Host Range, Prevalence, and Genetic Diversity of Adenoviruses in Bats (original) (raw)
Abstract
Bats are the second largest group of mammals on earth and act as reservoirs of many emerging viruses. In this study, a novel bat adenovirus (AdV) (BtAdV-TJM) was isolated from bat fecal samples by using a bat primary kidney cell line. Infection studies indicated that most animal and human cell lines are susceptible to BtAdV-TJM, suggesting a possible wide host range. Genome analysis revealed 30 putative genes encoding proteins homologous to their counterparts in most known AdVs. Phylogenetic analysis placed BtAdV-TJM within the genus Mastadenovirus, most closely related to tree shrew and canine AdVs. PCR analysis of 350 bat fecal samples, collected from 19 species in five Chinese provinces during 2007 and 2008, indicated that 28 (or 8%) samples were positive for AdVs. The samples were from five bat species, Hipposideros armiger, Myotis horsfieldii, M. ricketti, M yotis spp., and Scotophilus kuhlii. The prevalence ranged from 6.25% (H. armiger in 2007) to 40% (M. ricketti in 2007). Comparison studies based on available partial sequences of the pol gene demonstrated a great genetic diversity among bat AdVs infecting different bat species as well as those infecting the same bat species. This is the first report of a genetically diverse group of DNA viruses in bats. Our results support the notion, derived from previous studies based on RNA viruses (especially coronaviruses and astroviruses), that bats seem to have the unusual ability to harbor a large number of genetically diverse viruses within a geographic location and/or within a taxonomic group.
Members of the family Adenoviridae are nonenveloped, icosahedral viruses approximately 70 to 100 nm in size. The family is divided into four genera: Mastadenovirus, Aviadenovirus, Atadenovirus, and Siadenovirus (3, 6, 7). Adenoviruses (AdVs) contain a linear, nonsegmented, double-stranded DNA (dsDNA) with a genome size ranging from 30 to 36 kb for mastadenoviruses, 31 to 36 kb for atadenoviruses, and 26 to 45 kb for siadenoviruses (3).
AdV infection can be identified in mammals, birds, amphibians, reptiles, and fish, and live AdVs have been isolated from at least 40 vertebrate species (3, 6, 21, 25). A total of 52 human AdV (hAdV) serotypes have been identified and classified into seven groups, designated serotypes A through G. AdVs are highly prevalent in the human population and can cause human infections ranging from respiratory disease (mainly by AdV-B and -C) and conjunctivitis (AdV-B and -D) to gastroenteritis (AdV-F serotypes 40 and 41) (11, 24). In animals, canine AdV type 1 (CAV-1) and canine AdV type 2 (CAV-2) cause hepatitis and respiratory and enteric diseases in dogs (20, 30). The egg drop syndrome-1976 virus (EDS-76 virus), belonging to the aviadenoviruses, is the causative agent of an economically important disease characterized by a severe and sudden drop in egg production (17).
Bats are reservoirs of numerous new or emerging viruses, including henipavirus, Ebola virus, Marbourg virus, Menangle virus, rabies virus, coronavirus, and astrovirus, and most of the bat viral species reported to date are RNA viruses (4, 5, 14, 23, 28, 31). Although numerous virus species and strains were identified in recent years by PCR and sequencing, the isolation of live bat viruses remains rare and difficult, probably due to the lack of appropriate bat cell lines. Recently, two bat adenoviruses (bat AdV-FBV1 and bat AdV-2 PPV1) were isolated from fruit bat (Pteropus dasymallus yayeyamae) and common pipistrelles (P teropus pipistrellus), respectively. The agent was identified as novel adenovirus by partial sequencing (16, 22).
In this study, we report the isolation of a novel AdV from bat fecal samples using a newly established bat primary kidney cell culture. The isolated AdV, named bat adenovirus strain TJM (BtAdV-TJM), is capable of infecting several vertebrate cell lines and inducing a cytopathic effect (CPE). The near-full-length genome sequence (except the 5′- and/or 3′-terminal ends) of BtAdV-TJM is 31,681 bp and carries 30 putative genes. Our epidemiological investigation demonstrated that among the 19 bat species surveyed in this study, bat AdVs are prevalent mainly in Myotis species and Scotophilus kuhlii. This report represents a first detailed study of a DNA virus group in bats.
MATERIALS AND METHODS
Sample collection.
The bat fecal samples were collected during May 2007 and May 2008. The sampling of individual bats was performed as described previously (14). To collect fecal samples from grouped bats, clean plastic sheets measuring 2.0 by 2.0 m were placed under known bat roosting sites at about 18:00. Relatively fresh fecal samples were collected at approximately 5:30 to 6:00 the next morning and stored in liquid nitrogen. Samples were transported to the laboratory and stored at −80°C until analyzed.
Establishment of a bat primary kidney cell line.
The liver, spleen, gut, kidney, heart, and brain taken from a 1-week-old baby bat were washed with D-Hanks (0.3 mM Na2HPO4, 137 mM NaC1, 5.4 mM KCl, 0.4 mM KH2PO4 [pH 7.4]) solution on ice four to five times. Organs were cut into 0.5 to 1 mm2 in size and digested with 0.25% trypsin for 10 min at 37°C. The digested tissues were centrifuged at 500 × g for 2 min and cultured with RPMI 1640 medium containing 20% fetal bovine serum (FBS) (Gibco, Invitrogen), 100 U penicillin/ml, and 0.1 mg streptomycin/ml at 37°C in an incubator supplemented with 5% CO2. After 6 passages, the cells from kidney were growing very well and used for virus isolation. All animal work was conducted under conditions and with permits approved by animal ethics committees of the Wuhan Institute of Virology, Chinese Academy of Sciences.
Virus isolation, purification, and examination by electron microscopy.
All procedures dealing with live-virus isolation were performed in a biosafety cabinet in biosafety level 2 (BSL-2) laboratories. Bat primary kidney (BtMsK) cells were maintained in RPMI 1640 medium supplemented with 20% FBS. Aliquots of 100 mg of feces were homogenized with 500 μl of phosphate-buffered saline (PBS) and centrifuged at 1,000 × g (catalog no. 1-15; Sigma) for 5 min. The supernatant from each sample was diluted 1:10 in RPMI 1640 medium and filtered through a 0.45-μm filter (Millipore). One milliliter of the diluted supernatant was added to BtMsK cells in 35-mm dishes. After incubation at 37°C for 1 h, the inoculum was removed and replaced with fresh RPMI 1640 medium supplemented with 10% FBS. Cell cultures were checked daily for cytopathic effects (CPEs). At 72 h postinoculation, the cell supernatant was collected and inoculated onto monolayer BtMsK cells. After incubation at 37°C for 1 h, the inoculum was removed and replaced with fresh RPMI 1640 medium with 10% FBS. Cultures were blindly passed three times.
