Conserved nucleosome positioning defines replication origins (original) (raw)
Abstract
The origin recognition complex (ORC) specifies replication origin location. The Saccharomyces cerevisiae ORC recognizes the ARS (autonomously replicating sequence) consensus sequence (ACS), but only a subset of potential genomic sites are bound, suggesting other chromosomal features influence ORC binding. Using high-throughput sequencing to map ORC binding and nucleosome positioning, we show that yeast origins are characterized by an asymmetric pattern of positioned nucleosomes flanking the ACS. The origin sequences are sufficient to maintain a nucleosome-free origin; however, ORC is required for the precise positioning of nucleosomes flanking the origin. These findings identify local nucleosomes as an important determinant for origin selection and function.
Keywords: DNA replication, origin recognition complex (ORC), nucleosome, chromatin
Initiation of DNA replication occurs at multiple genomic loci, termed origins of replication. In Saccharomyces cerevisiae, replication origins were originally identified as short (∼150-base-pair [bp]) autonomously replicating sequence (ARS) elements that were sufficient for the maintenance of episomes. The origin recognition complex (ORC) binds the ARS consensus sequence (ACS), an 11-bp T-rich sequence that is necessary but not sufficient for origin activity. During G1, ORC coordinates the recruitment of several additional replication factors to load the replicative DNA helicase, the Mcm2–7 complex, onto origin DNA to form the prereplicative complex (pre-RC) (for review, see Sclafani and Holzen 2007).
The S. cerevisiae genome contains, by various metrics, 6000–40,000 potential ACS sequence matches, of which only a few hundred are bound specifically by ORC. Although active transcription of a sequence has been shown to prevent ORC binding and pre-RC formation (Mori and Shirahige 2007), the large majority of potential ACS matches are intergenic, suggesting that additional chromosomal features are required to define the subset of these sites that are bound by ORC and act as replication origins.
All cellular events involving genomic DNA must operate within their chromosomal context. Nucleosomes are the most basic elements of chromatin structure. Nearly 80% of S. cerevisiae DNA is incorporated into stable nucleosomes, and their position relative to regulatory elements is a critical component of gene regulation. Significant regions of the genome are not in complex with nucleosomes, and are referred to as nucleosome-free regions (NFRs). NFRs represent particularly accessible parts of the genome that are frequently the site of multiprotein assemblies that regulate or perform key DNA templated processes (for review, see Rando and Chang 2009).
The DNA replication program has been shown to be regulated by the local chromatin environment (Donaldson 2005). Although progress has been made in establishing that chromatin modifications impact the activation of replication origins (Vogelauer et al. 2002; Knott et al. 2009), it has also been shown that nucleosome positioning is critical for origin function. Early nucleosome mapping experiments on a plasmid containing the ARS1 origin revealed that the ACS needed to be nucleosome-free, presumably to facilitate ORC binding (Simpson 1990). Studies of nucleosome positioning at the endogenous ARS1 locus confirmed that the ACS was nucleosome-free, and revealed that the position of the nucleosome adjacent to the ACS was important for origin function; moving this nucleosome farther from the ACS did not interfere with ORC binding, but did inhibit pre-RC formation (Lipford and Bell 2001). Thus, at ARS1, the precise position of local nucleosomes facilitates ORC binding and helicase loading. Here, we used high-throughput sequencing to assess the organization of nucleosomes at ORC binding sites throughout the yeast genome. Our results indicate that sequence-defined NFRs and ORC-dependent nucleosome positioning are critical determinants of replication origins in S. cerevisiae.
Results and Discussion
Identification of functional ACSs
In the S. cerevisiae genome, there are ∼250–350 ORC-binding sites that function as origins of replication (Wyrick et al. 2001; Xu et al. 2006). Multiple genomic studies have used chromatin immunoprecipitation (ChIP) and genomic microarrays (ChIP–chip) to identify ORC-binding sites and refine the ACS motif. Despite these efforts, the degeneracy of the ACS (Breier et al. 2004) and the limited resolution of the genomic array data have made it difficult to identify the specific ACSs bound by ORC. For example, a recent study used ChIP–chip data to identify 396 ORC-binding sites, but assigned multiple potential ACSs to 86 of those peaks (Xu et al. 2006).
