A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis (original) (raw)
Ribosome-anchored proteins Jjj1 and Zuo1 function together with Hsp70 to mediate ribosome biogenesis (see also the companion paper from Koplin et al. in this issue).
Abstract
Molecular chaperones assist cellular protein folding as well as oligomeric complex assembly. In eukaryotic cells, several chaperones termed chaperones linked to protein synthesis (CLIPS) are transcriptionally and physically linked to ribosomes and are implicated in protein biosynthesis. In this study, we show that a CLIPS network comprising two ribosome-anchored J-proteins, Jjj1 and Zuo1, function together with their partner Hsp70 proteins to mediate the biogenesis of ribosomes themselves. Jjj1 and Zuo1 have overlapping but distinct functions in this complex process involving the coordinated assembly and remodeling of dozens of proteins on the ribosomal RNA (rRNA). Both Jjj1 and Zuo1 associate with nuclear 60S ribosomal biogenesis intermediates and play an important role in nuclear rRNA processing, leading to mature 25S rRNA. In addition, Zuo1, acting together with its Hsp70 partner, SSB (stress 70 B), also participates in maturation of the 35S rRNA. Our results demonstrate that, in addition to their known cytoplasmic roles in de novo protein folding, some ribosome-anchored CLIPS chaperones play a critical role in nuclear steps of ribosome biogenesis.
Introduction
Molecular chaperones are ubiquitous proteins that facilitate de novo protein folding (Frydman, 2001; Hartl et al., 2009; Kramer et al., 2009) as well as most other aspects of protein homeostasis (Balch et al., 2008), including assembly and disassembly of oligomeric complexes (McClellan et al., 2007) and quality control of misfolded or stress-denatured proteins (McClellan et al., 2005; Bukau et al., 2006). In eukaryotes, cytosolic chaperones are organized into two distinct but overlapping networks: stress-inducible heat shock proteins protect the cellular proteome from misfolding and stress, whereas chaperones linked to protein synthesis (CLIPS) cooperate with the translational apparatus (Albanèse et al., 2006). The transcriptional coregulation of CLIPS with the translational apparatus suggested that this chaperone network functions in protein biogenesis, most likely in de novo folding of newly made polypeptides (Albanèse et al., 2006). Indeed, many CLIPS chaperones, such as the Hsp70s Ssb1 and Ssb2 (herein referred to as SSB), the prefoldin GIMc (genes involved in microtubules complex), and the chaperonin TRiC/CCT associate with newly translated polypeptides (Wegrzyn and Deuerling, 2005; Albanèse et al., 2006; Kramer et al., 2009). However, not all ribosome-associated CLIPS bind nascent chains, as neither Zuo1 nor its Hsp70 partner Ssz1 interacts directly with nascent chains (Yam et al., 2005; Albanèse et al., 2006). The Zuo1–Ssz1 complex, termed ribosome-associated complex (RAC; Gautschi et al., 2001), stimulates the ATPase activity of the highly homologous CLIPS Hsp70s SSB via the N-terminal J domain of Zuo1 (Table S1; Huang et al., 2005). Another RAC-like protein, Jjj1, was characterized as a strictly cytosolic protein that binds to Rei1 and helps recycle the 60S ribosomal export factor Arx1 (Table S1; Demoinet et al., 2007; Meyer et al., 2007).
Despite their disparate proposed functions, Zuo1 and Jjj1 share a homology domain and have a similar domain arrangement (Fig. 1 A). Starting from the surprising observation that these chaperones exhibit an aggravating genetic interaction, in this study, we uncover that these chaperones assist ribosome biogenesis, a highly ordered and regulated ribonucleoprotein assembly process that begins in the nucleolus and proceeds through nuclear and cytoplasmic steps (for reviews see Tschochner and Hurt, 2003; Strunk and Karbstein, 2009). Jjj1 and Zuo1 are required to assist nuclear steps of ribosome biogenesis, a process hitherto not shown to require chaperone assistance. Both J domain proteins cooperate with Hsp70-type CLIPS in a chaperone network that binds to nuclear ribosomal biogenesis intermediates and facilitates distinct steps in the ribosomal RNA (rRNA) maturation pathway. Our results uncover a novel function for CLIPS chaperones and indicate that eukaryotes use two distinct ribosome-anchored J domain proteins to link chaperones to the ribosome assembly process.
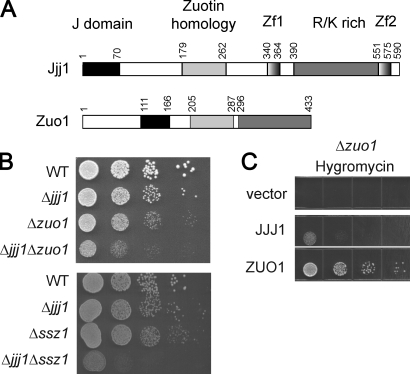
Figure 1.
Jjj1 and Zuo1–Ssz1 have overlapping biological functions. (A) Similar domain organization of Jjj1 and Zuo1. Jjj1 contains an N-terminal J domain, two zinc fingers (ZnF), and a C-terminal R/K-rich domain. Alignment with Zuo1 defines an additional homology domain (ZHD; Fig. S2). (B) Cells deleted for both Jjj1 and either Zuo1 (top) or Ssz1 (bottom) present a synthetic growth phenotype. An equal number of cells was spotted as a 10-fold dilution series on YPD plates and incubated at 30°C for 2 d. (C) Jjj1 overexpression does not suppress the antibiotics sensitivity of Δ_zuo1_ cells. Δ_zuo1_ cells were transformed with an empty p426 vector, the p426-ZUO1 and p426-JJJ1. The serial dilution was performed as in B.
Results
Genetic interactions of Jjj1 with Zuo1–Ssz1
The domain organization of Jjj1 bears striking similarities to that of Zuo1 (Fig. 1 A). Both proteins have an N-terminal J domain known to regulate the ATPase activity of Hsp70s (Mayer and Bukau, 2005) and a K/R-rich, positively charged C-terminal domain, which, in the case of Jjj1, is flanked by two zinc fingers (Fig. 1 A and Table S1). Alignment of Jjj1 and Zuo1 across species revealed an additional homology region that defines a novel conserved domain of unknown function that we called zuotin homology domain (ZHD; Fig. S1). Notably, this domain is unique to Jjj1 and Zuo1 homologues, pointing to a shared and unique function for these J domain CLIPS.
A possible functional overlap between Zuo1 and Jjj1 was revealed by their genetic interactions (Fig. 1 B). Single deletion of either JJJ1 or ZUO1 caused a slow growth phenotype, showing that both proteins are independently important for cell survival. The double-deletion Δ_zuo1_Δ_jjj1_ exhibited a strong synthetic growth phenotype, suggesting that these proteins also have partially redundant functions within the same process (Fig. 1 B; Forsburg, 2001). Synthetic growth interactions were similarly observed between Jjj1 and Ssz1, the partner of Zuo1 in RAC (Fig. 1 B). Thus, the entire RAC interacts functionally with Jjj1, with expression of at least one of these J domain proteins critical for cell survival. Of note, the function of either protein requires the presence of a functional J domain (Fig. S2 A), suggesting that their activity involves regulation of the downstream Hsp70s SSB (Huang et al., 2005) and SSA (Meyer et al., 2007). Indeed, the N terminus of Zuo1, containing its J domain (111–165) and the N-terminal extension upstream from the J domain (1–111; Fig. S3), binds directly to Ssz1 and SSB. In contrast, the J domain of Jjj1 directly binds to and activates the ATPase of the related Hsp70 SSA (Fig. S3; Meyer et al., 2007).
Although RAC and Jjj1 overlap functionally, these chaperones are not fully interchangeable. Δ_zuo1_, Δssz1, and Δssb1/2 cells are hypersensitive to hygromycin, whereas Δ_jjj1_ cells are not (Fig. S2 C; Albanèse et al., 2006). In addition, Zuo1 overexpression could not rescue the Δjjj1 phenotype (Fig. S2 C; Meyer et al., 2007). Interestingly, although overexpression of Jjj1 could rescue the slow growth of Δzuo1 cells (Fig. S2 C), it could not suppress their hygromycin sensitivity phenotype (Fig. 1 C). Thus, Jjj1 and the RAC–SSB CLIPS have overlapping but distinct functions in a common pathway essential for cell survival.
Loss of either Jjj1 or RAC–SSB impairs 60S ribosome biogenesis
The genetic interaction between Jjj1 and the RAC–SSB complex was puzzling given their very different proposed functions. The RAC–SSB network is thought to function in the cytoplasm by binding nascent chains emerging from ribosomes (Pfund et al., 1998; Gautschi et al., 2002; Yam et al., 2005). In contrast, Jjj1 is proposed to interact with the cytosolic protein Rei1 to assist the cytoplasmic recycling of ribosomal export factor Arx1 (Demoinet et al., 2007; Meyer et al., 2007, 2010). We next tried to elucidate how these CLIPS with apparently disparate functions may cooperate.
