C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 16.
Published in final edited form as: Science. 2009 Aug 7;325(5941):750–753. doi: 10.1126/science.1176325
Abstract
The catalytic engine of RNAi is the RNA-induced silencing complex (RISC), wherein the endoribonuclease Argonaute and single-stranded siRNA direct target mRNA cleavage. Here we have reconstituted long dsRNA- and duplex siRNA-initiated RISC activities using recombinant Drosophila Dicer-2, R2D2 and Ago2 proteins. We employ this core reconstitution system to purify an RNAi regulator-_c_omponent _3 p_romoter _o_f RISC (C3PO), a complex of Translin and Trax. C3PO is a Mg2+-dependent endoribonuclease that promotes RISC activation by removing siRNA passenger strand cleavage products. These studies establish an in vitro RNAi reconstitution system and identify C3PO as a key activator of the core RNAi machinery.
Keywords: RNAi, RISC, Dcr-2/R2D2, Ago2, C3PO, endoribonuclease
RNA interference (RNAi) is post-transcriptional gene silencing initiated by the RNase III Dicer that processes double-stranded (ds)RNA into 21- to 22-nucleotide (nt) small interfering RNA (siRNA) (1–3). Nascent siRNA duplex is assembled into the effector RNA-induced silencing complex (RISC), wherein single-stranded siRNA guides the endoribonuclease Argonaute (Ago) to catalyze sequence-specific cleavage of complementary mRNA(1–3). A minimal RISC can be reconstituted with recombinant Ago2 and single-stranded siRNA, but not duplex siRNA (4), suggesting that additional factors are required for loading nascent siRNA onto Ago2. In Drosophila, Dicer-2 (Dcr-2) and R2D2 coordinately recruit duplex siRNA to Ago2 to promote RISC assembly (5–7). Moreover, Dcr-2/R2D2 complex senses thermodynamic asymmetry of siRNA and facilitates the guide strand selection (8). It remains unclear as to what constitute holo-RISC, how RISC is assembled, and how RISC is regulated. These outstanding questions can be effectively addressed using a classic biochemical fractionation and reconstitution approach.
We took a candidate approach to reconstitute the core RISC activity using recombinant Dcr-2, R2D2 and Ago2 proteins, all of which are essential for Drosophila RISC assembly (6, 7, 9). Besides PAZ and PIWI domains, Drosophila Ago2 carries a long stretch of amino-terminal poly glutamine (Q) repeats that are absent in most Ago proteins. Here we generated an active truncated His-Flag-tagged Ago2 that removes most polyQ repeats and fully restores duplex siRNA-initiated RISC activity in ago2 mutant lysate (Fig. S1) (9). Furthermore, purified recombinant Dcr-2/R2D2 and Ago2 proteins could successfully reconstitute long dsRNA- and duplex siRNA-initiated RISC activities (Fig. 1A). The RISC activity was abolished when using catalytic mutant Ago2 (Fig. 1A), indicating that Ago2 was responsible for mRNA cleavage in this reconstituted system.
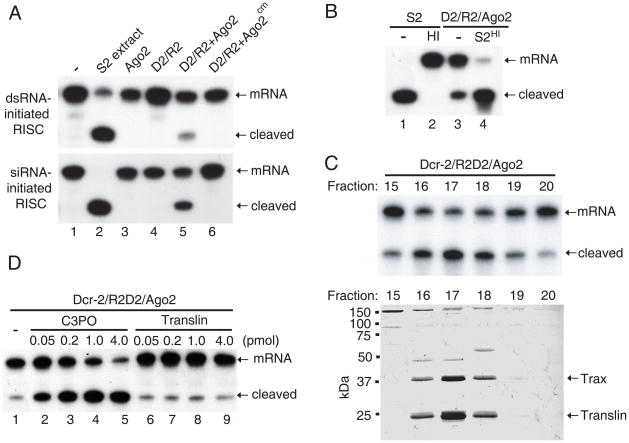
Fig. 1.