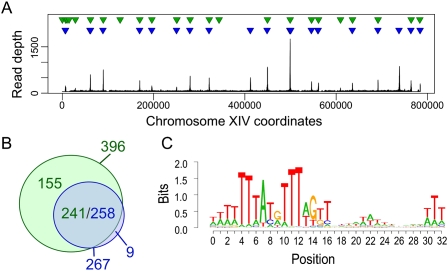
To identify functional ACS motif matches more precisely, we first used ChIP coupled with high-throughput sequencing (ChIP-seq) to identify sites of ORC localization across the yeast genome (Fig. 1A). We identified 267 peaks of ORC binding throughout the genome (Fig. 1A, blue triangles). Due to the very stringent criteria we imposed to minimize the potential for false positives, this represents 129 fewer ORC-binding sites than detected by a prior study (Fig. 1A, green triangles; Xu et al. 2006). Nevertheless, 258 of the 267 areas of ORC enrichment we identified overlapped with 241 previously identified ORC-binding sites (Fig. 1B). The difference in numbers (241 vs. 258) results from the identification of multiple discrete ORC-binding sites in regions the previous study had annotated as single ORC-binding sites.
Figure 1.
Precise localization of yeast origins. (A) Read depth of ORC-enriched sequence tags from chromosome XIV from G2-arrested cells. ORC-binding sites identified in this study (blue) and a prior study (green) (Xu et al. 2006) are indicated by triangles. (B) Venn diagram of the overlap between the 267 ORC peaks identified in this study (blue) and the 396 areas of ORC enrichment defined in Xu et al. (2006) (green) (see the Supplemental Material). Of the ORC peaks identified in this study, 258 overlapped 241 peaks from the previous study. (C) The position weight matrix of the ORC binding consensus built from the ORC–ACS set. Each nucleotide is represented by a discrete color (red [T], orange [G], blue [C], and green [A]).
We implemented a scoring system to identify the single most likely ACS associated with each ORC ChIP-seq peak, which we termed the ORC–ACS. Our analysis identified 253 likely ORC-binding locations at nucleotide resolution. The resulting consensus sequence (Fig. 1C) closely matches a previously published ACS motif (Xu et al. 2006). Comparison of the sites where both studies identified a common area of ORC enrichment (Supplemental Fig. 1) revealed that, in the majority (67%) of instances, we agreed on the ACS call, while in the remaining instances, we identified a novel ORC–ACS site.
Nucleosome positioning at origins of replication
The precise definition of ORC-binding sites provided by ChIP-seq allowed us to investigate the position of nucleosomes flanking origins of replication. Previous genome-wide studies of nucleosome positioning have suggested that S. cerevisiae origins are depleted of bulk nucleosomes (Albert et al. 2007; Field et al. 2008; Mavrich et al. 2008); however, these studies focused on previously annotated ARS elements (of ∼500 bp) rather than ORC's asymmetric binding site. This would be equivalent to analyzing the position of nucleosomes flanking promoters without considering the transcription start site (TSS) or the direction of transcription. Thus, we used the precisely defined ORC-binding sites described above and high-resolution mapping of nucleosomes across the yeast genome to comprehensively address the localization of nucleosomes relative to ORC binding at origins.
We identified and mapped the location of ∼67,500 nucleosomes from an asynchronous yeast population using high-throughput sequencing of mononucleosomal fragments. Our nucleosome mapping recapitulated previous findings demonstrating that nucleosomes are strongly positioned around TSSs (Supplemental Fig. 2; for review, see Rando and Chang 2009).