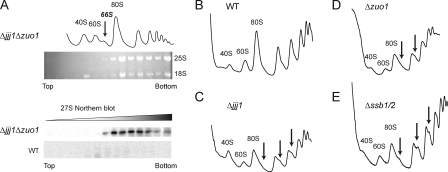
Insight into the shared function of Jjj1 and Zuo1 was provided by sucrose gradient analysis of doubly deleted Δjjj1Δzuo1 cells (Fig. 2 A). Loss of both chaperones led to a reduction in the levels of 80S ribosomes and translating polysomes (Fig. 2 A, top) as well as the appearance of a prominent extra peak between the 60S and the 80S ribosomal particles, which corresponded to the 66S particle, an intermediate in 60S ribosome biogenesis (Fig. 2 A, top). The 66S particle is normally nuclear and contains an immature 27S rRNA (for review see Tschochner and Hurt, 2003). This precursor would normally complete its processing and maturation in the nucleus to yield the 60S ribosomal subunit that contains the 25S rRNA. The identity of the 66S extra peak was confirmed by Northern blot analysis of the sucrose gradient fractions for the wild-type (WT) and Δjjj1Δzuo1 cells using a probe specific for the 27S rRNA (Fig. 2 A, bottom). This analysis revealed the accumulation of immature 27S rRNA precursors in Δjjj1Δzuo1 cells compared with WT extracts, where the low levels of 27S rRNA were below the detection limit (Fig. 2 A, bottom). Notably, the 27S rRNA was also present in heavier fractions in the gradient, which is suggestive of association with polysomes or formation of higher order aggregated species. This experiment suggested that the shared function of Jjj1 and Zuo1 was in ribosome biogenesis.
Figure 2.
Jjj1 and the CLIPS Zuo1, Ssz1, and SSB participate in ribosome biogenesis. (A) Joint deletion of JJJ1 and ZUO1 accumulates a 66S preribosomal particle containing the 27S rRNA precursor. (top) Lysates from WT and Δ_jjj1_Δ_zuo1_ cells were separated on a 7–47% sucrose gradient and the OD254 nm monitored. RNA from individual fractions was analyzed on formaldehyde agarose gels and ethidium bromide staining to visualize the 25S and 18S rRNAs. (bottom) Northern blot analysis for 27S rRNA of Δ_jjj1_Δ_zuo1_ and WT cell lysates. (B–E) Deletion of JJJ1, ZUO1, or SSB1/2 produces aberrant polysome profiles. 20 OD254 nm of yeast lysates from WT (A), Δ_jjj1_ (B), Δ_zuo1_ (C) or Δssb1/2 (D) were fractionated on a 7–47% sucrose gradient and the OD254 nm monitored. The position of 80S ribosomes and 40S and 60S ribosomal subunits are indicated. Arrows indicate the half-mers.
Previous sucrose gradient analysis of polysome profiles from Δ_jjj1_ cells showed that they contain half-mers within the polysomes (Fig. 2, B and C [arrows]; Demoinet et al., 2007; Meyer et al., 2007). These shoulders are hallmarks of defective 60S ribosomal subunit maturation: the lack of mature cytoplasmic 60S ribosomal subunits results in irreversible binding of abortive 48S preinitiation complexes on mRNAs (Demoinet et al., 2007; Meyer et al., 2007). Strikingly, the polysome profiles of both Δzuo1 and Δssb1/2 cells also exhibited half-mers in the polysomal fractions (Fig. 2, D and E). These anomalies indicate that the absence of the RAC–SSB chaperones also lead to a defect in 60S biogenesis.
Role of Jjj1 and RAC–SSB CLIPS in 60S ribosome maturation, export, and Arx1 recycling
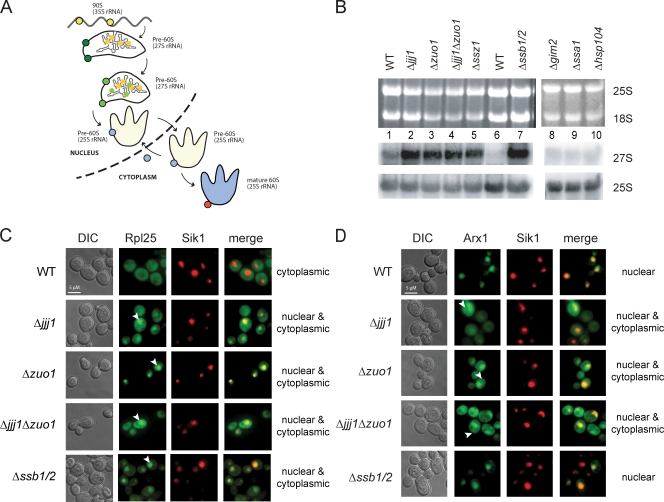
The results in Fig. 2 suggested that Jjj1 and RAC–SSB are both required for normal 60S ribosome biogenesis. In eukaryotes, ribosome biogenesis begins in the nucleolus and proceeds through nuclear steps of rRNA processing and ribonucleoprotein assembly, each mediated by a different set of nucleolar and nuclear factors leading to the formation of mature 60S and 40S particles (Fig. 3 A, yellow and green; and Table S2; for reviews see Fromont-Racine et al., 2003; Tschochner and Hurt, 2003; Strunk and Karbstein, 2009). These ribosomal subunits are exported to the cytoplasm with the help of shuttling factors, such as Arx1 for the 60S subunit (Fig. 3 A, blue), and the final cytoplasmic maturation is mediated by several factors, such as Rei1 for the 60S subunit (Fig. 3 A, red). We next examined how loss of Jjj1 and the RAC–SSB system affects three aspects of ribosome biogenesis, namely rRNA maturation (Fig. 3 B), ribosomal export from the nucleus (Fig. 3 C), and Arx1 recycling (Fig. 3 D). When compared with WT cells, we observed a strong accumulation of the 27S rRNA precursor in Δjjj1 and Δzuo1 strains as well as in Δssz1 and Δssb1/2 (Fig. 3 B). No accumulation was observed in strains deleted for other cytosolic chaperones, such as the CLIPS Gim2 or the stress-inducible Hsp104 (Fig. 3 B). Deletion of a single SSA Hsp70 homologue, SSA1, did not affect this process (unpublished data), most likely because of the presence of four redundant SSA genes in yeast (Frydman, 2001).
Figure 3.
Distinct effects of Jjj1, Zuo1, and SSB on 60S ribosomal export and Arx1 recycling. (A) Schematic representation of ribosome biogenesis of the 60S ribosomal subunit. Nucleolar ribosomal biogenesis factors are represented in yellow, nuclear ones in green, shuttling factors, like Arx1, in blue, and cytoplasmic factors in orange. (B) Jjj1 and Zuo1, Ssz1, and SSB are involved in 60S ribosome subunit biogenesis. Total RNA from the indicated yeast cells was extracted and separated on a formaldehyde agarose gel. The 27S rRNA was detected by Northern blotting. RNA loading was controled by ethidium bromide staining to visualize the 25S and 18S rRNAs (top) and by Northern blot analysis for the 25S rRNA (bottom). (C) Effects of CLIPS deletion on the export of the 60S ribosomal subunit. The export of the 60S subunit was monitored using Rpl25-GFP as a reporter (provided by D. Roser, University of Heidelberg, Heidelberg, Germany). Cells were grown to mid-log phase at 25°C, and the in vivo localization of Rpl25-GFP was monitored by fluorescence microscopy. The nucleolar marker Sik1-RFP was used to identify the position of the nucleus. (D) Zuo1 and Jjj1 but not SSB are required for the recycling of the shuttling factor Arx1. Cells were transformed with the Arx1-GFP (provided by A. Johnson, University of Texas at Austin, Austin, TX) plasmid and were grown at 25°C to mid-log phase. The in vivo localization of Arx1-GFP was monitored by fluorescence microscopy. DIC, differential interference contrast. See Results for description of arrowheads.
Defects in 60S ribosomal biogenesis often impair the export of the 60S particles to the cytoplasm, leading to their accumulation in the nucleus (Hurt et al., 1999). Thus, we examine whether 60S ribosomal export to the cytosol was affected in Δjjj1, Δzuo1, and Δssb1/2 cells using ribosomal protein Rpl25-GFP as a 60S marker (Fig. 3 C; Hurt et al., 1999) and Sik1-RFP as a nucleolar marker (Fig. 3 C; Sung and Huh, 2007). As expected, most 60S ribosomal particles in WT cells are cytoplasmic (Fig. 3 C). At 25°C, Δjjj1 cells accumulated Rpl25-GFP in the nucleus (Fig. 3 C, arrowheads; and note yellow overlap with the red nucleolar marker Sik1-RFP). The nuclear export defect in Δjjj1 cells was less marked at 30°C (unpublished data). Rpl25-GFP also accumulated in the nucleus in Δzuo1 and Δzuo1Δjjj1 cells (Fig. 3 C, arrowheads; yellow overlap in nucleus) and even more dramatically in Δssb1/2 cells (Fig. 3 C, bottom, arrowhead).
Jjj1 is proposed to bind to Rei1 and facilitate the recycling of Arx1 back to the nucleus (Demoinet et al., 2007; Meyer et al., 2007). Consistent with this role, we found that Arx1 accumulated in the cytoplasm of Δjjj1 and Δzuo1Δjjj1 cells (Fig. 3 D, arrowheads). Surprisingly, Arx1 recycling was also impaired in Δ_zuo1_ cells (Fig. 3 D, arrowheads). The defective recycling of Arx1 in these cells was also detected biochemically, as failure to dislodge Arx1 from cytoplasmic 60S subunits caused Arx1 to migrate with polysomes in both Δzuo1 and Δjjj1 cells (unpublished data). Interestingly, Arx1 recycling appeared unaffected in Δssb1/2 cells (Fig. 3 D), indicating that SSB is not required for Arx1 recycling. Collectively, these results indicate that both Jjj1 and the RAC–SSB network participate in the maturation of the 60S ribosomal subunit and normal 60S ribosomal export.
