Roles of the Lipid-binding Motifs of Atg18 and Atg21 in the Cytoplasm to Vacuole Targeting Pathway and Autophagy (original) (raw)
Abstract
Atg18 and Atg21 are homologous WD-40 repeat proteins that bind phosphoinositides via a novel conserved Phe-Arg-Arg-Gly motif and function in autophagy-related pathways. Atg18 is required for the cytoplasm to vacuole targeting (Cvt) pathway and autophagy, whereas Atg21 is only required for the Cvt pathway. Currently, the functions of both proteins are poorly understood. Here, we examined the relationship between the phosphatidylinositol 3-phosphate (PtdIns(3)P)-binding abilities of Atg18 and Atg21 and autophagy by expressing variants of these proteins that have mutations in their phosphoinositide-binding motifs. Cells expressing PtdIns(3)P-binding mutants of both these proteins showed highly reduced autophagy. Furthermore, the localization of components of two related ubiquitin-like protein conjugation systems, Atg8 and Atg16, to the phagophore assembly site is affected. Consistent with the aberrant localization of the above Atg proteins, precursor Ape1, a cargo of the Cvt pathway and autophagy, is partially protease-sensitive in starvation conditions. This finding suggests a requirement for the PtdIns(3)P binding capability of Atg18 and Atg21 in efficient completion of the sequestering autophagic vesicles. Finally, using a multiple knock-out strain, we found that Atg18 and Atg21 facilitate the recruitment of Atg8–PE to the site of autophagosome formation and protect it from premature cleavage by Atg4, which represents a key aspect of post-translational autophagy regulation. Taken together, our results suggest that PtdIns(3)P binding by at least Atg18 or Atg21 is required for robust autophagic activity and that the PtdIns(3)P-binding motifs of Atg18 and Atg21 can compensate for one another in the recruitment of Atg components that are dependent on PtdIns(3)P for their phagophore assembly site association.
Keywords: Autophagy, Lysosomes, Protein Targeting, Protein-Protein Interactions, Yeast, Stress, Vacuole
Introduction
Macroautophagy (hereafter referred to as autophagy) is an intracellular trafficking pathway wherein a double-membrane vesicle, the autophagosome, engulfs cytoplasmic components, including proteins and organelles. The autophagosome then fuses with a lysosome (or the vacuole in fungi and plants) where its cargo is degraded, and the breakdown products are recycled back into the cytosol. Autophagy plays a key role in cellular homeostasis and functions in diverse cellular processes such as development, programmed cell death, immune response, pathogenesis, tumor suppression, cardiac disorders, diabetes, and the prevention of neuronal degeneration (reviewed in Ref. 1). Autophagosome formation originates at a site known as the phagophore assembly site (PAS,4 also called the pre-autophagosomal structure (2, 3)). So far in yeast, more than 30 autophagy-related (Atg) proteins have been identified, and most of them localize at least transiently to the PAS. In yeast, apart from autophagy that is primarily a nonselective, degradative pathway that occurs during starvation conditions, there is another vesicular pathway called the cytoplasm to vacuole targeting (Cvt) pathway, which is a selective, biosynthetic pathway that takes place during vegetative growth (4). The Cvt pathway serves in the vacuolar transport of two hydrolases, α-mannosidase and the precursor form of aminopeptidase I (prApe1). Of the known Atg proteins, there are some that are exclusively involved in the Cvt pathway, others that are only needed for autophagy, and a subset of proteins that are required for both processes. For these pathways to function normally, various combinations of Atg proteins have to act in a concerted manner.
Atg18 and Atg21 are homologous Atg proteins; Atg18 functions in both autophagy and the Cvt pathway, whereas Atg21 is only required for the latter (5–8). There is a third homolog, Ygr223c, that is involved neither in Cvt nor autophagy (Ref. 9 and data not shown). Atg18, Atg21, and Ygr223c are predicted to be multiple WD-40 repeat proteins that fold to form seven bladed β-propellers (10). WD-40 repeat proteins are relatively common in all eukaryotes and are implicated in a wide variety of functions such as signal transduction, cytoskeleton assembly, transcriptional regulation, pre-mRNA processing, cell cycle control, and apoptosis (11–15). Repeated WD-40 motifs form β-propeller structures that act as sites for protein-protein interaction, and proteins containing these motifs serve as platforms for the assembly of protein complexes or as mediators of stable or transient interactions among other proteins (16, 17).
In addition to WD-40 motifs, Atg18, Atg21, and Ygr223c are able to bind both phosphatidylinositol 3-phosphate (PtdIns(3)P) and phosphatidylinositol (3,5)-bisphosphate (PtdIns(3,5)P2) (8, 10, 18). Both Atg18 and Atg21 bind phosphoinositides via conserved Phe-Arg-Arg-Gly (FRRG) motifs that lie within the proposed β-propellers; in the case of Atg18, these residues are 284FRRG287, and for Atg21 they are 342FRRG345. Mutations in the FRRG motif of Atg18 (FRRG to FTTG) result in a complete loss of phosphoinositide binding, whereas similar mutations in Atg21 result in highly reduced phosphoinositide binding, suggesting that this protein may have other lipid binding domains (8, 10, 18). The Atg18FTTG mutant shows a substantial block in the Cvt pathway and a partial decrease in autophagy (19). In contrast, the Atg21FTTG mutant is completely defective in the vacuolar transport of prApe1 via the Cvt pathway, thus underscoring the importance of phosphoinositide binding for both pathways (10, 18). The deletion of Fab1, an enzyme that converts PtdIns(3)P to PtdIns(3,5)P2, has no effect on either the Cvt pathway or autophagy (10, 19), suggesting that these processes require PtdIns(3)P but not PtdIns(3,5)P2. In addition to its roles in Cvt and autophagy, Atg18 mediates retrograde trafficking from the vacuole via the endosome to the Golgi, and this function depends on its ability to bind PtdIns(3,5)P2 (20). The role of the PtdIns(3,5)P2 binding function of Atg21 is currently unknown.
In yeast, PtdIns(3)P is generated by the specific phosphorylation of PtdIns at the D-3 position of the inositol ring by Vps34, the only PtdIns 3-kinase in this organism (21). Vps34 plays a role in a number of vesicular membrane trafficking processes such as endocytosis, multivesicular body formation, soluble vacuolar protein sorting, and autophagy (21–24). In growing cells, PtdIns(3)P localizes to endosomal and vacuolar membranes, whereas in starvation conditions it is enriched on autophagosomal membranes and is also transported into the vacuole by virtue of its presence on the membrane of autophagic bodies (25–27). Vps34 forms two distinct complexes, PtdIns 3-kinase complex I and PtdIns 3-kinase complex II, that function in autophagy and vacuolar protein sorting, respectively (23). Both of the PtdIns 3-kinase complexes have three common subunits as follows: Vps15, Vps34, and Vps30/Atg6. In addition to these common factors, each complex also contains one specific component; for the PtdIns 3-kinase complex I, this unique factor is Atg14, whereas Vps38 is specific to complex II (23). Atg14 mediates the localization of Vps30 and Vps34 to the PAS; however, Vps15 localizes to the PAS by an unknown mechanism that is independent of Atg14 (28).
The physiological significance of the presence of PtdIns(3)P on the autophagosome membrane and the functional relevance of Atg protein-PtdIns(3)P interaction is not well understood. Atg18 interacts with Atg2; however, the physiological significance of this interaction is currently unknown. Recent data show that one of the functions of PtdIns(3)P on autophagic membranes is the recruitment of the Atg2-Atg18 complex to the PAS, via the PtdIns(3)P binding activity of Atg18 (19). Although the Atg18FTTG mutant is able to interact with Atg2, it is defective in the localization of the Atg2-Atg18FTTG complex to the PAS (19). It is proposed that the localization of some other Atg components such as the Atg12–Atg5-Atg16 complex and Atg8 may be regulated by PtdIns(3)P, because in the absence of Atg14, the PAS localization of these proteins is abolished (29). These proteins, however, are not dependent on Atg18 for their PAS localization, suggesting an alternative and previously unidentified mechanism for their PAS recruitment. Here, we show that under nitrogen starvation conditions, the PAS localization of Atg16 and Atg8 does in fact depend on the PtdIns(3)P-binding motifs of Atg18 and Atg21; _atg18_Δ _atg21_Δ cells expressing Atg18FKKG and Atg21FKKG mutants are defective in the PAS recruitment of Atg16 and Atg8. Furthermore, as a result of the aberrant localization of these proteins, _atg18_Δ _atg21_Δ cells expressing the Atg18FKKG and Atg21FKKG mutants show a severe defect in autophagy, whereas either single mutant displays either no defect or a very negligible defect. Thus, our results suggest a model wherein the PtdIns(3)P-binding motifs of Atg18 and Atg21 can substitute for one another in the recruitment of Atg components that are dependent on PtdIns(3)P for their PAS association and in this manner allow autophagy to proceed at close to wild-type levels.
Finally, to further understand the role of Atg18 and Atg21 in Atg8 localization, we made use of a modified multiple knock-out (MKO) strain. In this strain, most of the ATG genes are deleted, and a subset of Atg proteins can therefore be expressed to reconstitute in vivo a particular step of autophagy or the Cvt pathway (30). In our modified MKO strain, we expressed GFP-Atg8ΔR along with the Atg3 and Atg7 proteins that conjugate Atg8ΔR with phosphatidylethanolamine (PE) (31, 32), the Atg12–Atg5-Atg16 complex that enhances Atg8–PE formation, the PtdIns 3-kinase components Atg6 and Atg14 that are required for the PAS localization of Atg8–PE, and the Atg4 cysteine protease required for the cleavage of PE from Atg8 prior to autophagosome completion. We found that under growing conditions in the presence of these proteins, GFP-Atg8ΔR was dispersed in the cytosol. The additional presence of wild-type Atg18 and Atg21 resulted in distinct GFP-Atg8ΔR puncta. Furthermore, puncta formation occurred in a mechanism dependent on the PtdIns(3)P-binding motifs of Atg18 and Atg21, suggesting a role for these proteins in the protection of Atg8–PE from unregulated cleavage by Atg4. These data provide the first evidence for an in vivo mechanism that regulates the cleavage of Atg8–PE.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The yeast Saccharomyces cerevisiae strains used in this study are listed in Table 1. Knock-out strains were constructed using the loxP/Cre system (33). Integration of the GFP tag at the 3′ end of the ATG16 open reading frame (ORF) was performed by a PCR-based procedure (34). GFP-Atg8ΔR(404) was linearized with BstBI and introduced into the ATG8 locus. The TRP1 marker in YCY146 was replaced with the Escherichia coli kanr marker using the marker exchange plasmid M3925 (trp1::kanMX3) digested with BamHI (35).
TABLE 1.
Yeast strains used in this study
| Strain | Descriptive name | Genotype | Source or Ref. |
|---|---|---|---|
| KTY148 | ATG16-GFP | SEY6210 ATG16-GFP::KAN | This study |
| KTY162 | _atg18_Δ ATG16-GFP | SEY6210 _atg18_Δ::HIS3 ATG16-GFP::KAN | This study |
| KTY164 | _atg21_Δ ATG16-GFP | SEY6210 _atg21_Δ::TRP1 ATG16-GFP::KAN | This study |
| SEY6210 | WT | _MAT_α ura3-52 leu2-3,112 his3_-Δ_200 trp1_-Δ_901 lys2-801 suc2_-Δ_9 GAL | 61 |
| TN121 | WT pho8_Δ_60 | MATa leu2-3,112 trp1 ura3-52 pho8::_pho8_Δ_60 pho13_Δ::URA3 | 43 |
| UNY82 | Atg18-3HA | SEY6210 ATG18-3HA::Kan | This study |
| UNY110 | _atg18_Δ _atg21_Δ _pep_4Δ _vps4_Δ | TN121 _atg18_Δ _atg21_Δ _pep4_Δ::_KAN vps4_Δ::BLE | This study |
| UNY120 | MKO (ATG3 ATG8_Δ_R) | SEY6210 _atg1_Δ, _2_Δ, _4_Δ, _5_Δ, _6_Δ, _7_Δ, _8_Δ, _9_Δ, _10_Δ, _11_Δ, _12_Δ, _13_Δ, _14_Δ, _16_Δ, _17_Δ, _18_Δ, _19_Δ, _20_Δ, _21_Δ, _23_Δ, _24_Δ, _27_Δ, _29_Δ ATG8_Δ_R::LYS2 | This study |
| UNY121 | _pep4_Δ | SEY6210 _pep4_Δ::LEU2 | This study |
| UNY122 | _atg8_Δ | SEY6210 _atg8_Δ::LEU2 | This study |
| UNY123 | _atg4_Δ | SEY6210 _atg4_Δ::LEU2 | This study |
| YCY14 | _atg21_Δ | SEY6210 _atg21_Δ::HIS3 | This study |
| YCY26 | _atg18_Δ | SEY6210 _atg18_Δ::KAN | This study |
| YCY28 | _atg18_Δ _atg21_Δ | SEY6210 _atg18_Δ::_KAN atg21_Δ::HIS3 | This study |
| YCY31 | _atg18_Δ _atg21_Δ pho8_Δ_60 | TN121 _atg18_Δ _atg21_Δ | This study |
| YCY38 | _atg18_Δ _atg21_Δ _pep4_Δ pho8_Δ_60 | TN121 _atg18_Δ _atg21_Δ _pep4_Δ::Kan | This study |
| YCY66 | _atg18_Δ _atg21_Δ ATG16-GFP | SEY6210 _atg18_Δ::HIS3 ATG16-GFP::_KAN atg21_Δ::URA3 | This study |
| YCY79 | _atg4_Δ::LEU ATG8-GFP | SEY6210 _atg4_Δ::LEU ATG8-GFP::LYS2 | This study |
| YCY146 | MKO (ATG3 GFP-ATG8_Δ_R) | SEY6210 _atg1_Δ, _2_Δ, _4_Δ, _5_Δ, _6_Δ, _7_Δ, _8_Δ, _9_Δ, _10_Δ, _11_Δ, _12_Δ, _13_Δ, _14_Δ, _16_Δ, _17_Δ, _18_Δ, _19_Δ, _20_Δ, _21_Δ, _23_Δ, _24_Δ, _27_Δ, _29_Δ GFP-ATG8_Δ_R::TRP1 | This study |
| YCY153 | MKO (ATG3 GFP-ATG8_Δ_R) | YCY146 GFP-ATG8_Δ_R::TRP1::KAN | This study |
| YCY155 | MKO (ATG3 ATG8-GFP) | YCY137 ATG8-GFP::LYS2 | This study |
| YCY156 | MKO (ATG3 GFP-ATG8_Δ_R) | YCY153 ATG8-GFP::LYS2 | This study |
Yeast cells were grown in rich medium (YPD: 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal medium (SMD: 0.67% yeast nitrogen base, 2% glucose, supplemented with the appropriate amino acids and vitamins). Cells were starved in synthetic medium lacking nitrogen (SD-N: 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose).
Plasmids
For constructing pATG18-PA(314) and pATG21-PA(314) (where PA is protein A), the full-length ATG18 and ATG21 ORFs with 1 kb of endogenous promoter were amplified from genomic DNA and cloned into the XhoI and XmaI sites of pNopPA(314) (36). To clone pATG21-GFP(416), the full-length ATG21 ORF with 1 kb of 5′ sequence, including the endogenous promoter, was PCR-amplified and cloned into the NotI and BamHI sites of pATG9-GFP(416) (pAPG9GFP(416) (37)). pATG18-GFP(416) was cloned by a two-step process. A silent mutation was first introduced into pATG18-PA(314) to remove an internal BamHI site in the ATG18 ORF, leading to pATG18ΔBamHI-PA(314); the full-length ATG18 ORF with 1 kb of 5′ sequence, including the endogenous promoter, was PCR-amplified from pATG18ΔBamHI-PA(314) and cloned into the same sites of pATG9-GFP(416). The full-length ATG21 ORF with 1 kb of 5′ sequence, including the endogenous promoter, and 1 kb of 3′ sequence, including the terminator, was amplified from genomic DNA and cloned as a BamHI and SalI fragment into pRS415 or pRS414 to generate pATG21(415) or pATG21(414). PCR-based site-directed mutagenesis was used to substitute two arginines (RR) with two lysines (KK) in the corresponding wild-type plasmids, leading to pATG18FKKG-PA(314), pATG21FKKG-PA(314), pATG21FKKG-GFP(416), pATG18FKKG-GFP(416), and pATG21FKKG(415).
pATG7-ATG10-ATG5-HA-ATG12(416), pATG16(415), and pATG16-ATG4(415) were made based on plasmids pATG7(414), pATG10(414), pATG5(416), pHA-ATG12(416), pATG16(416), and pATG4(414) (30). For constructing pATG7-ATG10-ATG5-HA-ATG12(416), an ATG7 fragment from pATG7(414) was first cloned into the XmaI site of pRS416 to generate pATG7(416); ATG10, ATG5, and HA-ATG12 fragments were cloned sequentially into pATG7(416) using a single restriction enzyme each time. KpnI was used to introduce ATG10, BamHI for ATG5, and XhoI for HA-ATG12. pATG16(415) was constructed from pATG16(416) by replacing its backbone with that of pRS415 using PvuI. An ATG4 fragment digested with SacI from pATG4(414) was cloned into the pATG16(415) vector linearized with the same enzyme, resulting in pATG16-ATG4(415).
To clone the multigene plasmids pATG18-ATG2(413) and pATG23-ATG27-ATG6-ATG14(414), single gene plasmids expressing an individual ATG gene with its endogenous promoter and terminator regions were first constructed and tested for functionality. XbaI and SacI sites were used to clone pATG18(413), XhoI and XmaI for pATG2(413), XhoI for pATG23(414), KpnI for pATG27(414), SacI and BamHI for pATG6(414), and SalI and PstI for pATG14(414). The promoter, ORF, and terminator inserts were then removed from the single gene plasmids and cloned sequentially to generate plasmids pATG18-ATG2(413) and pATG23-ATG27-ATG6-ATG14(414). To generate pATG6-ATG14(414), a fragment containing both the ATG6 and the ATG14 inserts was PCR-amplified from pATG23-ATG27-ATG6-ATG14(414) and cloned into the NotI site of pRS414. The same fragment was inserted into the NotI site of pATG21(414), leading to pATG21-ATG6-ATG14(414). Site-directed mutagenesis was performed to exchange the arginines (RR) in the PtdIns(3)P-binding motifs of Atg18 and Atg21 with lysines (KK), in the multigene plasmids.
pATG8-GFP(416) has been described previously (38). pATG8-GFP bearing the endogenous promoter of ATG8 was digested using NotI and HindIII, and a fragment containing the promoter of ATG8 and the ATG8 ORF fused to GFP were cloned into the pRS307 vector digested with the same enzymes. pAtg8-GFP(307) was linearized with BglII and integrated into the lys2-801 locus.
pATG8ΔR(414) bearing the endogenous ATG8 promoter was digested with SacI and XhoI, and cloned into the pRS307 vector digested with SacI and SalI. pATG8ΔR(307) was linearized with BglII and integrated into the lys2-801 locus. The pRS vectors have been described previously (39). pGFP-ATG8(316) was a kind gift from Dr. Yoshinori Ohsumi (Tokyo Institute of Technology, Yokohama, Japan).
Fluorescence Microscopy
Fluorescence microscopy was performed as described previously (30). Yeast cells were grown in YPD or SMD lacking the appropriate auxotrophic amino acids to mid-log phase before imaging. For starvation, cells were shifted to SD-N for 4 h. Images were captured with a ×100 objective lens for all microscopy experiments except for those to localize Atg16-GFP, in which a ×60 objective lens was used. Fluorescence signals were visualized on a fluorescence microscope (IX71; Olympus). The images were captured by a CCD camera (Photometrics CoolSNAP HQ; Roper Scientific) and deconvolved using DeltaVision software (Applied Precision).
Electron Microscopy
Sample preparation, image acquisition, and quantification of autophagic bodies were performed as described previously (40).
Yeast Two-hybrid Assay and Affinity Purification
For yeast two-hybrid screening, AD-Atg18 (pPS150) and BD-Atg21 (pPS153) plasmids were transformed into strain PJ69-4A (41). As negative controls, the strain PJ69-4A was transformed with BD-Atg21 and empty AD vector or AD-Atg18 and empty BD vector. Interaction was assayed by streaking transformants on SD plates lacking histidine and examining growth.
For affinity purification, the strain UNY82, expressing Atg18 chromosomally tagged with 3-hemagglutinin, was transformed with Atg21-PA (experimental strain) or with empty vector, pRS414 (negative control). As an additional negative control, strain SEY6210 transformed with Atg21-PA alone was used. Affinity purification was performed as described previously (42). The protein extracts were resolved by SDS-PAGE using an 8% gel and transferred to Immobilon polyvinylidene difluoride membrane (IPVH00010; Millipore, Billerica, MA). Immunoblotting was performed with monoclonal anti-hemagglutinin antibody (Sigma).
Other Assays
The protein extraction, Western blotting, prApe1 maturation, GFP-Atg8 processing, and alkaline phosphatase (Pho8Δ60) and protease protection assays have been described previously (43–45). For the protease protection assay, spheroplasts were resuspended in lysis buffer (20 mm PIPES, pH 6.8, 200 mm sorbitol, 5 mm MgCl2) and disrupted using a 25-mm Swin-Lok holder assembly fitted with a 3.0-μm Nucleopore Track-Etch membrane (Whatman).
RESULTS
Mutations in the PtdIns(3)P Binding Domains of Both Atg18 and Atg21 Result in a Strong Reduction of Autophagic Activity
To investigate the physiological role of PtdIns(3)P binding by Atg18 and Atg21, we generated mutations within the FRRG motifs of these proteins (FRRG to FKKG) and examined the effects of these mutations on the Cvt pathway and autophagy. Consistent with previous analyses (8, 18, 19), cells in rich medium expressing Atg18FKKG or Atg21FKKG showed a substantial or a complete block in prApe1 maturation, respectively (supplemental Fig. S1).
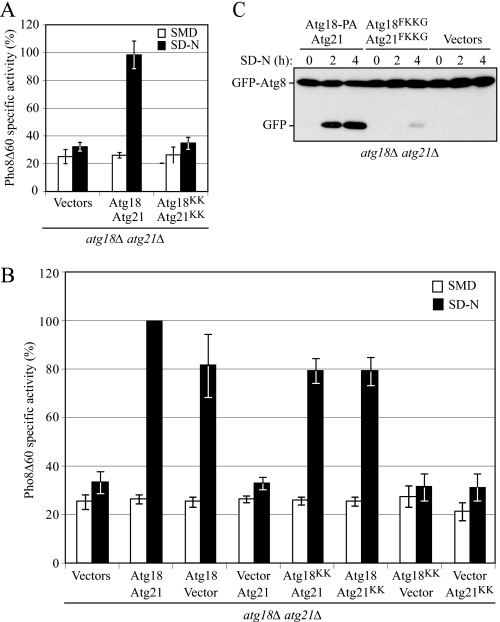
Previous reports indicate that _atg18_Δ cells expressing Atg18FKKG only show a slight reduction in autophagy (8), and in accordance with Atg21 not being involved in autophagy, Atg21FKKG also has essentially no effect on this process as determined by the Pho8Δ60 assay (Fig. 1A). In this assay, a truncated form of the vacuolar alkaline phosphatase precursor, Pho8Δ60, is expressed in the cytosol (43). To be proteolytically activated, precursor Pho8Δ60 must be transported to the vacuole by autophagy. S. cerevisiae only possesses two enzymes with alkaline phosphatase activity, Pho8 and Pho13. Thus, in a strain with a deletion of PHO13, where pho8_Δ_60 has replaced PHO8, the only alkaline phosphatase activity is derived from Pho8Δ60, which therefore serves as a marker for nonspecific autophagy. The enzymatic activity of mature Pho8Δ60 can be determined colorimetrically as a function of the cleavage of its substrate _p-_nitrophenyl phosphate. Because Atg18 and Atg21 are homologs, however, we considered the possibility that they were partially redundant. Therefore, we examined nonspecific autophagy in _atg18_Δ _atg21_Δ cells expressing both Atg18FKKG tagged with protein A and Atg21FKKG. Under nitrogen starvation conditions that induce autophagy, wild-type cells (double deletion cells expressing Atg18-PA and Atg21 from plasmids) showed a clear increase in Pho8Δ60-dependent alkaline phosphatase activity (Fig. 1A). In contrast, _atg18_Δ _atg21_Δ cells expressing the double mutants showed Pho8Δ60 activity levels that were almost similar to the background as observed in _atg18_Δ _atg21_Δ cells containing empty vectors. These results suggest that although Atg18 and Atg21 are not functionally identical, wild-type PtdIns(3)P binding activity of at least one of these proteins is required for autophagy in the absence of the other.
FIGURE 1.
Mutations in the PtdIns(3)P-binding motifs of both Atg18 and Atg21 result in a drastic reduction of autophagy. A, Pho8Δ60 activity is severely reduced in cells expressing Atg18FKKG-PA and Atg21FKKG. The _atg18_Δ _atg21_Δ pho8_Δ_60 (YCY31) cells were transformed with the following: empty vectors, centromeric plasmids expressing wild-type Atg18-PA and Atg21, or two plasmids bearing Atg18-PA and Atg21 constructs with mutations in their lipid binding domains (Atg18FKKG-PA and Atg21FKKG (denoted as Atg18KK and Atg21KK, respectively)). Cells were grown in nutrient-rich medium to mid-log phase and then shifted to starvation conditions for 4 h in SD-N medium. The Pho8Δ60-specific activity (nmol/min/mg) was measured as described under “Experimental Procedures.” The results represent the mean ± S.D. of three independent experiments. _B, atg18_Δ _atg21_Δ cells were transformed with the indicated plasmids and analyzed for Pho8Δ60-specific activity after 4 h in SD-N. The activity measured from cells expressing wild-type Atg18-PA and Atg21 was set to 100%, and the other values were normalized. C, GFP-Atg8 processing is severely affected in cells expressing Atg18 and Atg21 PtdIns(3)P-binding mutants. The _atg18_Δ _atg21_Δ (YCY28) strain was transformed with the combination of plasmids as in A. Cells were grown in nutrient-rich medium until mid-log phase (0-h time point) and then shifted to nitrogen-starvation conditions for 2 or 4 h. Aliquots were collected at the indicated time points and analyzed by immunoblotting using anti-yellow fluorescent protein antibody (which detects GFP). The positions of full-length GFP-Atg8 and free GFP are indicated.
Either single Atg18 or Atg21 PtdIns(3)P-binding mutant displayed autophagy activity that was close to the wild-type level (Fig. 1B), in agreement with previous results (7, 8, 46). Furthermore, both single mutants were competent for prApe1 maturation in starvation conditions (prApe1 can be delivered to the vacuole via the selective Cvt pathway in nutrient-rich conditions and by nonselective autophagy under starvation conditions; supplemental Fig. S2_A_). The defect in autophagy seen with the double mutants was not due to a decrease in stability resulting from the mutations, as both proteins displayed stability similar to their wild-type counterparts (supplemental Fig. S2_A_).
The drastic reduction in autophagy in _atg18_Δ _atg21_Δ cells expressing Atg18FKKG and Atg21FKKG was not previously examined, so we decided to confirm this result through another biochemical approach, GFP-Atg8 processing. Atg8 remains associated with the completed autophagosome in yeast and is degraded after delivery into the vacuole, whereas the GFP moiety is relatively stable (44); thus, the appearance of free GFP can serve as a measure of autophagy. _atg18_Δ _atg21_Δ cells were transformed with a plasmid-based GFP-Atg8, and either Atg18-PA and Atg21 (equivalent to wild type), empty vectors, or Atg18FKKG-PA and Atg21FKKG. The cells were grown in SMD medium until mid-log phase and then shifted to nitrogen starvation conditions. At various time points (0, 2, and 4 h), aliquots were removed, trichloroacetic acid-precipitated, and then subjected to Western blot using anti-yellow fluorescent protein antibody that recognizes GFP (Fig. 1C). In wild-type cells, the amount of free GFP increased over time during starvation, representing a functional autophagy pathway, whereas in autophagy-defective _atg18_Δ _atg21_Δ cells expressing only empty vectors, no free GFP was detected. In the presence of Atg18FKKG-PA and Atg21FKKG, only a very small amount of free GFP could be detected after 4 h of nitrogen starvation, representing a significant decrease in autophagy. This result is in agreement with the Pho8Δ60 analysis and further confirms the finding that _atg18_Δ _atg21_Δ cells expressing both the Atg18 and Atg21 lipid-binding mutants were defective in autophagy. Thus, although Atg18 and Atg21 are not functionally identical, wild-type PtdIns(3)P binding activity of at least one of these proteins is required for autophagy in the absence of the other. As expected based on these results, in _atg18_Δ _atg21_Δ cells expressing Atg18FKKG-PA and an empty vector, Pho8Δ60 activity levels were also similar to background (Fig. 1B).
We also examined whether the subcellular localization of the Atg18FKKG and Atg21FKKG mutants corresponded with their autophagy defects using GFP-tagged chimeras (supplemental Fig. S2, B and C). Consistent with previous results, we observed that Atg18 and Atg21 localize to several distinct structures in the cell such as the vacuolar membrane, the PAS, and endosomes (5–9, 19, 29). The Atg18FKKG-GFP and Atg21FKKG-GFP mutants, on the other hand, were not localized to membrane structures but instead were primarily dispersed throughout the cytosol in both growing and starvation conditions (supplemental Fig. S2, B and C) (8, 18, 19). Thus, the localization of Atg18 and Atg21 to the vacuolar membrane and other punctate structures is dependent on their PtdIns(3)P binding domains. We also determined that neither wild-type Atg18 nor Atg21 is dependent on the other for its localization, in rich medium or conditions of nitrogen deprivation (supplemental Fig. S2_D_).
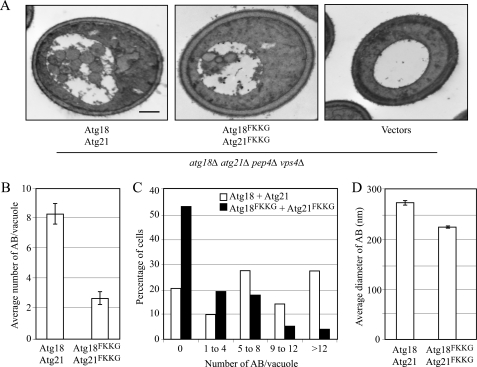
To determine the role for Atg18 and Atg21 PtdIns(3)P binding in autophagy, we examined the ultrastructure of autophagic bodies that accumulated under nitrogen starvation conditions in _atg18_Δ _atg21_Δ cells expressing both the Atg18FKKG and Atg21FKKG mutants, by electron microscopy (Fig. 2A). For the electron microscopy analysis, an _atg18_Δ _atg21_Δ strain was additionally deleted for the PEP4 and VPS4 genes. PEP4 encodes the vacuolar proteinase A, required for the degradation of the inner vesicles of autophagosomes (called autophagic bodies) in the vacuole. The deletion of PEP4 results in the preservation of autophagic bodies in the vacuole and allows them to be visualized by electron microscopy. We deleted the VPS4 gene to eliminate vesicles that are delivered into the vacuole via the multivesicular body pathway (47).
FIGURE 2.
Mutations in the Atg18 and Atg21 PtdIns(3)P-binding motifs resulted in fewer autophagosomes. A, wild-type (_atg18_Δ _atg21_Δ _pep4_Δ _vps4_Δ; UNY110 cells expressing Atg18-PA and Atg21), experimental (_atg18_Δ _atg21_Δ _pep4_Δ _vps4_Δ cells expressing Atg18FKKG-PA and Atg21FKKG), and negative control (UNY110 cells bearing empty vectors) strains were grown to early log phase in SMD lacking uracil and leucine, shifted to SD-N for 4 h to induce autophagy, fixed in potassium permanganate, and examined by electron microscopy. Scale bar, 0.5 μm. B, quantification of the average number of autophagic bodies per vacuole. The error bars represent standard error of the mean. C, quantification of autophagic body accumulation. The number of autophagic bodies accumulated was determined from cells containing similarly sized vacuoles. D, quantification of autophagic body size. The average diameters of cross-sections of autophagic bodies are shown; error bars represent mean ± S.E.; n > 175.
The _atg18_Δ _atg21_Δ _pep4_Δ _vps4_Δ strain was transformed with the following combinations of plasmids: Atg18-PA and Atg21, Atg18FKKG-PA and Atg21FKKG, or empty vectors. After 4 h of nitrogen starvation, no autophagic bodies were observed in the _atg18_Δ _atg21_Δ _pep4_Δ _vps4_Δ strain transformed with empty vectors (Fig. 2A). Under the same conditions, the average number of autophagic bodies in _atg18_Δ _atg21_Δ _pep4_Δ _vps4_Δ cells expressing Atg18-PA and Atg21 was 8.4 ± 0.7 (mean ± S.E., n = 97), whereas the same cells expressing Atg18FKKG-PA and Atg21FKKG showed a significantly smaller number (2.7 ± 0.44 (mean ± S.E., n = 74)) of autophagic bodies (Fig. 2B).
We also examined the distribution of autophagic bodies per vacuole and found that in cells expressing Atg18-PA and Atg21, 27% had more than 12 autophagic bodies per vacuole, compared with 3% in cells expressing Atg18FKKG-PA and Atg21FKKG (Fig. 2C). Additionally, 53% of the cells expressing the PtdIns(3)P-binding mutants of Atg18 and Atg21 showed no autophagic bodies (Fig. 2C). Finally, we also estimated that the average cross-sectional diameter of autophagic bodies in cells expressing the PtdIns(3)P-binding variants was slightly smaller (224.57 ± 0.33 (mean ± S.E., n = 194)) than in those expressing the wild-type Atg18-PA and Atg21 (274 ± 0.15 (mean ± S.E., n = 596)). Taken together, these results show that mutations in the PtdIns(3)P binding domains of both Atg18 and Atg21 result in highly reduced autophagy by affecting the number, and to some extent the size, of autophagic bodies.
Mutations in the PtdIns(3)P Binding Domains of Both Atg18 and Atg21 Result in the Incomplete Sequestration of prApe1 within Autophagosomes
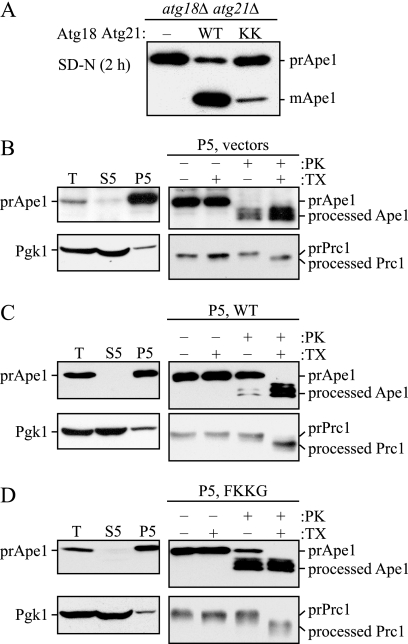
To gain further insight into the roles of PtdIns(3)P binding in the autophagy pathway, we wanted to identify the step that was affected in the _atg18_Δ _atg21_Δ strain expressing the lipid-binding mutants of Atg18 and Atg21. We first determined that consistent with a defect in autophagy in a strain expressing the FKKG variants of Atg18 and Atg21, there was a strong block in prApe1 maturation compared with wild-type cells after a shift to starvation conditions (Fig. 3A). In cells expressing the mutant Atg18 and Atg21 variants, the majority of the prApe1 protein was in the precursor form after a 2-h shift to medium lacking nitrogen, when compared with a wild-type strain where most of the Ape1 protein was in the mature form.
FIGURE 3.
Protease protection assay reveals that _atg18_Δ _atg21_Δ _pep4_Δ cells expressing the PtdIns(3)P-binding mutants of Atg18-PA and Atg21 are defective in autophagosome completion. A, mutations in the PtdIns(3)P-binding motifs of both Atg18 or Atg21 result in a reduction in prApe1 maturation during starvation conditions. _atg18_Δ _atg21_Δ pho8_Δ_60 (YCY31) cells bearing empty vectors, Atg18-PA and Atg21 (wild type (WT)), or Atg18FKKG-PA and Atg21FKKG (KK) centromeric plasmids were grown to mid-log phase in rich medium and then shifted to SD-N for 2 h. Protein extracts were prepared and examined by SDS-PAGE. The precursor and mature forms of Ape1 were examined by immunoblotting with anti-Ape1 antiserum. B–D, under starvation conditions, the majority of precursor Ape1 is protease-sensitive in _atg18_Δ _atg21_Δ _pep4_Δ cells expressing Atg18FKKG-PA and Atg21FKKG. _atg18_Δ _atg21_Δ _pep4_Δ (YCY38) cells bearing empty vectors (B), Atg18-PA and Atg21 (wild type; C), or Atg18FKKG-PA and Atg21FKKG (FKKG; D) were grown to mid-log phase in rich medium and converted to spheroplasts. To induce autophagy, the spheroplasts were starved for 1 h in SD-N medium supplemented with 1.2 m sorbitol. The spheroplasts were collected by centrifugation, resuspended in osmotically balanced lysis buffer, and then disrupted as described under “Experimental Procedures.” The lysate was clarified using a low speed centrifugation step to remove unbroken cells. The resulting total lysate (T) was further separated into 5,000 × g lysate (S5) and pellet (P5) fractions. The P5 fraction from each strain was divided into four parts and subjected to either no treatment, treatment with 0.4% Triton X-100 (TX), or treatment with proteinase K (PK) in the presence or absence of 0.4% Triton X-100 and analyzed by immunoblot using anti-Ape1 antiserum. Lysis conditions were verified by immunoblot analysis using the anti-Pgk1 and anti-Ape1 antisera. To verify that the lysis method did not disrupt the integrity of organellar membranes in the P5 fractions, maturation of the precursor form of Prc1 was examined by immunoblot analysis using anti-Prc1 antibody.
Next we analyzed the ability of _atg18_Δ _atg21_Δ cells expressing Atg18FKKG-PA and Atg21FKKG to completely sequester prApe1 in nitrogen starvation conditions by performing a protease protection assay to examine whether the cargo protein was enclosed within completed autophagosomes. In this assay, _atg18_Δ _atg21_Δ _pep4_Δ cells expressing empty vectors, wild-type Atg18-PA and Atg21, or Atg18FKKG-PA and Atg21FKKG were grown to mid-log phase and converted to spheroplasts. The spheroplasts were shifted for 1 h to osmotically supplemented nitrogen starvation medium to induce autophagosome formation. The cells were then collected by centrifugation and osmotically lysed. After the cell lysate was subjected to a low speed 300 × g centrifugation step to remove unbroken cells, the lysate was further separated into low speed supernatant and pellet fractions. The prApe1complex binds the membrane fraction in a MgCl2-dependent manner and can therefore be recovered in a low speed pellet fraction even when it is not enclosed in a completed autophagosome. The predominant separation of the cytosolic marker Pgk1 in the supernatant fraction indicated efficient lysis of spheroplasts (Fig. 3, B–D). The pellet fraction was divided into four parts as follows: the first was treated with no reagent; the second was treated with detergent; the third was treated with proteinase K; and the fourth was treated with proteinase K and detergent. The pellet fraction of a mutant that is defective in vesicle completion will accumulate prApe1 in a form sensitive to externally added protease; however, in a strain that is competent for autophagosome formation, prApe1 will be protected from proteinase K and will become sensitive to this protease only when autophagosome or vacuole membrane integrity is disrupted by detergent addition.
The pellet fraction of the _atg18_Δ _atg21_Δ _pep4_Δ strain bearing empty vectors was accessible to proteinase K both in the absence or presence of detergent (Fig. 3B), consistent with a complete block in autophagy in an _atg18_Δ _atg21_Δ mutant. In the _atg18_Δ _atg21_Δ _pep4_Δ strain expressing Atg18-PA and Atg21, prApe1 was predominantly protected from proteinase K alone, indicating efficient autophagosome completion. In the _atg18_Δ _atg21_Δ _pep4_Δ strain expressing the PtdIns(3)P-binding mutants of Atg18 and Atg21, however, prApe1 was largely sensitive to proteinase K, indicating that PtdIns(3)P-binding is required for efficient vesicle completion during autophagy.
To ascertain that our experimental conditions did not result in nonspecific membrane disruption, such as lysis of the autophagosome or vacuolar membranes, we examined the protease sensitivity of carboxypeptidase Y (Prc1), a vacuolar exopeptidase that is synthesized as a precursor and delivered to the vacuole via the secretory pathway where it is converted to the mature, active form by removal of a propeptide. Treatment of the pellet fraction with proteinase K in the absence of detergent did not yield the mature form of Prc1, whereas addition of both detergent and proteinase K resulted in processing of precursor Prc1 to its mature form (Fig. 3, B–D), thus confirming that the vacuolar membrane, and presumably autophagosomes, were intact in our pellet fractions.
atg18Δ atg21Δ Cells Expressing Atg18FKKG and Atg21FKKG Mutants Are Defective in the Recruitment of Atg8 in Rich Conditions and/or Dissociation of Atg8 from the PAS under Starvation Conditions
When expressed together, the Atg18FKKG and Atg21FKKG mutants result in a complete block in the Cvt pathway (data not shown) and a severe defect in autophagy (Fig. 1, A and C). Therefore, we wanted to determine the mechanism by which the mutants resulted in these phenotypes. We hypothesized that when expressed together, these mutant proteins may result in the mislocalization of some Atg proteins, and accordingly, we examined the localization of Atg8. The localization of Atg8 to the PAS under starvation conditions is dependent on at least 10 Atg proteins; the absence of seven of these proteins (Atg3, Atg4, Atg7, Atg10, and the Atg12–Atg5-Atg16 complex) results in the complete lack of Atg8 localization to the PAS, whereas the loss of the other three proteins (Atg9 and the PtdIns(3)-kinase complex components Atg14 and Atg6) results in a partial loss of Atg8 recruitment to the PAS (29). The loss of Atg18 does not affect the recruitment of Atg8–PE to the PAS under both exponential growth and starvation conditions (29). In contrast, Atg21 is required for the localization of GFP-Atg8 to the PAS under nutrient rich conditions; GFP-Atg8 displays a diffuse cytosolic staining in the absence of Atg21 (8).
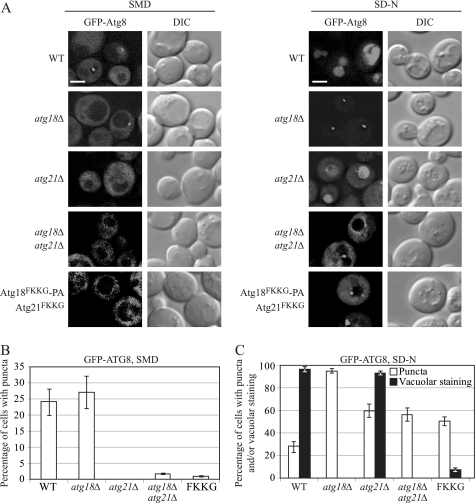
As with autophagy activity, previous studies did not examine the effect on Atg8 localization of mutating the PtdIns(3)P-binding sites on both Atg18 and Atg21 simultaneously. We observed the GFP-Atg8 localization pattern in exponentially growing cells in rich medium or after cells were shifted to starvation conditions for 4 h. In wild-type cells grown in rich medium, GFP-Atg8 showed cytosolic staining, and about 23% of the cells showed clear perivacuolar puncta as a result of the recruitment of GFP-Atg8 to the PAS (Fig. 4, A and B). Under starvation conditions, GFP-Atg8 is recruited to the PAS where it participates in autophagosome formation. GFP-Atg8 present on the inner membrane of the autophagosome is delivered to the vacuole via autophagy; the GFP moiety is cleaved from Atg8, and free GFP accumulates in the vacuole. This free GFP can be visualized by fluorescence microscopy. After 4 h in starvation conditions, in wild-type cells, ∼25% of the cells examined showed perivacuolar GFP-Atg8 puncta, and almost all the cells showed vacuolar staining (Fig. 4, A and C), consistent with normal autophagy. In agreement with previously published data, in an _atg18_Δ strain under both growing and starvation conditions, GFP-Atg8 was recruited to the PAS; however, free GFP was never visualized in the vacuolar lumen under starvation conditions (Fig. 4, A–C). Thus, Atg18 is not needed for the PAS recruitment of GFP-Atg8, but instead it has a role in the Cvt pathway and autophagy at a stage subsequent to the recruitment of Atg8.
FIGURE 4.
Atg18FKKG and Atg21FKKG mutants are defective in the recruitment of Atg8 to the PAS in nutrient-rich medium and in both its recruitment to and dissociation from the PAS under starvation conditions. A, GFP-Atg8 localization patterns were monitored by fluorescence microscopy in nutrient-rich conditions (SMD) and after a shift to starvation medium (SD-N) for 4 h in wild-type (WT; SEY6210), _atg18_Δ (YCY26), _atg21_Δ (YCY14), _atg18_Δ _atg21_Δ (YCY28), or _atg18_Δ _atg21_Δ cells expressing Atg18FKKG-PA and Atg21FKKG. DIC, differential interference contrast. Scale bar, 2.5 μm. B, quantification of perivacuolar GFP-Atg8 punctate dots under nutrient-rich conditions in the cells from A. C, percentage of cells showing perivacuolar, GFP-Atg8 punctate dots (white bars) and vacuolar GFP staining (black bars) under nitrogen starvation conditions in the cells from A. Three independent experiments with ∼100 cells for each strain were analyzed for scoring the percentage of cells with GFP-Atg8 puncta. Error bars represent mean ± S.D.
Also consistent with previous observations, in the absence of Atg21, GFP-Atg8 perivacuolar puncta were absent, and this protein showed diffuse cytosolic staining, in nutrient-rich conditions (Fig. 4, A and B) (8). As Atg21 does not have a role in starvation-induced autophagy, under these conditions, the localization pattern of GFP-Atg8 appeared similar to that in wild-type cells, although we detected an increase in the cytosolic GFP-Atg8 signal (Fig. 4, A and C). In _atg18_Δ _atg21_Δ cells, in nutrient-rich conditions, the GFP-Atg8 localization pattern appeared similar to that in an _atg21_Δ strain, showing a diffuse cytosolic fluorescence signal and a strong block in PAS recruitment (Fig. 4, A and B). These observations suggest that in growing conditions Atg21 is epistatic over Atg18 in the recruitment of Atg8 and that these two proteins do not have completely overlapping functions. After 4 h in starvation conditions, the GFP-Atg8 in _atg18_Δ _atg21_Δ cells showed very strong cytosolic staining, and although about 55% of the cells showed perivacuolar punctate dots, vacuolar GFP staining was not observed (Fig. 4, A and C). This phenotype suggests a defect in both the recruitment of GFP-Atg8 to the PAS and delivery of GFP-Atg8 to the vacuole; a defect in only recruitment would show cytosolic staining with no PAS puncta, whereas a defect in vacuolar delivery alone would be reflected by strong perivacuolar puncta, with the absence of vacuolar or cytosolic staining (for example, compare with _atg18_Δ cells that are not defective in recruitment). Similar results were observed in _atg18_Δ _atg21_Δ cells expressing both Atg18FKKG-PA and Atg21FKKG. In growing conditions, GFP-Atg8 in the double mutant was mostly cytosolic (Fig. 4, A and B), whereas in starvation conditions, in accordance with our Pho8Δ60 activity and GFP-Atg8 processing results (Fig. 1, A and C), vacuolar GFP-Atg8 fluorescence staining associated with autophagy was seldom seen; instead, this protein showed both strong cytosolic staining and perivacuolar punctate structures (Fig. 4, A and C). Atg18FKKG or Atg21FKKG alone did not affect GFP-Atg8 localization under starvation conditions (data not shown), consistent with either single mutation not affecting Pho8Δ60 activity (Fig. 1B). Overall, these results suggest that under starvation conditions, the wild-type localization of GFP-Atg8, and concomitantly normal autophagy, depends on the wild-type PtdIns(3)P binding ability of at least Atg18 or Atg21.