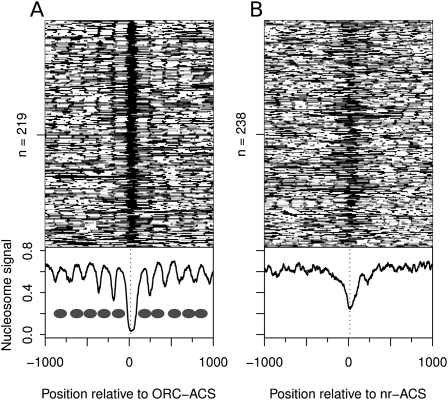
To examine the nucleosomal landscape at replication origins, we aligned the 219 members of the ORC–ACS set that were within nonrepetitive regions and were not within 10 kb of a chromosome end and plotted the nucleosome score as a heat map relative to the start of the ACS (Fig. 2). Positions fully occupied by a nucleosome are white (Fig. 2), and those lacking a nucleosome are black (Fig. 2). We aligned the origins using the T-rich strand of the ORC–ACS, and will subsequently define sequences 5′ of the T-rich strand of the ORC–ACS as “upstream” and sequences 3′ of the T-rich strand as “downstream.” Similarly, we will refer to the first nucleosome upstream of the ORC–ACS as the −1 nucleosome, and the first downstream nucleosome as the +1 nucleosome.
Figure 2.
Replication origins are associated with an asymmetric NFR flanked by well-positioned nucleosomes. (A) Heat map of nucleosome occupancy and average nucleosome signal from asynchronous cells for 219 ORC–ACS sites. Interpreted nucleosome positions are represented as ovals. (B) Nucleosome occupancy at 238 nr-ACS sites. Vertical dashed lines represent the center of the 33-bp ACS motif match. All ACS matches are oriented relative to the T-rich strand and are aligned at position 0.
Strikingly, we detected a pattern of nucleosome occupancy surrounding the ORC–ACS that is similar to that of open promoters (Fig. 2A; Supplemental Fig. 2). The ACS is located asymmetrically within an ∼125-bp NFR. As with the nucleosomes at promoters, both the −1 and +1 nucleosomes are stably positioned and set up an adjacent periodic pattern of nucleosome occupancy. The composite NFR is ∼90 bp larger than ORC's DNase I footprint, suggesting that ORC binding to the ACS is not solely responsible for maintaining the NFR. Importantly, the NFR is asymmetric with respect to the ACS (Figs. 2A, 3A), extending much farther downstream from than upstream of the ACS.
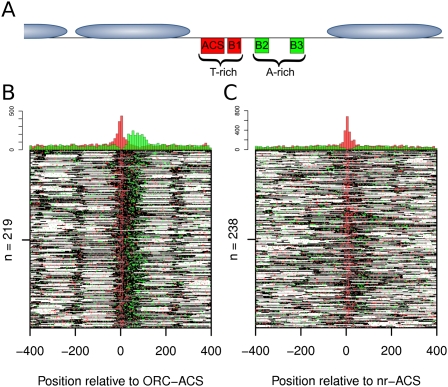
Figure 3.
Sequence polarity at replication origins. (A) Schematic of nucleosome positioning at the prototypical ARS1 replication origin. The positions of the ACS and B elements relative to the adjacent nucleosomes are indicated. (B) Heat map of nucleosome occupancy from asynchronous cells at the ORC–ACS sites. Red and green dots represent the bias in nucleotide composition (T-rich or A-rich, respectively) at each sequence (see the Materials and Methods). Histograms above the heat map show the cumulative incidence of T-rich or A-rich windows (red and green, respectively) over the region in 20-bp windows. (C) Nucleosome occupancy and sequence bias for the nr-ACS set.