Domain contribution of Jjj1 and Zuo1 to subcellular localization and ribosome association
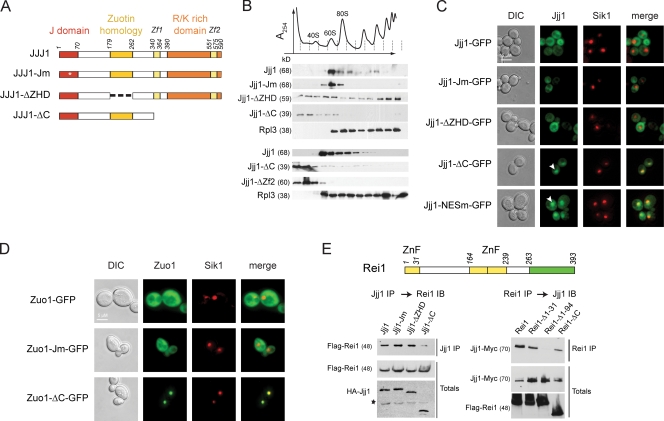
To understand how Jjj1 acts during ribosome biogenesis, we explored the contribution of its various domains to its association with ribosomes (Fig. 4) and its function (Fig. 5 and Table S3). Jjj1 has three clearly identifiable domains: an N-terminal J domain, the ZHD, and a C-terminal–charged domain flanked by two zinc fingers (Fig. 4 A). We initially examined how the various Jjj1 domains contribute to its association with ribosomal fractions (Fig. 4 A). Full-length WT Jjj1 comigrates with the 60S and 80S ribosomal fractions and to a lesser extent with polysomes (Fig. 4 B, middle [low expression plasmid] and bottom [chromosomal-tagged copy]; and Fig. S4 A, high copy plasmid). These interactions were largely abrogated by deletion of the charged domain (Jjj1-ΔC; Fig. 4 B, middle and bottom; and Fig. S4 A). A functional J domain, required for binding to SSA (Fig. S3), was not required for association with the 60S/80S ribosomal particles (Jjj1-Jm; Fig. 4 B and Fig. S4 A), although we noticed a consistent loss of association with polysomes even at high expression levels (Fig. S4 A). Notably, deletion of the ZHD domain had the opposite effect than the Jm mutation on Jjj1 sedimentation: Jjj1-ΔZHD was more weakly associated with the 60S and 80S fractions and enriched in the polysome fractions (Jjj1-ΔZHD; Fig. 4 B and Fig. S4). This suggests that the ZHD domain contributes to Jjj1 association with a subset of 60S ribosomal particles, whereas the C-terminal–charged domain mediates the stable association with ribosomes and polysomes. Collectively, our results reveal a complex pattern of interactions between Jjj1 and ribosomes, whereby different domains have distinct contributions to its interaction with ribosomes at different stages. A similar domain analysis for Zuo1 also indicated that the charged domain is required for polysome association (unpublished data) as previously reported (Yan et al., 1998).
Figure 4.
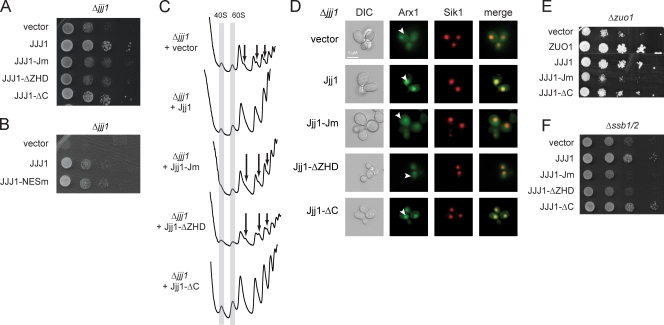
Jjj1 is a conserved modular protein with distinct functional domains. (A) Domain mutants of Jjj1. Jm, mutated in the canonical HPD motif of the J domain; ΔZHD, lacks the ZHD; ΔC, lacks the C-terminal K/R-rich domain. Asterisk indicates the point mutation in the HPD motif of the J domain. (B) Effect of Jjj1 mutations on the interaction with ribosomes. Yeast extracts from the indicated cells were fractionated on 7–47% sucrose gradients. The OD254 nm profile (top) identifies the polysomal fractions. Individual fractions were analyzed for the presence of Jjj1 mutants and the ribosomal protein Rpl3 by SDS-PAGE and immunoblotting. Similar results are obtained by expression of Jjj1 mutants from centromeric plasmids (middle), the endogenous chromosomal copy (bottom), or high copy number plasmids (Fig. S5). (C) Cellular localization of the different Jjj1 mutants. Δ_jjj1_ cells expressing the different mutants of Jjj1 in fusion with GFP were monitored by fluorescence microscopy. Sik1-RFP identifies the position of the nucleolus. The Jjj1-GFP fusion complements the slow growth phenotype of Δjjj1 cells (Fig. S5) and associates with ribosomes (not depicted). (D) Role of the Zuo1 C terminus and the J domain in its subcellular localization and function. Δzuo1 cells expressing the different mutants of Zuo1 in fusion with GFP were monitored by fluorescence microscopy. (E) Identification of Jjj1 domain mediating association with Rei1. (top) Domain structure of Rei1 indicating position of the three zinc fingers (yellow) and the C-terminal Rpl24-binding domain (green) are indicated (Lebreton et al., 2006). (bottom left) Yeast lysates were prepared from cells expressing HA-Jjj1 or the indicated Jjj1 domain variants together with Flag-tagged Rei1. After immunoprecipitation for Jjj1, the association with Rei1 was detected by immunoblotting with anti-Flag. Total Rei1 and Jjj1 protein was analyzed by immunoblotting against the respective tags. Asterisk indicates a background crossreacting band. (bottom right) Yeast lysates were prepared from cells carrying an endogenously tagged Jjj1 together with Flag-tagged Rei1 or Rei1 domain variants. After immunoprecipitation for Rei1, the association with Jjj1 was detected by immunoblotting with anti-Myc. Total Rei1 and Jjj1 protein was analyzed by immunoblotting. DIC, differential interference contrast; ZnF, zinc finger. See Results for description of arrowheads.
Figure 5.
A nuclear form of Jjj1 that does not bind Rei1 suffices to restore its function in ribosome biogenesis and Arx1 recycling. (A) Complementation of Δjjj1 by Jjj1 mutants. Cells expressing the Jjj1 variants were grown overnight at 30°C, and a dilution series was performed on −URA plates. (B) Jjj1-NESm is functional and complements the slow growth phenotype of the Δjjj1 cells. (C) Rescue of Δjjj1 aberrant polysome profile by Jjj1 domain mutants. Yeast lysates were fractionated on a 7–47% sucrose gradient, and the OD254 was monitored. The gray columns indicate the 40S and the 60S peaks. The arrows indicate the presence of half-mers containing extra 48S initiation complexes. (D) Rescue of defective Arx1 recycling in Δ_jjj1_ cells by Jjj1 domain mutants. Δ_jjj1_ cells were transformed with the plasmids expressing the different Jjj1 mutants and the Arx1-GFP plasmid, and localization of Arx1-GFP was monitored by fluorescence microscopy. Sik1-RFP protein was used to identify the nucleus. DIC, differential interference contrast. (E and F) Overexpression of Jjj1 can suppress the slow growth phenotype of the Δ_ssb1/2_ (E) and Δzuo1 (F) cells in a domain-specific manner. Cells were transformed with either WT or mutant Jjj1, and growth was assessed by a dilution series assay. White line indicates that intervening lanes have been spliced out. See Results for description of arrowheads.
We next examined the effect of the various Jjj1 domains on its subcellular localization (Fig. 4 C; and Fig. S4, B and D). As described previously, full-length Jjj1 is largely cytoplasmic (Fig. 4 C, top; Meyer et al., 2007). Similarly, Jjj1-Jm and Jjj1-ΔZHD were also cytoplasmic (Fig. 4 C). Surprisingly, the Jjj1-ΔC mutant showed a strong accumulation in the nucleus (Fig. 4 C, arrowheads). Thus, we searched for nuclear export signals (NESs; la Cour et al., 2004) and identified a canonical NES at the C terminus spanning amino acids 409–419 (LQALQAELAEI). Disruption of the NES by a single leucine to alanine mutation in the first leucine (Jjj1-NESm) led to the nuclear accumulation of Jjj1-NESm, which is similar to Jjj1-ΔC (Fig. 4 C, arrowheads). Thus, Jjj1 is largely cytosolic under steady-state conditions but shuttles between the cytoplasm and nucleus. Interestingly, Jjj1-ΔC was more tightly colocalized with the nucleolar Sik1 marker than Jjj1-NESm (Fig. 4 C, compare bottom merge panels), suggesting that Jjj1-ΔC may be concentrated in the nucleolus. Strikingly, similar results were obtained for Zuo1-ΔC, which also concentrated in the nucleolus (Fig. 4 D). Thus, Zuo1 and Jjj1 can localize to the nucleolus, which is the site of ribosome biogenesis.