Purification of C3PO as a RISC activator. (A) The dsRNA- and siRNA-initiated RISC assays were performed in buffer, S2 extract, recombinant Ago2, Dcr-2/R2D2 (D2/R2) complex, and D2/R2 plus wild-type or catalytic-mutant (cm/D965A) Ago2. Herein all siRNA-initiated RISC assays used duplex siRNA if not stated otherwise. (B) The siRNA-initiated RISC assays were performed with untreated or heat-inactivated (HI) S2 extract, or recombinant Dcr-2/R2D2/Ago2 in the absence or presence of S2HI extract. (C) Purification of C3PO through a seven-step chromatographic procedure. Following the final Mono Q step, individual fractions were assayed with recombinant Dcr-2/R2D2/Ago2 for the RISC-enhancing activity (top) or resolved by SDS-polyacrylamide gel (PAGE) followed by Colloidal-staining (bottom). (D) The siRNA-initiated RISC assays were performed using recombinant Dcr-2/R2D2/Ago2 alone or with increasing amount of recombinant C3PO or Translin.
However, recombinant Dcr-2/R2D2/Ago2 generated lower RISC activities than S2 extract (Fig. 1A), suggesting that additional factors are required to achieve maximal RISC activity. Therefore, we employed this core reconstitution system to search for new RISC-enhancing factors. We found that mild heat treatment (HI, 37°C for 30 minutes) abolished the RISC activity in S2 extract (Fig. S2) and that addition of S2HI extract greatly enhanced the RISC activity of recombinant Dcr-2/R2D2/Ago2 (Fig. 1B), suggesting the existence of an RNAi activator. We named this factor _c_omponent _3 p_romoter _o_f RISC (C3PO) because besides Dcr-2 and R2D2, this is the third component that promotes RISC activity.
We purified C3PO from S2 extract following a seven-step chromatographic procedure. At the final step, two proteins, ~27 kDa and ~37 kDa, showed close correlation with the RISC-enhancing activity (Fig. 1C). They were identified by mass spectrometry as the evolutionarily conserved Translin/TB-RBP and Translin-associated factor X (Trax). Translin is a single-stranded DNA/RNA-binding protein that co-purifies with siRNA after UV crosslinking (10, 11), whereas Trax has sequence similarity to and interacts with Translin (12). Consistently, recombinant C3PO complex, but not Translin, greatly enhanced the RISC activity of recombinant Dcr-2/R2D2/Ago2 (Fig. S3a and Fig. 1D). Maximal RISC activity was obtained only when Dcr-2/R2D2, C3PO and Ago2 were present (Fig. S3b).
Conversely, genetic depletion of C3PO diminished RISC activity in Drosophila ovary extract. Western blotting revealed that Translin and Trax were both missing in translin (trsn) mutant fly lysate (Fig. 2A) (11), suggesting that Trax is unstable without Translin. By contrast, the core RNAi components, i.e. Dcr-2, R2D2 and Ago2, remained at wild type levels in trsn mutant (Fig. 2A). While siRNA-generating activity was slightly higher in the mutant lysate (Fig. S4), duplex siRNA-initiated RISC activity was at ~25% of wild type level in trsn extract (Fig. 2B), and this defect was rescued by adding recombinant C3PO (Fig. 2C). Thus, C3PO is required for optimal RISC activity in vitro.
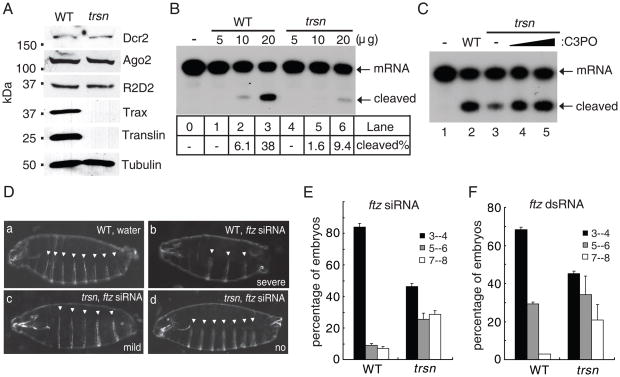
Fig. 2.
C3PO is required for efficient RNAi. (A) Western blots comparing the levels of Dcr-2, R2D2, Ago2, Translin, Trax, and β-tubulin between wild-type (WT) and trsn mutant lysates. (B) The siRNA-initiated RISC assays were performed in buffer, and WT or trsn mutant ovary extracts. The RISC activity was measured by the percentage of cleaved mRNA. (C) The siRNA-initiated RISC assays were performed in buffer, 20μg WT extract, or 20μg trsn mutant extract without or with recombinant C3PO. (D) Images showing segmentation phenotypes of WT and trsn mutant embryos: (a) WT embryo injected with water (8 abdominal cuticle belts); (b) WT embryo injected with _ftz_-siRNA (severe, 3–4 belts); (c) trsn embryo injected with _ftz-_siRNA (mild, 5–6 belts); (d) trsn embryo injected with _ftz_-siRNA (no phenotype, 7–8 belts). (E-F) Graphs showing distribution of WT or trsn mutant embryos with severe, mild, or no phenotype following injection of _ftz-_siRNA (E, n>100) or _ftz-_dsRNA (F, n>150).