To explore the potential role of the ACS and surrounding sequences in encoding an NFR, we examined the 238 highest-scoring ACS motifs that were not within genes, telomeres, or rDNA, or proximal to any ORC-binding sites identified in this or prior studies (Xu et al. 2006). The sequence composition of these 238 ACS motif matches, which we call nonreplicative ACS (nr-ACS) sites, is nearly indistinguishable from the ORC–ACS set (Supplemental Fig. 3). Despite their sequence similarity, the nr-ACSs show a markedly different nucleosomal organization (Fig. 2B). Although the nr-ACS motif tends to be nucleosome-free—not surprising for regions with such a high A/T composition—the nucleosome depletion is weaker and is centered on the ACS instead of extending downstream from the ACS, as seen for the ORC–ACS set. Finally, the adjacent upstream and downstream nucleosomes lack the precise periodic positioning found at the ORC–ACS sites.
There were a number of sites where the higher resolution of our ORC ChIP-seq experiments led us to identify a different ACS motif match as the likely ORC-binding site compared with a prior ChIP–chip study (Supplemental Fig. 1; Xu et al. 2006). For the ACSs jointly identified in both studies, we consistently found a pattern of asymmetric, well-positioned nucleosomes (Supplement Fig. 4). For the sites in which our findings disagreed with the previous study, we found that the ORC–ACS sites identified in this study more closely resembled the nucleosome profile of the jointly identified ACSs. Assuming the pattern of positioned nucleosomes we identified is a hallmark of yeast replication origins, which is well justified based on the sites that both studies agreed on, these data further underscore the low false-positive rate of our ORC–ACS identification.
Nucleosome depletion at origins is encoded by sequence
S. cerevisiae exhibits remarkable specificity in origin selection, despite the potential for ORC to interact with many genomic ACS matches. This selectivity could be the result of a subset of potential ORC-binding sites being obscured by nucleosomes, while others remain accessible. If so, we might expect that the wide, asymmetric NFR will not depend on ORC, but rather will be encoded in the origin DNA sequence.
To explore the role of primary DNA sequence in specifying the NFR and potentiating ORC binding, we examined the distribution of in vitro assembled nucleosomes surrounding both ORC–ACS and nr-ACS sites in the absence of competing _trans_-acting factors (Kaplan et al. 2009). A distinct NFR was maintained at the ORC–ACS sites in the absence of ORC or any other _trans_-acting factors (Supplemental Fig. 5A). Reminiscent of our in vivo nucleosome positioning data, we found that the NFR was positioned asymmetrically over the ORC–ACS, extending downstream from the ACS. Similarly, the in vitro NFR at nr-ACS matches also recapitulated the in vivo nucleosome mapping results, with a weaker NFR centered on the nr-ACS (Supplemental Fig. 5B). Despite maintaining an asymmetric NFR at ORC–ACS sites in the absence of _trans_-acting factors, the precise and periodic positioning of adjacent nucleosomes was not observed in the in vitro data. These findings imply that nucleosome-excluding signals are encoded in the sequence of the ORC–ACS set that do not exist in the nr-ACS set, but that additional _trans_-acting factors (e.g., ORC) are required for the precise and ordered nucleosome positioning found at origins.
In yeast, the ACS and additional downstream modular sequence elements (B elements) contribute to origin function (Fig. 3A). Unlike the ACS, the B elements are poorly defined, with minimal conservation of sequence or spacing between different origins (for review, see Bell 1995). A prior study found that the sequences downstream (50–100 bp) from the T-rich ACS were enriched for adenosine bases with no specific motifs identified (Breier et al. 2004). We posited that the A-rich sequences downstream from the ACS prevented the +1 nucleosome from encroaching on origin sequences. Consecutive 4- to 6-bp islands of As constitute a strong nucleosome excluding signal (for review, see Segal and Widom 2009), as poly-dA:dT islands resist bending around the histone octamer. We re-examined the sequence composition of replication origins to determine if A islands contributed to the asymmetric nature of the ORC–ACS-associated NFR. To visualize differences in base composition, we color-coded each genomic position: Shades of red (T-rich) or green (A-rich) in Figure 3, B and C, represent the bias in surrounding nucleotide composition. At both ORC–ACS (Fig. 3B) and nr-ACS (Fig. 3C) sites, the T-rich ACS match is visible as a red vertical line. Interestingly, we found a strong concentration of green A-rich islands near the downstream edge of the NFR that was present only in the ORC–ACS set (Fig. 3B,C). We displayed these differences graphically by plotting a histogram of A islands or T islands as a function of distance from the ACS (Fig. 3B,C, top).
The A-rich islands downstream from the ACS appear to be a determinant of ORC binding, as they are specific to the ORC–ACS set. These A-rich sequence elements are well beyond the known ORC footprint (Bell and Stillman 1992), and our ChIP-seq data do not support a robust interaction of ORC with these sites. Instead, we propose that these sequences maintain the large asymmetric NFR and thereby facilitate ORC binding. Consistent with this hypothesis, the density and position of these A islands varies with the width of the NFR (Supplemental Fig. 6). These findings strongly suggest that the large asymmetric NFR present at bona fide origins of replication is required for ORC association with origins in vivo, and is not simply a consequence of ORC binding.
It is interesting that start sites of bidirectional DNA replication have strongly polar sequence and chromatin compositions. If the role of the A-rich islands was simply to exclude nucleosomes downstream from the ACS, we would expect that T-rich islands could also serve a similar function. Interestingly, a similar polar bias in A-rich and T-rich sequence elements is found at the promoters of active genes residing in an open chromatin environment (Supplemental Fig. 7; Maicas and Friesen 1990). This may suggest that the transition from T-rich to A-rich sequences at both promoters and origins promotes the DNA unwinding that occurs at both sites. Alternatively, the T-rich/A-rich polarity of the ACS and surrounding sequence elements may facilitate transient long-range ORC interactions, or possibly pre-RC assembly.
ORC is necessary and sufficient for precise nucleosome positioning
Positioning of nucleosomes at ARS1 appeared to be dependent on ORC, as mutations of the ACS permitted the −1 nucleosome to encroach into the NFR in vitro (Lipford and Bell 2001). Because sequence is a determinant of nucleosome occupancy (Segal et al. 2006), and the ACS sequence is predicted to be resistant to nucleosome occupation, we re-evaluated the role of ORC in nucleosome positioning at origins of replication across the genome. To this end, we repeated the genome-wide nucleosome positioning assay in both wild type and a congenic strain with a temperature-sensitive ORC mutant at the restrictive temperature. Consistent with previous ChIP studies (Aparicio et al. 1997), the orc1-161 mutation eliminated DNA binding at 37°C (Supplemental Fig. 8). Biological replicates of each strain were arrested at the G2/M border and subsequently raised to the nonpermissive temperature for 2 h before isolating and sequencing the mononucleosomal fragments.
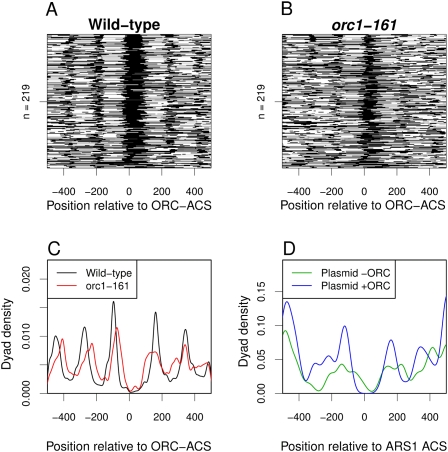
A comparison of the nucleosomal distribution around the set of ORC–ACSs in wild-type and orc1-161 cells revealed a clear effect of ORC on the positioning of the −1 and +1 nucleosomes (Fig. 4). At the nonpermissive temperature, the canonical positioning of the nucleosomes around the ORC–ACS was lost (Fig. 4B), although a smaller NFR was maintained (consistent with the in vitro nucleosome studies above). In contrast, the wild-type sample at the nonpermissive temperature (Fig. 4A) was nearly indistinguishable from the profile generated at 23°C (Fig. 2A; Supplemental Fig. 9A). To more easily view the differences in nucleosome positioning, we plotted the density of the wild-type (Fig. 4C, black) and orc1-161 (Fig. 4C, red) nucleosome dyads as a function of the distance from the ORC–ACS (Fig. 4C). In the absence of ORC binding, the nucleosomes on both sides of the ORC–ACS encroached into the NFR, and the adjacent nucleosomes lost their precise positioning. Thus, despite the substantial distance between the ACS and the +1 nucleosome, ORC binding strongly influenced the position of this nucleosome and its neighbors. Importantly, the nucleosome pattern at TSSs showed no change between the wild-type and the mutant samples, indicating that loss of ORC function did not result in a global remodeling of nucleosomes (Supplemental Fig. 9B).