Because Jjj1 is shown to associate with Rei1 through its zinc fingers (Meyer et al., 2010), we next examined the domains that mediate this interaction in both Jjj1 (Fig. 4 E, left) and Rei1 (Fig. 4 E, right). Cells expressing the indicated epitope-tagged Rei1 and Jjj1 domain variants were subjected to immunoprecipitation against one of the proteins followed by immunoblotting against the other (Fig. 4 E). As expected, the C terminus of Jjj1 was indeed required for interaction with Rei1. For Rei1, we find that the N terminus of Rei1, encompassing the first of the three zinc fingers of Rei1, mediates the interaction with Jjj1 (Fig. 4 E, right).
Nuclear Jjj1 can rescue all ribosome biogenesis defects in Δjjj1 cells
We next assessed the contribution of the Jjj1 domains to the Δjjj1 phenotype (Fig. 5 A; and Fig. S4, B and C). As expected, the slow growth phenotype was rescued by expression of WT Jjj1 (Fig. 5 A). The J domain mutant (Jjj1-Jm), which no longer interacts with the Hsp70 SSA (Fig. S4), does not rescue the Δjjj1 phenotype. Similarly, the ZHD domain (Jjj1-ΔZHD) is also required for Jjj1 function. Surprisingly, the slow growth phenotype is fully rescued by Jjj1 lacking the entire C-terminal–charged domain (Jjj1-ΔC; Fig. 5 A; and Fig. S4, B and D). This is independent of the expression levels, as WT growth rates are also observed upon deletion of the C-terminal domain or the Zf2 zinc finger from the chromosomal copy of JJJ1, where Jjj1 is expressed at endogenous low levels (Fig. S4 C). Thus, Jjj1 function requires both the J domain and the ZHD domain but, surprisingly, not the C-terminal–charged domain, which mediates association with polysomes and contains the Rei1-binding site. The Jjj1-NESm mutant was also able to rescue the slow growth phenotype of Δjjj1 (Fig. 5 B and Fig. S4 B). This further suggests that a mostly nuclear form of Jjj1 can rescue its growth phenotype.
We next examined the contribution of various Jjj1 domains to the 40S/60S balance (Fig. 5 C and Fig. S4 D). Expression of the full-length Jjj1 in Δjjj1 cells restored the normal polysome profiles (Fig. 5 C). Mutation of the J domain (Jm-Jjj1) or deletion of the ZHD domain could not restore the 40S/60S balance. In contrast, expression of Jjj1 lacking the C-terminal domain (Jjj1-ΔC) fully restored the normal polysome profile (Fig. 5 C). Importantly, restoration of the normal polysome profiles by Jjj1-ΔC was also observed at the low endogenous expression levels in cells carrying a deletion of the C-terminal domain or of the Zf2 zinc finger of the chromosomal copy of Jjj1 (Fig. S4 D). Furthermore, the Jjj1-NESm mutant also rescued the balance between 40S/60S ribosomal subunits (Fig. S4 E).
We next examined the role of Jjj1 domains in Arx1 recycling from the cytoplasm to the nucleus (Fig. 5 D). In cells expressing WT Jjj1, Arx1 was primarily in the nucleus (Fig. 5 D, Jjj1, yellow overlap with Sik1 marker), whereas Δ_jjj1_ cells contained a significant pool of cytoplasmic Arx1 (Fig. 5 D; Demoinet et al., 2007; Meyer et al., 2007). The Arx1 recycling defect was not rescued by either Jjj1-Jm or Jjj1-ZHD (Fig. 5 D). Strikingly, Jjj1-ΔC fully restored Arx1 nuclear localization, despite lacking the entire Rei1 interaction domain (Fig. 5 D, arrowheads). This result does not support the idea that Jjj1 facilitates Arx1 recycling via recruitment of Rei1. The observation that Jjj1-ΔC does not bind to Rei1, yet rescues every Δjjj1 defect we tested, suggests that Jjj1 plays an important role in normal growth and ribosome biogenesis that does not involve cytoplasmic interaction with Rei1. Although it is in principle possible that a small subpopulation of Jjj1-ΔC enters the cytosol, the predominantly nuclear localization of Jjj1-ΔC and Jjj1-NESm strongly argues for a nuclear function for Jjj1, such as assisting the correct conformational maturation of the 60S ribosomal particles in the nucleus (see Figs. 6 and 7).
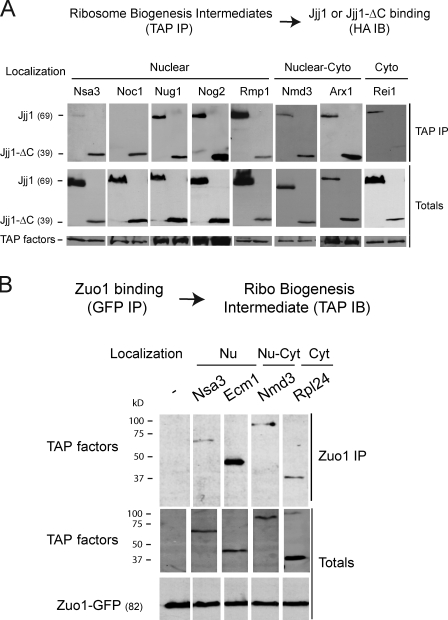
Figure 6.
Jjj1 and Zuo1 play early and distinct roles in ribosome biogenesis. (A) Association of Jjj1 and Jjj1-ΔC with nuclear and cytoplasmic (Cyto) ribosomal precursor particles. Ribosome biogenesis intermediates containing the chromosomally TAP-tagged version of the indicated biogenesis factors were isolated by TAP purifications from cells transformed with plasmids expressing HA-Jjj1 or HA–Jjj1-ΔC. After isolation and elution from the beads, proteins were analyzed by SDS-PAGE and immunoblotting (IB). (B) Association of Zuo1 with nuclear (Nu) and cytoplasmic (Cyt) ribosomal precursor particles. Zuo1-containing complexes were isolated from cells containing the chromosomally TAP-tagged version of the indicated biogenesis factors and transformed with a plasmid expressing Zuo1-GFP. The presence of ribosome biogenesis intermediates in the GFP immunoprecipitation (IP) was assessed and analyzed by SDS-PAGE, TAP, and GFP immunoblotting.
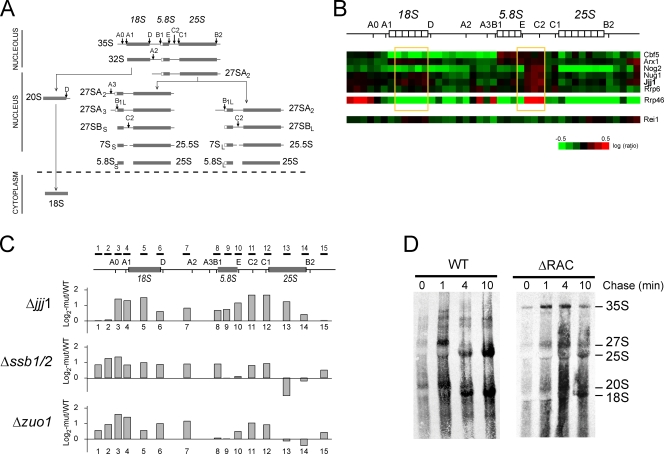
Figure 7.
Jjj1 and Zuo1 play early and distinct roles in ribosome biogenesis. (A) Schematic representation of biogenesis of eukaryotic ribosomal rRNA maturation particles. The position of endonucleolytic cleavage steps and the subcellular localization of each step are indicated. (B) Loss of Jjj1 blocks a nuclear step of rRNA processing. rRNA processing microarray data (obtained from Peng et al., 2003) extracted to show the genes clustering together with Jjj1. The microarray data from Rei1 and Arx1 deletions are also included for comparison. The regions enriched in the jjj1 deletion strain, reflecting slower processing respect to WT, are highlighted in yellow. (C) rRNA processing defects in Δjjj1, Δzuo1, and Δssb1/2 cells measured by microarray experiments as described previously (Peng et al., 2003) using the indicated rRNA probes. Log2 fold induction of the indicated mutants over WT was calculated from triplicate experiments. (D) Loss of Zuo1–Ssz1 (RAC) blocks a nuclear step of rRNA processing. Pulse-chase labeling with [3H]uracil was performed with the WT and ΔRAC cells. Cells were pulse labeled for 2 min and chased as indicated with an excess of cold uracil. Total labeled RNAs were purified, separated on a denaturing agarose gel, and autoradiographed. The positions of the intermediate and mature rRNAs are indicated.
The predominantly nuclear form of Jjj1 overlaps functionally with the RAC–SSB system
We next examined the Jjj1 domains required to rescue the growth defect of Δzuo1 cells (Fig. 5 E). Inactivation of the J domain or deletion of the ZHD in Jjj1 abolished the rescue of Δzuo1 slow growth (Fig. 5 E, JJJ1-Jm and JJJ1-ΔZHD). In contrast, JJJ1-ΔC restored the normal growth of Δzuo1 cells (Fig. 5 E, JJJ1-ΔC). Thus, the nuclear Jjj1-ΔC variant contains the region that overlaps functionally with Zuo1. Because Zuo1 regulates the activity of SSB, which also appears important for ribosome biogenesis, we next examined whether Jjj1 or its variants interact genetically with SSB (Fig. 5 F). Remarkably, Jjj1 overexpression fully restored the slow growth phenotype of Δssb1/2 (Fig. 5 F). Similar to Δzuo1 cells, the growth phenotype of Δssb1/2 was also fully restored by the nuclear Jjj1-ΔC variant (Fig. 5 F). In sum, the predominantly nuclear Jjj1 variant that does not interact with Rei1 can nonetheless rescue the Δjjj1, Δzuo1, and Δssb1/2 growth phenotypes. Collectively, our results suggest that the functional overlap between Jjj1 and the RAC–SSB chaperone system likely involves nuclear steps of ribosome biogenesis.