To determine if C3PO is required for RNAi in vivo, we injected wild type and trsn mutant embryos with fushi tarazu (ftz) siRNA or dsRNA that causes segmentation defects by silencing ftz expression (9). Following injection with ftz siRNA, more than 80% of wild type embryos displayed a severe segmentation phenotype, whereas a significant portion of trsn mutant embryos showed mild or no phenotype (Fig. 2D and 2E). A similar phenomenon was observed with ftz dsRNA injection (Fig. 2F). These experiments indicate that C3PO is required for efficient RNAi in vivo.
To distinguish if C3PO enhances RISC assembly or activity, we compared the amount of RISC activity generated by recombinant Dcr-2/R2D2/Ago2 with C3PO added before or after RISC assembly (Fig. S3c). In both cases, C3PO could enhance the core RISC activity, however, the RISC-enhancing effect was greatly diminished when C3PO was added late to pre-assembled RISC (Fig. S3d). Therefore, we conclude that C3PO primarily promotes RISC activation, but it also enhances the RISC-mediated target cleavage. Consistent with the latter, C3PO modestly enhanced single-stranded siRNA-initiated RISC activity (Fig. S3e).
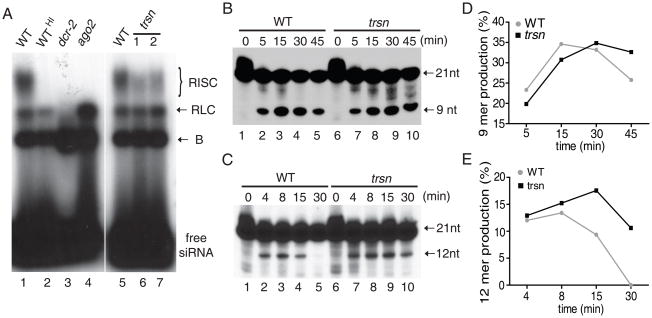
To further dissect the role of C3PO in RISC activation, we examined the stepwise process of RISC assembly by native siRNA gel-shift assay. As previously described (6, 9, 13), three siRNA-protein (siRNP) complexes, B, RISC loading complex (RLC) and RISC, were formed in wild-type ovary extract. RLC contains Dcr-2/R2D2 and siRNA, whose formation precedes and is required for RISC assembly (6, 13). While neither RLC nor RISC could form in dcr-2 mutant extract, only RISC was absent in ago2 mutant extract (Fig. 3A). By contrast, all three siRNP complexes could form in trsn mutant extract, but the amount of RISC was significantly less than that in wild-type extract (Fig. 3A). These results suggest that C3PO facilitates the transition from RLC to active RISC.
Fig. 3.
C3PO promotes RISC activation by removing siRNA passenger strand cleavage products. (A) Native siRNA gel-shift assays were performed using radiolabeled let-7 siRNA with untreated or heat-inactivated (HI) WT, dcr-2, ago2 mutant ovary extract (lanes 1–4), or 40μg of WT and two preps of trsn mutant ovary extract (lanes 5–7). (B-C) Passenger strand cleavage assays were performed with 40μg of WT or trsn mutant extract. The 9-nt or12-nt cleavage products were detected separately using siRNA whose passenger strand was radiolabeled at the 5′ (B) or 3′ (C) end. (D-E) The data in (B-C) was converted into graphs illustrating different stability of the 9-mer and 12-mer in WT and trsn extracts.
The central step of RISC activation is the unwinding of duplex siRNA and loading of the guide strand onto Ago2. Thus, we measured the efficiency of RISC assembly using the siRNA-unwinding assay (14). In the reconstitution system, recombinant C3PO enhanced the siRNA-unwinding activity of Dcr-2/R2D2/Ago2 (Fig. S5a). Conversely, the efficiency of siRNA unwinding was lower in trsn mutant extract than wild type control (Fig. S5b). Both results indicate that C3PO promotes siRNA unwinding and RISC activation.