Figure 4.
ORC is necessary and sufficient for precise nucleosome positioning. (A) Heat map of nucleosome occupancy around ORC–ACS sites in wild-type cells arrested in G2 at 37°C. (B) Heat map of nucleosome occupancy around ORC–ACS sites in orc1-161 cells arrested in G2 at 37°C. (C) Density of wild-type (black) and orc1-161 (red) nucleosome dyads surrounding ORC–ACS sites arrested in G2 at 37°C. (D) The density of nucleosome dyads assembled in vitro at ARS1 in the presence or absence of ORC.
We were able to recapitulate the ORC-dependent positioning of nucleosomes adjacent to origin DNA using purified proteins. We assembled nucleosomes on a plasmid containing the ARS1 origin of replication using the four purified S. cerevisiae histones and the ISWI nucleosome remodeling complex (Vary et al. 2004). Nucleosomes were then mapped by micrococcal nuclease digestion followed by high-throughput sequencing of the resulting mononucleosome-associated DNA. When this reaction was performed in the absence of ORC, we failed to observe precise positioning of nucleosomes around the ACS (Fig. 4D, green). Importantly, when nucleosomes were assembled on the plasmid in the presence of ORC, we observed clear positioning of the −1 and +1 nucleosomes, as well as adjacent nucleosomes on each side (Fig. 4D, blue). Thus, ORC alone (in the context of a nucleosome remodeling complex) is capable of positioning nucleosomes on either side of ARS1. Previous in vitro studies using chicken histones and a Drosophila embryo extract to assemble nucleosomes found that Abf1p positioned the +1 nucleosome at ARS1 (Lipford and Bell 2001). Consistent with these findings, we found that the addition of Abf1p to the purified reaction shifted the position of the +1 nucleosome relative to ORC alone (Supplemental Fig. 10A) almost perfectly recapitulating the nucleosome positioning observed in vivo at ARS1 (Supplemental Fig. 10B). Thus, at ARS1, ORC is sufficient to position nucleosomes in a manner analogous to that seen at most origins, but Abf1p acts to supersede this function at the +1 nucleosome. Importantly, most origins are not associated with a DNA-binding factor at the downstream position, suggesting that, consistent with the orc1-161 nucleosome mapping, ORC is responsible primarily for positioning nucleosomes on both sides of the origin.
Our findings show that ORC plays a key role in positioning nucleosomes on both sides of the origin NFR. The close proximity of the ACS to the −1 nucleosome strongly suggests that ORC DNA binding positions this nucleosome by DNA exclusion or by a direct interaction of the −1 nucleosome with ORC (Zhang et al. 2002). In contrast, the median distance from the ORC-binding site to the +1 nucleosome is ∼60 bp from the 3′ edge of the known ORC footprint, yet ORC clearly influences the positioning of this nucleosome. Because this positioning is also observed in the reconstituted reaction, ORC almost certainly acts directly on this nucleosome. It is possible that this effect is mediated by ORC wrapping the adjacent A-rich DNA. Such a model is consistent with electron microscopy studies of Drosophila ORC (Clarey et al. 2008) and DNase I footprinting studies that show that ORC can induce a 10-bp periodicity of cleavage, consistent with ORC wrapping DNA adjacent to the +1 nucleosome (Bell and Stillman 1992; Speck et al. 2005). Alternatively, ORC could directly bind the +1 nucleosome without interacting with the intervening DNA. In either case, ORC's interaction with the origin-proximal DNA/nucleosomes is not as simple as thought previously, and this more complex interaction may facilitate downstream events in DNA replication (e.g., by preparing origin DNA for Mcm2–7 loading). Genome-wide location analysis of the chromatin remodeler Ino80 revealed enrichment at a subset of origins (Shimada et al. 2008). Although ISWI is sufficient to position nucleosomes in an ORC-dependent manner in vitro, it is unclear if ISWI, Ino80, or another remodeling activity will facilitate the in vivo nucleosome positioning at origins of replication.