Jjj1 and Zuo1 interact with nuclear ribosome biogenesis intermediates
A possible nuclear function for Jjj1 and Zuo1 in ribosome biogenesis was further explored by assessing their interaction with ribosome assembly intermediates along the biogenesis pathway (Fig. 6 A and Table S2). For Jjj1, ribosome biogenesis intermediates were isolated via chromosomally integrated tandem affinity purification (TAP)–tagged assembly factors in cells expressing Jjj1 or Jjj1-ΔC (Fig. 6 A). Immunoblot analysis for Jjj1 indicated that this chaperone indeed associates with nucleolar and nuclear 60S ribosome biogenesis intermediates as well as with later intermediates in the biogenesis pathway. Interestingly, the nuclear Jjj1-ΔC interacted much more strongly than full-length Jjj1 with the very early 60S biogenesis factors Noc1 and Nsa3 (Fig. 6 A), perhaps because of the higher concentration of Jjj1-ΔC in the nucleus. A similar analysis for Zuo1 (Fig. 6 B) also revealed its association with nuclear ribosome biogenesis intermediates, most notably with particles containing Ecm1 (Fig. 6 B). These results support the role of both Zuo1 and Jjj1 in nuclear steps of ribosome biogenesis.
Jjj1 and RAC–SSB affect 35S and 27S rRNA maturation
Further insight into the function of these chaperones in ribosome biogenesis exploited the fact that eukaryotic ribosome biogenesis involves an ordered pathway of rRNA processing (Fig. 7 A; for review see Tschochner and Hurt, 2003). Ribosome biogenesis starts in the nucleolus with the formation of a 90S preribosomal particle containing the 35S rRNA precursor (Fig. 3 A and Fig. 7 A). Subsequent rRNA processing steps lead to the formation of 66S and 43S particles. The 66S particle, containing the 27S rRNA, completes its processing and maturation in the nucleus to form the 60S ribosomal subunit containing the 25S rRNA. The 43S particle, containing the 20S rRNA, is processed into the 40S small ribosomal subunit, containing the 18S rRNA. As perturbations in the processing and maturation steps lead to the accumulation of rRNA precursors that can be identified by hybridization with probes specific for different rRNA regions (Fig. 7, A–C), we next examined whether loss of Jjj1 or Zuo1 function led to specific blocks in the rRNA processing pathway.
Publicly available genomic data on ribosome biogenesis provided insight into Jjj1 function (Peng et al., 2003). A previous study had used oligonucleotide microarrays to examine defects in the biogenesis of noncoding RNAs in a large set of mutant strains (Peng et al., 2003). The microarray allowed detection of accumulation or depletion of specific RNA processing intermediates in a given mutant strain, thereby revealing defects in particular steps of rRNA maturation and ribosome biogenesis. The gene for Jjj1 (then a gene with unknown function called YNL227c) was included in this study. Clustering analysis linked JJJ1 to a subset of ribosome biogenesis proteins and exosome-associated factors required for processing of the 3′ end of 5.8S rRNA within the 27S rRNA, as well as for an earlier processing step 3′ of the 18S rRNA (Fig. 7 B; Peng et al., 2003). Nug1, but not Rei1 nor Arx1, clustered together with Jjj1 in this analysis (Peng et al., 2003). Interestingly, we find that Jjj1 physically interacts with Nug1-containing particles (Fig. 6 A).
Because information on other chaperones was absent from the public domain, we next performed a similar microarray-based analysis to examine a possible role for Zuo1 and SSB in nuclear steps of ribosome biogenesis (Fig. 7, A, B, and C [probes schematically indicated]; Peng et al., 2003). Our analysis of Δjjj1 yielded similar conclusions as those obtained previously (Fig. 7 B; Peng et al., 2003), confirming that Jjj1 plays a role in 27S rRNA processing and probably also in 35S rRNA processing. Analysis of Δzuo1 and Δssb1/2 cells indicated a block at very early 35S rRNA processing steps as well as a block in the 27S rRNA processing step, which is similar to that observed for Jjj1. The accumulation of 27S rRNA biogenesis intermediates in Δjjj1, Δzuo1, and Δssb1/2 cells was consistent with our previous Northern blot analyses and the polysome profiles (Figs. 2 and 3). We further examined the rRNA processing defect in cells lacking RAC by pulse-chase analysis of rRNA maturation using [3H]uracil (Fig. 7 D). This experiment further confirmed that loss of RAC function led to impaired rRNA maturation, which was very noticeable at the 35S rRNA cleavage (Fig. 7 D). Indeed, a function for Zuo1 and SSB in 35S rRNA maturation could explain the slight defect in 40S biogenesis observed in these cells (Fig. 2, D and E, low 40S peak; and not depicted).
Discussion
In this study, we show that ribosome biogenesis in the nucleus is assisted by a chaperone network consisting of two ribosome-anchored J-proteins, Jjj1 and Zuo1, that cooperate with different classes of Hsp70, SSA and Ssz1/SSB, respectively (Fig. 8). Our findings that some CLIPS chaperones play an important role in ribosome maturation in the nucleus, in addition to their previously reported cytosolic roles in cotranslational nascent chain binding, suggests a possible link between the cellular protein synthesis and folding machineries.
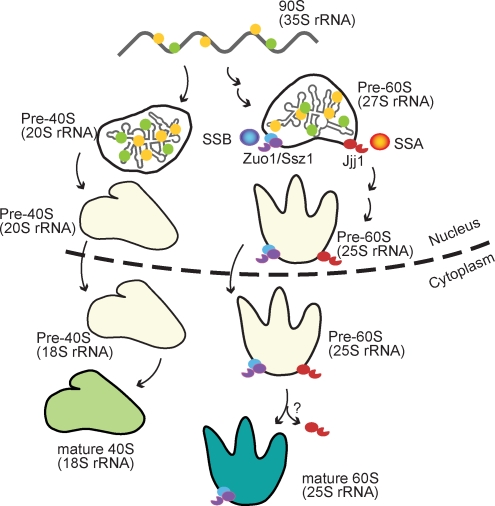
Figure 8.
Schematic representation of the role of Jjj1 and the Zuo1–Ssz1–SSB network in ribosome biogenesis. Major ribosomal maturation intermediates and rRNA processing steps are schematically indicated. Jjj1 and Zuo1 bind to nuclear assembly intermediates and regulate the activity of SSA and SSB, respectively. Jjj1 and Zuo1 bind first to nuclear 60S intermediates in the nucleus and stay bound to the mature cytoplasmic 60S ribosomal subunit. Red, Jjj1; orange, SSA; purple, Zuo1; blue, SSB. See Discussion for details.
Jjj1: a modular CLIPS chaperone that shares characteristics with Zuo1
Jjj1 exhibits a conserved modular domain structure, which bears striking parallels to the domain architecture of another CLIPS chaperone, Zuo1. In fact, JJJ1 is also a CLIPS chaperone, as it is also coregulated transcriptionally with translational components (Albanèse et al., 2006; unpublished data). The different Jjj1 domains contribute in a complex manner to its function and interaction with ribosomes (Table S3). The J domain is essential for the function of Jjj1 and Zuo1. The N-terminal extension and J domain of Zuo1 confer specificity for the Hsp70s Ssz1 and SSB, whereas the J domain of Jjj1 is specific for the Hsp70 SSA (Fig. S3). For both proteins, the C-terminal–charged domain is important for ribosome association (Fig. 4 and Fig. S4; Yan et al., 1998). Surprisingly, although the C-terminal domain is essential for Zuo1 function (Yan et al., 1998; unpublished data), it is dispensable for Jjj1 function. In contrast, the ZHD domain appears to play a key role in Jjj1 and Zuo1 function (Figs. 4 and 5; and not depicted). Our fractionation analyses suggest that this domain may play a role promoting binding to 60S ribosome biogenesis intermediates. This may account for the dispensability of the charged domain in Jjj1, which also promotes ribosome binding and appears to be important for the stable association of Jjj1 with ribosomes after export to the cytoplasm.
Jjj1 and the RAC–SSB network function in nuclear biogenesis of the 60S ribosomal particle
Our results indicate that Jjj1, Zuo1, and SSB also have important functions in ribosome biogenesis in the nucleus. Several lines of evidence support this conclusion: (a) Jjj1 and Zuo1 localize to the nucleus, (b) Jjj1 and Zuo1 interact with nuclear biogenesis factors, (c) there is an accumulation of 35S and 27S rRNA in Δjjj1 and Δzuo1 cells and of 66S ribosomal precursor in the Δjjj1Δzuo1 cells, and (d) genetic data indicate that Jjj1 and Zuo1 have overlapping but distinct functions within the ribosome biogenesis process. Our findings lead us to conclude that these chaperones are directly involved in assisting the ordered biogenesis of the ribosomal particle.
