Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers (original) (raw)
Abstract
Clustering of multiple transcription factor binding sites (TFBSs) for the same transcription factor (TF) is a common feature of _cis_-regulatory modules in invertebrate animals, but the occurrence of such homotypic clusters of TFBSs (HCTs) in the human genome has remained largely unknown. To explore whether HCTs are also common in human and other vertebrates, we used known binding motifs for vertebrate TFs and a hidden Markov model–based approach to detect HCTs in the human, mouse, chicken, and fugu genomes, and examined their association with _cis_-regulatory modules. We found that evolutionarily conserved HCTs occupy nearly 2% of the human genome, with experimental evidence for individual TFs supporting their binding to predicted HCTs. More than half of the promoters of human genes contain HCTs, with a distribution around the transcription start site in agreement with the experimental data from the ENCODE project. In addition, almost half of the 487 experimentally validated developmental enhancers contain them as well—a number more than 25-fold larger than expected by chance. We also found evidence of negative selection acting on TFBSs within HCTs, as the conservation of TFBSs is stronger than the conservation of sequences separating them. The important role of HCTs as components of developmental enhancers is additionally supported by a strong correlation between HCTs and the binding of the enhancer-associated coactivator protein Ep300 (also known as p300). Experimental validation of HCT-containing elements in both zebrafish and mouse suggest that HCTs could be used to predict both the presence of enhancers and their tissue specificity, and are thus a feature that can be effectively used in deciphering the gene regulatory code. In conclusion, our results indicate that HCTs are a pervasive feature of human _cis_-regulatory modules and suggest that they play an important role in gene regulation in the human and other vertebrate genomes.
The completion of the sequencing of the human genome (International Human Genome Sequencing Consortium 2001; Venter et al. 2001) and of several other vertebrates that followed (Aparicio et al. 2002; Mouse Genome Sequencing Consortium 2002; International Chicken Genome Sequencing Consortium 2004) has spurred the discovery of protein coding genes, but deciphering their regulation remains a challenging task. Significant progress has been made in understanding regulation of genes in nonvertebrate organisms, such as the unicellular eukaryote yeast (Wagner 1999; Balaji et al. 2006; Narlikar et al. 2007) or the invertebrate Drosophila (Berman et al. 2002; Lifanov et al. 2003; Boeva et al. 2007), but the regulatory code of vertebrates has proven much more complex. Extensive experimental and comparative genomic studies have shown that the regulation of genes in vertebrates likely depends on a complex interplay between proximal and distal enhancers (Winklehner-Jennewein et al. 1998; Grehan et al. 2001; Luster and Rizzino 2003), locus control regions (Li et al. 1999), repressors (Hammond et al. 2005), silencers, and other gene regulatory elements (Arnone and Davidson 1997), as well as the cellular context and epigenetic markers (Wilson et al. 2008). Numerous studies have also shown that many regulatory elements are evolutionarily conserved and their sequences are usually enriched in various combinations of transcription factor binding sites (TFBSs) (Arnone and Davidson 1997; Levy et al. 2001; Nobrega et al. 2003; Woolfe et al. 2005; Pennacchio et al. 2006; Pennacchio et al. 2007). While the co-occurrence of diverse TFBSs has been extensively studied and used for predicting novel regulatory elements and their function, grouping of binding sites for the same transcription factor (TF) into homotypic clusters of TFBSs (HCTs) has been studied and observed almost exclusively in invertebrates, especially in Drosophila (Lifanov et al. 2003). Evidence for functional contribution of HCTs to human regulatory elements remains rather limited (Murakami et al. 2004; Hu et al. 2007), despite the fact that HCTs present several mechanistic advantages. These include favoring lateral diffusion of a TF binding along a regulatory region (Kim et al. 1987; Khoury et al. 1990; Coleman and Pugh 1995), favoring high-affinity cooperative binding of some TFs (Hertel et al. 1997), and providing functional redundancy (Somma et al. 1991; Papatsenko et al. 2002). The lack of a genome-wide map of HCTs in vertebrates prompted us to test if HCTs might be associated with vertebrate _cis_-regulatory modules (CRMs). Indeed, the 126,045 HCTs defined in the human genome with 273 PWMs (see Methods) are strongly associated with promoters and distant-acting vertebrate enhancers, with half of all human promoters and known distant enhancers containing at least one HCT. In promoters, HCTs have a normal density distribution centered on the transcription start site (TSS), in agreement with binding density distributions of a few TFs experimentally determined by the ENCODE project (The ENCODE Project Consortium 2007). HCTs that are deeply conserved in the vertebrate lineage target preferentially TF-encoding genes, indicating an ancestral origin of the HCT-based regulation of TFs. HCTs are also 25-fold overrepresented in experimentally validated developmental enhancers, and are strongly associated with the transcriptional co-activator Ep300 protein, with the HCTs of certain TFs displaying significant tissue-specific biases. These findings support the hypothesis that HCTs are key components of vertebrate CRMs and play an important role in vertebrate development and gene regulation.
Results
Widespread distribution of HCTs in vertebrate genomes
To identify vertebrate HCTs, we initially screened human, mouse, chicken, and fugu genomes using known DNA binding motifs of vertebrate TFs, a total of 701 position weight matrices (PWMs) from TRANSFAC and JASPAR databases (see Methods). We used a hidden Markov model (HMM) (see Methods for details) trained for each PWM and genome independently to generate ranked lists of PWM HCTs, from which we retained, at most, 10,000 top scoring HCTs for each PWM for further analyses. After further eliminating HCTs longer than 3 kb and those with more than 25% of the TFBSs located in coding exons (see Methods), we found that the number of HCTs varied widely among different PWMs (for 189 PWMs we found more than 5000 HCTs), but we did not detect a correlation between the number of HCTs and either TFBS length or PWM information content (Supplemental Fig. S1). Assuming that evolutionary persistence of an HCT is an indicator of function, we further restricted our data set only to HCTs that are conserved between human and mouse (for conservation definition, see Methods). Examining intrafamilial TFBS clustering, we observed large fluctuations in the number of HCTs for TFs with similar PWMs from the same TF family, indicating that small variations in binding specificity can strongly modulate TF binding on a genome scale. For example, ZIC1, ZIC2, and ZIC3 have very similar PWMs as indicated by the intermotif distance (Narlikar et al. 2007), but the number of HCTs associated with each one of them varies over threefold (Supplemental Table S2). In addition, only a few ZIC HCTs overlap—only 40% of ZIC2 HCTs and 26.4% of ZIC3 HCTs overlap with ZIC1 HCTs, for example. The interfamilial variation in TFBS clustering prompted us to remove PWM redundancy for individual TFs, but not TF families. Among the multiple PWMs associated with the same TF, we retained that for which we found the most HCTs, resulting in a final data set of 273 PWMs (for details, see Supplemental Table S2). A total of 126,405 HCTs were retained, with a median length of 597 bp and a median of five TFBSs per HCT. The entire set of HCTs spans 50.4 Mb (1.6% of the human genome), a span that is comparable to that of protein coding exons. The majority of HCTs (77.6%) do not overlap protein-coding exons, while only 22.4% among HCTs are located in regions that also contain protein-coding exons (the vast majority of exons are located in between TFBSs, and only a limited number of TFBSs are allowed to overlap exons; see Methods). Most PWMs (208) are represented by less than 500 HCTs each, while 33 PWMs have more than 1000 HCTs (Fig. 1). The latter group of 12% TFs accounts for 53.4% of HCTs and corresponds to TFs such as E2F1, known to bind to the promoters of thousands of genes (Bieda et al. 2006). We further tested whether HCTs could provide increased specificity in predicting sequences that are likely to be bound by the corresponding TF relative to the presence of a single TFBS. We compared the fraction of evolutionarily conserved regions (ECRs) overlapping at least one TFBS with the fraction of ECRs overlapping at least one HCT (we used the entire set of human–mouse ECRs as reference sequences to account for the fact that our final set of HCTs is defined using human-mouse ECRs). We found that the number of ECRs with at least one TFBS was, on average, 372-fold higher than the number of ECRs overlapping HCTs (for the 273 PWMs considered), indicating that HCTs provide a much more specific measure for the presence of TFBSs in conserved regions.
Figure 1.

The distribution of the HCTs abundance for the 273 PWMs. The number of PWMs with abundant HCTs (>1000) is limited to 33, the best represented TFs being E2F1 (PWM: V$E2F_Q2; 5194 HCTs), ZFP161 (V$ZF5_01; 4844), and TEAD2 (V$ETF_Q6; 4382).