To study the relative contribution of different RISC activation mechanisms, we supplemented heat-inactivated S2 (S2HI) extract, which displayed no siRNA-unwinding activity due to Ago2 inactivation (Fig. S6b and Fig. S2), with recombinant wild-type or catalytic mutant Ago2. Only wild-type, but not mutant, Ago2 could effectively rescue siRNA unwinding in S2HI extract (Fig. S6c). This result, together with previous studies (15–17), strongly supports that the catalytic activity of Ago2 is indispensable for siRNA unwinding and RISC activation.
In the “slicer” model, Ago2 cleaves the passenger strand of siRNA into 9-nt and 12-nt fragments that simply melt away due to low binding energy, leaving the guide strand behind to form an active RISC with Ago2. By passenger strand cleavage assay(15, 17), we observed that both 9-nt and 12-nt fragments displayed longer half-life in trsn mutant than wild-type extract (Fig. 3B-3E and Fig. S7). Moreover, addition of C3PO complex, but not Translin, resulted in rapid degradation of the 9-nt fragment in the reconstituted system (Fig. S8a). Together, these findings suggest that C3PO promotes RISC activation by removing siRNA passenger strand cleavage products.
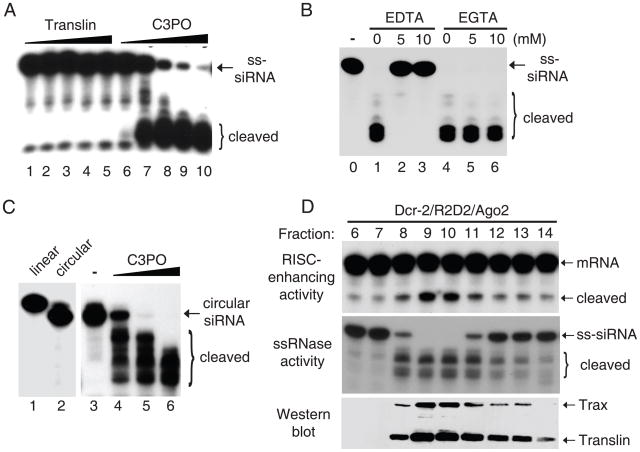
Further supporting this, recombinant C3PO displayed potent ribonuclease (RNase) activity towards single-stranded (ss)siRNA, but showed little or no activity towards double-stranded siRNA or ssDNA (Fig. 4A and Fig. S8b-c). The RNase activity of C3PO is Mg2+-dependent and could be blocked by EDTA, but not by EGTA (Fig. 4B). In addition, C3PO acts as an endonuclease for it could degrade circular as well as linear RNA (Fig. 4C). Moreover, the endogenous C3PO complex closely correlated with the RISC-enhancing activity as well as a ssRNase activity following sequential chromatography (Fig. 4D and Fig. S8d).
Fig. 4.
C3PO is an Mg2+-dependent endoribonuclease. (A) 5′-radiolabeled single-stranded (ss)-siRNA was incubated with increasing amounts of recombinant Translin or C3PO complex. (B) The RNase assays were performed with recombinant C3PO in the absence or presence of EDTA or EGTA. (C) The RNase assays were conducted by incubating circular ss-siRNA with increasing amounts of recombinant C3PO. (D) Following sequential chromatography, fractions were assayed for the RISC-enhancing activity (top), RNase activity (middle), or Western blotting to detect Translin and Trax (bottom).
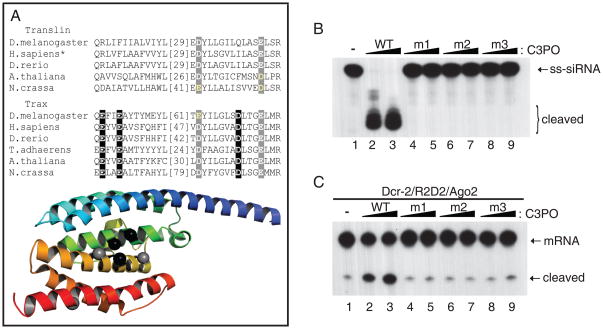
C3PO is a new class of endoribonuclease because neither subunit shows similarity to any known RNase by bioinformatics and structural analyses (18). To identify the catalytic sites of C3PO, we performed a multi-sequence alignment of Translin/Trax and observed three acidic residues (E123, E126, D204) that were invariant in Trax, but missing in all Translin (Fig. 5A and Fig. S9). Furthermore, modeling the structure of Drosophila Trax after crystal structure of human Translin (18) revealed that these residues existed in close spatial proximity, suggesting that they may coordinate Mg2+ ion for catalysis (Fig. 5A).