Our analyses suggest that Zuo1 and Jjj1 function in the nucleus, where they associate with nucleolar and nuclear ribosomal biogenesis intermediates (Fig. 6). A predominantly nuclear Jjj1-ΔC rescued every phenotype of Δjjj1 tested (Fig. 4), although it lost its association with Rei1 (Fig. 3). The presence of Jjj1 inside the nucleus is corroborated by the presence of a functional NES (Fig. 4 and Fig. S4). Interestingly, Jjj1-NESm is nuclear, whereas Jjj1-ΔC exhibits a more restricted nucleolar localization (Fig. 4 C). We interpret these results as indicating that the J and ZHD domains of Jjj1 specify for nucleolar localization or interaction with nucleolar ribosome biogenesis intermediates. Because the ribosome-binding C-terminal–charged domain is still present in Jjj1-NESm, it allows for continued binding to ribosomes upon exit from the nucleolus to the nucleoplasm. A functional NES signal is then required for efficient export of Jjj1 into the cytoplasm. Thus, although Jjj1 is largely cytoplasmic under steady-state conditions (Meyer et al., 2007), Jjj1 associates with ribosomal precursors in the nucleus and likely fluxes out of the nucleus with mature ribosomes.
Similar to Jjj1, Zuo1 also appears to cycle between the cytoplasm and the nucleus. Indeed, deletion of the C-terminal–charged domain in Zuo1 also confers a mostly nucleolar localization (Fig. 4 D). However, unlike Jjj1, the nuclear Zuo1-ΔC variant does not rescue the growth phenotype (Yan et al., 1998), perhaps because this chaperone serves a dual role in the cell and also promotes cytosolic protein biogenesis (Gautschi et al., 2002; Huang et al., 2005; Albanèse et al., 2006).
Of note, SSB contains a functional NES signal and also cycles between the cytoplasm and the nucleus (Shulga et al., 1999). The association of both SSA and SSB with ribosome biogenesis intermediates has often been noted (Horsey et al., 2004); however, these interactions are usually disregarded. Together, our data suggest that the J domain proteins Jjj1 and Zuo1 and the Hsp70s they regulate play important roles assisting ribosome biogenesis in the nucleus in addition to their previously known functions in the cytoplasm.
Jjj1 and RAC–SSB function at several nuclear steps in ribosome biogenesis
The synthetic aggravating interaction between Jjj1 and Zuo1 suggested that both chaperones function in overlapping but different steps within the same process, ribosome biogenesis. Microarray-based analysis of the rRNA processing pathway revealed that Δjjj1, Δzuo1, and Δssb1/2 cells exhibit blockage in overlapping nuclear steps in the rRNA processing pathway (Fig. 7). Given the complexity of the ribosome assembly pathway, further mechanistic work will be required to define the precise steps requiring chaperone assistance.
The hitherto unrecognized role of the Zuo1–Ssz1–SSB chaperone network in ribosome biogenesis is consistent with several previous observations. SSB and RAC have been linked to ribosomal frameshifting (Muldoon-Jacobs and Dinman, 2006). A genome-wide screen of Δzuo1 interactions suggested that Zuo1 does not play a very general role in cellular folding (Bobula et al., 2006) but identified interactions with three factors involved in 60S ribosome biogenesis: Rrp1, Rrp15, and Nop4 (Bobula et al., 2006). Consistent with our microarray-based analysis of Zuo1 function (Fig. 7 B), Rrp1 is involved in 27S rRNA processing (Horsey et al., 2004), Rrp15 is involved in 35S rRNA processing (De Marchis et al., 2005), and Nop4 facilitates localization to the nucleolus of the U3 snoRNA, which is required for rRNA processing of both 35S and 27S rRNA (Sun and Woolford, 1994; Qiu et al., 2008). Thus, our results resonate with several genetic and biochemical observations.
CLIPS chaperones with dual cytosolic and nuclear function
Jjj1, RAC, and SSB were reported to have very different functions in the cytosol. SSB is known to bind nascent chains, presumably with the help of RAC (Gautschi et al., 2002; Huang et al., 2005; Albanèse et al., 2006). We propose that these CLIPS serve as dual-function chaperones that first bind to and assist the biogenesis of ribosomal precursor particles and then stay on the ribosome to facilitate nascent chain folding. Both RAC components as well as SSB are very abundant. RAC concentration is estimated at 90,000 per cell, whereas SSB, estimated at 280,000 per cell, is approximately equimolar with ribosomes (310,000/cell) and probably binds to most if not all ribosomes (Huh et al., 2003; Raue et al., 2007).
Jjj1 must act transiently during ribosome biogenesis given the low abundance of this chaperone (2,300 copies per cell; Huh et al., 2003). Although we do find a fraction of Jjj1 associated with translating polysomes even under the low endogenous expression levels (Fig. 4), most of it must either be recycled to return to the nucleus or degraded in the cytoplasm (Fig. 8). Perhaps the previously described interaction with Rei1 and other cytoplasmic factors plays a role in these processes (Demoinet et al., 2007; Meyer et al., 2007). Although deletion of the Rei1 interaction domain did not affect the phenotypes examined in this study, Jjj1 likely has additional functions in the cytoplasm (Demoinet et al., 2007; Meyer et al., 2007) in addition to its function in nuclear ribosome biogenesis. In support of this idea, deletion of Arx1 alleviates the cold sensitivity of the Jjj1 deletion (Fig. S5 A; Demoinet et al., 2007; Meyer et al., 2007), although it does not rescue its aberrant polysome profile (Fig. S5 B, note the presence of half-mers in polysome profiles).
Eukaryotic ribosome biogenesis: a multistep process requiring chaperone assistance
Eukaryotic ribosome biogenesis is a highly complex process starting in the nucleolus and ending in the cytoplasm. Although chaperones had not been previously implicated in the process of eukaryotic ribosome assembly, this role is not surprising given its complexity. Interestingly, ribosome biogenesis in prokaryotes also requires the assistance of chaperones (El Hage et al., 2001; Maki et al., 2002; El Hage and Alix, 2004). The precise mechanism and steps assisted by prokaryotic chaperones have not been elucidated.
Our identification of Jjj1 and RAC–SSB as promoting ribosome biogenesis raises many questions regarding their mechanism of action. RNA folding and RNA chaperones operate through very different principles than protein folding and protein chaperones (Russell, 2008). Thus, it is unlikely that these CLIPS chaperones act by remodeling the rRNA structure (Russell, 2008). More likely, Jjj1 and Zuo1 bind to specific locations within the ribosomal precursors and by virtue of the recruitment of the Hsp70s SSA or SSB serve to remodel the ribosome biogenesis intermediates, either by facilitating the ordered recruitment and/or the dissociation of ribosomal proteins or assembly factors. Another possible function for SSA and SSB is that of maintaining the highly unstructured ribosomal proteins, many of which cannot fold until incorporated into the ribosome, in a conformation that can be assembled into ribosomal particles. However, our data suggest that the role of these Hsp70s may not be limited to this function because Jjj1, which does not bind nascent chains (unpublished data), can suppress the slow growth phenotype of Δssb1/2. Furthermore, Zuo1, Ssz1, and Jjj1 do not bind directly to nascent chains, and thus, it would be unlikely to bring them into the nucleus. Clearly, future studies are required to identify the targets and precise steps mediated by these chaperones. It will also be interesting to define their interplay with previously described ribosome assembly factors, which include AAA ATPases and prolyl isomerases, which may have protein-remodeling functions (for review see Strunk and Karbstein, 2009). The finding that chaperones involved in cytosolic nascent chain binding also assist early steps in nuclear ribosome biogenesis provides an intriguing mechanism to link the capacity for protein synthesis and that of protein folding.
One of the major differences between prokaryotic and eukaryotic de novo folding is the emergence in eukaryotic cells of machinery capable of efficient cotranslational folding (Agashe et al., 2004). Notably, DnaK and GroEL, the prokaryotic chaperones facilitating ribosome biogenesis, do not remain stably associated with ribosomes (Young et al., 2004) and assist de novo folding in bacteria primarily in a posttranslational manner (Agashe et al., 2004). In contrast, we show that eukaryotic ribosome biogenesis involves ribosome-anchored chaperones that remain associated with the mature ribosome. It is tempting to speculate that the primordial role of these ribosome-bound eukaryotic chaperones was to facilitate the assembly of the ribosome itself and that they later evolved additional functions such as facilitating cotranslational folding and perhaps even the dynamic ribosomal rearrangements occurring during translation. Thus, our findings provide a plausible rationale for the evolution in eukaryotic cells of cotranslationally acting chaperone machinery.
Materials and methods
Yeast strains and plasmids
All the yeast strains used in this study were derived from the Saccharomyces Genome Deletion Project (Winzeler et al., 1999). Double-deleted strains were obtained by mating the corresponding haploid cells followed by sporulation and tetrad dissections. Chromosomally integrated mutants of Jjj1, including 9myc-tagged Jjj1 deletions (ΔC, Δ390–590; ΔZF2, Δ551–590; and ΔZHD, Δ179–262) and Zuo1-GFP were generated by homologous recombination using PCR fragments in the BY4742 strain.