Computationally predicted HCTs agree with experimentally determined TFBSs
To verify whether HCTs correspond to regions actively bound by TFs, we used chromatin immunoprecipitation with microarray hybridization (ChIP-chip) and chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) experimental data available for several TFs: REST (Johnson et al. 2007), YY1 (Xi et al. 2007), STAT1 (Robertson et al. 2007), and E2F1, E2F4, MYC (The ENCODE Project Consortium 2007). In the case of E2F1, E2F4, and MYC, experimental data, and therefore our analysis, were limited to the ENCODE regions (The ENCODE Project Consortium 2007). We evaluated the fraction of HCTs that correspond to regions bound by the corresponding TF by overlapping the genomic coordinates of HCTs with those corresponding to regions bound by TFs. In all cases we observed a highly significant enrichment of experimentally determined binding sites at the respective computationally identified HCTs (Table 1). The strongest association was observed for E2F1, for which 65% of the HCTs overlapped experimentally validated binding sites, representing a 13-fold enrichment relative to the expectation of random overlaps. These results indicate that a large fraction of predicted HCTs are bound by the corresponding TF in cells.
Table 1.
Regions experimentally found to be bound by TFs overlap significantly with regions covered by HCTs
Additionally, the function of enhancers, and implicitly the binding of the corresponding TFs, has been associated with various epigenetic modifications, such as the acetylation of the core histone 3 (H3) and the monomethylation of H3K4 (Roh et al. 2005, 2007; Heintzman et al. 2007). To test if HCT-containing regions reflect epigenetic modifications specific to regulatory elements, we compared (see Methods) the location of HCTs with that of the histone modification patterns associated with gene activation and repression in CD4+ T cells (Barski et al. 2007). We found that 128 PWMs (47%) are significantly overrepresented in regions associated with gene activation (Supplemental Fig. S3), suggesting that these HCTs play an active role in gene regulation. We also found that HCTs of 60 PWMs (22%) overlap significantly regions linked to gene repression, however to a much smaller extent (Supplemental Fig. S3). These data suggest that HCTs play a significant role in gene regulation, although the specific overlap pattern observed in CD4+ T cells, for which the histone methylation patterns were recorded (Barski et al. 2007), is likely to be different in other tissues.
HCTs are predominantly located in promoters
To investigate how HCTs are positioned relative to genes, we surveyed the positions of all HCTs and found that most of them (38.6%) are located in promoter regions, while the rest were evenly distributed between intergenic (30.7%) and genic regions excluding promoters (30.7%). Given the span of promoters relative to other genomic regions, the promoter occupancy is more than 10-fold higher than expected (P = 0, binomial test), indicating a strong association of HCTs with proximal promoters. By constructing HCT gene coverage profiles (Fig. 2), we observed a symmetric well-defined peak in the HCT density centered on the TSS. This is consistent with the ENCODE project TF-binding data, which showed a similar symmetrical binding profile around the TSS for E2F1, E2F4, and MYC (The ENCODE Project Consortium 2007).
Figure 2.
Coverage of protein-coding genes by E2F HCTs. The coverage profile reveals a clear peak with a symmetric distribution around the transcription start site (TSS). Gene features are represented proportional to the median values of all human protein coding genes: 5′ UTR, 181 nt; first coding exon, 124; first intron, 2682; internal coding exon, 123 (multiple internal exons, as well as introns, are pooled together); internal intron, 1419; last coding exon, 149; 3′ UTR, 751. The promoter region is considered to be 1.5 kb in all cases. Depending on the length of the various gene features at a particular locus, HCTs could occasionally reach internal introns and exons. In the case of genes with split 5′ UTR regions, the first intron is likely to be covered by a promoter-based HCT, resulting, as in this case, in a higher coverage relative to the first coding exon.
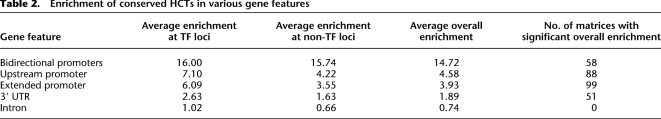
We further analyzed the enrichment of HCTs in promoters, UTRs, and introns. We found that HCTs of 99 out of 273 PWMs (36.3%) are significantly enriched in promoters (Table 2), 30 among them having more than half of their HCTs located in promoters (e.g., 75% for TEAD2, 73% for E2F1). The HCTs of 99 promoter-associated PWMs overlap 53% of all annotated promoters in the human genome, indicating the extensive use of HCTs by proximal gene regulatory elements. We observed that the most pronounced fold-enrichment (>14-fold) occurs in bidirectional promoters, with 61% of bidirectional promoters containing at least one HCT. This indicates that the genes under the control of a bidirectional promoter rely on HCTs significantly more than unidirectional promoters (P = 5 × 10−4, Fisher's exact test). Additionally, we found no enrichment in intronic regions, and only a moderate enrichment in 3′ UTR regions (less than twofold).
Table 2.
Enrichment of conserved HCTs in various gene features
The preference of many PWMs toward promoters should be reflected in the sequence composition of the HCTs themselves. Specifically, their sequence should have a high GC content, as it is known that promoter regions of many genes are GC-rich and enriched in CpG islands (CGIs) (Larsen et al. 1992). Indeed, we found that many HCTs are located in GC-rich regions, especially those of PWMs with abundant (>1000) HCTs (Fig. 3; Supplemental Table S2). These all have GC content higher than 60%, with the exception of PDX1 HCTs, which have 33.6% GC content. While the GC content is relatively constant throughout the span of HCTs, it differs strikingly from the GC content of flanking sequences. The high GC content appears to be specific only to the location of HCTs, and drops sharply to the genome average of ∼45% within 1 kb of flanking sequence (Fig. 4). To test whether the enrichment of HCTs in promoter regions simply reflects a TFBS recognition bias due to the PWM sequence composition, we constructed a set of HCTs defined with binding sites recognized by randomized PWMs that preserve the GC content, but distort the sequence recognition of the original PWM (see Methods). We found that only four out of 32 PWMs with GC-rich and abundant HCTs have a CGI preference of randomized PWMs not significantly different from the real HCTs (Supplemental Table S1). Furthermore, we tested (see Methods) whether the enrichment in promoter regions is due to the enrichment of CGIs themselves near promoters (Adachi and Lieber 2002). We found that the presence of CGIs alone cannot completely explain the enrichment observed in promoters for any of the 99 PWMs with HCTs enriched in promoters, even though the HCTs of some PWMs are tightly linked to the presence of CGIs (Supplemental Table S2). Similarly to the case of promoters, we tested whether the enrichment observed in bidirectional promoters is simply due to the presence of CGIs, and found that CGIs cannot completely explain the enrichment observed for any of the 58 PWMs with HCTs enriched in bidirectional promoters. These results indicate that the promoter preference observed for certain HCTs is not due to the GC content of the defining PWM or the number of CpG dinucleotides in their consensus sequence, and the alternative hypothesis of PWMs recognizing functional binding sites, which are enriched in promoter regions, should be favored.
Figure 3.
Bimodal distribution of the HCT count for 273 PWMs with respect to the GC content of HCTs. PWMs with more than 400 HCTs (28.6%, black) have HCTs that are either AT-rich (5.5%) or GC-rich (23.1%). The GC content was calculated for the entire span of the HCT, but avoiding coding and repetitive sequences.
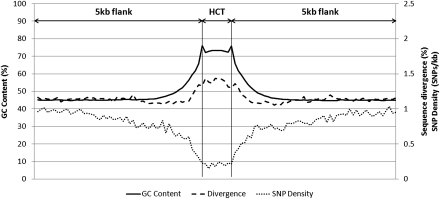
Figure 4.
GC content, SNP density, and human–chimp divergence profiles of 5194 E2F1 HCTs. The GC content and the divergence from chimp increase, while the SNP density decreases sharply in HCTs as compared to flanking regions, which show values similar to the genome average. The increased GC content at the beginning and end of the HCT is due to the first and last E2F1 TFBSs that define the HCT, whereas the position of the other TFBSs is variable. All values were computed by avoiding coding and repetitive sequences.
TFBSs in conserved HCTs evolve under purifying selection
Our assumption of gene regulatory activity associated with regions containing HCTs implies active binding to and functional relevance of individual TFBS within HCTs. To test this hypothesis, we investigated if TFBSs in HCTs are subject to an elevated evolutionary constraint compared to the rest of the HCT sequence. We compared the mutational pattern in TFBSs with that in sequences located in between TFBSs (intersites) within HCTs (see Methods). First, we found that the sequence divergence observed in TFBSs is lower than the divergence observed in intersites for the majority (83%) of PWMs (Fig. 5), providing evidence of negative selection acting specifically on TFBS and, thus, supporting the hypothesis that identified TFBSs are biologically active. Additionally, we found that HCTs display an overall divergence different from that observed in flanking sequences, higher in HCTs with high GC content (Fig. 5), and lower in HCTs with lower GC content (Supplemental Fig. S4).
Figure 5.
Divergence in TFBSs and intersite sequences within HCTs for 273 PWMs. The sequence divergence in human–chimp comparisons indicates that TFBSs tend to be more conserved than intersite sequences for HCTs of most PWMs (86.4%). For 31 of them (red) the difference is significant (after correction for multiple testing).
In addition to the GC content and divergence, we also analyzed the single nucleotide polymorphism (SNP) density in HCTs. First, we found that similarly to the GC content and divergence, the SNP density in HCTs is roughly constant throughout the HCT, but is strikingly different from the flanking sequences. For example, the overall SNP density in HCTs for E2F1 (0.2 SNPs/kb) is comparable to the SNP density in ultraconserved regions in the human genome (Ovcharenko 2008). This value is fivefold lower than that observed in flanking sequences (Fig. 4) and the overall SNP density in promoter regions (0.96 SNPs/kb). We attributed this rapid decrease in SNP density to the SNP ascertainment bias resulting in SNP density being strongly correlated with GC content, so that the GC content of HCTs is sufficient to explain the drop in SNP density (Supplemental Fig. S5). To provide an unbiased assessment of SNP density, we had to restrict the analysis to regions with very similar sequence composition. Therefore, similarly to the sequence divergence analysis, we compared SNP density in TFBS to sequences within HCTs located between TFBS. We found that the SNP density is lower in TFBSs than in intersites for most PWMs (68%) (Supplemental Fig. S6). However, because of the low SNP counts in TFBSs (146 out of 273 PWMs have less than 10 SNPs in TFBSs) we failed to detect any significant shift in derived allele frequencies (DAF), which would have pointed to negative selection if a shift toward a rare allele is observed, or to positive selection in the case of a shift toward common alleles (Fay et al. 2001). Nonetheless, the combined evidence from lower sequence divergence and decreased SNP density indicates that TFBSs within HCTs evolve under purifying selection.
Regulation of transcription factors relies on HCTs
To address the functional specificity of genes relying on HCTs for their regulation, we tested whether particular Gene Ontology (GO) categories (Ashburner et al. 2000) are enriched in the set of 9599 genes with HCTs in promoters. “Protein binding” and “transcription factor activity” were identified as the two most significantly enriched molecular function GO categories (P = 8.5 × 10−54 and 3.98 × 10−29 after multiple testing correction, respectively; significance of enrichment for GO categories was evaluated using the hypergeometric test, as described in Methods). Twelve other GO categories related to TF activity were also significantly enriched in this gene set, including “nucleotide binding” (P = 3.49 × 10−22), “sequence-specific DNA binding” (P = 1.36 × 10−18), and “regulation of transcription, DNA dependent” (P = 2.5 × 10−13). A significant fraction of human TF genes (62%, P = 2 × 10−25) have at least one HCT in the promoter region, indicating that TFs use HCTs extensively for their regulation, consistent with recent ChIP-chip binding analyses of individual TFs (Rabinovich et al. 2008). By separating TFs from the remaining human genes, we found that the HCT enrichment in promoters of TF gene loci is, on average, twice as high as for other loci (Table 2), reinforcing the strong association between promoter-based HCTs and TFs.
We further investigated the connection between HCTs and TFs at deeper levels of conservation, using HCTs that are also conserved in chicken and fugu. This analysis confirmed a significant association between TFs and HCTs with the increase of evolutionary separation of species—not only that TFs are significantly overrepresented in all target gene sets (Table 3), but also that their participation increases in the genes targeted by deeply conserved clusters. This suggests an ancestral nature of HCT-based regulation of TFs. For example, in the case of HCTs conserved between human, mouse, chicken, and fugu, the fraction of TFs in the target gene set is more than doubled (31%) compared to that of HCTs conserved between human and mouse (15.1%). Table 4, which contains all significant GO categories, illustrates that in the case of deeply conserved clusters the single and most important overrepresented category of target genes is transcription factors, indicating that the regulation of TFs during vertebrate evolution is strongly associated with the presence of HCTs at their promoters.
Table 3.
Enrichment of TFs in the set of genes harboring conserved HCTs at their promoters
Table 4.
GO categories with significant enrichment (after Bonferroni correction for multiple testing) in the set of 128 genes with HMCF-conserved clusters in their promoters
Abundant HCTs in developmental enhancers
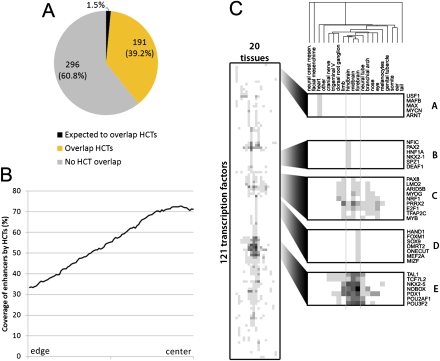
To assess whether conserved HCTs are also found at distal regulatory elements, we analyzed the relationship between conserved HCTs and distant-acting developmental enhancers for which reproducible tissue-specific activity has been shown in vivo in E11.5 mouse embryos (Pennacchio et al. 2006). This data set consists of 487 human noncoding sequences, the majority of which are conserved between human, mouse, chicken, frog, and fish. By overlapping their chromosomal coordinates with HCTs, we observe that 191 (39.2%) enhancers contain at least one conserved HCT, a fraction more than 25-fold larger than expected by chance (Fig. 6A). We note that HCTs are located toward the center of the enhancer elements (Fig. 6B). This agrees with the active core of regulatory elements being observed in the center of a conserved element (Ovcharenko et al. 2004b; Prabhakar et al. 2006), and with the fact that flanking sequences of up to several hundred base pairs were added to the elements tested for enhancer activity in mouse transgenic assays (Pennacchio et al. 2006).
Figure 6.
Developmental enhancers overlap conserved HCTs. (A) The fraction of enhancers functional in mouse (487) that overlap with at least three sites in HCTs (39.2%, yellow) is significantly higher (P < 8.5 × 10−102) than what is expected by chance (1.5%, black). The fraction increases to above 50% if a lower number of TFBSs in an HCT are allowed for an HCT-enhancer overlap. (B) Coverage of positive enhancers by HCTs shows that HCTs are located toward the center of the enhancer. (C) HCTs of different transcription factors are associated with enhancers that show tissue specific, e.g., heart (A), hindbrain (B), or forebrain (D), or a more ubiquitous expression pattern (C,E) in E11.5 mouse embryos. The shade of gray is proportional to the number of tissue-specific enhancers overlapping HCTs for a specific TF, with the lightest shade of gray indicating one, and black indicating 10 (only observed for NOBOX and forebrain-specific enhancers) overlap instances. The tissues and the corresponding number of enhancers found to be active in them (indicated in parentheses) are: Mesenchyme derived from neural crest (1), facial mesenchyme (6), heart (20), other (22), cranial nerve (25), trigeminal V (ganglion, cranial) (18), dorsal root ganglion (34), limb (79), hindbrain (rhombencephalon) (142), midbrain (mesencephalon) (148), forebrain (156), neural tube (103), branchial arch (22), nose (20), eye (32), melanocytes (3), genital tubercle (3), somite (7), ear (6), and tail (2).
To test whether the HCT enrichment observed in enhancer elements is simply due to elevated conservation of tested enhancers across several vertebrate species, we compared the presence of conserved HCTs in nonrepetitive ECRs across the entire human genome with that observed for ECRs located in enhancer elements. We performed separate analyses for the sets of human–mouse, human–chicken, and human–fugu ECRs. In each case we found that the fraction of ECRs outside of enhancer elements that overlap with conserved HCTs is significantly lower than the fraction of ECRs located within enhancer elements that overlap conserved HCTs (mouse: sixfold, P = 2.1 × 10−80; chicken: 2.7-fold, P = 1.3 × 10−41; fugu: 1.6-fold, P = 1.05 × 10−7, Fisher's exact test). This indicates that the HCT enrichment we observe in enhancer elements is not due to the presence of conserved sequences, but it is likely due to the biological role of HCTs.