Finally, we wanted to test whether the aberrant localization of GFP-Atg8 in _atg18_Δ _atg21_Δ cells was due to a defect in the conjugation of Atg8 with PE. Atg8 is present in a cytosolic pool, and it is processed by the proteolytic removal of a C-terminal arginine by the cysteine protease Atg4. After Atg4-dependent processing, Atg8 is conjugated to PE via two enzymes as follows: Atg7, the E1-like activating enzyme, and Atg3, an E2-like conjugating enzyme (48–51). The conjugation of Atg8 with PE, which is required for its PAS localization, is facilitated by a multimeric complex that may function as an E3-like enzyme and is composed of three proteins, Atg12, Atg5, and Atg16 (3, 52). Atg8–PE can be resolved from Atg8 by carrying out SDS-PAGE in the presence of urea and performing immunoblot analysis with the Atg8 antibody (53). Under growing conditions, the amount of conjugated Atg8 was significantly less in an _atg18_Δ _atg21_Δ strain expressing empty vectors or the Atg18FKKG-PA and Atg21FKKG mutants compared with that in the same strain expressing wild-type Atg18 and Atg21 (supplemental Fig. S3). After starvation for 4 h, even though there was a considerable accumulation of the unconjugated form of Atg8 in _atg18_Δ _atg21_Δ cells expressing empty vectors, or expressing the Atg18FKKG-PA and Atg21FKKG mutants, there was no discernible difference in the amount of Atg8–PE relative to the wild type (supplemental Fig. S3). Therefore, the autophagy defect in _atg18_Δ _atg21_Δ cells expressing the Atg18FKKG-PA and Atg21FKKG mutants was not simply due to the inability of the cells to form Atg8–PE.
atg18Δ atg21Δ Cells Expressing the Atg18FKKG and Atg21FKKG Mutants Are Defective in the PAS Recruitment of Atg16 under Starvation Conditions
Having determined that the Atg18 and Atg21 FKKG mutants affect the localization pattern of Atg8, we wanted to extend our analysis to examine the localization of other autophagy-related proteins. We specifically chose to examine the localization of Atg16 in a strain expressing the PtdIns(3)P-binding mutants of Atg18 and Atg21. There are two reasons for examining the localization of Atg16. First, Atg16 affects the recruitment of Atg8 to the PAS; and second, the PAS localization of Atg16 itself requires PtdIns(3)P (27). Indeed, the PAS localization of the Atg12–Atg5-Atg16 complex is blocked in the absence of the PtdIns 3-kinase (_vps34_Δ strain) and in the absence of Atg14, which is required for targeting the PtdIns 3-kinase to the PAS. However, because Atg16 does not have an obvious PtdIns(3)P-binding motif, and the deletion of Atg18 alone does not affect its PAS localization (29), we reasoned that in the absence of Atg18, Atg21 may have a role in the PAS recruitment of this protein.
To examine the localization pattern of Atg16 by fluorescence microscopy, we chromosomally tagged Atg16 with GFP. In nutrient-rich conditions, the fluorescence signal of Atg16 in wild-type cells was very weak and could be detected in less than 10% of the cells (data not shown). Accordingly, we chose to study its localization only under starvation conditions. After 4 h in nitrogen-depleted medium, Atg16-GFP showed cytosolic staining and a clear punctate perivacuolar dot in ∼40% of the wild-type cells (Fig. 5, A and B). In an _atg18_Δ background, more than 60% of the cells showed Atg16-GFP puncta. These results are consistent with Atg18 having a role in the dissociation of proteins such as Atg8 from the PAS. In accordance with Atg21 not having a role in autophagy, the loss of this protein had no appreciable effect on the PAS localization of Atg16-GFP after 4 h in starvation conditions (Fig. 5, A and B). The deletion of both Atg18 and Atg21, however, resulted in less than 20% of cells that showed Atg16-GFP perivacuolar puncta (Fig. 5, A and B). Next, we examined the possible contribution of Atg18 and Atg21 lipid binding in the PAS localization of Atg16. _atg18_Δ _atg21_Δ cells transformed with Atg18-PA and Atg21 or Atg18FKKG-PA and Atg21FKKG showed ∼50 or 22% Atg16-GFP puncta, respectively (Fig. 5, C and D). Taken together, these results indicated that under starvation conditions the recruitment of Atg16 to the PAS was compromised in the absence of Atg18 and Atg21 and additionally that the lipid-binding motifs of these proteins were required for their role in the PAS recruitment of Atg16.
FIGURE 5.
Atg18 and Atg21 lipid-binding mutants are defective in the recruitment of Atg16 to the PAS under starvation conditions. Each of the following strains was chromosomally tagged with Atg16-GFP. A and B, wild-type (WT, SEY6210), _atg18_Δ (YCY26), _atg21_Δ (YCY14), and _atg18_Δ _atg21_Δ (YCY28). C and _D, atg18_Δ _atg21_Δ strain was additionally transformed with plasmids expressing wild-type Atg18-PA and Atg21, the FKKG mutants, or empty vectors. A and C, Atg16-GFP localization pattern was examined by fluorescence microscopy; B and D, percentage of cells showing Atg16-GFP puncta in each of these strains after a 4-h shift to starvation conditions was quantified. The quantification shown here is from three independent experiments. Approximately 500 cells for each strain were analyzed for scoring the percentage of cells with fluorescent Atg16-GFP PAS puncta. Error bars represent the mean ± S.E. DIC, differential interference contrast. Scale bar, 2.5 μm.
Atg18 and Atg21 Protect Atg8 from Atg4-mediated Cleavage
Our current data suggest that Atg18 and/or Atg21 play a role in the PAS recruitment to, and dissociation from, the PAS of Atg8 and Atg16. To dissect out the role(s) of Atg18 and Atg21 in the PAS localization of Atg8 under nutrient-rich conditions, we utilized an MKO strain (30). In this strain, most of the autophagy-related genes are deleted, and therefore it is deficient in the assembly of a functional PAS. The advantage of the MKO system is that it allows us to investigate a particular function or contribution of a subset of Atg proteins in the complete absence of the other Atg proteins that are not being examined.
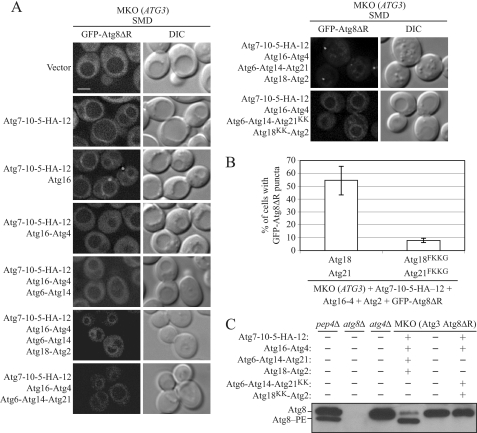
Our MKO strain has endogenous, wild-type copies of the VPS34 and VPS15 genes but not VPS30/ATG6, a component of PtdIns 3-kinase complexes I and II. Therefore, although it is not defective in PtdIns(3)P synthesis, it has a reduced amount of this lipid (54). We started with an MKO strain expressing Atg3 (MKO (ATG3)) and also expressing GFP-Atg8ΔR in which the C-terminal arginine residue has been removed; the latter allowed us to bypass the initial Atg4-mediated processing of Atg8. When we examined the localization of GFP-Atg8ΔR in this strain, we found that its fluorescence signal was evenly distributed in the cytosol (Fig. 6, Vector). In the presence of additional components needed for Atg8–PE conjugation and/or stability (Atg7, Atg10, Atg5, and Atg12 (30, 52, 55)), we observed that GFP-Atg8ΔR was localized to the vacuolar rim, in addition to being distributed in the cytosol. When the MKO (ATG3) strain bearing these components was supplemented with Atg16, GFP-Atg8ΔR was recruited to distinct puncta in addition to being present around the vacuolar rim, presumably because the presence of Atg16 further enhances the efficiency of Atg8–PE conjugation and/or directs conjugation to the PAS, or its equivalent in the MKO strain (56, 57). When the cysteine protease Atg4 was additionally expressed in the MKO (ATG3) strain containing Atg7, Atg10, Atg5, Atg12, and Atg16, GFP-Atg8ΔR-containing puncta and vacuolar rim staining disappeared, and the signal was dispersed in the cytosol, presumably because of the deconjugation of the PE moiety from Atg8. The addition of the PtdIns 3-kinase complex subunits Atg14 and Atg6 to the abovementioned MKO system did not result in the relocation of the GFP-Atg8ΔR fluorescence back into puncta or onto the vacuolar membrane. In wild-type cells, GFP-Atg8 can be detected as a punctum at the PAS even in the presence of Atg4, suggesting that the MKO strain expressing the Atg8 and Atg12 conjugation proteins and PtdIns 3-kinase complex subunits lacks some additional component(s) that regulate the second cleavage by Atg4.
FIGURE 6.