Our studies strongly support a model in which the characteristic nucleosome pattern surrounding origins of replication is generated in two steps. First, the large and asymmetric NFR found at origins of replication is established in a DNA sequence-dependent manner and is associated with a subset of ACS matches that are bound by ORC. We propose that the asymmetry of the NFR provides ORC access to the ACS and the adjacent A-rich sequences. In contrast, ACS matches not bound by ORC exhibit weak symmetric NFRs. These data argue that the differential affinity for nucleosomes encoded by the primary origin DNA sequence is a specificity factor for the initial recruitment of ORC to the origin. In a second step, we show that ORC bound at the origin precisely positions the flanking upstream and downstream nucleosomes to establish the conserved nucleosomal signature evident at origins of replication.
The large size of the NFR at origins of replication is likely to provide space for the association of other replication factors in addition to ORC. The origin NFR is ∼90 bp larger than the ORC DNase I footprint, which, based on the size of the archeal MCM complex (100 Å) (Pape et al. 2003), would provide room for the assembly of two to three Mcm2–7 complexes. We considered the possibility that the size of the NFR at origins may be dynamic throughout the cell cycle, possibly expanding upon loading of multiple Mcm2–7 complexes during G1 (Remus et al. 2009). To test this hypothesis, we examined nucleosome positioning in G1 (pre-RCs present) and G2 (pre-RCs absent) cells (Supplemental Fig. 11). We found no statistically significant differences in nucleosome organization at origins between G1 and G2 cell populations, suggesting that the size of the NFR is independent of pre-RC formation and Mcm2–7 loading. Instead, we hypothesize that the large NFR that is predictive for ORC binding is also sufficient to allow subsequent Mcm2–7 loading. Consistent with this hypothesis, our analysis of the distribution of Mcm2–7 proteins relative to the ACS reveal that the bulk of the Mcm2–7 signal is distributed downstream from the ACS in the NFR (WM Lam, S Kang, ML Eaton, DM MacAlpine, and SP Bell, in prep.).
The conserved positioning of nucleosomes at almost all ORC-binding sites identified by this study (including those associated with silencers at the mating type loci) suggests that nucleosome organization is an essential and critical determinant of origin function. Consistent with the hypothesis that precise nucleosome positioning is essential for DNA replication, prior work at ARS1 has demonstrated that moving the −1 nucleosome further upstream of the ACS results in a loss of origin function and inhibition of Mcm2–7 loading (Lipford and Bell 2001). As almost all of the replication origins identified in this study share a common nucleosomal profile, it is not surprising that we found no correlation between nucleosome organization and origin efficiency or timing. Instead, we suggest that this nucleosome organization at origins is a fundamental determinant required at all origins, and that other chromatin-associated features, such as histone modifications (Knott et al. 2009), will control the efficiency and timing of replication initiation. Given that ORC exhibits little, if any, sequence specificity in higher eukaryotes (Vashee et al. 2003; Remus et al. 2004), and that, in Drosophila, ORC localizes to open chromatin (MacAlpine et al. 2010), we propose that nucleosome organization will be a defining feature of ORC binding in metazoans.