For virus purification, infected cells were harvested at 24 h postinfection when strong CPEs appeared. After three freeze-thaw cycles, cell debris were clarified by centrifugation at 3,000 × g for 10 min and filtered through a 0.45-μm filter. Viruses in the supernatant were purified by ultracentrifugation through a 30% sucrose cushion at 40,000 rpm for 3 h by using a Ty70 rotor (Beckman). The pelleted viruses were dissolved with 400 μl of PBS and stored at −70°C in aliquots.
Purified viruses were checked by electron microscopy using Formvar- and carbon-coated copper grids (200 mesh), negatively stained with 2% phosphotungstic acid (pH 7.0), and examined at 75 kV with a Hitachi H-7000FA transmission electron microscope.
Cell susceptibility study.
Cells of different origins (Table 1) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated FBS. Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. Monolayer cells were inoculated with the newly isolated bat AdV at 2 PFU/cell. After 1 h of adsorption at 37°C, the inoculum was removed and replaced with fresh DMEM containing 2% FBS. Cell cultures were checked daily for CPEs. Infection was further confirmed by PCR using primers derived from the genome sequence (see below) from 10−6-diluted cell supernatants.
TABLE 1.
Susceptibilities of cells of different mammalian species to BtAdV-TJM
| Cell line | Species origin | Organ origin | Susceptibility |
|---|---|---|---|
| BHK-21 | Hamster | Kidney | − |
| MDCK | Dog | Kidney | + |
| FK | Cat | Kidney | − |
| PK-15 | Pig | Kidney | + |
| HEK 293 | Human | Kidney | + |
| Vero | Monkey | Kidney | + |
| Vero E6 | Monkey | Kidney | + |
| Tb1-Lu | Bat (Tadarida brasiliensis) | Lung | − |
| BtMsK | Bat (Myotis chinensis) | Kidney | + |
Genome characterization by random PCR.
Since the identity of the genome was not completed resolved at this stage of the study, a procedure that covers both RNA and DNA was employed. Purified virus was treated with 10 ng/μl RNase A and 100 U DNase I in a total volume of 140 μl at 37°C for 30 min. The viral nucleic acid (either DNA or RNA) was extracted with a QIAamp viral RNA minikit (Qiagen, Germany) according to the manufacturer's protocols. Viral nucleic acid was eluted in 60 μl AVE buffer and stored at −70°C.
Viral cDNA synthesis was performed by incubation of the extracted viral nucleic acid at 65°C for 5 min, followed by quenching on ice for 2 min. A 20-μl reaction mixture containing the following ingredients was prepared: 10 pmol of universal primer-dN6 (5′-GCC GGA GCT CTG CAG AAT TCN NNN NN-3′) (8), 10 μl of denatured nucleic acid, 0.6 mM each deoxynucleoside triphosphate (dNTP), 20 U of RNase inhibitor, and 200 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega). The mixture was incubated at 25°C for 10 min, followed by 37°C for 1 h. To synthesize second-strand cDNA/DNA, the reaction mixture was boiled for 2 min and cooled rapidly on ice, followed by incubation at 37°C for 30 min in the presence of 5 U Klenow fragment (exo−; New England Biolabs, Beverly, MA) and 10 pmol of universal primer-dN6.
PCR was conducted with a 50-μl reaction mixture volume containing 10 μl PCR buffer, 1 mM MgSO4, 0.2 mM each dNTP, and 40 pmol universal primer (5′-GCC GGA GCT CTG CAG AAT TC-3′) (8), 1 U KOD-Plus DNA polymerase (Toyobo, Japan), and 2 μl of the second-strand cDNA/DNA mixture described above. The reaction was conducted for 40 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 2 min followed by incubation for 10 min at 68°C. The products were analyzed by agarose gel electrophoresis. PCR products larger than 500 bp in size were purified with an EZNA gel extraction kit (Omega Bio-Tek) and cloned into the pGEM-T Easy vector (Promega) after adding adenine (A) at the 3′ termini of the PCR products using Taq polymerase. Insert-containing plasmids were sequenced by using the M13 forward and reverse primers with an ABI Prism 3730 DNA analyzer (Applied Biosystems). The sequences of the PCR products were compared with sequences in the GenBank database by using BLAST.
Full-genome sequencing.
The purified viral DNA was used as a template for PCR amplification with a combination of primer sets (available upon request) designed either from published AdV genome sequences in GenBank or from BtAdV-TJM, obtained by random PCR sequencing as described above. PCR products were either purified with an EZNA gel extraction kit (Omega Bio-Tek) and sequenced directly or cloned into the pGEM-T Easy vector (Promega) before being sequenced. For each PCR fragment cloned, at least three independent clones were subjected to sequencing to obtain a consensus sequence. The Roche 3′ random amplification of cDNA ends (RACE) kit (Roche Diagnostics) was used to determine genome terminal sequences of both the sense and antisense strands according to the manufacturer's instructions.
Molecular epidemiology.
Viral DNA was extracted from individual bat fecal swabs with QIAamp DNA blood minikits (Qiagen, CA) according to the manufacturer's instructions. For screening the presence of AdVs and studying their genetic diversity, a partial sequence of the AdV DNA polymerase (pol) gene was amplified with nested PCR. For the first round amplification, the 25-μl reaction mix contained 2 μl of extracted DNA, 2.5 μl PCR buffer, 20 pmol each primer (pol-F [5′-CAGCCKCKGTTRTGYAGGGT-3′] and pol-R [5′-GCHACCATYAGCTCCAACTC-3′]), 0.2 mM dNTP, and 0.5 U Taq DNA polymerase (Promega). After an initial incubation step at 94°C for 5 min, 30 cycles of amplification were carried out, consisting of denaturation at 94°C for 30 s, annealing at 48°C for 30 s, and extension at 72°C for 30 s and a final extension step at 72°C for 10 min. For the second round of amplification, 1 μl of first-round PCR products was used as a template and amplified with forward primer pol-nF (5′-GGGCTCRTTRGTCCAGCA-3′) and reverse primer pol-nR (5′-TAYGACATCTGYGGCATGTA-3′) with the same cycling parameters as those for the first round. Standard precautions were taken to avoid PCR contamination, and a negative control was included in every PCR assay. The expected products were gel purified by using an EZNA gel extraction kit (Omega Bio-Tek) and sequenced with the two nested-PCR primers.
Sequence and phylogenetic analysis.
Routine sequence management and analysis were carried out by using DNAStar (DNAStar Inc.). The identification of open reading frames (ORFs) was performed by a translated BLAST search (BLASTx at http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Splice site predictions were performed by using software available at the Berkeley Drosophila Genome Project website (http://www.fruitfly.org). Protein identity comparisons were performed by aligning sequences with BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Sequence alignment was performed by using T-coffee and corrected manually (18). The phylogenetic trees were constructed by using the neighbor-joining (NJ) method with a bootstrap of 1,000 replicates with the MEGA, version 4.1, program (13). Gaps were regarded as a complete deletion unless otherwise specified.
Nucleotide sequence accession numbers.
The following published AdV genome sequences were retrieved from GenBank and included in the analysis in this study: human adenovirus A (accession no. NC_001460), human adenovirus B1 (accession no. NC_011203), human adenovirus B2 (accession no. NC_011202), human adenovirus C (accession no. NC_001405), human adenovirus D (accession no. NC_010956), human adenovirus E (accession no. NC_003266), human adenovirus F (accession no. NC_001454), ovine adenovirus A (accession no. NC_002513), ovine adenovirus D (accession no. NC_004037), porcine adenovirus C (accession no. NC_002702), bovine adenovirus B (accession no. NC_001876), bovine adenovirus D (accession no. NC_002685), canine adenovirus 1 (accession no. NC_001734), canine adenovirus 2 (accession no. AC_000020), murine adenovirus A (accession no. NC_000942), tree shrew adenovirus (accession no. NC_004453), snake adenovirus (accession no. NC_009989), frog adenovirus (accession no. NC_002501), turkey adenovirus A (accession no. NC_001958), fowl adenovirus A (accession no. NC_001720), fowl adenovirus D (accession no. NC_000899), duck adenovirus A (accession no. NC_001813), and bat adenovirus FBV1 (accession no. AB303301). All partial pol gene sequences of bat AdVs sequenced in this study were submitted to GenBank (accession no. GU226951 to GU226969).
RESULTS
Isolation of a novel adenovirus using bat primary kidney cells.
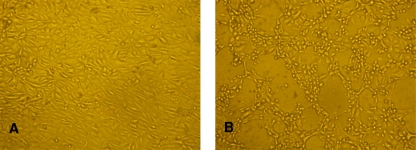
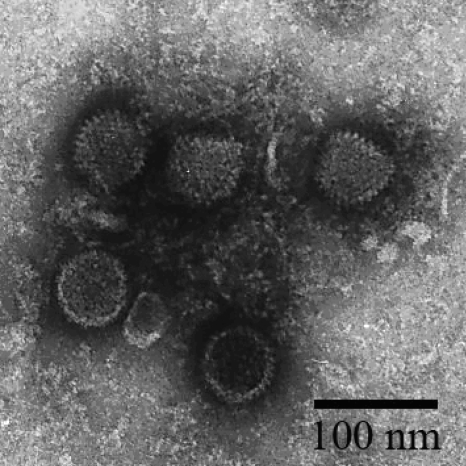
Bat primary kidney cells were derived from Myotis chinensis bats as described in Materials and Methods. After five continuous passages, the growth of bat primary kidney cells became stable (Fig. 1A). The cells were then used to isolate bat viruses from fecal samples collected from Myotis bats at different locations in China in 2007 and 2008. After three passages, one sample (M. ricketti, collected from Tianjin City in July 2007) exhibited an evident CPE, as shown in Fig. 1B. After further passage and purification, the virus morphology was examined by transmission electron microscopy, which revealed an average viral particle size of 60 to 70 nm in diameter (Fig. 2). Although the size distribution was below the average size of 90 nm for most known AdVs, the electron micrograph images suggested that the virus is an AdV.
FIG. 1.
Isolation of bat adenovirus in bat primary kidney cells. (A) Noninfected bat primary kidney cells. (B) The same cells as those shown in A after infection with BtAdV-TJM at 2 PFU/cell, showing CPEs observed at 24 h postinfection.
FIG. 2.
Examination of viral particles by electron microscopy. See Materials and Methods for a description of the preparation of the samples.
Infection of cell lines of different origins by the newly isolated virus.
To test the potential host range of the putative bat AdV, cell lines of different origins were inoculated with purified bat AdV at 2 PFU/cell. At 24 h postinfection, an evident CPE in MDCK, PK-15, HEK 293, Vero, and Vero E6, but not in BHK-21, FK, or Tb1-Lu, cells was observed. The productive infection of these cells was later confirmed by specific PCR amplification of viral genomic DNA (data not shown).
Molecular identification of the putative bat adenovirus.
Although the electron microscopy analysis suggested that the isolated virus could be an AdV, a random sequencing strategy that covers both RNA and DNA viral genomes was used to ensure successful genome characterization in the absence of a confident classification of the virus. As detailed in Materials and Methods, concentrated viral particles were filtered and treated with both RNase and DNase before viral nucleic acid extraction and random amplification, followed by cloning and sequencing analysis. Out of 15 sequenced clones, 3 clones (inserts at 364, 425, and 415 nt, respectively) were homologous to the gene coding for the AdV DNA polymerase (Pol), 3 clones (1300, 770, and 770 nt) were homologous to the DNA binding protein (DBP), 1 clone (607 nt) was homologous to ORFC of the E4 region (E4-ORFC), and the other 8 clones were contaminations of cellular genomic fragments. All fragments randomly sequenced demonstrated the best match to the homologous genome regions of canine adenovirus type 2 (CAdV-2) strain Toronto A26/61 (GenBank accession no. CAU77082). The amino acid sequence identities of Pol, DBP, and E4-ORFC to the counterparts of CAdV-2 were 83%, 78%, and 30%, respectively. These results confirmed that the newly isolated bat virus is a member of the family Adenoviridae. The virus isolated form bat fecal samples was named bat adenovirus strain TJM (BtAdV-TJM), after the name of the city and bat species from which the sample was collected.
Characterization of the genome coding structure.
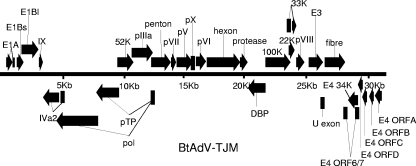
The full-length genome (GenBank accession no. GU226970) of BtAdV-TJM was sequenced by using primers designed using the randomly obtained genome fragment sequences and published AdV genome sequences and a 3′ RACE strategy. The genome size is 31,681 bp or larger (see below). The genome organization of BtAdV-TJM is similar to that of most known members in the genus Mastadenovirus (Fig. 3). The average G+C content is 56.8%, ranging from 43.6 to 69.4% in different genes, with slightly higher values in the center of the genome than at the genome terminal regions. CpG dinucleotide analysis of BtAdV-TJM using FUZZNUC software (http://bioweb.pasteur.fr/seqanal/interfaces/fuzznuc.html) revealed 1,885 CpG dinucleotides located within the genome. Despite several attempts, we were not able to identify inverted terminal repeats (ITRs) present in all known AdVs. This suggests that (i) one or two ends of the genome are not completely sequenced and (ii) the genome most likely contains more than 31,681 bp.
FIG. 3.
Genome organization of BtAdV-TJM. The viral genome is represented by the double line in the center marked with 5-kbp intervals. Protein-coding regions are shown as filled arrows. Arrows above the black line represent open reading frames (ORFs) encoded on the forward (top) strand oriented from left to right. Arrows underneath the genome line represent ORFs that are carried on the complementary (bottom) strand. ORFs were named according to the nomenclature used for homologous proteins of other AdVs.
Ninety-nine open reading frames were identified in both strands of the BtAdV-TJM genome using a search strategy defining ATG as the start codon and a cutoff size of 60 aa. Among them, 30 predicted protein products had significant sequence identity to other known AdV homologs. Eleven of these genes were carried by the complementary strand (Fig. 3 and Table 2). Similar to other members of the genus Mastadenovirus, the middle part of the BtAdV-TJM genome was predicted to contain 18 genus-common genes. These include genes coding for DNA replication (Pol, pTP, and DBP), DNA encapsidation (52K and IVa2); the formation and structure of the virion (pIIIa, penton, pVII, pX, pVI, hexon, protease, 100K, 33K, pVIII, and fiber); 22K, which originates from a lack of splicing in 33K; and the U exon. No gene coding for VA RNA was identified.
TABLE 2.
Predicted gene products encoded by the BtAdV-TJM genome
| Gene product | Location(s) (nt)a | Length (aa) | Feature(s) of homologous AdV products | G+C content (%) |
|---|---|---|---|---|
| E1A | 391-883, 970-1130 | 217 | Early E1A 25-kDa protein | 53.5 |
| E1Bs | 1270-1818 | 182 | E1B protein, small T antigen | 53.6 |
| E1Bl | 1632-3011 | 459 | E1B protein, large T antigen | 53.4 |
| IX | 3077-3397 | 106 | Adenovirus hexon-associated protein (IX) | 57.0 |
| IVa2 | 3421-4736, 5036-5048c | 442 | Adenovirus IVa2 protein, maturation protein | 55.2 |
| Pol | 4521-7940, 12472-12480c | 1142 | DNA polymerase | 58.5 |
| pTP | 7796-9616, 12472-12480c | 609 | Adenoviral DNA terminal protein | 62.8 |
| 52K | 9543-10859 | 438 | Late L1 52-kDa protein, encapsidation protein 52K | 64.3 |
| pIIIa | 10735-12459 | 574 | IIIa protein precursor, hexon-associated protein (IIIa) | 63.8 |
| Penton | 12359-13936 | 525 | Adenovirus penton base protein | 60.6 |
| pVII | 13968-14372 | 134 | Adenoviral core protein VII | 69.4 |
| pV | 14438-15739 | 433 | Adenovirus minor core protein pV, L2 minor core protein | 68.4 |
| pX | 15765-15974 | 69 | Mu peptide precursor, late L2 mu core protein, pX | 66.2 |
| pVI | 16023-16844 | 273 | Capsid protein precursor pVI, minor capsid protein VI | 65.7 |
| Hexon | 16914-19640 | 908 | Hexon capsid protein | 50.4 |
| Protease | 19652-20272 | 206 | 23K endopeptidase, protease | 58.5 |
| DBP | 20315-21733 | 472 | Early E2A DNA binding protein, single-stranded DNA binding protein | 57.6 |
| 100K | 21746-23794 | 682 | L4 100-kDa hexon assembly protein | 58.3 |
| 33K | 23676-23820, 23933-24282 | 164 | Protein, 33K | 60.6 |
| 22K | 23676-24143 | 155 | Encapsidation protein, 22K | 62.6 |
| pVIII | 24286-24954 | 222 | Adenovirus hexon-associated protein, protein VIII | 62.0 |
| E3 | 25321-26469 | 382 | E3-ORFA | 58.5 |
| U exon | 26442-26645c | 67 | U exon | 51.0 |
| Fiber | 26644-28311 | 555 | Adenoviral fiber protein | 53.5 |
| E4-ORF6/7 | 28325-28567, 29303-29350c | 96 | Control protein E4-ORF6/7 | 51.2 |
| E4-34K | 28568-29350c | 260 | Adenovirus early E4 protein, E4-34K | 47.9 |
| E4-ORFD | 29352-29744c | 130 | E4-ORFD | 44.5 |
| E4-ORFC | 29723-30097c | 124 | E4-ORFC | 46.7 |
| E4-ORFB | 30300-30659c | 119 | E4-ORFB | 43.6 |
| E4-ORFA | 30751-31314c | 187 | E4-ORFA | 48.4 |
A summary of the sequence similarity comparison between homologous proteins of BtAdV-TJM and selected members of the genus Mastadenovirus is presented in Table 3. The most conserved protein is the hexon protein (908 aa), which showed high sequence identity (86%) to that of CAdV-2. The penton protein is less conserved, with a sequence identity of 55 to 84%. The Arg-Gly-Asp (RGD) motif, which plays a key role in the interaction of the virus with the cell surface integrins αvβ3 and αvβ5 and is the main target of virus neutralization (27), is missing in the predicted penton protein sequence of BtAdV-TJM. Neither RGD sequence was found in other viral structural proteins, including pIIIa, penton, pVII, pX, pVI, hexon, protease, 100K, 33K, pVIII, and fiber. Similarly, the motif leucine-aspartic acid-valine (LDV), which interacts with integrin α4β1 (12), is also missing in the BtAdV-TJM penton protein. On the other hand, the AdV fiber knob, which is responsible for attachment to cellular receptors, was identified in BtAdV-TJM with an amino acid identity of 24 to 50% compared to that of other mastadenoviruses.
TABLE 3.
Amino acid sequence comparison of selected BtAdV-TJM proteins with their homologs from other members of the genus Mastadenovirusa
| Virus | Amino acid identity (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1A | E1Bl | IVa2 | Pol | pTP | 52K | Penton | pVII | pV | Hexon | DBP | Protease | Fiber | E4 34K | |
| HAdV-A | NA | 28 | 60 | 63 | 63 | 57 | 62 | 44 | 36 | 71 | 37 | 64 | 25 | 28 |
| HAdV-B1 | 24 | 29 | 61 | 65 | 63 | 57 | 56 | 42 | 36 | 69 | 35 | 66 | 27 | 27 |
| HAdV-B2 | 24 | 26 | 61 | 65 | 63 | 53 | 56 | 38 | 35 | 68 | 37 | 67 | 26 | 29 |
| HAdV-C | 23 | 28 | 60 | 64 | 62 | 52 | 60 | 40 | 33 | 67 | 37 | 66 | 24 | 30 |
| HAdV-D | 30 | 28 | 62 | 64 | 63 | 54 | 58 | 41 | 35 | 69 | 36 | 67 | 25 | 28 |
| HAdV-E | 27 | 30 | 61 | 65 | 64 | 52 | 58 | 43 | 36 | 69 | 39 | 67 | 26 | 29 |
| HAdV-F | NA | 28 | 61 | 65 | 63 | 57 | 61 | 42 | 34 | 70 | 39 | 64 | 25 | 30 |
| SAdV-1 | 29 | 28 | 63 | 66 | 63 | 57 | 62 | 38 | 38 | 71 | 43 | 66 | 27 | 31 |
| SAdV-A | NA | 28 | 60 | 65 | 63 | 54 | 64 | 44 | 37 | 70 | 36 | 65 | 29 | 29 |
| CAdV-1 | 47 | 51 | 73 | 78 | 85 | 75 | 84 | 71 | 58 | 86 | 64 | 79 | 45 | 50 |
| CAdV-2 | 47 | 55 | 75 | 79 | 86 | 78 | 83 | 73 | 68 | 86 | 64 | 82 | 50 | 51 |
| TsAdV | NA | 27 | 60 | 66 | 61 | 65 | 69 | 35 | 33 | 75 | 50 | 69 | 28 | 30 |
| OAdV-A | NA | 29 | NI | 67 | 61 | NI | 63 | NI | 32 | 73 | NI | 66 | NI | 27 |
| PAdV-A | NA | 27 | 60 | 62 | 63 | 55 | 71 | 38 | 29 | 70 | 52 | 58 | 31 | 29 |
| PAdV-C | NA | 27 | 66 | 68 | 64 | 66 | 66 | 36 | 31 | 75 | 58 | 65 | 29 | 30 |
| BAdV-A | 25 | 30 | 60 | 68 | 62 | 67 | 66 | NI | 32 | 75 | 59 | 69 | 29 | NA |
| BAdV-B | NA | 27 | 63 | 62 | 58 | 60 | 67 | 42 | 34 | 69 | 51 | 69 | 27 | 28 |
| MAdV-3 | NA | 26 | 59 | 57 | 41 | 51 | 55 | NA | NA | 67 | 41 | 61 | 26 | 25 |
| MAdV-A | NA | 25 | 59 | 56 | 41 | 52 | 55 | 39 | 23 | 66 | 32 | 61 | 26 | 28 |
With the exception of one genus-specific gene (pV) that is located between pVII and pX, most genus-specific genes are located near the ends of the genome. The two ends of the BtAdV-TJM genome are predicted to encode the E1 and E4 regions. E1 is encoded by the top or forward strand. One putative E1A protein (23.3 kDa), the product of a splicing event, was identified. The protein sequence of E1A is highly divergent and has only 23 to 47% sequence identity to its counterpart of human and canine AdVs. Two coding sequences, E1Bs (20.7 kDa, small T antigen) and E1Bl (50.4 kDa, large T antigen), were identified in the E1B region. Similar to human AdV and CAdV-2, the BtAdV-TJM genome E4 region contains six genes encoded by the complementary strand that are named E4-ORFA, E4-ORFB, E4-ORFC, E4-ORFD, E4-34K, and E4-ORF6/7. The ORF6/7 mRNA results from further splicing between the 5′ end of 34K and the region immediately downstream of 34K. The E4-34K protein has an amino acid sequence identity of 27 to 51% to its homolog of other genus members. Only one ORF was identified in the E3 region, which is located between pVIII and the U exon, whereas five to nine genes were identified in this region in human AdVs and two were identified in CAdV-2.
Phylogeny.
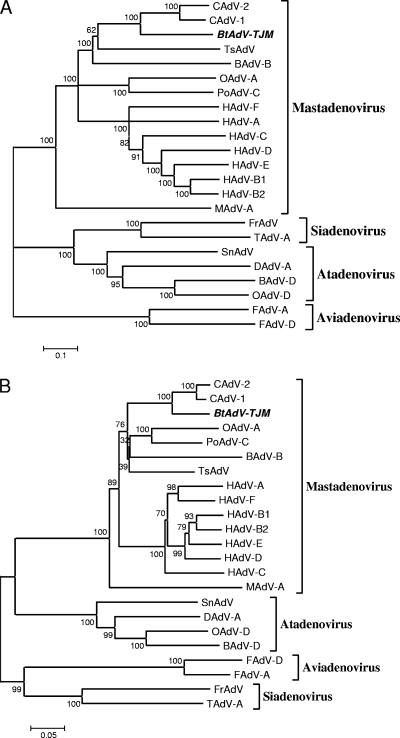
Phylogenetic analysis was conducted to establish the taxonomy of BtAdV-TJM. Phylogenetic trees based on the full-length genome sequences (Fig. 4A) and the hexon protein sequences (Fig. 4B) both unequivocally indicated that BtAdV-TJM is a member of the genus Mastadenovirus. These trees also suggest that BtAdV-TJM is most closely related to CAdV and tree shrew AdV.
FIG. 4.
Phylogenetic analysis of bat adenoviruses. The phylogenetic trees were constructed based on the alignment of full-length AdV genome nucleotide sequences (A), the amino acid sequences of hexon proteins (B), and partial amino acid sequences of the polymerases (C) of bat adenoviruses by using the neighbor-joining (NJ) method with a bootstrap of 1,000 replicates with the MEGA 4.1 program. Gaps were regarded as a complete deletion unless otherwise specified. A condensed tree is presented in C. The scale bar is in units of nucleotide (A) or amino acid (B and C) residue substitutions per site. Bootstrap values (percent) are indicated at the branch nodes of the phylogenetic trees. Abbreviations are as follows (with GenBank accession numbers in parentheses): HAdV-A, human adenovirus A (accession no. NC_001460); HAdV-B1, human adenovirus B1 (accession no. NC_011203); HAdV-B2, human adenovirus B2 (accession no. NC_011202); HAdV-C, Human adenovirus C (NC_001405); HAdV-D, human adenovirus D (accession no. NC_010956); HAdV-E, human adenovirus E (accession no. NC_003266); HAdV-F, human adenovirus F (accession no. NC_001454); OAdV-A, ovine adenovirus A (accession no. NC_002513); OAdV-D, ovine adenovirus D (accession no. NC_004037); PoAdV-C, porcine adenovirus C (accession no. NC_002702); BAdV-B, bovine adenovirus B (accession no. NC_001876); BAdV-D, bovine adenovirus D (accession no. NC_002685); CAdV-1, canine adenovirus 1 (accession no. NC_001734); CAdV-2, canine adenovirus 2 (accession no. AC_000020); MAdV-A, murine adeno- virus A (accession no. NC_000942); TsAdV, tree shrew adenovirus (accession no. NC_004453); SnAdV, snake adenovirus (accession no. NC_009989); FrAdV, frog adenovirus (accession no. NC_002501); TAdV-A, turkey adenovirus A (accession no. NC_001958); FAdV-A, fowl adenovirus A (accession no. NC_001720); FAdV-D, fowl adenovirus D (accession no. NC_000899); DAdV-A, duck adenovirus A (accession no. NC_001813); bat AdV-FBV1, bat adenovirus FBV1 (accession no. AB303301). The bat AdV detected in this study is named by the sample number followed by the abbreviation of the sampling location and species name. The sequences derived from different locations are highlighted with different symbols. HB, Hubei; HN, Hainan; GD, Guangdong; YN, Yunnan; TJ, Tianjin; Ha, Hipposideros armiger; Md, Myotis daubentoni; Mh, Myotis horsfieldii; Mspp, unidentified Myotis species; Mr, Myotis ricketti; SK, Scotophilus kuhlii.
Prevalence of adenoviruses in bats.
To gain more knowledge about the prevalence and distribution of AdVs in bats, 350 individual bat fecal samples from 19 bat species collected in five provinces of China in 2007 and 2008 were examined by nested PCR, which targets a 261-bp region of the polymerase gene. A total of 28 (8%) samples tested positive for AdV in five bat species, including Hipposideros armiger, M. horsfieldii, M. ricketti, M yotis spp., and Scotophilus kuhlii. The prevalence ranged from 6.25% to 40% (Table 4). It is notable that AdV was detected in M. ricketti bats in four out of five provinces; thus, members of the genus Myotis are the most represented AdV hosts among the different bat species surveyed in this study.
TABLE 4.
Detection of adenovirus in bats by PCR
| Bat species | No. of positive samples/no. of tested samples (% positive) at sampling location and date | |||||
|---|---|---|---|---|---|---|
| Tianjin 2007 | Hubei 2007 | Guangdong 2007 | Yunnan 2007 | Hainan 2007 | Hubei 2008 | Total |
| Rousettus leschenaulti | 0/1 | 0/1 | ||||
| Eptesicus serotinus | 0/1 | 0/1 | ||||
| Rhinolophus pusillus | 0/1 | 0/1 | ||||
| Rhinolophus affinis | 0/11 | 0/4 | 0/45 | 0/60 | ||
| Rhinolophus luctus | 0/1 | 0/1 | ||||
| Rhinolophus sinicus | 0/14 | 0/11 | 0/7 | 0/14 | 0/26 | |
| Rhinolophus pearsoni | 0/9 | 0/1 | 0/10 | |||
| Rhinolophus macrotis | 0/1 | 0/3 | 0/4 | |||
| Hipposideros armiger | 0/8 | 1/16 (6.25) | 0/12 | 1/36 (2.78) | ||
| Hipposideros larvatus | 0/13 | 0/13 | ||||
| Hipposideros pomona | 0/9 | 0/18 | 0/27 | |||
| Myotis daubentoni | 0/18 | 0/18 | ||||
| Myotis horsfieldii | 2/8 (25) | 2/8 (25) | ||||
| Myotis ricketti | 3/36 (8.33) | 2/5 (40) | 7/33 (21.2) | 1/16 (6.25) | 13/90 (14.4) | |
| Myotis davidii | 0/2 | 0/2 | ||||
| Myotis chinensis | 0/3 | 0/3 | ||||
| Myotis spp. | 3/13 (23.1) | 3/13 (23.1) | ||||
| Scotophilus kuhlii | 9/26 (34.6) | 9/26 (34.6) | ||||
| Minipopterus schreibersii | 0/10 | 0/10 |
Genetic diversity of adenoviruses in bats.
BtAdV-TJM was compared with two other recently isolated bat AdVs, AdV-FBV1 and AdV-2 PP1, using available pol gene sequences. BtAdV-TJM shares amino acid sequence identities of 66% and 78% to AdV-FBV1 and AdV-2 PP1, respectively, in the sequences compared, indicating that these three bat AdVs are not genetically closely related. When the sequences of the 261-bp pol gene fragments from bat AdVs detected in this study were analyzed, it became even more obvious that bat AdVs are a group of genetically diverse AdVs, with nucleotide and amino acid sequence identities of 66 to 72% and 68 to 75%, respectively. Interestingly, genetic diversity was also observed among AdVs identified in the same bat species, including M. ricketti (with nucleotide and amino acid sequence identities of 70 to 98% and 74 to 100%, respectively) and M. horsfieldii (61% identity in amino acid sequence). AdVs from S. kuhlii are more conserved, with a nucleotide sequence identity of 99 to 100% and an identical amino acid sequence. The phylogenetic tree based on the aligned amino acid sequences of partial pol genes indicated that bat AdVs that come from the same bat species cluster together, with the exception of samples 1285 and 1213, obtained from two M. horsfieldii isolates (Fig. 4C).
DISCUSSION
For the last decade or so, an increasing number of emerging RNA viruses has been associated with bats as a natural reservoir. However, there has been much less reporting on the identification and isolation of DNA viruses from bats. Two bat adenovirus, AdV-FBV1 and AdV-2 PPV1, were recently isolated, but only partial sequence information was available (16, 22). In this study, we have conducted a more comprehensive investigation of the host range, prevalence, and genetic diversity of bat AdVs in Chinese bat populations.
Using a newly established Myotis bat primary kidney cell line developed by our group, we succeeded in isolating a novel adenovirus (BtAdV-TJM) from an M. ricketti fecal sample that is substantially different from bat AdV-FBV1 and bat AdV-2 PP1. This new bat AdV is capable of infecting and causing CPEs in several mammalian cells of different species origins, from dog to human, indicating a potential wide host range.
Determination of the full-length genome indicated that BtAdV-TJM has a coding structure similar to that of most known AdVs. Despite several trials of RACE experiments, we failed to detect inverted terminal repeats (ITRs), which are present in all known AdVs. This suggests that one or both ends of the genome were not successfully sequenced in this study. This dilemma will be resolved later. Phylogenetic analysis based on the full coding sequence of the genome and the deduced penton protein sequence placed BtAdV-TJM within the genus Mastadenovirus, and it is most closely related to CAdV-2. The average GC content of BtAdV-TJM is 56.8%, higher than those of CAdV-1 (47%) and CAdV-2 (50.3%). The most conserved genes are the E1A, E1Bl, IVa2, Pol, pTP, 52K, penton, pVII, pV, hexon, DBP, protease, fiber, and E4-34K genes, which have 47 to 86% amino acid sequence identity to those of the CAdV-2 homologs.
There are several unusual features revealed for the BtAdV-TJM genome and its encoded proteins. In the E3 region of the genome, only one ORF was identified for BtAdV-TJM, whereas there are two for CAdV-2 and five for hAdVs in the same region. Recently, Klempa et al. showed that E3 was missing in a novel murine adenovirus (MAdV-3) (10), suggesting that genes carried within the E3 region may be dispensable for animal AdV. This notion is supported by findings that genes in the E3 region from hAdVs play a role in modulating the host immune response to infection but are not essential for virus growth in vitro (9, 15). The other unexpected finding was that two important amino acid motifs in the predicted penton protein sequence of the BtAdV-TJM were missing. They are the RGD (Arg-Gly-Asp) and LDV (leucine-aspartic acid-valine) motifs, which are important for interactions with the cell surface integrins αvβ3 and αvβ5 (27) and integrin α4β1, respectively (12). It is not clear whether bat AdV no longer requires these functional motifs or whether they are replaced by other yet-to-be-identified structural components.
When partial sequences of the pol genes of different bat AdVs were examined by PCR surveillance of 350 bat fecal samples, two conclusions were drawn. First, bat AdVs are present in a wide range of different bat species, ranging from microbats to fruit bats. Among the species surveyed, AdVs appeared to be most abundant in the genera Myotis and S. kuhlii. Second, bat AdVs present in different bat species display great genetic diversity. This is true for sequences detected in different members of the genus Myotis and among different individuals of the same species, M. horsfieldii, at a single location. The great genetic diversity of viruses among the same species of bats at the same location has been observed for RNA viruses in the past as one of the unexplained features of bat viruses that is rarely seen for other animal viruses (4, 23, 29, 31). Here we have demonstrated this again for a DNA virus. The genetic diversity of bat viruses is an interesting and important observation that requires further investigation.
Based on this discovery from our limited surveillance work, it can be predicted that bat AdVs are prevalent in more bat species and in more geographic locations. Further epidemiology work is required to fully appreciate the genetic diversity and distribution of bat AdVs in China and elsewhere.
Human AdV 5 (Ad5) is widely used as a vector for gene therapy or a vaccine. A major hurdle to the successful clinical use of the AdV vector is the preexisting neutralizing antibodies to the viral vector (2). Thus, AdV vectors derived from immunologically distinct viruses could potentially be advantageous in that regard. A recent report suggested that CAV-2 and recombinant HAd5/CAV-2 vectors may be the antithesis of Adenoviridae immunogenicity and that each one may have specific clinical advantages (19). The main structural proteins (penton and hexon) of BtAdV-TJM share amino acid sequence identities of 56 to 62% and 68 to 71%, respectively, with those of human AdVs. In addition, the GC content, which is recognized by the innate immune system (1), is high in the bat AdV genome. Both of these features suggest that bat AdV might be an ideal substitute for existing AdV vectors applied in gene therapy and vaccine delivery for humans.
Bats have been reported to be natural reservoirs of many viruses, including henipaviruses, lyssaviruses, filoviruses, coronaviruses, and astroviruses. Most bat viruses discovered to date are RNA viruses. Here we have extended this trend of bat virus discovery into a group of highly diverse DNA viruses. In addition, we have also discovered a group of genetically diverse adeno-associated viruses (AAVs) in the same sample collection used in this study (our unpublished data), which provided further support to our results presented in this paper, as most AAVs are associated with AdVs. Our findings, together with the discovery of bat herpesvirus (26) and AdVs (16, 22) by others, revealed that bats carry diverse DNA viruses as well. Considering the wide host range and geographic locations of bat AdVs and more contact between humans and bats, there is a need to conduct future research into the potential spillover of these virus into human and domestic animal populations and their pathogenic potentials in spillover hosts.
Acknowledgments
We thank Mária Benkő (Veterinary Medical Research Institute, Hungarian Academy of Sciences, Hungary) for suggestions on sequence annotation. We give great thanks to April Davis (Wadsworth Center, New York State Department of Health) for revising the manuscript.
This work was jointly funded by State Key Program for Basic Research grant 2005CB523004 from the Chinese Ministry of Science, National Natural Science Foundation (grant 30970137), and Technology and the Knowledge Innovation Program Key Project administered by the Chinese Academy of Sciences (grant KSCX1-YW-R-07).
Footnotes
▿
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Aderem, A., and D. A. Hume. 2000. How do you see CG? Cell 103**:**993-996. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., M. G. Pau, J. H. Custers, W. Koudstaal, S. Kostense, M. J. Havenga, D. M. Truitt, S. M. Sumida, M. G. Kishko, J. C. Arthur, B. Korioth-Schmitz, M. H. Newberg, D. A. Gorgone, M. A. Lifton, D. L. Panicali, G. J. Nabel, N. L. Letvin, and J. Goudsmit. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172**:**6290-6297. [DOI] [PubMed] [Google Scholar]
- 3.Benkö, M., and B. Harrach. 2003. Molecular evolution of adenoviruses. Curr. Top. Microbiol. Immunol. 272**:**3-35. [DOI] [PubMed] [Google Scholar]
- 4.Chu, D. K., L. L. Poon, Y. Guan, and J. S. Peiris. 2008. Novel astroviruses in insectivorous bats. J. Virol. 82**:**9107-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K. B., G. Crameri, A. Hyatt, M. Yu, M. R. Tompang, J. Rosli, J. McEachern, S. Crameri, V. Kumarasamy, B. T. Eaton, and L. F. Wang. 2007. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 104**:**11424-11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, A. J., K. M. Wright, and B. Harrach. 2000. DNA sequence of frog adenovirus. J. Gen. Virol. 81**:**2431-2439. [DOI] [PubMed] [Google Scholar]
- 7.Farkas, S. L., M. Benko, P. Elo, K. Ursu, A. Dan, W. Ahne, and B. Harrach. 2002. Genomic and phylogenetic analyses of an adenovirus isolated from a corn snake (Elaphe guttata) imply a common origin with members of the proposed new genus Atadenovirus. J. Gen. Virol. 83**:**2403-2410. [DOI] [PubMed] [Google Scholar]
- 8.Froussard, P. 1993. rPCR: a powerful tool for random amplification of whole RNA sequences. PCR Methods Appl. 2**:**185-190. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz, M. S. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 6(Suppl. 1)**:**S172-S183. [DOI] [PubMed] [Google Scholar]
- 10.Klempa, B., D. H. Kruger, B. Auste, M. Stanko, A. Krawczyk, K. F. Nickel, K. Uberla, and A. Stang. 2009. A novel cardiotropic murine adenovirus representing a distinct species of mastadenoviruses. J. Virol. 83**:**5749-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojaoghlanian, T., P. Flomenberg, and M. S. Horwitz. 2003. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13**:**155-171. [DOI] [PubMed] [Google Scholar]
- 12.Komoriya, A., L. J. Green, M. Mervic, S. S. Yamada, K. M. Yamada, and M. J. Humphries. 1991. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 266**:**15075-15079. [PubMed] [Google Scholar]
- 13.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5**:**150-163. [DOI] [PubMed] [Google Scholar]
- 14.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310**:**676-679. [DOI] [PubMed] [Google Scholar]
- 15.Lichtenstein, D. L., K. Toth, K. Doronin, A. E. Tollefson, and W. S. Wold. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23**:**75-111. [DOI] [PubMed] [Google Scholar]
- 16.Maeda, K., E. Hondo, J. Terakawa, Y. Kiso, N. Nakaichi, D. Endoh, K. Sakai, S. Morikawa, and T. Mizutani. 2008. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae). Emerg. Infect. Dis. 14**:**347-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFerran, J. B., R. M. McCracken, E. R. McKillop, M. S. McNulty, and D. S. Collins. 1978. Studies on a depressed egg production syndrome in Northern Ireland. Avian Pathol. 7**:**35-47. [DOI] [PubMed] [Google Scholar]
- 18.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302**:**205-217. [DOI] [PubMed] [Google Scholar]
- 19.Perreau, M., F. Mennechet, N. Serratrice, J. N. Glasgow, D. T. Curiel, H. Wodrich, and E. J. Kremer. 2007. Contrasting effects of human, canine, and hybrid adenovirus vectors on the phenotypical and functional maturation of human dendritic cells: implications for clinical efficacy. J. Virol. 81**:**3272-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prier, J. E. 1962. Canine hepatitis virus and human adenovirus. Public Health Rep. 77**:**290-292. [PMC free article] [PubMed] [Google Scholar]
- 21.Schrenzel, M., J. L. Oaks, D. Rotstein, G. Maalouf, E. Snook, C. Sandfort, and B. Rideout. 2005. Characterization of a new species of adenovirus in falcons. J. Clin. Microbiol. 43**:**3402-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonntag, M., K. Muhldorfer, S. Speck, G. Wibbelt, and A. Kurth. 2009. New adenovirus in bats, Germany. Emerg. Infect. Dis. 15**:**2052-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang, X. C., J. X. Zhang, S. Y. Zhang, P. Wang, X. H. Fan, L. F. Li, G. Li, B. Q. Dong, W. Liu, C. L. Cheung, K. M. Xu, W. J. Song, D. Vijaykrishna, L. L. Poon, J. S. Peiris, G. J. Smith, H. Chen, and Y. Guan. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80**:**7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M. P. de Bethune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cecchini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77**:**8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellehan, J. F., A. J. Johnson, B. Harrach, M. Benko, A. P. Pessier, C. M. Johnson, M. M. Garner, A. Childress, and E. R. Jacobson. 2004. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 78**:**13366-13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wibbelt, G., A. Kurth, N. Yasmum, M. Bannert, S. Nagel, A. Nitsche, and B. Ehlers. 2007. Discovery of herpesviruses in bats. J. Gen. Virol. 88**:**2651-2655. [DOI] [PubMed] [Google Scholar]
- 27.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73**:**309-319. [DOI] [PubMed] [Google Scholar]
- 28.Wong, S., S. Lau, P. Woo, and K. Y. Yuen. 2007. Bats as a continuing source of emerging infections in humans. Rev. Med. Virol. 17**:**67-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo, P. C., M. Wang, S. K. Lau, H. Xu, R. W. Poon, R. Guo, B. H. Wong, K. Gao, H. W. Tsoi, Y. Huang, K. S. Li, C. S. Lam, K. H. Chan, B. J. Zheng, and K. Y. Yuen. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81**:**1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright, N. G., H. Thompson, and H. J. Cornwell. 1971. Canine adenovirus pneumonia. Res. Vet. Sci. 12**:**162-167. [PubMed] [Google Scholar]
- 31.Zhu, H. C., D. K. Chu, W. Liu, B. Q. Dong, S. Y. Zhang, J. X. Zhang, L. F. Li, D. Vijaykrishna, G. J. Smith, H. L. Chen, L. L. Poon, J. S. Peiris, and Y. Guan. 2009. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 90**:**883-887. [DOI] [PubMed] [Google Scholar]