The plasmids for Zuo1 and SSB yeast expression have been described previously (Albanèse et al., 2006). For Jjj1-S, an S tag was inserted at the SmaI and ClaI sites to generate a C terminus tagging by using oligonucleotides. The point mutation in the HPD motif of the J domain and the NES mutation were generated by PCR mutagenesis. In addition to chromosomally integrated versions, the Jjj1 variants were cloned into the BamH1–Sma1 sites of high copy number p426 plasmid (Labbé and Thiele, 1999) and the centromeric p416 plasmid (Labbé and Thiele, 1999). The Jjj1-GFP fusions were obtained using the same strategy but in a p426 plasmid containing an in-frame C-terminal GFP moiety. The different mutants were generated by PCR. The chromosomally TAP-tagged strains were obtained from Thermo Fisher Scientific.
Lysate preparation and ribosome fractionation
400 ml yeast in exponential growth phase was treated with 100 µg/ml cycloheximide, harvested, washed with cold lysis buffer, resuspended in 1 ml of buffer A (20 mM Hepes, 50 mM KCl, 10 mM MgCl2, 1% Triton X-100, 1 mM DTT, and protease inhibitor cocktail), and frozen as drops in liquid nitrogen. The cells were ground using a grinder (MM301; Retsch). 1 ml of buffer A was added to the powder, and the lysates were clarified by centrifugation at 14,000 g for 10 min. 20 OD lysate was applied on a 12 ml 7–47% sucrose gradient in buffer A and centrifuged in a rotor (SW41; Beckman Coulter) for 150 min at 39,000 rpm at 4°C. Fractions were collected using a UA/6 detector (ISCO, Inc.). When indicated, polysomes were dissociated by treatment with 25 mM EDTA. The fractions were TCA precipitated, separated by SDS-PAGE, and subjected to immunoblot analysis.
Immunoprecipitation and tandem affinity purification
For the yeast GST pull-downs, 1 mg of total proteins was incubated with 10 µl Sepharose gluthatione beads (GE Healthcare) for 1 h at 4°C. The beads were washed two times in buffer A, one time with buffer A containing 500 mM KCl and 1% Triton X-100, and then one time with buffer A. The beads were eluted with 50 µl of lysis buffer containing 25 mM reduced glutathione.
Immunoprecipitation of endogenously TAP-tagged proteins and subsequent tobacco etch virus cleavage was performed as described previously (Puig et al., 2001). In brief, logarithmic growth cells were harvested, washed in cold water, resuspended, and lysed in 3 ml of lysis buffer B (20 mM Hepes, pH 7.5, 50 mM KCl, 10 mM MgCl2, 0.1% Nonidet-P40, 1 mM DTT, 1 mM PMSF, and 100 µg/ml cycloheximide) supplemented with protease inhibitor cocktail and 50U/ml superasin (Applied Biosystems). Lysates were incubated with 1 ml strepavidin magnetic beads coupled with biotinylated IgG (Dynabeads M280; Invitrogen) for 2 h with gentle rotation at 4°C. The magnetic beads were washed three times for 2 min in cold buffer B and resuspended in 100 µl of buffer B containing 80 U AcTEV protease (Invitrogen) and incubated for 2 h at 16°C. The final eluate was frozen in liquid nitrogen, and 20–50 µl was used to perform the Western blotting.
Recombinant protein purification
The J domain and the J domain harboring a point mutation in the J domain (HPD to QPD) were cloned into the pGEX4T1 vector. The plasmids were transformed into the BL21 Rosetta. The cells were grown at 37°C to OD0.6 and induced with 0.5 mM IPTG for 4 h. The cells were harvested and washed in 1 vol PBS. The cell pellet was resuspended in 2 ml PBS with PMSF and protease inhibitors and lysed by French Press. The cell lysate was centrifuged for 30 min at 10,000 rpm. The supernatant was loaded on a 1 ml column of Sepharose glutathione beads. The column was washed with 100 ml PBS and eluted with 5× 1 ml of elution buffer (50 mM Hepes, pH 8, 50 mM NaCl, and 25 mM GSH). The fractions containing the fusion proteins were pooled and dialyzed overnight against 5 liters of 20 mM Hepes, 50 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM DTT, and 1 mM PMSF. After dialysis, the samples were concentrated on Vivaspin column, aliquoted, and frozen in liquid nitrogen. Ydj1 was purified from Escherichia coli as described previously (Caplan et al., 1992; Cyr et al., 1994), and Ssa1 was purified from yeast as described previously (McClellan and Brodsky, 2000).
Fluorescence microscopy
Overnight cultures were diluted with fresh medium to OD600 of 0.2–0.4, and cells were grown for another 4 h at 25 or 30°C as indicated. Cells were mounted in liquid minimal culture medium on glass slides. The images were taken at room temperature. Fluoresence was visualized on a microscope (Axiovert; Carl Zeiss, Inc.) equipped with a 100× NA 1.3 oil immersion objective lens and a digital camera controlled with AxioVision software. Images were prepared using Photoshop (version 7.0; Adobe). Proteins used in this study were tagged with GFP or RFP. The nucleolus was localized using the Sik1-RFP plasmid (provided by W.-K. Huh, Seoul National University, Seoul, Korea; Sung and Huh, 2007).
ATPase assay
ATP hydrolysis activity was measured at 30°C in the following buffer: 20 mM Hepes/NaOH, pH 7.4, 50 mM NaCl, 10 mM MgCl2, 10% glycerol, and 1 mM DTT in the presence of 1 mM α[32P]ATP, essentially as described previously (Reissmann et al., 2007). After 5 min of preincubation, the reaction was started by mixing 2 µl α[32P]ATP (0.1 µCi/µl) solution with 8 µl of 1.25× reaction mix. After 5-, 10-, 15-, and 20-min incubation at 30°C, 2 µl samples were taken and transferred onto cellulose F TLC plastic sheets (PEI; EMD). The plates were developed in a solvent system containing 0.5 M LiCl and 1 M formic acid in H2O, air dried, and exposed to a phosphor screen (Kodak). After scanning the screen in an imager (Typhoon 9410; GE Healthcare), the amount of free α[32P]ADP was quantified using Image Quant (version 5.2; Molecular Dynamics).
Dilution plating
Strains were grown overnight in appropriate media at 30°C and diluted to yield an OD600 of .08. This sample was subjected to 10-fold serial dilutions for subsequent spotting onto plates.10 µl of each dilution sample, i.e., with an OD600 of .08, .008, .0008, and .00008, was spotted onto YPD plates or YPD containing drugs and allowed to grow at 30°C: YPD + 50 µg/ml hygromycin for 2 d.
RNA isolation, Northern blotting, and microarray experiments
Northern blot analysis was performed as described previously (Albanèse et al., 2006), starting from 60 ODs of cells. For RNA isolation, cell pellets were washed in sterile water and resuspended in 420 µl of lysis buffer (50 mM sodium acetate, pH 5.3, 10 mM EDTA, and 1% SDS). RNA was extracted by phenol/chloroform, and the top phase was precipitated with 0.3 M final sodium acetate and ethanol. 10 µg total RNA was denatured in formaldehyde and formamide at 65°C for 10 min and cooled to 4°C. The RNA were separated on a 1% agarose gel containing 2.2 M formaldehyde and transferred onto a Hybond N+ membrane using a semi-dry electrophoretic transfer cell (Bio-Rad Laboratories, Inc.) as recommended by the manufacturer’s protocol. The membrane was UV cross-linked and prehybridized overnight at 42°C in 5 ml of hybridization buffer (50% formamide, 50 mM phosphate buffer, pH 7.0, 0.8 M NaCl, 2.5× Denhardt’s solution, 1 mM EDTA, 0.1% SDS, and 0.250 mg/ml denatured salmon sperm DNA). 106 cpm/ml of the probe was added to the hybridization buffer, and the hybridization was performed overnight at 42°C. The membrane was washed for 15 min in buffer I (50 mM NaCl, 20 mM phosphate buffer, pH 7.0, 1 mM EDTA, and 0.1% SDS) at room temperature, 10 min at room temperature in buffer II (25 mM NaCl, 20 mM phosphate buffer, pH 7, 1 mM EDTA, and 0.1% SDS), and 10 min at room temperature in 0.1× SSC with 0.1% SDS. The membrane was exposed overnight to a phosphor screen (Kodak). The screen was scanned in an imager (Typhoon 9410).
For microarray analysis, 5 µg RNA was reversed transcribed in the presence of 5–3-aminoallyl-dUTP and natural dNTPs exactly as described previously in Hogan et al. (2008). The labeling, hybridization, and analysis of the microarrays as well as the microarray design, printing, and sequence of probes were performed as described previously (Hogan et al., 2008; http://cmgm.stanford.edu/pbrown/protocols/index.html). Fold changes between mutant and WT cells are the mean of three experiments.
Pulse-chase RNA labeling
Metabolic labeling of RNA was performed as described previously (Fatica and Tollervey, 2002). The strains were transformed with the empty p426 plasmid containing the URA3 gene and grown in glucose minimal medium lacking uracil. 50 ml of cells at 0.6 OD600 was centrifuged, resuspended in 4 ml of medium, and labeled with 100 µCi 5,6[3H] uracil for 1 min followed by a chase with excess unlabeled uracil for 0, 1, 4, and 10 min. RNA of each time point was extracted following standard procedures and loaded on 1.2% agarose/formaldehyde gels. After transfer on a nitrocellulose membrane, the membrane was cross-linked and incubated in amplify solution (GE Healthcare) before exposure on a film (BioMax MR; Kodak) for 7–14 d.