Materials and methods
Yeast strains and growth conditions
Wild-type (W303) and mutant (MdVy200, orc1-161) yeast strains were grown in rich medium for all experiments. Cells were arrested at the G2/M transition by 10 μg/mL nocodazole or at G1 with α-factor. For nucleosome positioning in the presence or absence of ORC, wild-type and mutant strains were grown at the permissive temperature (23°C), arrested at G2/M for 2 h, and then shifted for 2 h to the restrictive temperature (37°C).
ChIP and mononucleosome preparation
Mononucleosomes were prepared from 1.5 × 109 cells that were cross-linked with 0.1% formaldeyhde for 30 min and quenched with 125 mM glycine. After cross-linking, the cells were spheroplasted and treated with 80 U of micrococcal nuclease to generate mononucleosome fragments.
ChIP extracts were prepared from 5 × 108 cells arrested at the G2/M transition with 10 μg/mL nocodazole as described previously (Aparicio et al. 1997). An anti-ORC polyclonal antibody was used for the ChIP as described previously (Tanny et al. 2006).
Reconstituted chromatin assembly
Chromatin was assembled on an _ARS1_-containing plasmid using purified recombinant S. cerevisiae histones expressed in Escherichia coli and the ISWI complex (Vary et al. 2004) in the presence or absence of recombinant ORC and/or Abf1p (see the Supplemental Material for details). Nucleosome positions were determined by micrococcal nuclease digestion and high-throughput multiplex sequencing of the mononucleosomal fragments.
High-throughput sequencing
Libraries of mononucleosomal and ChIP fragments were prepared for sequencing using the genomic DNA sample preparation kit (Illumina) according to the manufacturer's protocol. The library was sequenced using the Illumina 1G Genome Analyzer.
Data analysis
See the Supplemental Material for detailed descriptions of all data analysis. All genomic data and ORC-binding and ORC-ACS locations have been deposited at GEO with accession number GSE16926.
Acknowledgments
We thank Daniel Lew and members of the MacAlpine and Bell laboratories for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute (S.P.B.), Whitehead Foundation Scholar Award (D.M.M.), and National Institutes of Health grant HG004279 (D.M.M.). K.G. is a Fellow of The Leukemia and Lymphoma Society.
Footnotes
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Bell SP 1995. Eukaryotic replicators and associated protein complexes. Curr Opin Genet Dev 5: 162–167 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Breier A, Chatterji S, Cozzarelli N 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22 http://genomebiology.com/2004/5/4/R22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarey MG, Botchan M, Nogales E 2008. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol 164: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD 2005. Shaping time: Chromatin structure and the DNA replication programme. Trends Genet 21: 444–449 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett A, Tillo D, Field Y, LeProust E, Hughes T, Lieb J, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes & Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J, Bell S 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas E, Friesen JD 1990. A sequence pattern that occurs at the transcription initiation region of yeast RNA polymerase II promoters. Nucleic Acids Res 18: 3387–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Shirahige K 2007. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem 282: 4447–4452 [DOI] [PubMed] [Google Scholar]
- Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S 2003. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep 4: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY 2009. Genome-wide views of chromatin structure. Annu Rev Biochem 78: 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall E, Botchan M 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. Poly(da:dt) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang J-PZ, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18: 566–575 [DOI] [PubMed] [Google Scholar]
- Simpson RT 1990. Nucleosome positioning can affect the function of a _cis_-acting DNA element in vivo. Nature 343: 387–389 [DOI] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B 2005. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny R, MacAlpine D, Blitzblau H, Bell S 2006. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol Biol Cell 17: 2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary JC Jr, Fazzio TG, Tsukiyama T 2004. Assembly of yeast chromatin using ISWI complexes. Methods Enzymol 375: 88–102 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly T, Walter J 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes & Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer B, Grunstein M 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Wyrick J, Aparicio J, Chen T, Barnett J, Jennings E, Young R, Bell S, Aparicio O 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavare S 2006. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276 doi: 10.1186/1471-2164-7-0276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21: 4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]