Fig. 5.
The RNase activity of C3PO is required for RISC activation. (A) A partial Translin/Trax multi-sequence alignment (top) and a modeled structure of Drosophila Trax based on crystal structure of human Translin from 1j1j (bottom). The comprehensive alignment of Translin/Trax is shown in Figure S9. The putative catalytic D/E residues are colored in black. (B) The RNase activity was compared between WT and three catalytic mutant (m1=E123, m2=E126, m3=D204) C3PO complexes. (C) The RISC-enhancing activity was compared between WT and catalytic mutant C3PO by assaying together with recombinant Dcr-2/R2D2/Ago2.
To test this hypothesis, we individually mutated E123, E126 and D204 residues of Trax to alanine. Recombinant mutant C3PO complexes were generated by co-expressing His-tagged wild-type Translin and untagged mutant Trax (Fig. S10a). These point mutations did not affect protein folding or complex formation because wild type and mutant C3PO displayed same column behaviors and bound single-stranded siRNA equally well in a non-cleaving condition (Fig. S10b). Consistently, mutating each putative catalytic residue abolished the RNase activity and the RISC-enhancing activity of C3PO (Fig. 5B-C and Fig. S10c). By contrast, mutation of other highly conserved D/E residues in Translin or Trax did not significantly affect the RNase or RISC-enhancing activity of C3PO (Fig. S10d-e). These studies suggest that the intrinsic RNase activity of C3PO is required for its RISC-enhancing activity.
We establish the first in vitro reconstitution system for dsRNA and duplex siRNA-initiated RISC activities, demonstrating that Dcr-2/R2D2/Ago2 comprise the catalytic core of Drosophila RNAi. This reconstitution system enables us to purify C3PO, a multimeric complex of Translin and Trax, as a key activator of the core RNAi machinery. Our biochemical studies indicate that the “slicer” mechanism plays a dominant role in Drosophila RISC assembly. C3PO, a Mg2+-dependent endoribonuclease for which Trax is the catalytic subunit, promotes RISC activation by removing siRNA passenger strand cleavage products. The RNase activity of C3PO may be stimulated by Ago2-mediated nick in duplex siRNA and/or fraying of the ends of cleavage products. The exonuclease QIP may function in a similar manner in Neurospora crassa (19). Finally, this robust and progressive reconstitution system should greatly facilitate in-depth mechanistic studies of the assembly, function and regulation of holo-RISC, the catalytic engine of RNAi.
Supplementary Material
supplementary data
Acknowledgments
We thank Drs. R. Koch, B. Suter, R. Carthew, M. Siomi, H. Siomi for reagents, and Drs. Y. Liu, H. Yu, D. Corey, Z. Paroo for discussion and reading the manuscript. Y.L. is supported by the Sara and Frank McKnight fellowship. N.V.G. is a Howard Hughes Medical Institute investigator. The work is supported by a Welch grant (I-1608) and National Institute of Health grants awarded to J.P. (AG025688) and Q.L. (GM078163 and GM084010).
References and notes
- 1.Carthew RW, Sontheimer EJ. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siomi H, Siomi MC. Nature. 2009;457:396. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 3.Paroo Z, Liu Q, Wang X. Cell Res. 2007;17:187. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, et al. Science. 2003;301:1921. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 6.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. Cell. 2004;117:83. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Rna. 2006;12:1514. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. Science. 2004;306:1377. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 9.Okamura K, Ishizuka A, Siomi H, Siomi MC. Genes Dev. 2004;18:1655. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Boja ES, Oubrahim H, Chock PB. Biochemistry. 2004;43:13424. doi: 10.1021/bi048847l. [DOI] [PubMed] [Google Scholar]
- 11.Claussen M, Koch R, Jin ZY, Suter B. Genetics. 2006;174:1337. doi: 10.1534/genetics.106.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suseendranathan K, et al. Eur J Cell Biol. 2007;86:173. doi: 10.1016/j.ejcb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Tomari Y, et al. Cell. 2004;116:831. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 14.Nykanen A, Haley B, Zamore PD. Cell. 2001;107:309. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 15.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Cell. 2005;123:607. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Genes Dev. 2005;19:2837. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rand TA, Petersen S, Du F, Wang X. Cell. 2005;123:621. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Sugiura I, et al. Acta Crystallogr D Biol Crystallogr. 2004;60:674. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 19.Maiti M, Lee HC, Liu Y. Genes Dev. 2007;21:590. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary data