. A. 2001a. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 273, 259–265. ( 10.1016/S0378-1119(01)00584-4) [DOI] [PubMed] [Google Scholar]
- Stenström C. M., Jin H., Major L. L., Tate W. P., Isaksson L. A. 2001b. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 263, 273–284. ( 10.1016/S0378-1119(00)00550-3) [DOI] [PubMed] [Google Scholar]
- Studer S. M., Joseph S. 2006. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 22, 105–115. ( 10.1016/j.molcel.2006.02.014) [DOI] [PubMed] [Google Scholar]
- Suzuki H., Brown C. J., Forney L. J., Top E. M. 2008. Comparison of correspondence analysis methods for synonymous codon usage in bacteria. DNA Res. 15, 357–365. ( 10.1093/dnares/dsn028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R. P., Noller H. F. 2005. mRNA helicase activity of the ribosome. Cell. 120, 49–58. ( 10.1016/j.cell.2004.11.042) [DOI] [PubMed] [Google Scholar]
- Thanaraj T. A., Argos P. 1996. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 5, 1594–1612. ( 10.1002/pro.5560050814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J., Sauna Z. E., Kimchi-Sarfaty C., Ambudkar S. V., Gottesman M. M., Nussinov R. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 383, 281–291. ( 10.1016/j.jmb.2008.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyo K. E., Alper H. S., Stephanopoulos G. N. 2007. Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol. 25, 132–137. ( 10.1016/j.tibtech.2007.01.003) [DOI] [PubMed] [Google Scholar]
- Varenne S., Lazdunski C. 1986. Effect of distribution of unfavourable codons on the maximum rate of gene expression by an heterologous organism. J. Theor. Biol. 120, 99–110. ( 10.1016/S0022-5193(86)80020-0) [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. 1984. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180, 549–576. ( 10.1016/0022-2836(84)90027-5) [DOI] [PubMed] [Google Scholar]
- Venter J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 304, 66–74. ( 10.1126/science.1093857) [DOI] [PubMed] [Google Scholar]
- Villalobos A., Ness J. E., Gustafsson C., Minshull J., Govindarajan S. 2006. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinform. 7, 285 ( 10.1186/1471-2105-7-285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J. D., Lancaster L., Hodges C., Zeri A. C., Yoshimura S. H., Noller H. F., Bustamante C., Tinoco I. 2008. Following translation by single ribosomes one codon at a time. Nature. 452, 598–603. ( 10.1038/nature06716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W., Peske F., Beringer M., Gromadski K. B., Savelsbergh A., Rodnina M. V. 2004. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem. Soc. Trans. 32, 733–737. ( 10.1042/BST0320733) [DOI] [PubMed] [Google Scholar]
- Wu G., Dress L., Freeland S. J. 2007a. Optimal encoding rules for synthetic genes: the need for a community effort. Mol. Syst. Biol. 3, 134 ( 10.1038/msb4100176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Nie L., Freeland S. J. 2007b. The effects of differential gene expression on coding sequence features: analysis by one-way ANOVA. Biochem. Biophys. Res. Commun. 358, 1108–1113. ( 10.1016/j.bbrc.2007.05.043) [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25, 568–579. ( 10.1093/molbev/msm284) [DOI] [PubMed] [Google Scholar]
- Yarus M., Folley L. S. 1985. Sense codons are found in specific contexts. J. Mol. Biol. 182, 529–540. ( 10.1016/0022-2836(85)90239-6) [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y., Weiss R., Arnold F. H. 2002. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 99, 16 587–16 591. ( 10.1073/pnas.252535999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Cohen R. E., Pickart C. M. 1999. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli. Biotechniques. 27, 950–954. [DOI] [PubMed] [Google Scholar]