Online supplemental material
Fig. S1 shows that sequence alignment reveals the evolutionary conservation of the ZHD domain between Zuo1 and Jjj1 homologues. Fig. S2 shows the requirement of functional J domains in Jjj1 and Zuo1 for complementation of Δjjj1Δzuo1 phenotype, that cells deleted for ZUO1 but not JJJ1 are hypersensitive to hygromycin, and that overexpression of Jjj1 can suppress the slow growth phenotype of Δ_zuo1_, but overexpression of Zuo1 cannot suppress the Δ_jjj1_ phenotype. Fig. S3 shows that J domains of Zuo1 and Jjj1 suffice to confer specificity for different Hsp70s. Fig. S4 shows that ribosome association and biological function of Jjj1 and Jjj1 domain mutants is independent of expression levels. Fig. S5 shows that deletion of Arx1 rescues the cold sensitivity of Δjjj1 cells but not the ribosome biogenesis defect. Table S1 shows CLIPS chaperones analyzed in this study and previously proposed function. Table S2 shows selected features and functions of ribosome biogenesis factors used in this study. Table S3 shows summary of function of different domains in JJJ1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001054/DC1.
Acknowledgments
We thank Drs. Jonathan Warner, Jeff Brodsky, Elizabeth Craig, Won-Ki Huh, Arlen Johnson, and Daniela Roser for their generous gifts of antibodies and plasmids. We also thank Pat Brown and Dan Hogan for their generous help and advice with the microarray experiments. We thank Marta del Alamo and Stefanie Duttler for help with microarray experiments, Charles Parnot for developing scripts for polysome analysis and PhotoShop microscopy processing, and Gabriela Arroyo for help with polysome analysis. We also thank Raul Andino, Charles Parnot, and Stephanie Escusa for useful discussions and comments on the manuscript.
This work was supported by the National Institutes of Health (grant GM56433). V. Albanèse was a recipient of postdoctoral fellowships from the Fondation pour la Recherche Médicale and the Association pour la Recherche contre le Cancer.
S. Reissmann’s present address is Max Planck Institute for Terrestrial Microbiology, Marburg, Germany.
Abbreviations used in this paper:
CLIPS
chaperones linked to protein synthesis
NES
nuclear export signal
RAC
ribosome-associated complex
rRNA
ribosomal RNA
TAP
tandem affinity purification
WT
wild type
ZHD
zuotin homology domain
References
- Agashe V.R., Guha S., Chang H.C., Genevaux P., Hayer-Hartl M., Stemp M., Georgopoulos C., Hartl F.U., Barral J.M. 2004. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 117:199–209 10.1016/S0092-8674(04)00299-5 [DOI] [PubMed] [Google Scholar]
- Albanèse V., Yam A.Y., Baughman J., Parnot C., Frydman J. 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 124:75–88 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Bobula J., Tomala K., Jez E., Wloch D.M., Borts R.H., Korona R. 2006. Why molecular chaperones buffer mutational damage: a case study with a yeast Hsp40/70 system. Genetics. 174:937–944 10.1534/genetics.106.061564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. 2006. Molecular chaperones and protein quality control. Cell. 125:443–451 10.1016/j.cell.2006.04.014 [DOI] [PubMed] [Google Scholar]
- Caplan A.J., Cyr D.M., Douglas M.G. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 71:1143–1155 10.1016/S0092-8674(05)80063-7 [DOI] [PubMed] [Google Scholar]
- Cyr D.M., Langer T., Douglas M.G. 1994. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19:176–181 10.1016/0968-0004(94)90281-X [DOI] [PubMed] [Google Scholar]
- De Marchis M.L., Giorgi A., Schininà M.E., Bozzoni I., Fatica A. 2005. Rrp15p, a novel component of pre-ribosomal particles required for 60S ribosome subunit maturation. RNA. 11:495–502 10.1261/rna.7200205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E., Jacquier A., Lutfalla G., Fromont-Racine M. 2007. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA. 13:1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A., Alix J.H. 2004. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 51:189–201 10.1046/j.1365-2958.2003.03813.x [DOI] [PubMed] [Google Scholar]
- El Hage A., Sbaï M., Alix J.H. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 264:796–808 10.1007/s004380000369 [DOI] [PubMed] [Google Scholar]
- Fatica A., Tollervey D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313–318 10.1016/S0955-0674(02)00336-8 [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. 2001. The art and design of genetic screens: yeast. Nat. Rev. Genet. 2:659–668 10.1038/35088500 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603–647 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- Gautschi M., Lilie H., Fünfschilling U., Mun A., Ross S., Lithgow T., Rücknagel P., Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA. 98:3762–3767 10.1073/pnas.071057198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M., Mun A., Ross S., Rospert S. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA. 99:4209–4214 10.1073/pnas.062048599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M., Hayer-Hartl M., Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16:574–581 10.1038/nsmb.1591 [DOI] [PubMed] [Google Scholar]
- Hogan D.J., Riordan D.P., Gerber A.P., Herschlag D., Brown P.O. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:e255 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsey E.W., Jakovljevic J., Miles T.D., Harnpicharnchai P., Woolford J.L., Jr 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 10:813–827 10.1261/rna.5255804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Gautschi M., Walter W., Rospert S., Craig E.A. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497–504 10.1038/nsmb942 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., Simos G. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144:389–401 10.1083/jcb.144.3.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G., Boehringer D., Ban N., Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16:589–597 10.1038/nsmb.1614 [DOI] [PubMed] [Google Scholar]
- la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527–536 10.1093/protein/gzh062 [DOI] [PubMed] [Google Scholar]
- Labbé S., Thiele D.J. 1999. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306:145–153 10.1016/S0076-6879(99)06010-3 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Saveanu C., Decourty L., Rain J.C., Jacquier A., Fromont-Racine M. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173:349–360 10.1083/jcb.200510080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki J.A., Schnobrich D.J., Culver G.M. 2002. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell. 10:129–138 10.1016/S1097-2765(02)00562-2 [DOI] [PubMed] [Google Scholar]
- Mayer M.P., Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62:670–684 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A.J., Brodsky J.L. 2000. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics. 156:501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A.J., Tam S., Kaganovich D., Frydman J. 2005. Protein quality control: chaperones culling corrupt conformations. Nat. Cell Biol. 7:736–741 10.1038/ncb0805-736 [DOI] [PubMed] [Google Scholar]
- McClellan A.J., Xia Y., Deutschbauer A.M., Davis R.W., Gerstein M., Frydman J. 2007. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 131:121–135 10.1016/j.cell.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Meyer A.E., Hung N.J., Yang P., Johnson A.W., Craig E.A. 2007. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA. 104:1558–1563 10.1073/pnas.0610704104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.E., Hoover L.A., Craig E.A. 2010. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J. Biol. Chem. 285:961–968 10.1074/jbc.M109.038349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon-Jacobs K.L., Dinman J.D. 2006. Specific effects of ribosome-tethered molecular chaperones on programmed -1 ribosomal frameshifting. Eukaryot. Cell. 5:762–770 10.1128/EC.5.4.762-770.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.T., Robinson M.D., Mnaimneh S., Krogan N.J., Cagney G., Morris Q., Davierwala A.P., Grigull J., Yang X., Zhang W., et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell. 113:919–933 10.1016/S0092-8674(03)00466-5 [DOI] [PubMed] [Google Scholar]
- Pfund C., Lopez-Hoyo N., Ziegelhoffer T., Schilke B.A., Lopez-Buesa P., Walter W.A., Wiedmann M., Craig E.A. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981–3989 10.1093/emboj/17.14.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Qiu H., Eifert J., Wacheul L., Thiry M., Berger A.C., Jakovljevic J., Woolford J.L., Jr., Corbett A.H., Lafontaine D.L., Terns R.M., Terns M.P. 2008. Identification of genes that function in the biogenesis and localization of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:3686–3699 10.1128/MCB.01115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U., Oellerer S., Rospert S. 2007. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282:7809–7816 [DOI] [PubMed] [Google Scholar]
- Reissmann S., Parnot C., Booth C.R., Chiu W., Frydman J. 2007. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat. Struct. Mol. Biol. 14:432–440 10.1038/nsmb1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. 2008. RNA misfolding and the action of chaperones. Front. Biosci. 13:1–20 10.2741/2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., James P., Craig E.A., Goldfarb D.S. 1999. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J. Biol. Chem. 274:16501–16507 10.1074/jbc.274.23.16501 [DOI] [PubMed] [Google Scholar]
- Strunk B.S., Karbstein K. 2009. Powering through ribosome assembly. RNA. 15:2083–2104 10.1261/rna.1792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Woolford J.L., Jr 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.K., Huh W.K. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255–263 10.1016/S0962-8924(03)00054-0 [DOI] [PubMed] [Google Scholar]
- Wegrzyn R.D., Deuerling E. 2005. Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell. Mol. Life Sci. 62:2727–2738 10.1007/s00018-005-5292-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Yam A.Y., Albanèse V., Lin H.T., Frydman J. 2005. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280:41252–41261 10.1074/jbc.M503615200 [DOI] [PubMed] [Google Scholar]
- Yan W., Schilke B., Pfund C., Walter W., Kim S., Craig E.A. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17:4809–4817 10.1093/emboj/17.16.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781–791 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]