Atg18-Atg2 and Atg21 protect Atg8 from Atg4-mediated cleavage. A, MKO strain expressing Atg3 and GFP-Atg8ΔR, which lacks the C-terminal arginine residue of Atg8, was transformed with plasmids expressing different combinations of Atg proteins as indicated. The GFP-Atg8ΔR localization pattern in these different strains was monitored in nutrient-rich conditions by fluorescence microscopy. Similar results were observed with cells shifted to starvation conditions for 4 h. DIC, differential interference contrast. Scale bar, 2.5 μm. B, percentage of cells with fluorescent GFP-Atg8ΔR puncta were scored from three independent experiments. Error bars represent mean ± S.D. C, lipidation of Atg8 is affected in the MKO strain expressing FKKG mutants of Atg18 and Atg21. Atg8ΔR was expressed from the lys2 locus of an MKO strain bearing ATG3. This strain was transformed with empty vectors or the indicated plasmids. _pep4_Δ, _atg8_Δ, or _atg4_Δ strains were used as controls. Cells were grown to mid-log phase and collected, and protein extracts were subjected to SDS-PAGE followed by immunoblotting with anti-Atg8 antibody.
Under nutrient-rich conditions, the absence of ATG18 and ATG21 resulted in the cytosolic dispersal of GFP-Atg8 (Fig. 4A); therefore, we hypothesized that Atg18 and Atg21 may play a role in the punctate localization of GFP-Atg8 by protecting it from unregulated cleavage by Atg4. The addition of either Atg18-Atg2 or Atg21 to the MKO strain containing Atg7-Atg10-Atg5-Atg12, Atg16-Atg4, and Atg6-Atg14 resulted in the partial relocation of some of the GFP-Atg8ΔR fluorescence back onto the vacuolar membrane (Fig. 6); however, neither Atg18 nor Atg21 alone restored the PAS localization of Atg8. In contrast, in the presence of both Atg18-Atg2 and Atg21, in addition to the components from the two conjugation systems and the PtdIns 3-kinase complex I, GFP-Atg8ΔR appeared as puncta in ∼55% of the cells examined, in the presence of Atg4, suggesting that in wild-type cells, Atg18-Atg2 and Atg21 may play a role in protecting Atg8–PE from premature cleavage by Atg4 before autophagosome formation is complete (Fig. 6, A and B). In this case, the vacuolar rim staining was not apparent, which may indicate that the inhibition of the second Atg4 cleavage reaction was restricted to the punctate GFP-Atg8ΔR. Next, we examined whether we could reconstitute GFP-Atg8ΔR puncta formation in the MKO system in the presence of the Atg18- and Atg21-PtdIns(3)P-binding mutants. We found that in the MKO strain expressing Atg18FKKG and Atg21FKKG mutants, only ∼6% of the cells examined showed GFP-Atg8ΔR puncta; however, almost all the cells examined showed weak but discernible vacuolar rim staining (Fig. 6, A and B).
We carried out one additional analysis to verify that the punctate GFP-Atg8ΔR seen in the presence of Atg18-Atg2 and Atg21 did not reflect inhibition of Atg4 enzymatic activity in the MKO strain expressing only a subset of Atg proteins. We transformed the MKO (ATG3) strain with a plasmid expressing Atg8 with GFP fused at the C terminus, which is normally subject to cleavage by Atg4 in wild-type cells (36). The generation of free GFP can be used to monitor Atg4 activity. We carried out this analysis in the strain expressing GFP-Atg8ΔR, which we used in the previous analyses, and also in the MKO (ATG3) strain without GFP-Atg8ΔR to avoid the possibility that the free GFP was generated from the latter construct. In either case, the MKO (ATG3) strain retained normal Atg4 protease activity, and the presence of the additional Atg components did not interfere with cleavage of GFP from Atg8 when fused at the C terminus (supplemental Fig. S4). Thus, lack of Atg4 enzymatic activity could not explain the restoration of GFP-Atg8ΔR puncta.
Finally, we wanted to test the hypothesis that the GFP-Atg8ΔR puncta formation seen in the presence of Atg18-Atg2 and Atg21 is due to the protection of Atg8–PE cleavage by Atg4. For this purpose, we constructed an MKO strain expressing Atg3 and Atg8ΔR. We transformed this strain with empty vectors or with components from the two conjugation systems and the PtdIns 3-kinase complex I in the presence of wild-type Atg18-Atg2 and Atg21 or Atg2-Atg18FKKG and Atg21FKKG mutants, and we analyzed Atg8–PE formation by immunoblot analysis. We found that in the MKO strain expressing wild-type Atg18 and Atg21, Atg8ΔR was predominantly conjugated to PE, whereas in the same strain expressing the PtdIns(3)P-binding mutants of Atg18 and Atg21, Atg8ΔR was found in the PE-unconjugated form (Fig. 6C). As expected, Atg8ΔR was not PE-conjugated in our MKO strain transformed with empty vectors. Taken together with our fluorescence microscopy results, these data suggest a role for Atg18 and Atg21 in the protection of Atg8–PE puncta from unregulated cleavage by Atg4, in a mechanism dependent on their ability to bind PtdIns(3)P.
DISCUSSION
In this work we analyzed the physiological significance of the PtdIns(3)P binding domains of two homologous proteins, Atg18 and Atg21, in the Cvt pathway and autophagy. Although the PtdIns 3-kinase Vps34, and components of the kinase complex, are required for autophagy-related processes (21, 27, 58), the role of PtdIns(3)P is still unclear, but it is absolutely required for both the Cvt and autophagy pathways.
To analyze the PtdIns(3)P-binding roles of Atg18 and Atg21, we generated point mutations within their conserved Phe-Arg-Arg-Gly (FRRG) motifs. Neither Atg18FKKG nor Atg18FTTG (19) mutants completely abolished Pho8Δ60 activity (Fig. 1A), suggesting that the role of Atg18 in autophagy depends on more than its ability to bind PtdIns(3)P. However, it cannot be ruled out that these mutants still retain some level of PtdIns(3)P binding. One important finding in this study was that in _atg18_Δ _atg21_Δ cells expressing Atg18FKKG and Atg21FKKG, autophagy was severely reduced, suggesting that robust autophagy requires the PtdIns(3)P binding activity of at least one of these two proteins (Figs. 1 and 2). It is important to note that although the PtdIns(3)P binding activity of Atg21 can compensate for that of Atg18, the two proteins are not completely redundant, as the deletion of Atg18 cannot be rescued by Atg21, suggesting that in autophagy Atg18 has a function that is distinct from its ability to bind PtdIns(3)P.
Similar to the defect in nonspecific autophagy, we found that the majority of the Cvt pathway cargo protein prApe1 was sensitive to exogenously added protease in cells expressing Atg18FKKG and Atg21FKKG following spheroplast lysis (Fig. 3), suggesting that PtdIns(3)P binding by at least Atg18 or Atg21 is required for some stage of autophagosome assembly such as vesicle expansion or vesicle sealing.
In contrast to their wild-type counterparts, the Atg18FKKG-GFP and Atg21FKKG-GFP mutants were not localized to membrane structures but instead were primarily dispersed throughout the cytosol in both growing (supplemental Fig. S2_B_) and starvation conditions (supplemental Fig. S2_C_) (8, 18, 19), even in the presence of the other wild-type protein. This finding seems to disagree with the observation that either single mutant is largely normal for autophagy activity (Fig. 1); however, we found that Atg18 and Atg21 interact based on yeast two-hybrid studies and affinity isolation (supplemental Fig. S5). Thus, we hypothesize that some amount of PAS binding does occur with the single FKKG mutants, but the level may be below detection by fluorescence microscopy. Clearly, the efficient localization of Atg18 and Atg21 to the vacuolar membrane and other punctate structures is dependent on their respective PtdIns(3)P binding domains. The lipid-binding FRRG motif is also conserved in the human Atg18 homolog WIPI-1. WIPI-1 binds PtdIns(3)P and forms puncta in an autophagy-dependent manner in several cell lines, and the deletion of this four amino acid motif in WIPI-1 results in a mutant that has reduced PtdIns(3)P-binding and is incompetent for puncta formation (59).
Atg8 is a ubiquitin-like protein subject to lipid conjugation, whose steady state levels are significantly elevated during autophagy-inducing starvation conditions (32). Atg8–PE is one of the last phagophore markers to be recruited to the PAS during autophagosome formation (29), and the amount of Atg8 at the PAS controls the size of the autophagosome (40). Ag8-PE associates with the inner and outer membranes of the expanding phagophore; upon autophagic vesicle completion, Atg8–PE on the outer membrane of the autophagosome is cleaved by the action of the cysteine protease Atg4, whereas the Atg8 trapped in the inner membrane is degraded in the vacuole (40). Atg18 and Atg21 are required for the efficient recruitment to, and dissociation from, the PAS of Atg8 (Fig. 4). Although at this point the reason for the block in autophagosome formation in _atg18_Δ _atg21_Δ cells expressing the Atg18 FKKG and Atg21FKKG mutants is not clear, the autophagy defect is not simply due to the inability of these cells to form Atg8–PE (supplemental Fig. S3). Similarly, we found that Atg18 and Atg21, via their PtdIns(3)P-binding motifs, regulate the PAS recruitment of Atg16 (Fig. 5). Prior to this work, it was unclear how Atg16 is targeted to the PAS, as it requires PtdIns(3)P for its PAS localization but is reported to be independent of Atg18 (29).