The experimental validation of the developmental enhancers recorded not only the binary outcome (positive or negative) of the transgenic assay for each element, but it also recorded the tissues where each element was observed to drive expression (Pennacchio et al. 2006). This allowed us to link each TF to enhancer elements containing its HCTs, and further to the tissues where those elements are active. Using this data, we grouped TFs into clusters of similar tissue-specific expression (Fig. 6C). Seventy-two percent of enhancers overlapping HCTs exhibit activity in either brain (forebrain, midbrain, hindbrain) or neural tube. This tissue specificity is characteristic to the entire enhancer data set, as a similar association with brain activity was observed in the set of elements that do not overlap HCTs (67.6%, P = 0.31, Fisher's exact test). For some TFs, such as POU3F2, the expression pattern of HCT targets agrees with the known function of the TF. POU3F2 is known to play an important role in mammalian neurogenesis (Atanasoski et al. 1995), and we observe POU3F2 HCTs in enhancers driving expression almost exclusively in neural tissues (forebrain, midbrain, hindbrain, neural tube, dorsal root ganglion, and nose). For some TFs, such as E2F1, E2F4, and PDX1, we observe a broad expression pattern in several tissues (Fig. 7C,E), while for others we observe expression activity restricted to only one tissue; e.g., MYCN, ARNT, USF1, and MAX are associated only with expression in heart (Fig. 6C,A), in agreement with known expression patterns for some of these TFs, such as MYCN (Jakobovits et al. 1985). However, the strongest TF-tissue-specificity association was observed between NOBOX and forebrain expression (P = 0.01, Fisher's exact test). Overall, the unexpectedly high overlap between positive enhancer elements and HCTs indicates that the latter may play an important role in the development by being an integral part of distal enhancers.
Figure 7.
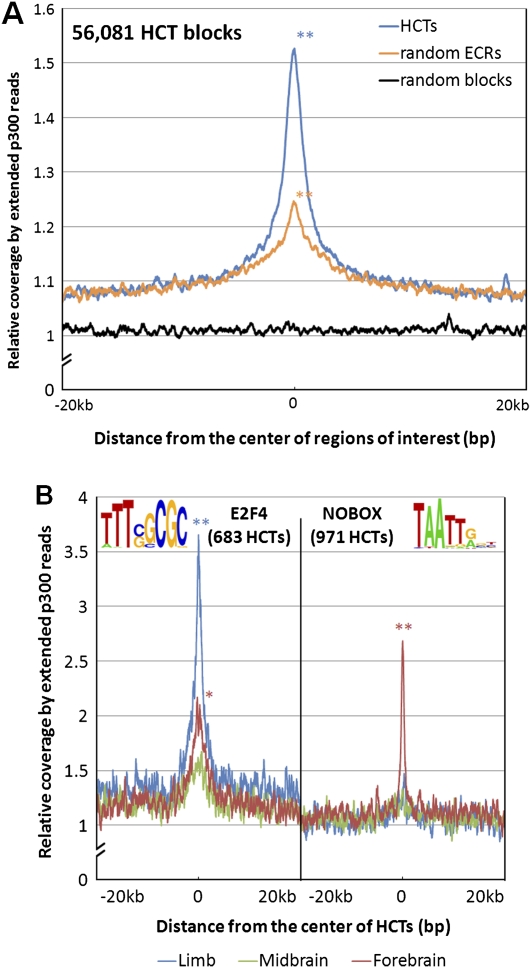
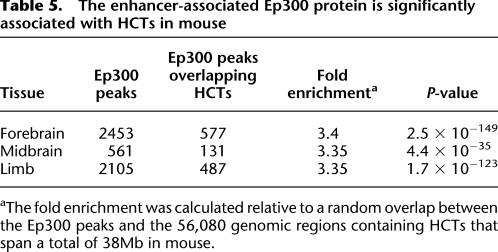
The enhancer activity of HCTs is supported by their association with the enhancer coactivator protein Ep300 in mouse. (A) Mouse HCTs corresponding to all human HCTs for 273 PWMs were combined into 56,081 regions (median size, 549 bp). Their Ep300 coverage profile was constructed by overlapping a total of 9,519,543 Ep300 reads with 40-kb regions centered on the middle point of HCTs. Even though ECRs are known to be significantly associated with Ep300 (P = 0), HCTs show an even stronger association by the means of a significantly higher Ep300 peak (P = 0). (B) Ep300 coverage profiles for HCTs of specific TFs and tissues reveal tissue-specific activity for certain TFs. For example, the coverage profile of 683 E2F4 HCTs reveals a peak significantly higher in limb than in either forebrain (P = 1.4 × 10−21) or midbrain (P = 1.2 × 10−40), while the difference between the forebrain and midbrain coverage is only marginally significant (P = 9.7 × 10−6). In the case of the 971 NOBOX HCTs, their coverage is significantly higher in forebrain than in either limb (P = 2.7 × 10−27) or midbrain (P = 7.3 × 10−43), while limb and midbrain coverage are not significantly different from each other (P = 0.07). These data strongly suggest a limb-specific activity for E2F4 HCTs, and a forebrain-specific activity for NOBOX HCTs. Statistical significance was evaluated with Fisher's exact test, as in Visel et al. (2009);*P < 0.01; **P < 10−20.
The important HCT contribution to enhancer elements is further reinforced by the significant colocalization of HCTs with the enhancer-associated Ep300 protein. The genomic locations where Ep300 was found to bind were recently described in mouse for three types of embryonic tissue (forebrain, midbrain, and limb) (Visel et al. 2009). By overlapping mouse Ep300 peaks with the mouse homologs of the human HCTs we found an enrichment that is threefold higher than expected (Table 5). Also, the extended Ep300 ChIP-seq read coverage profile of all HCTs (Fig. 7A) indicates a stronger association between HCTs and Ep300 than between other ECRs and Ep300. Individual coverage profiles with the tissue-specific Ep300 reads reveal potential tissue-specific activity for certain TFs, such as limb-specific activity for E2F4 HCTs, or forebrain-specific activity for NOBOX HCTs (Fig. 7B). The latter reinforces the association between NOBOX and forebrain specificity observed with the set of human enhancers. In fact, the association between NOBOX and forebrain is the strongest among any TF-tissue pair when estimated only using Ep300 peaks (25-fold difference, P = 2.99 × 10−50, Fisher's exact test). Given that Ep300 is a protein associated with enhancers, this strongly supports the hypothesis that HCTs are an integral part of developmental enhancers.
Table 5.
The enhancer-associated Ep300 protein is significantly associated with HCTs in mouse
Experimental testing of the HCT enhancer activity
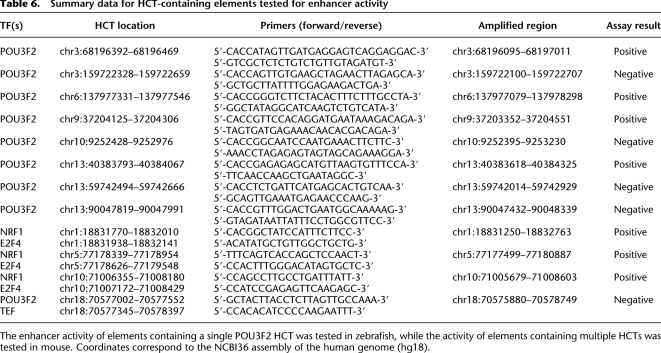
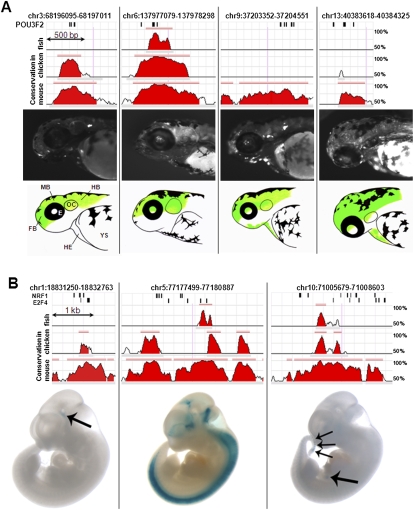
The strong association between HCTs and experimentally validated enhancers suggests that HCTs have the potential to predict the location of enhancers. To test this hypothesis, we used an in vivo reporter assay in zebrafish to record the enhancer activity of eight putative enhancers (Table 6) containing HCTs for POU3F2 (also known as OCT7). POU3F2 is the TF most highly represented among the TFs with more than five HCTs overlapping experimentally validated enhancers (13 HCTs out of 536, 2.4%). Moreover, all 13 enhancers that contain POU3F2 HCTs are active in the central nervous system (forebrain, midbrain, hindbrain, or neural tube), in agreement with the known function of the POU TFs. Therefore, POU3F2 represents a suitable candidate for testing the enhancer prediction potential of HCTs. Among the POU3F2 HCTs that do not overlap promoters, HCTs for other TFs, or elements previously tested for enhancer activity, we selected the HCTs with the highest HMM clustering score. Additionally, to assess the influence of distant evolutionary conservation, we selected four elements that are conserved in chicken, and four that are not (Table 6). Zebrafish was chosen as a testing system because developing zebrafish embryos are abundantly available and transparent, allowing direct visualization of reporter gene expression throughout development, and therefore are an ideal system for enhancer properties screening of sequences of unknown spatial and temporal specificities (Lieschke and Currie 2007). Each element was PCR-amplified from the human genome (Table 6) and cloned in an EGFP reporter cassette, driven by a minimal mouse Fos promoter, as described previously (Fisher et al. 2006a,b). Each construct was injected in 100–200 two-cell stage zebrafish embryos. We evaluated the injected G0 developing embryos for GFP expression at 24, 48, 72, and 96 h post-fertilization. For a construct to be classified as an enhancer we required a minimum of 20% of developing fish expressing GFP in a consistent spatial pattern, comparable with rates previously reported (McGaughey et al. 2009). Of the eight elements tested, four displayed consistent enhancer properties, with GFP expression throughout various domains of the central nervous system and, for one construct, the developing heart (Fig. 8A), in agreement with the known role of POU TFs in the development of the nervous system (Schreiber et al. 1993; Atanasoski et al. 1995; Jin et al. 2009) and the function of the 13 enhancers experimentally validated in mouse that contain POU3F2 HCTs (Fig. 6C). This suggests that HCTs do have the potential to predict the location of enhancers, even though larger experimental samples will be required to quantify the success rate of HCT-based prediction more accurately. We also observed that among the four HCTs that had enhancer activity in zebrafish, two were conserved in chicken and two were not. The same 1:1 ratio of elements deeply conserved and nondeeply conserved was observed for the set of elements that did not have enhancer activity in zebrafish. This suggests that the potential of HCTs to predict enhancers correctly does not strongly correlate with the presence of deep evolutionary conservation.
Table 6.
Summary data for HCT-containing elements tested for enhancer activity
Figure 8.
Experimental validation of predicted enhancers. (A) Four constructs containing POU3F2 HCTs that produced reproducible expression patterns of GFP in zebrafish. The sequence conservation profiles for mouse, chicken, and zebrafish represented as ECR Browser screen shots, correspond to the entire amplified regions (Table 6). Positions of the POU3F2 binding sites are represented by short vertical black bars above the conservation profile. All four elements are represented at the same scale. Pictures of 48–72-h post-fertilization zebrafish embryos with representative GFP expression patterns are shown below each element. The corresponding spatial domains of expression of each enhancer are also diagrammatically illustrated (regions where enhancer activity was recorded are shown in green). FB, forebrain; MB, midbrain; HB, hindbrain; E, eye; OC, otic capsule; HE, heart; YS, yolk sac. (B) Three constructs containing HCTs for NRF1 and E2F4 that produced reproducible LacZ expression pattern in transgenic mice. All three elements are represented at the same scale (this is different from the scale of elements tested in zebrafish). Arrows point to specific organs where the activity of these enhancers was observed, namely diencephalon for the enhancer on chromosome 1, and pancreas and caudal somites for the enhancer located in chromosome 10. Additional replicates for all elements presented here are included in the Supplemental material.
In contrast to the eight POU3F2 HCTs, which do not overlap HCTs for other TFs, the majority (69.6%) of HCTs are located in regions where other HCTs could be detected, raising the possibility of gene expression coactivation when a particular combination of cognate TFs is present and bound, and thus could imply tissue specific activity. To test the enhancer activity associated with co-occurring HCTs, we used an in vivo transgenic mouse assay (Pennacchio et al. 2006), a validation system complementary to zebrafish, which benefits from the shorter evolutionary distance between human and mouse. We decided to investigate the enhancer activity of regions containing HCTs for the nuclear respiratory factor (NRF1) and the E2F transcription factor 4 (E2F4), whose HCTs show a moderate co-occurrence (21% of the E2F4 HCTs overlap with 6% of the NRF1 HCTs). While NRF1 plays a role in metabolism, nuclear respiration, and cellular growth, E2F4 is associated with tumor suppression, and recent evidence indicates that together they might target a common set of genes (Cam et al. 2004). Among the 148 genome-wide occurrences of combinatorial E2F4/NRF1-HCTs, we randomly selected three regions that do not overlap promoters and regions previously tested for enhancer activity. One HCT pair is located in the first intron of the paired box 7 gene (PAX7) on chromosome 1, the second one is located in an intergenic region more than 67 kb upstream of the TBCA gene on chromosome 5, and the third is located 2.6 kb upstream of the NEUROG3 gene on chromosome 10 (Fig. 8B; Table 6). The mouse transgenic assays revealed that all three elements show enhancer activity with high reproducibility. The enhancer located in the first intron of the PAX7 gene showed reproducible expression in the same subregion of the brain (Fig. 8B) in seven out of eight embryos, suggesting that transcriptional activation by this element contributes to the previously reported PAX7 activity in the brain (Ziman and Kay 1998). The second element showed a reproducible compound pattern that includes subregions of the forebrain, midbrain, and hindbrain, as well as much of the neural tube in four out of five embryos. Finally, the element located upstream of NEUROG3 showed highly reproducible expression in pancreas and caudal somites in four out of five embryos, indicating that this element, in combination with a minimal promoter, is sufficient to drive pancreas-specific gene expression in embryos and does not require (but might complement or enhance) the proximal NEUROG3 promoter region used in previous studies to maintain this activity (Lee et al. 2001). Overall, these results indicate that the presence of multiple HCTs in the same region appear to be a strong predictor for enhancer function. However, we tested an additional element, which contains HCTs for POU3F2 (whose HCTs have enhancer capacity as shown by zebrafish transgenic assays) and TEF. This element is located in the second intron of the ZNF407 gene on chromosome 18, and did not show reproducible reporter expression at E11.5, indicating that the presence of multiple HCTs does not guarantee the enhancer capability of the tested regions. We also note that despite a positive outcome, the three tested enhancers containing HCTs for NRF1 and E2F4 have very different expression patterns, suggesting that the tissue-specific activity depends on more than the presence of a specific combination of HCTs, as TFBSs for additional TFs could be present in those regions without being identified by our approach as forming HCTs.
While large-scale in vivo studies will be required to validate the overall effectiveness of HCT-based enhancer predictions, the limited number of examples studied here, together with data from previous studies, suggests that the presence of HCTs may provide a viable computational strategy to identify high confidence enhancer candidate regions in genomes.
Discussion
In this study, we analyzed the distribution of evolutionarily conserved HCTs in vertebrate genomes and found that similarly to invertebrates, HCTs are also numerous in the human genome. Our analysis indicates that HCTs are an important component of proximal promoter regulatory elements, as well as of distal developmental enhancers. This suggests that the use of HCTs in gene regulation is widely spread not only in invertebrates, but also in the vertebrate clade, possibly due to their functional advantages as they facilitate recruitment of TFs and are functionally redundant (Somma et al. 1991; Papatsenko et al. 2002; Lifanov et al. 2003).
We observe that abundant HCTs are located in regions that differ sharply from flanking regions in terms of their sequence composition (Fig. 5), which is consistent with the idea that a contrasting sequence environment is beneficial in the process of TF recruitment (Lifanov et al. 2003). At the same time, regions with reduced sequence complexity, such as GC-rich and GC-poor regions, which characterize abundant HCTs (Fig. 4), could be regarded as a constant source of TFBSs that can be obtained through relatively few mutations. This could be a factor contributing to the evolutionary stability of HCTs in such regions. A previous study by Sethupathy et al. (2008) indicates that human- and primate-specific TFBSs in promoter regions are likely to be subject to positive selection, which is consistent with the idea of TFBSs being created in these regions. Our analysis indicates that TFBSs within an evolutionarily conserved HCT are more conserved than sequences separating them; thus, they appear to be evolving under negative selection. These two complementary views suggest that the evolutionary stability of HCTs is the consequence of two opposing forces acting on TFBSs. On the one hand, TFBSs in the center of HCTs are preserved by negative selection, as indicated by a higher incidence of ECRs toward the center of HCTs (Supplemental Fig. S7). This might be due to the need to preserve a certain distance between the HCT and the gene, most notably the TSS. On the other hand, the well-known phenomenon of site turnover (Dermitzakis and Clark 2002) maintains the presence of TFBSs in the flanks of HCTs. Since most PWMs are relatively short, new TFBSs could be easily created by a few random mutations (Florea et al. 2000; Ludwig et al. 2000), facilitated by the reduced sequence complexity that results from an elevated or decreased GC content of the sequence of abundant HCTs (Figs. 3, 4; Supplemental Fig. S4).
Our analysis also indicates that the regulation of a significant fraction of TFs (62%) and developmental genes (66%) depends on HCTs. This implies that the regulation of those genes was likely conserved during vertebrate evolution, and it is thus conceivable that a top-level regulatory network responsible for the development of vertebrate organisms has been maintained across great evolutionary distances. The important contribution of HCTs to vertebrate development is supported by two independent lines of evidence: HCTs are part of nearly half of known human developmental enhancers, and HCTs are strongly associated with the enhancer co-activator protein Ep300. While the overlap between these sets is highly significant (Figs. 6, 7; Table 5), it is certainly arguable that the fraction of developmental enhancers utilizing HCTs could be, in fact, higher, because we are effectively missing TF binding information due to limitations in TFBS databases (existing TRANSFAC and JASPAR 3 vertebrate matrices only cover less than 25% of all human TFs). It is also reasonable to think that among the 567 elements not found to drive expression in mouse embryos at E11.5 and recorded as “negatives,” there are elements that could drive specific expression at other time points, and be in fact “positive” enhancers. Supporting this idea is the fact that a significantly lower fraction (34%, P = 0.046, Fisher's exact test) among the “negative” elements also overlap HCTs. It is also possible that some of them are repressors, and therefore their activity would not be revealed in an enhancer assay. A surprising finding was the strong association between NOBOX and forebrain activity in both human and mouse, despite the fact that NOBOX is not known to be active in brain. A possible explanation could be that our data indicate a novel function for NOBOX. Alternatively, it is possible that, rather than NOBOX, a TF with a binding motif similar to that of NOBOX, but with known brain activity binds in fact to the sites recognized by the NOBOX PWM. Such a candidate could be LHX3, which has a PWM that is not significantly different from that of NOBOX as indicated by an intermotif distance of 0.126 (Narlikar et al. 2007). LHX3 is known to be expressed in several regions of the brain during early embryogenesis (Sloop et al. 1999), but further experiments are required to clarify the nature of the strong association between the PWM for NOBOX/LHX3 and forebrain activity. The tissue-specific activity of the HCTs for any TF can be verified with the overlap with the enhancer-associated protein Ep300, which has been shown to have a high accuracy in predicting the tissues where certain enhancers are active (Visel et al. 2009). Our data indicate that HCTs have significant tissue-specific activity differentiation, and thus they could be used in the future to help predict the tissue specificity of the regulatory elements of which they are part. It is, however, noteworthy that tissue specificities likely depend on a complex interplay of TFs and additional regulatory cues that are present in the HCT-containing modules. This is strikingly demonstrated by the very different activity pattern that we observed for the three elements containing HCTs for both NRF1 and E2F4 TFs and that were tested for enhancer activity in mouse transgenic assays (Fig. 8B). These differences in tissue specificity are consistent with the presence of HCTs for distinct sets of additional TFs in the regions tested for enhancer activity, such as one HCT for E2F1 in the case of the enhancer located on chromosome 1, and HCTs for multiple TFs for the other two enhancers (VSX2, PDX1, PRRX2, NKX2-5 for the enhancer on chromosome 5, and MYOG, OVOL2, TCF3, SP3, SPZ1, WT1, ZFP161 for the enhancer on chromosome 10). Moreover, additional singular copies of TFBSs not detected by our HCT screen, such as those previously reported in the case of the NEUROG3 enhancer (Lee et al. 2001), might also contribute to defining the tissue specificity. In contrast, the elements with a single HCT for POU3F2 yielded consistent expression patterns in zebrafish (throughout various domains of the central nervous system), in agreement with the known function of POU TFs. This not only suggests that the presence of HCTs could be an indicator of the enhancer activity for noncoding elements, but it also confirms that the presence of an HCT for a given TF might dictate the function of an enhancer (such as in the case of enhancers containing only HCTs for POU3F2). At the same time, it remains clear that quantifying the functional predictive power of combinatorial HCTs will require experimental testing of substantially larger sample sizes.
In summary, we find that the regulation of genes in vertebrate organisms relies heavily on HCTs. Their strong association with the regulation of TFs and developmental genes indicates that HCTs play an important role in vertebrate development, and suggests that homotypic clustering as a general organization principle of _cis_-regulatory regions is conserved between vertebrates and invertebrates. More importantly, a detailed knowledge about the function of HCTs for specific TFs, such as the association with tissue-specific developmental enhancers, could be used to find new regulatory elements and possibly refine the knowledge about the function of the known ones, effectively contributing to the deciphering of the gene regulatory code.
Methods
Identification of TFBSs and homotypic clusters of TFBS
For the purpose of identifying the location of potential binding sites, we used the profiles of binding sites for vertebrate TFs stored in the form of 601 and 100 position-specific weight matrices in the TRANSFAC (Matys et al. 2003) Professional database (release 11.4), and JASPAR 3 database (Sandelin et al. 2004), respectively. We trained an in-house developed software called tfSearch (Pennacchio et al. 2007) on random sequence to create optimized position-specific scoring matrices (PSSM) to maintain the rate of false-positive discoveries in real genomic sequence to about five false-positives in 10 kb of sequence. We then used tfSearch and the optimized vertebrate PSSMs to scan the sequences of four vertebrate genomes available from the UCSC Genome Browser website (http://genome.cse.ucsc.edu): human (hg18), mouse (mm9), chicken (galGal3), and fugu (fr2). We discarded from our analysis TFBSs located in low complexity and simple repeat regions in order to avoid a high rate of false-positives. As a result, this inevitably eliminated known functional binding sites, such as a cluster of four SP1 binding sites located in a simple repeat region that is required for the efficient expression of the human insulin receptor gene (Araki et al. 1991). We also eliminated from our analysis regions occupied by transposable elements in order to avoid detecting lineage-specific elements that could be introduced, for example, by mammalian-specific MIR or primate-specific Alu elements. By avoiding these two types of repetitive regions, our analysis should provide a conservative view of the extent that HCTs conserved in the vertebrate lineage occupy in the human genome. For the purpose of defining clusters of TFBS, we developed a two-state four-parameter HMM that takes into account the genome-wide background distribution of individual TFBSs and generates a ranked list of HCTs (Supplemental Fig. S8). The HMM was trained independently for each PWM–genome pair. The minimum distance between sites in a cluster was set to 5 to avoid redundancy in sites, and the maximum distance was set to 500 bp, in line with previous studies that suggest this to be a reasonable distance between TFBSs within a CRM (Berman et al. 2002; Kel et al. 2008). A cluster was required to contain at least four sites, to minimize the rate of false-positives. For each PWM and genome, we retained at most the 10,000 top scoring HCTs (a total of 2,719,773), from which we eliminated those longer than 3 kb (this was set to match the size of the extended promoter region—see below) and those with more than 25% of their TFBSs located in coding exons (2,540,832 were retained) in order to avoid detecting genes with a repetitive domain structure (one should note that the exon-based filtering does not eliminate HCTs that contain coding exons within their span unless those coding exons contain more than 25% of the TFBSs).
HCT conservation and overlap with other genomic features.
For determining the conservation status for each HCT, we used the sets of independently defined HCTs for the corresponding PWM in human and every other vertebrate species (mouse, chicken, fugu) and three sets of pairwise evolutionarily conserved regions, or ECRs (Ovcharenko et al. 2004a), linking the genomic segments in the human genome to homologous regions in each of the mouse, chicken, and fugu genomes, respectively. An HCT is determined to be conserved if it spans an ECR that links to a genomic region in the second species that contains an HCT determined independently in that species (Supplemental Fig. S2). For determining the HCT conservation status, only ECR containing less than 50% repetitive sequence were considered. To minimize the rate of false discoveries, we restricted our data set only to those HCTs that are conserved between human and mouse (a total of 9.2%), under the assumption that evolutionary conservation is indicative of function. Furthermore, among the PWMs associated with the same TF, we retained only the PWM with the most conserved HCTs for each TF (a total of 273 PWMs). The fraction of conserved HCTs for these 273 PWM (10.1%) is similar to that of the original set, while the majority of HCTs (89.9%) do not have a corresponding HCT at the orthologous mouse position. Our primary data set consists of a total of 126,405 HCTs conserved between human and mouse that were defined for the 273 selected PWMs. For these, we estimated a false discovery rate by overlapping them with HCTs obtained using PWMs with shuffled columns, so that the PWM GC content is maintained, but the biological relevance of consecutive positions is disrupted. The process of defining HCTs from the set of binding sites defined by the shuffled PWMs was identical to defining real HCTs, except that we did not check for the presence of similarly defined HCTs at the orthologous position in mouse (this allows for an upper bound estimate). The false discovery rate was then estimated from the number of real HCTs that overlap with at least 50% of their size HCTs obtained using shuffled PWMs. We found that only 28 out of 273 PWMs (10.3%) have false discovery rates defined this way higher than 10% (details for each PWM are provided in Supplemental Table S2), with PWMs illustrated throughout the manuscript having low false discovery rates (E2F1, 9.8%; NOBOX, 6.7%; E2F4, 0%; ZNF5, 20.1%). Analyses of overlaps between HCTs and other genomic features, such as gene loci, regions experimentally determined to be bound by TFs, and distant enhancers, were performed with custom Perl scripts. The significance of overlaps between HCTs and other genomic features was evaluated with the binomial test, where the number of trials is equal to the total number of HCTs for a given TF, and the probability of success (e.g., an HCT overlaps a promoter) is equal to the fraction of the genome (blocks of Ns and repeats larger than 500 bp were subtracted from the total genome size) occupied by the feature of interest. In order to obtain a conservative estimate of the significance of difference, we maximized the probability of success by considering all features of interest regardless of their conservation status (as opposed to our set of HCTs, which are defined as overlapping human–mouse ECRs), and subtracting the HCT span from the total genome size.
Gene annotation
For defining gene loci in the human genome, we used the RefSeq annotation track of April 29, 2008 available from the UCSC Genome Browser website. This contains information for 26,280 transcripts, which we combined into a set of 18,649 gene loci. Each locus is defined by one or more transcripts that are located on the same strand and share at least 30 amino acids, allowing different gene loci to overlap (e.g., genes located on opposite strands, or genes located on the same strand, but do not share any coding exons). The transcription start site for each gene locus is defined as the genomic start coordinate of the most 5′ transcript among the transcripts that contribute to a gene locus. The upstream promoter for each gene locus was defined as a region of up to 1.5 kb upstream of the gene itself, being limited only by the presence of another gene locus at a distance smaller than 1.5 kb from the TSS. The extended promoter region comprises the upstream promoter and 1.5 kb downstream of the TSS, thus having a size of maximum 3 kb. Bidirectional promoters are defined as genomic regions (75 nt–3 kb in size) flanked by two gene loci located on opposite strands in a head-to-head relative orientation.
The influence of CpG islands on promoter enrichment
CpG islands (CGIs) are defined as relatively short (>200 nt) DNA regions characterized by a high GC content (>50%), and a high frequency of CpG dinucleotides (the ratio of observed-to-expected > 0.6). The location of CGIs was obtained from the “cpgIslandExt” UCSC annotation track of the hg18 assembly of the human genome (a total of 27,639). To test whether the enrichment of CGIs in promoter regions determines the enrichment of HCTs for GC-rich PWMs in promoter regions, we tested whether the distribution of HCTs is uniform across CGIs located in and outside promoters. If this was true, it would support the hypothesis that promoter enrichment observed for certain TFs is due to the presence of CGIs alone, with the alternative that CGIs cannot completely explain the enrichment observed. For this purpose we selected only 19,599 CGIs that overlap human–mouse ECRs to account for the HCT bias toward conserved regions, and counted how many of the 10,826 promoter-based CGIs and how many of the 8773 CGIs located outside of promoters contain at least one HCT. The significance of difference was calculated using the Fisher's exact test as implemented in the R package v2.7.1. An identical approach was used in the case of the limited number of 782 CGIs located in bidirectional promoters.
For testing whether the HCT preference for promoters is due to the GC content of the PWM, we constructed a new set of PWMs by shuffling the columns of the original ones. This maintains the GC content of the shuffled PWMs identical to the real PWMs, but disrupts the co-occurrence of nucleotides that is derived from biological relevant sequences. We then overlapped these new HCTs with the 11,384 CGIs found in promoters. Fisher's exact test was used to calculate the significance of the difference between the overlaps of CGIs with HCTs defined using read and shuffled PWMs.
Gene Ontology annotation and analysis
We assigned to each gene locus all GO categories assigned to all transcripts in that locus. For this purpose we used the April 29, 2008 versions of gene2refseq and gene2go annotation files available from the NCBI website. The enrichment analysis of GO categories among genes in a set of interest was limited only to those GO categories assigned to at least 10 genes, and was evaluated using the hypergeometric distribution as implemented in the R package v2.7.1, and the Bonferroni correction for multiple testing was then applied to the _P_-value for each GO category tested.
External data sets
For the purpose of verifying the overlap between homotypic clusters of TFBS and experimental data, we used the coordinates of peak regions as provided in the Supplementary materials of the respective papers for the following TFs: REST (Johnson et al. 2007); YY1 (Xi et al. 2007); STAT1 (Robertson et al. 2007); E2F1, E2F4, MYC (The ENCODE Project Consortium 2007). If coordinates provided corresponded to an assembly other than hg18, they were converted with the help of the liftOver tool and corresponding chain files available from the UCSC Genome Browser website.
Histone methylation data were obtained from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspx. Specifically, we used the summary bed files and the definition of “tag islands” in Barski et al. (2007) to define the genomic regions characterized by each histone methylation type. We further defined the regions associated with gene activation and repression by combining all the tag islands associated with the two states: monomethylation of H3K27, H3K9, H4K20, H3K79, and H2BK5 for gene activation, and trimethylation of H3K27, H3K9, and H3K79 for gene repression, respectively.
The set of 849 distant enhancers tested in vivo (Pennacchio et al. 2006) was downloaded from http://enhancer.lbl.gov, and their coordinates were converted from hg17 to hg18 with the liftOver tool and chain files available from the UCSC Genome Browser website. Clustering of TFs and tissue-specific expression was performed with Cluster v2.11 software (Eisen et al. 1998), and the result was visualized with the TreeView v1.6 (Eisen et al. 1998).
The overlap of HCTs with regions targeted by the enhancer-associated Ep300 protein was evaluated using the mouse HCTs corresponding to human HCTs and the Ep300 peaks included in Supplemental Tables 2–4 of Visel et al. (2009). For constructing the Ep300 coverage profiles of HCTs, extended Ep300 reads from the Gene Expression Omnibus (GEO) series GSE13845 were used. Only reads mapped to mouse chromosomes 1–19, X, and Y were considered from each of the three samples: GSM348064: forebrain, 3,602,919 reads; GSM348065: midbrain, 3,504,753 reads; GSM348066: limb, 2,411,871 reads. The coverage values were normalized to the value of Ep300 genome-wide coverage (1.12×) for Figure 7A, and with the genome-wide coverage of tissue-specific reads (forebrain, 0.42×; midbrain, 0.41×; limb, 0.28×) for Figure 7B. Significance of coverage peaks was evaluated with the Fisher's exact test.
Analysis of sequence divergence
For this purpose we utilized human–chimp pairwise alignments available from the UCSC Genome Browser website (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/vsPanTro2/). Only syntenic alignments included in the “hg18.panTro2.syn.net.gz” file were utilized, and alignments from the highest-scoring chain were favored when multiple alignments are defined for the same region. Divergence in all regions was calculated after eliminating the coding sequences corresponding to coding exons in the knownGene and RefSeq annotation tracks, and repetitive sequences included in the simpleRepeat and RepeatMasker tracks.
Testing for purifying selection acting on TFBSs
For this purpose we compared the sequence divergence between human and chimp and SNP density observed in TFBSs to that observed in sequences separating the TFBSs (intersites) within HCTs. The significance of difference was tested with the Fisher's exact test. The use of intersite sequence as reference minimizes the likelihood of biased inference due to difference in GC content, mutation rate, ascertainment bias or position relative to gene features, as we found HCTs to differ strikingly from flanking sequences (Fig. 5).
Testing for in vivo enhancer activity
Zebrafish were raised and bred in accordance with standard conditions (Kimmel et al. 1995; Whitlock and Westerfield 2000). Embryos were obtained from natural crosses, incubated at 30.0°C and staged (Warga and Kimmel 1990). Sequences of interest were amplified with specific attB-containing primers (Table 6) and cloned into a donor vector (pDONR 221) of the Gateway cloning system (Invitrogen). Plasmid DNAs for microinjection were purified on QIAprep Mini-prep (Qiagen) spin columns. Transposase RNA was transcribed in vitro using the mMessage mMachine Sp6 kit (Ambion). Injection solutions were made with 25 ng/mL of transposase RNA, and 15–25 ng/mL of circular plasmid, in water. DNA was injected into the yolk of wild-type embryos at the two-cell stage. Embryos were analyzed in a Leica MZ16F stereomicroscope and imaged using the QCAPTURE Pro 6.0 package. In the case of transgenic mouse assays, the regions containing combinations of HCTs were extended so that they can be amplified (for primers and coordinates, see Table 6) and cloned according to our standard mouse transgenic assay (Pennacchio et al. 2006). The generation of transgenic mice and embryo staining was done as previously described (Poulin et al. 2005).
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health; National Library of Medicine (V.G., I.O.); National Institutes of Health grants HG004428 (National Human Genome Research Institute) and HL088393 (National Heart, Lung and Blood Institute) (M.A.N., J.M.W.); and the Department of Energy Contract DE-AC02-05CH11231, University of California, E.O. Lawrence Berkeley National Laboratory (A.V., L.A.P.). L.A.P. was also supported by grant HL066681, Berkeley-Program for Genomic Applications, under the Programs for Genomic Applications, funded by the National Heart, Lung and Blood Institute, and HG003988 funded by the National Human Genome Research Institute.
Footnotes
References
- Adachi N, Lieber MR 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807–809 [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 [DOI] [PubMed] [Google Scholar]
- Araki E, Murakami T, Shirotani T, Kanai F, Shinohara Y, Shimada F, Mori M, Shichiri M, Ebina Y 1991. A cluster of four Sp1 binding sites required for efficient expression of the human insulin receptor gene. J Biol Chem 266: 3944–3948 [PubMed] [Google Scholar]
- Arnone MI, Davidson EH 1997. The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasoski S, Toldo SS, Malipiero U, Schreiber E, Fries R, Fontana A 1995. Isolation of the human genomic brain-2/N-Oct 3 gene (POUF3) and assignment to chromosome 6q16. Genomics 26: 272–280 [DOI] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L 2006. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J Mol Biol 360: 213–227 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB 2002. Exploiting transcription factor binding site clustering to identify _cis_-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci 99: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ 2006. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res 16: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Clement J, Regnier M, Roytberg MA, Makeev VJ 2007. Exact p-value calculation for heterotypic clusters of regulatory motifs and its application in computational annotation of _cis_-regulatory modules. Algorithms Mol Biol 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 16: 399–411 [DOI] [PubMed] [Google Scholar]
- Coleman RA, Pugh BF 1995. Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem 270: 13850–13859 [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG 2002. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI 2001. Positive and negative selection on the human genome. Genetics 158: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS 2006a. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312: 276–279 [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS 2006b. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Florea L, Li M, Riemer C, Giardine B, Miller W, Hardison RC 2000. Validating computer programs for functional genomics in gene regulatory regions. Curr Genomics 1: 11–27 [Google Scholar]
- Grehan S, Tse E, Taylor JM 2001. Two distal downstream enhancers direct expression of the human apolipoprotein E gene to astrocytes in the brain. J Neurosci 21: 812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Crable SC, Anderson KP 2005. Negative regulatory elements are present in the human LMO2 oncogene and may contribute to its expression in leukemia. Leuk Res 29: 89–97 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Maniatis T 1997. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol 9: 350–357 [DOI] [PubMed] [Google Scholar]
- Hu Z, Hu B, Collins JF 2007. Prediction of synergistic transcription factors by function conservation. Genome Biol 8: R257 doi: 10.1186/gb-2007-8-12-r257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Jakobovits A, Schwab M, Bishop JM, Martin GR 1985. Expression of N-myc in teratocarcinoma stem cells and mouse embryos. Nature 318: 188–191 [DOI] [PubMed] [Google Scholar]
- Jin Z, Liu L, Bian W, Chen Y, Xu G, Cheng L, Jing N 2009. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J Biol Chem 284: 8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kel AE, Niehof M, Matys V, Zemlin R, Borlak J 2008. Genome wide prediction of HNF4α functional binding sites by the use of local and global sequence context. Genome Biol 9: R36 doi: 10.1186/gb-2008-9-2-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury AM, Lee HJ, Lillis M, Lu P 1990. Lac repressor-operator interaction: DNA length dependence. Biochim Biophys Acta 1087: 55–60 [DOI] [PubMed] [Google Scholar]
- Kim JG, Takeda Y, Matthews BW, Anderson WF 1987. Kinetic studies on Cro repressor-operator DNA interaction. J Mol Biol 196: 149–158 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H 1992. CpG islands as gene markers in the human genome. Genomics 13: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, German MS 2001. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 50: 928–936 [DOI] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S, Workman C 2001. Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics 17: 871–877 [DOI] [PubMed] [Google Scholar]
- Li Q, Harju S, Peterson KR 1999. Locus control regions: Coming of age at a decade plus. Trends Genet 15: 403–408 [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD 2007. Animal models of human disease: Zebrafish swim into view. Nat Rev Genet 8: 353–367 [DOI] [PubMed] [Google Scholar]
- Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA 2003. Homotypic regulatory clusters in Drosophila. Genome Res 13: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567 [DOI] [PubMed] [Google Scholar]
- Luster TA, Rizzino A 2003. Regulation of the FGF-4 gene by a complex distal enhancer that functions in part as an enhanceosome. Gene 323: 163–172 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Stine ZE, Huynh JL, Vinton RM, McCallion AS 2009. Asymmetrical distribution of non-conserved regulatory sequences at PHOX2B is reflected at the ENCODE loci and illuminates a possible genome-wide trend. BMC Genomics 10: 8 doi: 10.1186/1471-2164-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Murakami K, Kojima T, Sakaki Y 2004. Assessment of clusters of transcription factor binding sites in relationship to human promoter, CpG islands and gene expression. BMC Genomics 5: 16 doi: 10.1186/1471-2164-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar L, Gordan R, Hartemink AJ 2007. A nucleosome-guided map of transcription factor binding sites in yeast. PLoS Comput Biol 3: 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I 2008. Widespread ultraconservation divergence in primates. Mol Biol Evol 25: 1668–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L 2004a. ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32: W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Stubbs L, Loots GG 2004b. Interpreting mammalian evolution using Fugu genome comparisons. Genomics 84: 890–895 [DOI] [PubMed] [Google Scholar]
- Papatsenko DA, Makeev VJ, Lifanov AP, Regnier M, Nazina AG, Desplan C 2002. Extraction of functional binding sites from unique regulatory regions: The Drosophila early developmental enhancers. Genome Res 12: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I 2007. Predicting tissue-specific enhancers in the human genome. Genome Res 17: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin F, Nobrega MA, Plajzer-Frick I, Holt A, Afzal V, Rubin EM, Pennacchio LA 2005. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85: 774–781 [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA 2006. Close sequence comparisons are sufficient to identify human _cis_-regulatory elements. Genome Res 16: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ 2008. E2F in vivo binding specificity: Comparison of consensus versus nonconsensus binding sites. Genome Res 18: 1763–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev 19: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K 2007. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res 17: 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B 2004. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Tobler A, Malipiero U, Schaffner W, Fontana A 1993. cDNA cloning of human N-Oct3, a nervous-system specific POU domain transcription factor binding to the octamer DNA motif. Nucleic Acids Res 21: 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Giang H, Plotkin JB, Hannenhalli S 2008. Genome-wide analysis of natural selection on human _cis_-elements. PLoS One 3: e3137 doi: 10.1371/journal.pone.0003137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloop KW, Meier BC, Bridwell JL, Parker GE, Schiller AM, Rhodes SJ 1999. Differential activation of pituitary hormone genes by human Lhx3 isoforms with distinct DNA binding properties. Mol Endocrinol 13: 2212–2225 [DOI] [PubMed] [Google Scholar]
- Somma MP, Pisano C, Lavia P 1991. The housekeeping promoter from the mouse CpG island HTF9 contains multiple protein-binding elements that are functionally redundant. Nucleic Acids Res 19: 2817–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A 1999. Genes regulated cooperatively by one or more transcription factors and their identification in whole eukaryotic genomes. Bioinformatics 15: 776–784 [DOI] [PubMed] [Google Scholar]
- Warga RM, Kimmel CB 1990. Cell movements during epiboly and gastrulation in zebrafish. Development 108: 569–580 [DOI] [PubMed] [Google Scholar]
- Whitlock KE, Westerfield M 2000. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development 127: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT 2008. Species-specific transcription in mice carrying human chromosome 21. Science 322: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklehner-Jennewein P, Geymayer S, Lechner J, Welte T, Hansson L, Geley S, Doppler W 1998. A distal enhancer region in the human β-casein gene mediates the response to prolactin and glucocorticoid hormones. Gene 217: 127–139 [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi: 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Yu Y, Fu Y, Foley J, Halees A, Weng Z 2007. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res 17: 798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman MR, Kay PH 1998. Differential expression of four alternate Pax7 paired box transcripts is influenced by organ- and strain-specific factors in adult mice. Gene 217: 77–81 [DOI] [PubMed] [Google Scholar]