Sulfonylurea pharmacogenomics in Type 2 diabetes: the influence of drug target and diabetes risk polymorphisms (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 1.
Published in final edited form as: Expert Rev Cardiovasc Ther. 2010 Mar;8(3):359–372. doi: 10.1586/erc.09.154
Abstract
The sulfonylureas stimulate insulin release from pancreatic β cells, and have been a cornerstone of Type 2 diabetes pharmacotherapy for over 50 years. Although sulfonylureas are effective antihyperglycemic agents, interindividual variability exists in drug response (i.e., pharmacodynamics), disposition (i.e., pharmacokinetics) and adverse effects. The field of pharmacogenomics has been applied to sulfonylurea clinical studies in order to elucidate the genetic underpinnings of this response variability. Historically, most studies have sought to determine the influence of polymorphisms in drug-metabolizing enzyme genes on sulfonylurea pharmacokinetics in humans. More recently, polymorphisms in sulfonylurea drug target genes and diabetes risk genes have been implicated as important determinants of sulfonylurea pharmacodynamics in patients with Type 2 diabetes. As such, the purpose of this review is to discuss sulfonylurea pharmacogenomics in the setting of Type 2 diabetes, specifically focusing on polymorphisms in drug target and diabetes risk genes, and their relationship with interindividual variability in sulfonylurea response and adverse effects.
Keywords: drug target, KCNJ11, pharmacogenomic, sulfonylurea, SUR1, TCF7L2, Type 2 diabetes
Overview of Type 2 diabetes pharmacotherapy
The prevalence of diabetes is increasing at an alarming rate, affecting over 250 million individuals worldwide [101]. In adults, Type 2 diabetes is the most prevalent form, accounting for 90–95% of the worldwide cases of diabetes [101]. In terms of pathophysiology, Type 2 diabetes is characterized by decreased insulin secretion from pancreatic β cells and decreased tissue responsiveness to the normal action of insulin (i.e., insulin resistance). Over time, declining β-cell function and insulin resistance lead to the inability to maintain normoglycemia, and hyperglycemia ensues [1].
Chronic hyperglycemia, as a result of Type 2 diabetes, has many long-term adverse consequences. Chronic hyperglycemia results in microvascular complications such as retinopathy, nephropathy and neuropathy, and macrovascular complications such as cardiovascular disease, stroke and peripheral vascular disease. Given these long-term complications, the goal of Type 2 diabetes treatment in men and nonpregnant women is to obviate periods of poor glycemic control and maintain glycated hemo globin (HbA1c) levels of less than 7% [2]. Lowering glucose to near-nondiabetic levels in patients with Type 2 diabetes has been shown to reduce the risk of microvascular complications [3-5]. In terms of the prevention of macrovascular complications, the United Kingdom Prospective Diabetes Study (UKPDS) showed a nonsignificant 16% reduction in the risk of myocardial infarction in the group of Type 2 diabetes patients receiving intensive glucose control [3]. Furthermore, in the intensive glucose control group, 10-year follow-up showed a reduction in the risk of myocardial infarction and death from any cause over time [6]. Recently, some trials have investigated the effect of more intensive glucose control (e.g., HbA1c ≤6.0 or ≤6.5%) on cardiovascular events in patients with Type 2 diabetes [7-9]. However, the results have largely shown no benefit on major cardiovascular end points, and in the case of the Action to Control Cardiovascular Risk in Diabetes (ACCORD) study, an increased risk of mortality associated with intensive glucose control [7-9]. By contrast, two recently published meta-analyses showed that, compared with conventional glucose control, intensive glucose control reduced the risk of some cardiovascular end points (e.g., nonfatal myocardial infarction) [10,11]. However, intensive glucose control was also associated with a higher incidence of weight gain and severe hypoglycemia compared with the conventional glucose control groups [10,11].
Based on existing data at the time regarding the association between glucose control and the prevention of microvascular and macrovascular complications, the 2009 American Diabetes Association Standards for Medical Care in Diabetes recommends a HbA1c goal of less than 7% in most patients with Type 2 diabetes. There are numerous nonpharmacologic and pharmacologic treatment modalities to achieve and maintain normoglycemia. Lifestyle interventions, such as diet and exercise, are effective; however, long-term adherence is typically poor. Furthermore, lifestyle interventions have a modest durability in the face of worsening glycemia over time [12]. As such, most patients with Type 2 diabetes will require one or more antidiabetic drug therapies to manage their hyperglycemia. Available pharmacologic agents are listed in Table 1 and include biguanides, insulin, sulfonylureas, thiazolidinediones, glucagon-like peptide 1 analogs, α-glucosidase inhibitors, nonsulfonylurea insulin secretagogues, amylin agonists and dipeptidyl peptidase (DPP)-4 inhibitors [12].
Table 1. Available pharmacologic agents for the treatment of Type 2 diabetes.
| Drug class | Role in Type 2 diabetes therapy |
|---|---|
| Biguanides (metformin) | Tier 1, step 1 |
| Insulin | Tier 1, step 2 |
| Sulfonylureas (tolbutamide, tolazamide, chlorpropamide, acetohexamide, glyburide [also known asglibenclamide], glipizide, gliclazide and glimepiride) | Tier 1, step 2 |
| Thiazolidinediones† (pioglitazone and rosiglitazone) | Tier 2 |
| Glucagon-like peptide 1 analogs (exenatide) | Tier 2 |
| α-glucosidase inhibitors (acarbose and miglitol) | Other therapy |
| Nonsulfonylurea insulin secretagogues (repaglinide and nateglinide) | Other therapy |
| Amylin agonists (pramlintide) | Other therapy |
| DPP-4 inhibitors (sitagliptin) | Other therapy |
Given the variety of pharmacologic treatment options, the American Diabetes Association and the European Association for the Study of Diabetes recently published a consensus statement providing guidance on the management of hyperglycemia in adults with Type 2 diabetes [12]. In this consensus statement, therapies are divided into tiers, and the tiers are further divided into steps. Tier 1 represents well-validated core therapies that are established, clinically effective and cost effective. Tier 1 therapies include lifestyle interventions to decrease weight and increase physical activity, metformin, sulfonylureas and insulin. Tier 2 represents less well-validated therapies such as thiazolidinediones, pioglitazone and the glucagon-like peptide 1 analog, exenatide. Medications that fall into the ‘other therapy’ category are those not recommended in tier 1 or 2 approaches, and include α-glucosidase inhibitors, nonsulfonylurea insulin secretagogues, pramlintide and DPP-4 inhibitors.
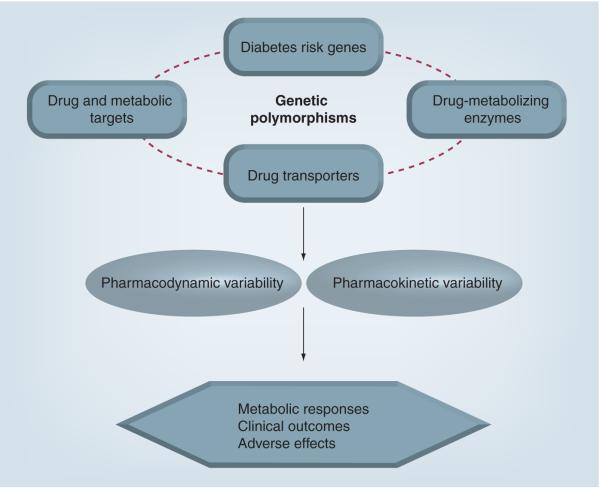
The tiered drug treatment approach serves as a useful guidance for the pharmacotherapeutic management of Type 2 diabetes. However, it does not solve the problem that substantial interindividual variability exists in antidiabetic drug disposition and response. As such, the field of pharmacogenomics has recently been applied to the treatment of Type 2 diabetes. The goal of pharmacogenomics is to elucidate the relationship between variants in the human genome (i.e., polymorphisms) and variability in the effects of drugs [13]. It is anticipated that pharmacogenomics will improve the use of drug therapy in Type 2 diabetes by identifying genetic predictors of metabolic response, clinical outcomes and adverse effects (Figure 1). Sulfonylureas are in the tier 1 category and have been a mainstay of Type 2 diabetes pharmacotherapy for over 50 years. It is well recognized that interindividual variability exists in sulfonylurea response (i.e., pharmacodynamics), disposition (i.e., pharmacokinetics) and adverse effects. The field of pharmacogenomics has been applied to sulfonylurea clinical studies in order to elucidate the genetic underpinnings of this response variability. Historically, most studies have sought to determine the influence of polymorphisms in drug-metabolizing enzyme genes on sulfonylurea pharmacokinetics in humans. More recently, polymorphisms in sulfonylurea drug target genes and diabetes risk genes have been implicated as important determinants of sulfonylurea pharmacodynamics in patients with Type 2 diabetes. As such, the purpose of this article is to discuss sulfonylurea pharmacogenomics in the setting of Type 2 diabetes, specifically focusing on polymorphisms in drug target and diabetes risk genes, and their relationship with interindividual variability in sulfonylurea response and adverse effects. For comprehensive reviews of the influence of drug-metabolizing enzyme and drug-transporter polymorphisms on variability in sulfonylurea pharmacokinetics, the reader is referred to previous articles by Kirchheiner and colleagues [14] and Pacanowski and colleagues, respectively [15]. In addition, the focus of this present review is on sulfonylurea response in polygenic Type 2 diabetes. For a comprehensive discussion of the use and efficacy of sulfonylureas in patients who possess mutations that cause monogenic forms of diabetes, the reader is referred to articles by Murphy and colleagues [16] and Gloyn and colleagues [17].
Figure 1. Application of pharmacogenomics to sulfonylurea therapy.
Adapted and reprinted with permission from [15].
Sulfonylurea clinical pharmacology
Mechanistically, the sulfonylureas bind to ATP-sensitive potassium (KATP) channels on pancreatic β cells. The KATP channel exists as a complex comprised of the sulfonylurea receptor 1 (SUR1), which is the regulatory subunit, and the inward-rectifier potassium ion channel (Kir6.2), which forms the pore of the channel. Four SUR1 subunits and four Kir6.2 subunits make up the KATP channel. The binding of sulfonylureas to SUR1 results in closure of the KATP channel, increased concentrations of intracellular potassium, depolarization of the β-cell membrane, and the subsequent opening of voltage-gated calcium channels. As calcium moves into the β cell, it stimulates the movement of insulin-containing secretory granules to the cell surface. These insulin-containing secretory granules are ultimately released from the β cell and into the circulation [18]. In short, sulfonylureas stimulate insulin release from pancreatic β cells in a glucose-independent manner.
The sulfonylurea drug class has evolved as different generations of agents. The first-generation sulfonylureas are the oldest, and include tolbutamide, tolazamide, chlorpropamide and acetohexamide. The second-generation sulfonylureas include glyburide (also known as glibenclamide), glipizide, gliclazide and glimepiride. All sulfonylureas have the same mechanism of action; however, the second-generation agents are more potent than the first-generation agents on a weight basis. Most sulfonylureas are extensively metabolized in the liver, primarily by the cytochrome P450 (CYP) 2C9 isoenzyme. The half-life of most sulfonylureas is relatively short, with the exception of chlorpropamide, which has a half-life of 24–48 h [19]. In terms of clinical response, the sulfonylureas, on average, lower HbA1c by 1–2% [12]. As insulin secretagogues, the pharmacodynamic effects of sulfonylureas are dependent on functioning β cells; thus they are typically used earlier in the course of the disease process. In later stages of the disease, when β-cell dysfunction becomes most prevalent, sulfonylurea efficacy may decline and may necessitate an increase in drug dosage or the switch to other non-insulin secretagogue therapy. More recently, studies in isolated rodent and human islets showed that sulfonylureas induce β-cell apoptosis [20-23]. As such, concern has been raised as to whether sulfonylureas may exacerbate the pathogenesis of Type 2 diabetes, particularly β-cell dysfunction in the later stages of the disease. However, at this time, no definitive in vivo conclusions have been drawn [23].
Given their mechanism of action, the most common adverse effect of sulfonylureas is hypoglycemia. Sulfonylurea-induced hypoglycemia is more likely to occur with the longer acting agents (e.g., chlorpropamide and glibenclamide), in patients with irregular eating habits, and in those who consume alcohol excessively [24]. Sulfonylureas should also be used with caution in patients who are unable to recognize the symptoms of hypoglycemia (e.g., those on concomitant β-blocker therapy or elderly patients). Weight gain, which is typically 1–4 kg, is another concern of sulfonylurea therapy, particularly given that many Type 2 diabetes patients are already overweight or obese [12,24]. Rare adverse effects include: cholestatic jaundice, skin rash, hemolytic anemia, thrombocytopenia, agranulocytosis, flushing (chlorpropamide) and hyponatremia (chlorpropamide) [24]. Sulfonylureas are susceptible to drug–drug interactions. Pharmacological interactions that may increase sulfonylurea plasma concentrations and glucose-lowering effects include drugs that displace sulfonylureas from plasma proteins (e.g., salicylates, sulfonamides, warfarin, phenylbutazone and fibric acid derivatives), decrease the hepatic metabolism of sulfonylureas (e.g., warfarin, monoamine oxidase inhibitors, chloramphenicol and phenylbutazone) and decrease the renal excretion of sulfonylureas (e.g., salicylates, probenecid and allopurinol) [24]. In addition, inducers of hepatic drug metabolism (e.g., rifampin) may decrease sulfonylurea plasma concentrations and efficacy.
There has been a long-standing debate regarding the potential of sulfonylureas to increase cardiovascular risk. This debate was spurred by the results of the University Group Diabetes Program study, which showed a higher incidence of cardiovascular mortality in patients treated with tolbutamide as compared with patients treated with insulin or placebo [25]. By binding to and closing pancreatic KATP channels, sulfonylureas promote insulin release. However, sulfonylureas also close myocardial KATP channels, which is proposed to abolish the protective effects of myocardial ischemic preconditioning [26]. Further complicating this issue, sulfonylureas are thought to have different selectivities for pancreatic versus cardiac KATP channels, which may confer differing effects on cardiovascular pathophysiology and risk [27]. Despite these mechanistic hypotheses, an increase in cardiovascular risk was not observed in UKPDS, where the myocardial infarction case–fatality rate was similar between sulfonylurea users and nonusers [28]. Along the same lines, there was no evidence of increased cardiovascular risk in the Action in Diabetes and Vascular Disease: Preterax and Diamicron Modified Release Controlled Evaluation (ADVANCE) trial, which included an intensive glucose control strategy involving gliclazide (modified release) and other drugs as required [8]. Thus, while it has been documented that sulfonylureas have negative effects on the myocardium, these mechanistic findings have not consistently translated into an increased cardiovascular risk in large-scale clinical trials.
Interindividual variability in sulfonylurea response
Although the sulfonylureas are effective antihyperglycemic agents, interindividual variability exists in drug response. Definitions of sulfonylurea failure vary between clinical studies. However, it is estimated that 10–20% of patients will have less than a 20-mg/dl reduction in fasting plasma glucose following the initiation of sulfonylurea therapy (i.e., primary sulfonylurea failure) [29]. Approximately 50–60% of patients will initially have a greater than 30 mg/dl reduction in fasting plasma glucose, but will fail to reach the desired glycemic treatment goals [29]. For patients who have a good initial response to therapy, the rate of secondary sulfonylurea failure is approximately 5–7% per year [29]. Variability in sulfonylurea response has also been demonstrated in large-scale randomized trials. For example, A Diabetes Outcome Progression Trial (ADOPT) showed that the incidence of sulfonylurea monotherapy failure (as defined by a fasting plasma glucose >180 mg/dl) at 5 years was 34% compared with 15% for rosiglitazone and 21% for metformin [30]. While some patients fail sulfonylurea therapy, other patients appear to be uniquely sensitive to the hypoglycemic effects of these agents. In the UKPDS study, 31% of patients experienced mild hypoglycemia during the first year of glibenclamide treatment [3]. Severe hypoglycemia with sulfonylurea therapy is less common, with a reported incidence of 1% per year in UKPDS [3]. Clinical factors, such as declining β-cell function, long-standing diabetes, high baseline glucose levels and a high degree of insulin resistance are known to predispose patients to sulfonylurea failure. Along the same lines, sulfonylurea-induced hypoglycemia may result from the duration of sulfonylurea action (e.g., the incidence of hypoglycemia is higher with chlorpropamide and glyburide), mild baseline hyperglycemia, irregular eating patterns, excessive alcohol intake and age.