Finally, we resolved an apparent discrepancy between in vitro and in vivo results concerning the regulation of the Atg4 protease. Previous in vitro studies suggest that Atg7 and Atg3 are sufficient for correct conjugation of Atg8 (60). Those analyses, however, rely on the use of Atg8ΔR and do not include Atg4, which is normally present in the cell. In contrast, Atg8–PE is not detected in an MKO strain expressing these same components (30). The latter system, which included Atg4, suggested that additional components prevent the second Atg4-dependent cleavage of Atg8–PE from occurring prematurely, an aspect of autophagy regulation that has not been understood. We now show that in the presence of Atg4, wild-type Atg18, and Atg21, but not Atg18FKKG and Atg21FKKG, mutants (along with the Atg12–Atg5-Atg16 conjugation system and the Atg6-Atg14 PtdIns 3-kinase components) protected the formation of punctate GFP-Atg8 (Fig. 6). Furthermore, under these conditions, we did not detect GFP-Atg8 localization on the vacuole rim, which is otherwise seen in the absence of Atg4. This may indicate that Atg16 by itself does not dictate the correct localization of Atg8 conjugation, rather, a combination of Atg16 and cleavage by Atg4 may be responsible for limiting Atg8–PE to the PAS. However, in wild-type cells lacking Atg18 and Atg21, GFP-Atg8 is present in puncta under starvation conditions (Fig. 4), suggesting that other components may also participate in protection of Atg8–PE from premature cleavage by Atg4.
In conclusion, we think that the second Atg4-dependent cleavage event, which removes Atg8 from PE, may be highly regulated and may ultimately determine the efficiency of the Cvt and autophagy pathways. One of the primary conclusions from these studies is that one mechanism for preventing premature cleavage of Atg8 from PE is by Atg18 and Atg21 regulating access to the substrate rather than by controlling Atg4 activity (Fig. 7). This role, in addition to their function in recruitment and release of Atg8 and Atg16 from the PAS, is dependent on the ability of Atg18 and Atg21 to bind PtdIns(3)P. In addition, although Atg18 and Atg21 do not have absolutely identical functions, their ability to bind PtdIns(3)P allows them to partially substitute for one another during autophagosome formation. Further information on the nature of phosphoinositide binding will be of interest because these proteins lack known lipid-binding motifs. Structural studies may also provide insight into the mechanism through which they regulate Atg4 cleavage of Atg8–PE.
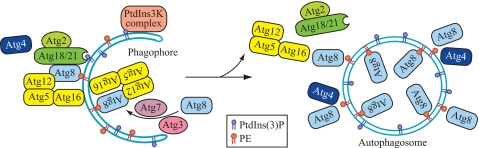
FIGURE 7.
Model for the function of Atg18 and Atg21 in regulating Atg4 access to Atg8–PE. Atg4 initially cleaves the C-terminal arginine of Atg8 (not depicted). Atg8 is subsequently conjugated to PE through the action of Atg7 and Atg3. The PtdIns 3-kinase complex generates PtdIns(3)P that allows recruitment of Atg18 (and its binding partner Atg2) and Atg21, which in turn are needed for efficient recruitment of Atg8 and Atg16; Atg18 and Atg21 also regulate the dissociation of these proteins from the PAS/phagophore. Atg16 at the PAS and subsequent cleavage by Atg4 at other sites may limit Atg8–PE to the PAS and the forming phagophore; the presence of Atg18 and Atg21 blocks access of Atg4 to Atg8–PE at the PAS/phagophore, preventing a premature second cleavage event. Following completion of the autophagosome, Atg18 and Atg21, along with the Atg12–Atg5-Atg16 complex (which is present as a tetramer) dissociate in a process that is probably regulated by Atg1, allowing Atg4 to cleave Atg8 from PE on the surface of the autophagosome.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Clinton Bartholomew, Wei-Lien Yen, Jiefei Geng, and Zhifen Yang for reagents and helpful suggestions.
*
This work was supported, in whole or in part, by National Institutes of Health Grant GM53396 (to D. J. K.).
4
The abbreviations used are:
PAS
phagophore assembly site
Cvt
cytoplasm to vacuole targeting
PtdIns(3)P
phosphatidylinositol 3-phosphate
PtdIns(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
ORF
open reading frame
PIPES
1,4-piperazinediethanesulfonic acid
GFP
green fluorescent protein
PE
phosphatidylethanolamine
MKO
multiple knock-out.
REFERENCES
- 1.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) J. Biol. Chem. 277, 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky D. J., Cueva R., Yaver D. S. (1992) J. Cell Biol. 119, 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth H., Meiling-Wesse K., Epple U. D., Thumm M. (2002) FEBS Lett. 512, 173–179 [DOI] [PubMed] [Google Scholar]
- 6.Barth H., Meiling-Wesse K., Epple U. D., Thumm M. (2001) FEBS Lett. 508, 23–28 [DOI] [PubMed] [Google Scholar]
- 7.Guan J., Stromhaug P. E., George M. D., Habibzadegah-Tari P., Bevan A., Dunn W. A., Jr., Klionsky D. J. (2001) Mol. Biol. Cell 12, 3821–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strømhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. (2004) Mol. Biol. Cell 15, 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krick R., Henke S., Tolstrup J., Thumm M. (2008) Autophagy 4, 896–910 [DOI] [PubMed] [Google Scholar]
- 10.Dove S. K., Piper R. C., McEwen R. K., Yu J. W., King M. C., Hughes D. C., Thuring J., Holmes A. B., Cooke F. T., Michell R. H., Parker P. J., Lemmon M. A. (2004) EMBO J. 23, 1922–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisenko O. N., Bomsztyk K. (1997) Mol. Cell. Biol. 17, 4707–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamura T., Sato S., Haque D., Liu L., Kaelin W. G., Jr., Conaway R. C., Conaway J. W. (1998) Genes Dev. 12, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Suprenant K. A. (1994) J. Biol. Chem. 269, 31777–31784 [PubMed] [Google Scholar]
- 14.Achsel T., Ahrens K., Brahms H., Teigelkamp S., Lührmann R. (1998) Mol. Cell. Biol. 18, 6756–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y., Benedict M. A., Wu D., Inohara N., Núñez G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4386–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D., Roberts R. (2001) Cell. Mol. Life Sci. 58, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith T. F., Gaitatzes C., Saxena K., Neer E. J. (1999) Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 18.Krick R., Tolstrup J., Appelles A., Henke S., Thumm M. (2006) FEBS Lett. 580, 4632–4638 [DOI] [PubMed] [Google Scholar]
- 19.Obara K., Sekito T., Niimi K., Ohsumi Y. (2008) J. Biol. Chem. 283, 23972–23980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efe J. A., Botelho R. J., Emr S. D. (2007) Mol. Biol. Cell 18, 4232–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. (1993) Science 260, 88–91 [DOI] [PubMed] [Google Scholar]
- 22.Katzmann D. J., Stefan C. J., Babst M., Emr S. D. (2003) J. Cell Biol. 162, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munn A. L., Riezman H. (1994) J. Cell Biol. 127, 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burd C. G., Emr S. D. (1998) Mol. Cell 2, 157–162 [DOI] [PubMed] [Google Scholar]
- 26.Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obara K., Noda T., Niimi K., Ohsumi Y. (2008) Genes Cells 13, 537–547 [DOI] [PubMed] [Google Scholar]
- 28.Obara K., Sekito T., Ohsumi Y. (2006) Mol. Biol. Cell 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 30.Cao Y., Cheong H., Song H., Klionsky D. J. (2008) J. Cell Biol. 182, 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W. P., Scott S. V., Kim J., Klionsky D. J. (2000) J. Biol. Chem. 275, 5845–5851 [DOI] [PubMed] [Google Scholar]
- 32.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 35.Voth W. P., Jiang Y. W., Stillman D. J. (2003) Yeast 20, 985–993 [DOI] [PubMed] [Google Scholar]
- 36.He C., Song H., Yorimitsu T., Monastyrska I., Yen W. L., Legakis J. E., Klionsky D. J. (2006) J. Cell Biol. 175, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noda T., Kim J., Huang W. P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. (2000) J. Cell Biol. 148, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Huang W. P., Klionsky D. J. (2001) J. Cell Biol. 152, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheong H., Nair U., Geng J., Klionsky D. J. (2008) Mol. Biol. Cell 19, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 44.Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. (2005) Mol. Biol. Cell 16, 5843–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meiling-Wesse K., Barth H., Voss C., Eskelinen E. L., Epple U. D., Thumm M. (2004) J. Biol. Chem. 279, 37741–37750 [DOI] [PubMed] [Google Scholar]
- 47.Reggiori F., Wang C. W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. (2004) Mol. Biol. Cell 15, 2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 49.Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. (1999) Mol. Biol. Cell 10, 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanida I., Tanida-Miyake E., Komatsu M., Ueno T., Kominami E. (2002) J. Biol. Chem. 277, 13739–13744 [DOI] [PubMed] [Google Scholar]
- 51.Tanida I., Tanida-Miyake E., Ueno T., Kominami E. (2001) J. Biol. Chem. 276, 1701–1706 [DOI] [PubMed] [Google Scholar]
- 52.Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. (2007) J. Biol. Chem. 282, 37298–37302 [DOI] [PubMed] [Google Scholar]
- 53.Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burda P., Padilla S. M., Sarkar S., Emr S. D. (2002) J. Cell Sci. 115, 3889–3900 [DOI] [PubMed] [Google Scholar]
- 55.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. (1998) Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 56.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. (2008) Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N., Noda T., Ohsumi Y. (1999) EMBO J. 18, 3888–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kametaka S., Okano T., Ohsumi M., Ohsumi Y. (1998) J. Biol. Chem. 273, 22284–22291 [DOI] [PubMed] [Google Scholar]
- 59.Proikas-Cezanne T., Ruckerbauer S., Stierhof Y. D., Berg C., Nordheim A. (2007) FEBS Lett. 581, 3396–3404 [DOI] [PubMed] [Google Scholar]
- 60.Ichimura Y., Imamura Y., Emoto K., Umeda M., Noda T., Ohsumi Y. (2004) J. Biol. Chem. 279, 40584–40592 [DOI] [PubMed] [Google Scholar]
- 61.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data