Beyond these clinical variables, data suggest that genetic polymorphisms may contribute to the observed interindividual variability in sulfonylurea response, disposition and side effects. In terms of drug-metabolizing enzyme polymorphisms, CYP2C9*3 (Ile359Leu) and to a lesser extent CYP2C9*2 (Arg144Cys), influence the pharmacokinetics of many sulfonylureas [14]. Oral clearance of tolbutamide, glyburide (glibenclamide), glipizide and glimepiride is reduced in patients carrying a CYP2C9*3 allele, resulting in decreased clearance and increased plasma drug exposure of most of these agents. For example, the oral clearance of tolbutamide was approximately 6.5-fold lower in CYP2C9*3 homozygotes compared with wild-type homozygotes [31]. Similarly, glimepiride oral clearance in CYP2C9*3 homozygotes was only half that of CYP2C9 wild-type homozygotes [32]. The majority of CYP2C9 sulfonylurea pharmacogenomic studies have been conducted in healthy volunteers; therefore, the extent to which changes in plasma sulfonylurea exposure, as a result of CYP2C9 polymorphisms, affect sulfonylurea response and adverse effects in patients with Type 2 diabetes has not been comprehensively studied. However, one study in Type 2 diabetes patients showed that carriers of a CYP2C9*3 allele required significantly lower tolbutamide doses than wild-type homozygotes [33]. A recent population-based study of incident sulfonylurea users found that Type 2 diabetes patients with CYP2C9*2/*2, *2/*3 or *3/*3 genotypes were 3.4-times more likely to achieve a treatment HbA1c of less than 7% compared with CYP2C9 wild-type homozygotes [34]. Furthermore, patients with at least one copy of the CYP2C9*2 or *3 allele were less likely to experience sulfonylurea monotherapy treatment failure. CYP2C9 polymorphisms may also serve as useful predictors of adverse effects. For example, a different study showed that sulfonylurea-treated patients who possessed the CYP2C9*3/*3 or *2/*3 genotype had 5.2-times the odds of a severe hypoglycemic event than the other CYP2C9 genotype groups [35]. While the findings that CYP2C9 polymorphisms influence sulfonylurea response and adverse effects are intriguing, the utility of CYP2C9 genotyping prior to initiating sulfonylurea therapy is unclear. Prospective studies in patients with Type 2 diabetes are warranted in order to discern whether CYP2C9 genotyping is a useful tool to guide sulfonylurea dosing and treatment. In recent years, the field of sulfonylurea pharmacogenomics has expanded beyond just variation in drug-metabolism genes and now includes polymorphisms in drug target and diabetes risk genes, which will be the focus of this review hereafter.
Sulfonylurea drug target pharmacogenomics
As previously discussed, SUR1 and Kir6.2 are subunits that make up KATP channels. The SUR1 regulatory subunit is encoded by the ATP-binding cassette, subfamily C, member 8 (ABCC8) gene, while the Kir6.2 pore subunit is encoded by the potassium inwardly-rectifying channel, subfamily J, member 11 (KCNJ11) gene. The ABCC8 and KCNJ11 genes are located in 4.5-kb proximity to each other on chromosome 11 [36]. Interest in ABCC8 and KCNJ11 as candidate pharmacogenomic genes in polygenic Type 2 diabetes has evolved from clinical experience with monogenic forms of diabetes [16,17]. In transient and permanent neonatal diabetes mellitus, activating mutations in KCNJ11 and ABCC8 cause an open KATP channel state, which promotes membrane hyperpolarization and impaired insulin release [37]. Sulfonylureas are useful in patients with some of these activating mutations as they promote KATP channel closure, which leads to calcium influx and insulin release [38,39]. By contrast, loss-of-function mutations in KCNJ11 and ABCC8 promote channel closure and insulin hypersecretion, and are associated with hyperinsulinemic hypoglycemia of infancy [37]. Mutations causing monogenic forms of diabetes are rare; thus, the search for common polymorphisms that explain variability in sulfonylurea response in polygenic Type 2 diabetes has been an emerging area of research in the field of Type 2 diabetes pharmacogenomics. Given that sulfonylureas interact with KATP channels, both ABCC8 and KCNJ11 are logical candidate drug-target genes to investigate in terms of drug response. A summary of the published ABCC8 and KCNJ11 sulfonylurea pharmacogenomic studies in patients with Type 2 diabetes is shown in Table 2.
Table 2. Summary of sulfonylurea drug target Type 2 diabetes pharmacogenomic studies.
| Study author(year) | Study population | Sulfonylurea | Primary responsephenotype | Polymorphism(s)studied | Primary results | Ref. |
|---|---|---|---|---|---|---|
| Meirhaeghe_et al._ (2001) | Population-based French study;122 Type 2 diabetes patientswere selected based on medi-cal diagnosis and treatment(n = 70, sulfonylurea; n = 52,other treatmentsexcluding insulin) | Various | Single fasting blood samplefor glucose, insulin, totalcholesterol and triglycerides | ABCC8 intron/exon 16 −3C>T | No influence of the intron/exon 16−3 C>T polymorphism on glucoseor insulin levels, regardlessof treatment.In the sulfonylurea group, the−3C allele was associated withlower triglyceride levels than theT/T genotype | [41] |
| Zychma et al.(2002) | Type 2 diabetes ≤5 years andtreated with insulin (n = 68);or Type 2 diabetes for at least15 years and treated with oralagents (sulfonylurea alone orin combination with metforminor acarbose, n = 99). Patientswere selected from twoprevious cohort studies | Various | Differences in genotype andallele frequencies betweenthe two patient groups | ABCC8 intron/exon 16 −3C>T | No significant differences ingenotype distributions or allelefrequencies between groups | [42] |
| Zhang et al.(2007) | Type 2 diabetes with FPG≥7.8 mmol/l and treatedwith diet and exercise, butno hypoglycemic agents for2 months before the study(n = 115) | Gliclazide 40 mg twicedaily for 8 weeksDosage adjustmentsat weeks 2 and 4 wereallowed based on FPG | FPG, HbA1c | ABCC8 Ser1369Ala | Greater decrease in HbA1c in Alaallele carriers compared with Serhomozygotes | [44] |
| Feng et al.(2008) | Type 2 diabetes with onset afterthe age of 35 and all of the fol-lowing (n = 1268): diabetes di-agnosed within the past 5 yearsand no antidiabetic treatmentwithin the past 2 months; BMI<28 kg/m2; and FPG between7.8 and 15 mmol/l | Gliclazide 40 mg twicedaily for 8 weeksDosage adjustments wereallowed based on FPG atweeks 2 and 4 | Continuous variables: FPG,2-h glucose post 75 gOGTT, HbA1cLogistic regression:gliclazide response asdefined by week 8 FPG<7.8 mmol/l | 25 SNPs in 11 candidategenes, including ABCC8and KCNJ11 | ABCC8 variant Ala1389 carriershad greater decreases in FPG and2-h glucose compared with SerhomozygotesReductions in HbA1c were greaterin Ala/Ala (−1.7%) and Ser/Ala(−1.5%) compared with Ser/Ser(−1.2%), although this was notsignificantThe odds of gliclazide responsewere 1.4-times greater in Ser/Alaand 2.2-times greater in Ala/Alacompared with Ser/Ser_KCNJ11_ rs5210 was alsoassociated with the percentreduction in FPG | [45] |
| Gloyn et al.(2001) | Patients from the UKPDS studywho were randomly allo-cated to sulfonylurea therapy(n = 364) | Chlorpropamide orglipizide | Change in FPG in the firstyear from randomization;patients on sulfonylureatherapy at 1 year post-randomization wereclassified as ‘successes’,while patients who requiredadditional therapy whenFPG rose to >15 mmol wereclassified as ‘failures’ | KCNJ11 E23K and L270V | The E23K and L270Vpolymorphisms were notsignificantly associated withsulfonylurea response | [46] |
| Sesti et al.(2006) | Type 2 diabetes patients(n = 525) recruited consecutive-ly with the following criteria:onset of diabetes >35 yearsof age, absence of ketonuriaat diagnosis and negative forantiglutamic acid decarboxylaseantibody | Glibenclamide 15 mgper day. Metforminwas added if FPG was>300 mg/dl on twoseparate occasions | Secondary sulfonylureafailure: those requir-ing insulin due to FPG>300 mg/dl despitesulfonylurea and metformintherapy, appropriate diet,and absence of conditionsthat cause hyperglycemia | KCNJ11 E23K | The odds of secondary sulfonylureafailure were 1.69-times higherin variant K23 carriers comparedwith E/E | [52] |
| Holstein et al.(2009) | Case–control study of 43 Type 2diabetes patients admitted tothe emergency departmentowing to severe hypoglycemia(patients treated with insulinwere excluded) and 54 Type 2diabetes control patientswithout a history of severehypoglycemia | Glibenclamide andglimepiride | Severe hypoglycemiadefined as a symptomaticevent requiring treatmentwith intravenous glucoseand confirmed by a glucosemeasurement <50 mg/dl | KCNJ11 E23K | The frequency of the variant K23allele was higher in the controlgroup (46%) versus the case group(31%). K/K homozygotes hadhigher HbA1c than the E/K or E/Egenotypes | [53] |
SUR1 (ABCC8)
Some of the first data to suggest that the polymorphisms in ABCC8 may alter the mechanism of action of sulfonylureas in patients with Type 2 diabetes came from a healthy volunteer study. Following mutational analysis of ABCC8, the combination of a synonymous polymorphism in exon 18 (Thr759Thr) and a C>T polymorphism at position −3 of the intron/exon 16 splice acceptor was associated with a 40% decrease in insulin secretion and a 50% decrease in serum C-peptide secretion following tolbutamide injection. There were no genotype differences in insulin or C-peptide secretion following intravenous glucose administration, which suggested that the observed diminished β-cell response following tolbutamide administration was possibly due to altered interactions of the ligand with its drug target [40]. When this hypothesis was tested in patients with Type 2 diabetes, the intron/exon 16 −3 polymorphism was not associated with differences in plasma glucose or insulin concentrations in patients receiving sulfonylureas compared with those on other therapies [41]. This population-based study did have limitations, with only one fasting blood sample drawn per patient. Thus, pre- and postglucose and insulin concentrations following sulfonylurea therapy were not available. Another observational study evaluated whether the frequency of the intron/exon 16 −3T allele differed in patients who had early sulfonylurea failure versus patients who had diabetes for at least 15 years and who were treated with sulfonylureas alone or in combination with metformin or acarbose [42]. No significant differences in intron/exon 16 −3 C>T genotype distributions or allele frequencies were observed between groups. Potential limitations of this study were a small sample size and the lack of a uniform definition of sulfonylurea failure. In summar, these two published studies in patients with Type 2 diabetes suggest that the intron/exon 16 −3 polymorphism does not influence sulfonylurea response. However, given the limitations of these study designs, this polymorphism merits investigation in additional cohorts of Type 2 diabetes patients receiving sulfonylurea therapy. Other polymorphisms in ABCC8, namely Ser1369Ala (T>G, exon 33), have been more consistently associated with differences in sulfonylurea response in patients with Type 2 diabetes. In terms of functional significance, the Ser1389Ala polymorphism is located in the second nucleotide-binding fold, and the Ala/Ala genotype is associated with decreased insulin secretion [43]. Zhang and colleagues prospectively evaluated the effects of the Ser1369Ala polymorphism on response to gliclazide 40 mg twice daily for 8 weeks in 115 patients with Type 2 diabetes [44]. The decrease in HbA1c from baseline to week 8 was greater in Ala allele carriers (−1.6%) compared with Ser/Ser homozygotes (−0.76%; p = 0.044). Of note, the change in fasting plasma glucose did not differ significantly between genotype groups. The authors hypothesized that the Ala allele conferred hypersensitivity to sulfonylurea therapy, resulting in more efficacious HbA1c reductions. The reason for the discrepancy between the HbA1c and fasting plasma glucose results is unclear, but is possibly due to differences in postprandial glucose levels between groups, which is captured by HbA1c measurements.
The ABCC8 Ser1369Ala polymorphism was also identified as an important determinant of sulfonylurea response in a large prospective study. Feng and colleagues investigated whether Type 2 diabetes candidate gene variants influenced response to gliclazide 40 mg twice daily following 8 weeks of therapy [45]. A total of 25 polymorphisms in 11 candidate genes were studied (Table 2). The study population (n = 1268) consisted of two cohorts based on the geographic location of the hospitals. In a pooled analysis of both cohorts, patients with the Ser/Ala and Ala/Ala genotypes had 2.8 and 7.7% greater decreases in fasting plasma glucose than Ser/Ser homozygotes. Decreases in 2-h glucose following an oral glucose tolerance test were also greater in the Ser/Ala (−10.8%) and Ala/Ala (−11.9%) genotype groups compared with Ser/Ser. Reductions in HbA1c from baseline to week 8 were 1.2, 1.5 and 1.7% in the Ser/Ser, Ser/Ala and Ala/Ala genotype groups, respectively, although these differences were not significant between groups. The odds of responding to gliclazide therapy were higher in the Ser/Ala (OR: 1.4, 95% CI: 1.0–2.1) and Ala/Ala groups (OR: 2.2; 95% CI: 1.4–3.6) compared with the Ser/Ser genotype. There was no association between genotype and change in insulin levels. Of note, only one other polymorphism of the 25 studied was significantly associated with the percent reduction in fasting plasma glucose. This SNP, rs5210, is located in the 3′ untranslated region of the KCNJ11 gene.
Although this prospective study had a large sample size, well-defined phenotypic end points and a replication cohort, there were some limitations that merit consideration. The treatment duration was short and the study excluded obese patients and patients who had diabetes for more than 5 years. Furthermore, it is unclear if these results can be applied to other racial groups, as this population was solely comprised of Chinese patients.
KCNJ11
As the pore of the KATP channel, polymorphisms in KCNJ11 have been studied regarding their relationship with sulfonylurea response in patients with Type 2 diabetes. The most widely studied polymorphism has been Glu23Lys (E23K). KCNJ11 E23K has emerged as a Type 2 diabetes risk allele [46-48]. Furthermore, in vitro studies have demonstrated that the variant K23 allele is associated with KATP channel overactivity (i.e., decreased ATP inhibitory sensitivity) and in some clinical studies, the variant K allele is associated with decreased insulin secretion in humans [47-49]. Of note, E23K is in strong linkage disequilibrium with the ABCC8 Ser1389Ala polymorphism; therefore, most carriers of a KCNJ11 K23 risk allele also carry an ABCC8 Ala1369 risk allele [47,50,51]. Thus, like ABCC8 Ala1369, it has been hypothesized that the KCNJ11 variant K23 allele may be associated with interindividual variability in sulfonylurea response.
This hypothesis was tested in a group of newly diagnosed diabetes patients treated with sulfonylureas who were participating in the UKPDS study [46]. In this study, the KCNJ11 E23K variant was not significantly associated with response to sulfonylurea therapy. In another study, Sesti and colleagues investigated whether the E23K variant was associated with an increased risk of secondary sulfonylurea failure in patients with Type 2 diabetes [52]. The study group was comprised of 525 Caucasian patients with Type 2 diabetes who were treated with glibenclamide up to 15 mg per day. The frequency of the K23 allele was 66.8% in patients with secondary sulfonylurea failure compared with 58% in patients who did not fail sulfonylurea therapy (OR: 1.45; 95% CI: 1.01–2.09). After adjusting for potential covariates, the odds of secondary sulfonylurea failure in K allele carriers were higher than in E/E homozygotes (OR: 1.69; 95% CI: 1.02–2.78). Of note, in the patient group without secondary sulfonylurea failure, the age of diabetes onset was lower and glucose levels were higher in patients who carried a K allele. However, there were no significant genotype differences in age of diabetes onset or glucose levels in the secondary sulfonylurea failure group. The reasons for an increased risk of secondary sulfonylurea failure with the E23K variant in the face of similar diabetes duration and glucose levels are unclear. Also, the reason why the K23 allele was associated with increased sulfonylurea failure in this study, whereas previous studies have shown improved sulfonylurea response with the linked ABCC8 Ala1369 allele, is unclear. The investigators also studied the effects of the E23K genotype on ex vivo human pancreatic islet cell function. Islets of carriers of the K allele demonstrated reduced insulin secretion following glibenclamide administration, although this did not reach statistical significance. Following 24 h of high glucose exposure, islets of carriers of the K allele had significantly lower glibenclamide-stimulated insulin secretion than E23 homozygotes.
Recently, the KCNJ11 E23K variant has been evaluated for its association with severe sulfonylurea-induced hypoglycemia [53]. Holstein and colleagues hypothesized that Type 2 diabetes patients who possess the variant K allele would have a decreased risk of severe hypoglycemia following sulfonylurea therapy. They conducted a case–control study of 43 patients with Type 2 diabetes taking sulfonylurea therapy (either glimepiride or glibenclamide) who were admitted to the hospital owing to severe hypoglycemia. The study included 54 matched control patients who also had Type 2 diabetes and who were also treated with sulfonylurea therapy, but who had not experienced severe hypoglycemia. The doses of sulfonylureas and the use of metformin were comparable between the case and control groups. The frequency of the K variant allele was higher in the control group compared with cases (46 vs 31%; OR: 0.54; 95% CI: 0.30–0.98; p = 0.04). In univariate logistic regression analysis, the presence of the K variant was associated with a decreased risk of sulfonylurea-induced hypoglycemia. However, after controlling for covariates, this genotype association was no longer significant. Additional analyses revealed that K/K homozygotes had significantly higher HbA1c than patients with the E/K or E/E genotypes (7.57, 6.89 and 6.77%, respectively; p = 0.04). The major limitation of this small observational study was that it was probably underpowered to assess the primary end point.
It is important to note that none of the KCNJ11 clinical studies prospectively set out to determine the influence of the E23K variant on sulfonylurea response. Thus, variability exists in the definition of sulfonylurea failure, choice and dose of sulfonylurea used, and the baseline demographics of the respective populations. In addition, most studies focused exclusively on the E23K polymorphism. Recently, a large, prospective study showed that another KCNJ11 polymorphism, rs5210, located in the 3′ untranslated region, was significantly associated with the percentage decrease in fasting glucose after 8 weeks of gliclazide therapy [45]. However, replication of this finding in other study populations has yet to be performed. Last, the strong linkage disequilibrium between the KCNJ11 K23 and ABCC8 Ala1369 risk alleles has prompted investigators to determine the functional consequences of this risk haplotype (i.e., K23/A1369). Using recombinant human KATP channels, Hamming and colleagues found that variant K23/A1369 KATP channels have increased sensitivity to gliclazide as compared with wild-type E23/S1369 KATP channels [54]. Furthermore, they found that the increased sensitivity to gliclazide with the K23/A1369 haplotype was a result of the ABCC8 A1369 risk allele. These data provide intriguing mechanistic hypotheses regarding the K23/A1369 risk haplotype and merit further investigation in prospective clinical trials of sulfonylurea response in patients with Type 2 diabetes.
Disease-risk polymorphisms & sulfonylurea response
Type 2 diabetes is a polygenic disease, and the biological pathways underlying the pathophysiology of Type 2 diabetes are numerous and complex. Genes hypothesized to predispose individuals to Type 2 diabetes are those involved in pancreatic β-cell function and development, insulin secretion, signaling and resistance, and glucose transport [55]. To date, candidate gene-association studies and genome-wide association studies have identified over 15 Type 2 diabetes risk genes [55]. In terms of sulfonylurea response, polymorphisms in genes that influence insulin, glucose or β-cell physiology could potentially confer differential responses to these agents in Type 2 diabetes. A summary of published studies evaluating the associations between Type 2 diabetes disease risk polymorphisms and sulfonylurea response is shown in Table 3.
Table 3. Summary of association studies evaluating polygenic Type 2 diabetes risk polymorphisms and sulfonylurea response in patients with Type 2 diabetes.
| Study author(year) | Study population | Sulfonylurea | Primary responsephenotype | Polymorphism(s)studied | Primary results | Ref. |
|---|---|---|---|---|---|---|
| Pearson et al.(2007) | Type 2 diabetes (with age ofdiagnosis >40 years and no use ofinsulin within 6 months of diagnosis)and incident sulfonylurea use(n = 901) in the GoDARTS data-base. Incident sulfonylurea use wasdefined as no previous diabetestreatment for at least 6 months priorto index sulfonylurea prescription.In addition, incident users wererequired to have at least one HbA1crecorded in the 3–12 months follow-ing sulfonylurea initiation | Various | Failure to attain HbA1c ≤7%in the 3–12 months follow-ing the index sulfonylureaprescription | TCF7L2 rs12255372G>T and rs7903146C>T | The odds of sulfonylurea failure were2.16-times higher in rs12255372T/T vs wild-type G/G, and 1.9-timeshigher in rs7903146 T/T vswild-type C/C | [62] |
| Sesti et al. (2004) | Type 2 diabetes patients (n = 477)recruited consecutively with thefollowing criteria: onset of diabetes>35 years of age, absence ofketonuria at diagnosis andnegative for antiglutamic aciddecarboxylase antibody | In most patients,glibenclamide 15 mgper dayMetformin wasadded if FPG was>300 mg/dl on twoseparate occasions | Secondary sulfonylurea fail-ure: those requiring insulindue to FPG >300 mg/dldespite sulfonylurea andmetformin therapy, appro-priate diet and the absenceof conditions that causehyperglycemia | IRS1 Gly972Arg | Arg972 variant carriers had two-timesthe risk of secondary sulfonylureafailure compared with wild-typeGly homozygotes | [68] |
| Becker et al.(2008) | Population-based cohort study (Rot-terdam). Incident sulfonylurea users(n = 619) during 1 January 1991and 1 January 2005 were includedin the analysis | Glibenclamide,tolbutamide, gliclazideand glimepiride | Change in prescribed dailydose; mortality | _NOS1AP_rs10494366 T>G | T/G genotype was associated with agreater change in prescribed gliben-clamide dose than the T/T genotype.In glibenclamide users, G allelecarriers had 2.8-times increased riskof mortality compared with T/T. Intolbutamide or glimepiride users,G allele carriers had 0.3-times the riskof mortality compared with T/T | [73] |
TCF7L2
TCF7L2 is a key transcription factor in the WNT signaling pathway that is involved in glucose homeostasis and lipid metabolism, proliferation and function of pancreatic β cells, and the production of glucagon-like peptide 1 [56]. Altered regulation of this signaling pathway has been implicated in the development of Type 2 diabetes, and TCF7L2 has emerged as a Type 2 diabetes susceptibility gene. Two intronic TCF7L2 polymorphisms, rs7903146 C>T and rs12255372 G>T, have been the strongest and most consistent predictors of increased Type 2 diabetes risk [56-59]. Meta-analysis data reveal that homozygosity for these variant alleles is associated with an approximately twofold increase in the risk for Type 2 diabetes compared with wild-type homozygotes [58]. These risk alleles were also associated with a fivefold increase in TCF7L2 protein expression in pancreatic islets from patients with Type 2 diabetes, which, in turn, was associated with reduced insulin secretion from these cells in vivo and in vitro [60,61]. Other hypothesized mechanisms by which TCF7L2 polymorphisms confer an increased risk of Type 2 diabetes include decreased β-cell mass, impaired insulin processing or release, impaired GLP-1 signaling in pancreatic β cells, decreased glucagon secretion and hepatic insulin resistance [59].
Given the putative role of TCF7L2 polymorphisms in β-cell function, Pearson and colleagues set out to determine the impact of TCF7L2 rs12255372 G>T and rs7903146 C>T on early response to sulfonylurea therapy in Scottish patients with Type 2 diabetes [62]. Using the Genetics of Diabetes Audit and Research Tayside (GoDARTS) database, the investigators identified 901 incident sulfonylurea users. The primary outcome of this study was the failure to attain an HbA1c 7% or less in the 3–12 months following the index sulfonylurea prescription. For rs1225372, 57% of patients with the T/T genotype failed to attain an HbA1c of 7% or less, compared with 41 and 40% of patients with the G/T and G/G genotypes, respectively (p = 0.006). For rs7903146, 53% of patients with the variant T/T genotype failed to attain an HbA1c of 7% or less compared with 42 and 40% of patients with the C/T and C/C genotypes, respectively (p = 0.035). In logistic regression analysis, the odds of sulfonylurea failure were higher in patients with the rs12255372 T/T genotype compared with the G/G genotype (OR: 1.94; 95% CI: 1.23–3.06; p = 0.005) and higher in patients with the rs7903146 T/T genotype compared with the C/C genotype (OR: 1.73; 95% CI: 1.11–2.70; p = 0.015). When baseline HbA1c was included as a covariate in the regression models, the associations between TCF7L2 genotype and sulfonylurea failure were strengthened. The odds of sulfonylurea failure for rs12255372 T/T vs G/G were 2.16 (95% CI: 1.21–3.86; p = 0.009), and for rs7903146, 1.90 (95% CI: 1.09–3.33; p = 0.024).
This study also investigated whether TCF7L2 polymorphisms influenced metformin response in incident metformin users in the GoDARTS database. Unlike the sulfonylurea findings, TCF7L2 polymorphisms were not associated with metformin treatment failure in this population. The finding that sulfonylurea, but not metformin response, was affected by TCF7L2 polymorphism lends indirect support to the hypothesis that the functional consequences of TCF7L2 polymorphisms are to alter β-cell function. To date, this is the only study that has investigated the association between TCF7L2 polymorphisms and sulfonylurea response, and there are limitations associated with observational studies (e.g., nonrandomized design, lack of rigorous compliance assessment measures and potential prescriber bias). Nonetheless, these findings are intriguing, particularly since the frequency of variant homozygotes is common in the population (i.e., genotype frequency of 12%).
Insulin receptor substrate-1
Insulin receptor substrate (IRS)-1 is an important signal-transduction protein that mediates the metabolic effects of insulin. A common polymorphism, Gly972Arg, in the IRS-1 gene was associated with an increased risk of Type 2 diabetes in one meta-analysis [63], but these results were not replicated by other large-scale genetic association studies [64,65]. The variant Arg allele has been associated with decreased glucose-stimulated insulin secretion and decreased insulin response to sulfonylureas in ex vivo and in vitro studies [66,67]. Sesti and colleagues investigated whether the IRS-1 Gly972Arg polymorphism was associated with an increased risk of secondary sulfonylurea failure in patients with Type 2 diabetes (n = 477) [68]. The frequency of the Arg972 allele was higher in patients with secondary sulfonylurea failure compared with those well controlled on sulfonylurea therapy (16.7 vs 8.7%, respectively). In logistic regression analysis, Arg972 allele carriers had two-times the risk of secondary sulfonylurea failure as compared with noncarriers. This finding was independent of age, sex, BMI, metabolic control, age at diagnosis, duration of diabetes and the _PPAR_γ Pro12Ala genotype.
Nitric oxide synthase 1 adaptor protein
Nitric oxide synthase 1 adaptor protein (NOS1AP) binds to and regulates neuronal nitric oxide synthase. Neuronal nitric oxide synthase is hypothesized to reduce intracellular calcium levels through its effects on voltage-gated calcium channels. Subsequently, this enzyme plays a role in the electrical current of the heart and in insulin release from pancreatic β cells [69-72]. Given the relationship between NOS1AP and insulin release, Becker and colleagues set out to determine if NOS1AP polymorphisms are associated with efficacy and mortality risk in sulfonylurea-treated patients with Type 2 diabetes [73]. In this population-based cohort study of incident sulfonylurea users, patients with the rs10494366 T/G genotype had a greater change in the prescribed dose of glibenclamide compared with the wildtype T/T genotype. However, rs10494366 genotype was not associated with differences in prescribed doses of tolbutamide, gliclazide or glimepiride. In glibenclamide users, variant G allele carriers had an increased risk of mortality compared with the T/T genotype (HR: 2.8; 95% CI: 1.09–7.22). By contrast, mortality risk was lower in tolbutamide and glimepiride users who carried a G allele compared with the T/T genotype (tolbutamide, HR: 0.30; glimepiride, HR: 0.18). Of note, no genotype differences in mortality were observed in metformin or insulin users. The mechanisms through which this polymorphism influenced mortality risk and the reason why this association differed based on the type of sulfonylurea used are unclear. The authors hypothesized that it may be due to differences in the effect of sulfonylureas on other potassium channels, such as calcium-dependent potassium channels, or other sulfonylurea receptor subtypes, such as SUR2A.
Expert commentary
Sulfonylurea efficacy is intrinsically linked to β-cell physiology; therefore, the future study of sulfonylurea pharmacogenomics will be a challenging and complex process. Genes encoding the direct targets of sulfonylurea action (i.e., ABCC8 and KCNJ11) are the most logical candidate genes for investigation. To date, the ABCC8 Ser1369Ala polymorphism has been significantly associated with gliclazide response in two prospective studies. Both of these studies were conducted in Asian populations, therefore the applicability of these findings to patients of other races is unclear. Furthermore, both of these studies included only an 8-week treatment period. Therefore, the impact of these polymorphisms on long-term sulfonylurea response and durability outcomes is not known. The KCNJ11 polymorphism E23K is in strong linkage disequilibrium with the ABCC8 Ser1369Ala polymorphism. Recent data suggest that the K23/A1369 risk haplotype confers increased sensitivity to gliclazide in vitro, and this effect is primarily governed by the A1369 risk allele. In terms of Type 2 diabetes risk genes, polymorphisms in TCF7L2 and IRS-1 have been implicated as predictors of sulfonylurea response in two studies. However, to date, neither of these findings have been investigated in replication cohorts. Recently, a polymorphism in NOS1AP has been associated with changes in prescribed glibenclamide doses. Interestingly, this polymorphism also conferred differences in mortality risk based on the type of sulfonylurea used. These findings have also yet to be investigated in additional cohorts. Taken together, a number of notable drug target and Type 2 diabetes disease risk polymorphisms have been implicated as important determinants of sulfonylurea response; however, the clinical utility of this information is uncertain as most of these findings have yet to be replicated.
Currently, there are some limitations associated with sulfonylurea pharmacogenomic data. Published studies differ substantially in their designs (e.g., prospective vs population-based cohorts), inclusion criteria, choice of sulfonylurea, treatment duration and outcomes of interest. With the exception of a few investigations, most of the existing sulfonylurea pharmacogenomic studies have been retrospective. In addition, some studies have failed to exclude patients who potentially have monogenic forms of diabetes, such as maturity-onset diabetes of the young, or Type 1.5 diabetes. This is important, as mutations causing monogenic forms of diabetes (e.g., hepatocyte nuclear factor-1α) have been shown to influence response to sulfonylurea therapy [74]. Another challenge with interpreting the sulfonylurea pharmacogenomic literature is the lack of formal definitions of sulfonylurea failure. Heterogeneity in the definitions of sulfonylurea response and failure may lead to inconsistencies in the observed findings between studies for a given polymorphism. Last, while many studies have identified genetic associations with sulfonylurea response, none of the studies have been designed to elucidate how this genetic information can be used to select therapy and guide sulfonylurea dosing. It is not known whether a priori genetic knowledge will decrease the chance of long-term adverse clinical outcomes, particularly microvascular complications, in patients with Type 2 diabetes.
Five-year view
To date, only a limited number of polymorphisms in sulfonylurea drug target genes and Type 2 disease risk genes have been studied, and most of the results have been limited to small, observational studies. Given the complex pathophysiology of Type 2 diabetes and the role of the sulfonylureas in modulating this disease process (i.e., promoting insulin secretion), future studies will likely need to take a genome-wide association approach, or be conducted as association studies with hundreds of biologically relevant genes in order to identify the polymorphisms that are most predictive of sulfonylurea response. Future studies will also need to differentiate between the influence of polymorphisms on early sulfonylurea response and secondary sulfonylurea failure, as different genes are likely to affect sulfonylurea response as Type 2 diabetes progresses. In conclusion, the field of sulfonylurea pharmacogenomics has made some notable findings in recent years. In the coming years, more comprehensive assessments of these existing sulfonylurea pharmacogenomic associations will be necessary in order to determine if genetic information has clinical utility in improving the pharmacotherapeutic management of Type 2 diabetes.
Key issues.
- The sulfonylureas are oral agents used in the treatment of Type 2 diabetes. Sulfonylureas lower glycated hemoglobin levels by 1–2%.
- Sulfonylureas bind to ATP-sensitive potassium channels on pancreatic β cells and stimulate insulin release in a glucose-independent manner.
- Although the sulfonylureas are effective antihyperglycemic agents, interindividual variability exists in drug response and disposition.
- Recently, polymorphisms in drug target genes (i.e., ATP-binding cassette, subfamily C, member 8 [_ABCC8_] and potassium inwardly-rectifying channel, subfamily J, member 11 [_KCNJ11_]) and diabetes risk genes (e.g., TCF7L2 and insulin receptor substrate [_IRS-1_]) have been associated with variability in sulfonylurea response in patients with Type 2 diabetes.
- ABCC8 encodes the regulatory subunit of the sulfonylurea receptor, and the ABCC8 Ser1369Ala polymorphism has been associated with differential response to sulfonylurea therapy in patients with Type 2 diabetes. Clinical studies have shown that patients carrying the variant Ala allele have greater sulfonylurea-mediated reductions in fasting plasma glucose and glycated hemoglobin levels than wild-type homozygotes.
- KCNJ11 encodes Kir6.2, the pore subunit of the sulfonylurea receptor, and the KCNJ11 E23K polymorphism is associated with interindividual variability in sulfonylurea response and adverse effects in patients with Type 2 diabetes.
- The KCNJ11 E23K and ABCC8 Ser1369Ala polymorphisms are in strong linkage disequilibrium. Recent data suggest that the K23/Ala1369 risk haplotype confers increased sensitivity to gliclazide in vitro. This finding is primarily governed by the effects of the Ala1369 risk allele.
- TCF7L2 is a transcription factor in the WNT signaling pathway and it is a Type 2 diabetes risk gene. Polymorphisms in TCF7L2 have been associated with differential response to sulfonylurea therapy in patients with Type 2 diabetes.
- IRS-1 is a signal transduction protein that mediates the metabolic effects of insulin. The IRS-1 Gly972Arg polymorphism is associated with an increased risk of Type 2 diabetes and an increased risk of secondary sulfonylurea failure in patients with Type 2 diabetes.
- While the field of sulfonylurea pharmacogenomics has made some notable findings in recent years, few replication studies have been conducted. In the future, additional studies are also needed to determine how this genetic information can be used to select therapy and guide dosing, or if a priori genetic knowledge can be used to decrease the chance of long-term clinical outcomes, particularly microvascular complications, in patients with Type 2 diabetes.
Acknowledgments
Christina Aquilante currently holds investigator-initiated research grants from the NIH (K23 DK073197), the American College of Clinical Pharmacy and Tibotec Therapeutics.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365(9467):1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.American Diabetes Association Standards of medical care in diabetes – 2009. Diabetes Care. 2009;32(Suppl. 1):S13–S61. doi: 10.2337/dc09-S013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UK Prospective Diabetes Study (UKPDS) group Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with Type 2 diabetes (UKPDS 33) Lancet. 1998;352(9131):837–853. No authors listed. [PubMed] [Google Scholar]
- 4.UK Prospective Diabetes Study (UKPDS) group Effect of intensive blood-glucose control with metformin on complications in overweight patients with Type 2 diabetes (UKPDS 34) Lancet. 1998;352(9131):854–865. No authors listed. [PubMed] [Google Scholar]
- 5.Ohkubo Y, Kishikawa H, Araki E, et al. Intensive insulin therapy prevents the progression of diabetic microvascular complications in Japanese patients with non-insulin-dependent diabetes mellitus: a randomized prospective 6-year study. Diabetes Res. Clin. Pract. 1995;28(2):103–117. doi: 10.1016/0168-8227(95)01064-k. [DOI] [PubMed] [Google Scholar]
- 6.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in Type 2 diabetes. N. Engl. J. Med. 2008;359(15):1577–1589. doi: 10.1056/NEJMoa0806470. [DOI] [PubMed] [Google Scholar]
- 7.Gerstein HC, Miller ME, Byington RP, et al. Effects of intensive glucose lowering in Type 2 diabetes. N. Engl. J. Med. 2008;358(24):2545–2559. doi: 10.1056/NEJMoa0802743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A, MacMahon S, Chalmers J, et al. Intensive blood glucose control and vascular outcomes in patients with Type 2 diabetes. N. Engl. J. Med. 2008;358(24):2560–2572. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 9.Duckworth W, Abraira C, Moritz T, et al. Glucose control and vascular complications in veterans with Type 2 diabetes. N. Engl. J. Med. 2009;360(2):129–139. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 10.Kelly TN, Bazzano LA, Fonseca VA, Thethi TK, Reynolds K, He J. Systematic review: glucose control and cardiovascular disease in Type 2 diabetes. Ann. Intern. Med. 2009;151(6):394–403. doi: 10.7326/0003-4819-151-6-200909150-00137. [DOI] [PubMed] [Google Scholar]
- 11.Ray KK, Seshasai SR, Wijesuriya S, et al. Effect of intensive control of glucose on cardiovascular outcomes and death in patients with diabetes mellitus: a meta-analysis of randomised controlled trials. Lancet. 2009;373(9677):1765–1772. doi: 10.1016/S0140-6736(09)60697-8. [DOI] [PubMed] [Google Scholar]
- 12.Nathan DM, Buse JB, Davidson MB, et al. Medical management of hyperglycemia in Type 2 diabetes: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2009;32(1):193–203. doi: 10.2337/dc08-9025. • Provides an up-to-date overview of Type 2 diabetes treatment recommendations.
- 13.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann. Intern. Med. 2006;145(10):749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchheiner J, Roots I, Goldammer M, Rosenkranz B, Brockmoller J. Effect of genetic polymorphisms in cytochrome p450 (CYP) 2C9 and CYP2C8 on the pharmacokinetics of oral antidiabetic drugs: clinical relevance. Clin. Pharmacokinet. 2005;44(12):1209–1225. doi: 10.2165/00003088-200544120-00002. • Comprehensive review of reported associations between drug-metabolizing enzyme polymorphisms and sulfonylurea pharmacokinetics.
- 15.Pacanowski MA, Hopley CW, Aquilante CL. Interindividual variability in oral antidiabetic drug disposition and response: the role of drug transporter polymorphisms. Expert Opin. Drug Metab. Toxicol. 2008;4(5):529–544. doi: 10.1517/17425255.4.5.529. • Provides a review of the role of drug transporters and drug transporter polymorphisms on sulfonylurea disposition.
- 16.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic β-cell diabetes. Nat. Clin. Pract. Endocrinol. Metab. 2008;4(4):200–213. doi: 10.1038/ncpendmet0778. • Comprehensive review of monogenic forms of diabetes.
- 17.Gloyn AL, Ellard S. Defining the genetic aetiology of monogenic diabetes can improve treatment. Expert Opin. Pharmacother. 2006;7(13):1759–1767. doi: 10.1517/14656566.7.13.1759. • Comprehensive review of monogenic forms of diabetes and how this information can be used to guide pharmacologic management of the disease.
- 18.Reis AF, Velho G. Sulfonylurea receptor-1 (SUR1): genetic and metabolic evidences for a role in the susceptibility to Type 2 diabetes mellitus. Diabetes Metab. 2002;28(1):14–19. [PubMed] [Google Scholar]
- 19.Marchetti P, Navalesi R. Pharmacokinetic–pharmacodynamic relationships of oral hypoglycaemic agents. An update. Clin. Pharmacokinet. 1989;16(2):100–128. doi: 10.2165/00003088-198916020-00004. [DOI] [PubMed] [Google Scholar]
- 20.Efanova IB, Zaitsev SV, Zhivotovsky B, et al. Glucose and tolbutamide induce apoptosis in pancreatic β-cells. A process dependent on intracellular Ca2+ concentration. J. Biol. Chem. 1998;273(50):33501–33507. doi: 10.1074/jbc.273.50.33501. [DOI] [PubMed] [Google Scholar]
- 21.Maedler K, Carr RD, Bosco D, Zuellig RA, Berney T, Donath MY. Sulfonylurea induced β-cell apoptosis in cultured human islets. J. Clin. Endocrinol. Metab. 2005;90(1):501–506. doi: 10.1210/jc.2004-0699. [DOI] [PubMed] [Google Scholar]
- 22.Del Guerra S, Marselli L, Lupi R, et al. Effects of prolonged in vitro exposure to sulphonylureas on the function and survival of human islets. J. Diabetes Complicat. 2005;19(1):60–64. doi: 10.1016/j.jdiacomp.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Del Prato S, Pulizzi N. The place of sulfonylureas in the therapy for Type 2 diabetes mellitus. Metabolism. 2006;55(5 Suppl. 1):S20–S27. doi: 10.1016/j.metabol.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Krentz AJ, Bailey CJ. Oral antidiabetic agents: current role in Type 2 diabetes mellitus. Drugs. 2005;65(3):385–411. doi: 10.2165/00003495-200565030-00005. [DOI] [PubMed] [Google Scholar]
- 25.Meinert CL, Knatterud GL, Prout TE, Klimt CR. A study of the effects of hypoglycemic agents on vascular complications in patients with adult-onset diabetes. II. Mortality results. Diabetes. 1970;19(Suppl.):789–830. [PubMed] [Google Scholar]
- 26.Thisted H, Johnsen SP, Rungby J. Sulfonylureas and the risk of myocardial infarction. Metabolism. 2006;55(5 Suppl. 1):S16–S19. doi: 10.1016/j.metabol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Kar P, Holt RI. The effect of sulphonylureas on the microvascular and macrovascular complications of diabetes. Cardiovasc. Drugs Ther. 2008;22(3):207–213. doi: 10.1007/s10557-008-6090-2. [DOI] [PubMed] [Google Scholar]
- 28.Stevens RJ, Coleman RL, Adler AI, Stratton IM, Matthews DR, Holman RR. Risk factors for myocardial infarction case fatality and stroke case fatality in Type 2 diabetes: UKPDS 66. Diabetes Care. 2004;27(1):201–207. doi: 10.2337/diacare.27.1.201. [DOI] [PubMed] [Google Scholar]
- 29.DeFronzo RA. Pharmacologic therapy for Type 2 diabetes mellitus. Ann. Intern. Med. 1999;131(4):281–303. doi: 10.7326/0003-4819-131-4-199908170-00008. [DOI] [PubMed] [Google Scholar]
- 30.Kahn SE, Haffner SM, Heise MA, et al. Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N. Engl. J. Med. 2006;355(23):2427–2443. doi: 10.1056/NEJMoa066224. [DOI] [PubMed] [Google Scholar]
- 31.Kirchheiner J, Bauer S, Meineke I, et al. Impact of CYP2C9 and CYP2C19 polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics. 2002;12(2):101–109. doi: 10.1097/00008571-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Kirchheiner J, Brockmoller J, Meineke I, et al. Impact of CYP2C9 amino acid polymorphisms on glyburide kinetics and on the insulin and glucose response in healthy volunteers. Clin. Pharmacol. Ther. 2002;71(4):286–296. doi: 10.1067/mcp.2002.122476. [DOI] [PubMed] [Google Scholar]
- 33.Becker ML, Visser LE, Trienekens PH, Hofman A, van Schaik RH, Stricker BH. Cytochrome P450 2C9 *2 and *3 polymorphisms and the dose and effect of sulfonylurea in Type II diabetes mellitus. Clin. Pharmacol. Ther. 2008;83(2):288–292. doi: 10.1038/sj.clpt.6100273. [DOI] [PubMed] [Google Scholar]
- 34.Zhou K, Donnelly L, Burch L, et al. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in Type 2 diabetes: a Go-DARTS study. Clin. Pharmacol. Ther. 2009;87(1):52–56. doi: 10.1038/clpt.2009.176. [DOI] [PubMed] [Google Scholar]
- 35.Holstein A, Plaschke A, Ptak M, et al. Association between CYP2C9 slow metabolizer genotypes and severe hypoglycaemia on medication with sulphonylurea hypoglycaemic agents. Br. J. Clin. Pharmacol. 2005;60(1):103–106. doi: 10.1111/j.1365-2125.2005.02379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flanagan SE, Clauin S, Bellanne-Chantelot C, et al. Update of mutations in the genes encoding the pancreatic β-cell KATP channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum. Mutat. 2009;30(2):170–180. doi: 10.1002/humu.20838. • Review of key ABCC8 and KCNJ11 mutations and their role in monogenic forms of diabetes.
- 37.Sattiraju S, Reyes S, Kane GC, Terzic A. KATP channel pharmacogenomics: from bench to bedside. Clin. Pharmacol. Ther. 2008;83(2):354–357. doi: 10.1038/sj.clpt.6100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rafiq M, Flanagan SE, Patch AM, Shields BM, Ellard S, Hattersley AT. Effective treatment with oral sulfonylureas in patients with diabetes due to sulfonylurea receptor 1 (SUR1) mutations. Diabetes Care. 2008;31(2):204–209. doi: 10.2337/dc07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson ER, Flechtner I, Njolstad PR, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N. Engl. J. Med. 2006;355(5):467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- 40.Hansen T, Echwald SM, Hansen L, et al. Decreased tolbutamide-stimulated insulin secretion in healthy subjects with sequence variants in the high-affinity sulfonylurea receptor gene. Diabetes. 1998;47(4):598–605. doi: 10.2337/diabetes.47.4.598. [DOI] [PubMed] [Google Scholar]
- 41.Meirhaeghe A, Helbecque N, Cottel D, et al. Impact of sulfonylurea receptor 1 genetic variability on non-insulin-dependent diabetes mellitus prevalence and treatment: a population study. Am. J. Med. Genet. 2001;101(1):4–8. doi: 10.1002/ajmg.1297. [DOI] [PubMed] [Google Scholar]
- 42.Zychma MJ, Gumprecht J, Strojek K, et al. Sulfonylurea receptor gene 16–13 polymorphism – association with sulfonylurea or insulin treatment in Type 2 diabetic subjects. Med. Sci. Monit. 2002;8(7):CR512–CR515. [PubMed] [Google Scholar]
- 43.Florez JC, Jablonski KA, Kahn SE, et al. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes. 2007;56(2):531–536. doi: 10.2337/db06-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Liu X, Kuang H, Yi R, Xing H. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in Type 2 diabetes. Diabetes Res. Clin. Pract. 2007;77(1):58–61. doi: 10.1016/j.diabres.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Feng Y, Mao G, Ren X, et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese Type 2 diabetic patients. Diabetes Care. 2008;31(10):1939–1944. doi: 10.2337/dc07-2248. • The largest sulfonylurea pharmacogenomic study to date in terms of the number of patients evaluated and the number of polymorphisms and genes studied.
- 46.Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, Turner RC. Association studies of variants in promoter and coding regions of β-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53) Diabet. Med. 2001;18(3):206–212. doi: 10.1046/j.1464-5491.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 47.Florez JC, Burtt N, de Bakker PI, et al. Haplotype structure and genotype–phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53(5):1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen EM, Hansen L, Carstensen B, et al. The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of Type 2 diabetes. Diabetes. 2003;52(2):573–577. doi: 10.2337/diabetes.52.2.573. [DOI] [PubMed] [Google Scholar]
- 49.Schwanstecher C, Meyer U, Schwanstecher M. K(IR)6.2 polymorphism predisposes to Type 2 diabetes by inducing overactivity of pancreatic β-cell ATP-sensitive K+ channels. Diabetes. 2002;51(3):875–879. doi: 10.2337/diabetes.51.3.875. [DOI] [PubMed] [Google Scholar]
- 50.Inoue H, Ferrer J, Warren-Perry M, et al. Sequence variants in the pancreatic islet β-cell inwardly rectifying K+ channel Kir6.2 (Bir) gene: identification and lack of role in Caucasian patients with NIDDM. Diabetes. 1997;46(3):502–507. doi: 10.2337/diab.46.3.502. [DOI] [PubMed] [Google Scholar]
- 51.Barroso I, Luan J, Middelberg RP, et al. Candidate gene association study in Type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS Biol. 2003;1(1):E20. doi: 10.1371/journal.pbio.0000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sesti G, Laratta E, Cardellini M, et al. The E23K variant of KCNJ11 encoding the pancreatic β-cell adenosine 5′-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with Type 2 diabetes. J. Clin. Endocrinol. Metab. 2006;91(6):2334–2339. doi: 10.1210/jc.2005-2323. [DOI] [PubMed] [Google Scholar]
- 53.Holstein A, Hahn M, Stumvoll M, Kovacs P. The E23K variant of KCNJ11 and the risk for severe sulfonylurea-induced hypoglycemia in patients with Type 2 diabetes. Horm. Metab. Res. 2009;41(5):387–390. doi: 10.1055/s-0029-1192019. [DOI] [PubMed] [Google Scholar]
- 54.Hamming KS, Soliman D, Matemisz LC, et al. Coexpression of the Type 2 diabetes susceptibility gene variants KCNJ11 E23K and ABCC8 S1369A alter the ATP and sulfonylurea sensitivities of the ATP-sensitive K+ channel. Diabetes. 2009;58(10):2419–2424. doi: 10.2337/db09-0143. • Elucidates the molecular mechanisms of the K23/A1369 risk haplotype.
- 55.Grant RW, Moore AF, Florez JC. Genetic architecture of Type 2 diabetes: recent progress and clinical implications. Diabetes Care. 2009;32(6):1107–1114. doi: 10.2337/dc08-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin T. The WNT signalling pathway and diabetes mellitus. Diabetologia. 2008;51(10):1771–1780. doi: 10.1007/s00125-008-1084-y. [DOI] [PubMed] [Google Scholar]
- 57.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of Type 2 diabetes. Nat. Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 58.Tong Y, Lin Y, Zhang Y, Yang J, Liu H, Zhang B. Association between TCF7L2 gene polymorphisms and susceptibility to Type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med. Genet. 2009;10:15. doi: 10.1186/1471-2350-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearson ER. Translating TCF7L2: from gene to function. Diabetologia. 2009;52(7):1227–1230. doi: 10.1007/s00125-009-1356-1. [DOI] [PubMed] [Google Scholar]
- 60.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of Type 2 diabetes. J. Clin. Invest. 2007;117(8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hattersley AT, Pearson ER. Minireview: pharmacogenetics and beyond: the interaction of therapeutic response, β-cell physiology, and genetics in diabetes. Endocrinology. 2006;147(6):2657–2663. doi: 10.1210/en.2006-0152. [DOI] [PubMed] [Google Scholar]
- 62.Pearson ER, Donnelly LA, Kimber C, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56(8):2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 63.Jellema A, Zeegers MP, Feskens EJ, Dagnelie PC, Mensink RP. Gly972Arg variant in the insulin receptor substrate-1 gene and association with Type 2 diabetes: a meta-analysis of 27 studies. Diabetologia. 2003;46(7):990–995. doi: 10.1007/s00125-003-1126-4. [DOI] [PubMed] [Google Scholar]
- 64.Zeggini E, Parkinson J, Halford S, et al. Association studies of insulin receptor substrate 1 gene (IRS1) variants in Type 2 diabetes samples enriched for family history and early age of onset. Diabetes. 2004;53(12):3319–3322. doi: 10.2337/diabetes.53.12.3319. [DOI] [PubMed] [Google Scholar]
- 65.Florez JC, Sjogren M, Burtt N, et al. Association testing in 9,000 people fails to confirm the association of the insulin receptor substrate-1 G972R polymorphism with Type 2 diabetes. Diabetes. 2004;53(12):3313–3318. doi: 10.2337/diabetes.53.12.3313. [DOI] [PubMed] [Google Scholar]
- 66.Porzio O, Federici M, Hribal ML, et al. The Gly972-->Arg amino acid polymorphism in IRS-1 impairs insulin secretion in pancreatic β cells. J. Clin. Invest. 1999;104(3):357–364. doi: 10.1172/JCI5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchetti P, Lupi R, Federici M, et al. Insulin secretory function is impaired in isolated human islets carrying the Gly(972)-->Arg IRS-1 polymorphism. Diabetes. 2002;51(5):1419–1424. doi: 10.2337/diabetes.51.5.1419. [DOI] [PubMed] [Google Scholar]
- 68.Sesti G, Marini MA, Cardellini M, et al. The Arg972 variant in insulin receptor substrate-1 is associated with an increased risk of secondary failure to sulfonylurea in patients with Type 2 diabetes. Diabetes Care. 2004;27(6):1394–1398. doi: 10.2337/diacare.27.6.1394. [DOI] [PubMed] [Google Scholar]
- 69.Schulz R, Rassaf T, Massion PB, Kelm M, Balligand JL. Recent advances in the understanding of the role of nitric oxide in cardiovascular homeostasis. Pharmacol. Ther. 2005;108(3):225–256. doi: 10.1016/j.pharmthera.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 70.Massion PB, Pelat M, Belge C, Balligand JL. Regulation of the mammalian heart function by nitric oxide. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005;142(2):144–150. doi: 10.1016/j.cbpb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 71.Lajoix AD, Reggio H, Chardes T, et al. A neuronal isoform of nitric oxide synthase expressed in pancreatic β-cells controls insulin secretion. Diabetes. 2001;50(6):1311–1323. doi: 10.2337/diabetes.50.6.1311. [DOI] [PubMed] [Google Scholar]
- 72.Gunawardana SC, Rocheleau JV, Head WS, Piston DW. Mechanisms of time-dependent potentiation of insulin release: involvement of nitric oxide synthase. Diabetes. 2006;55(4):1029–1033. doi: 10.2337/diabetes.55.04.06.db05-1532. [DOI] [PubMed] [Google Scholar]
- 73.Becker ML, Aarnoudse AJ, Newton-Cheh C, et al. Common variation in the NOS1AP gene is associated with reduced glucose-lowering effect and with increased mortality in users of sulfonylurea. Pharmacogenet. Genomics. 2008;18(7):591–597. doi: 10.1097/FPC.0b013e328300e8c5. [DOI] [PubMed] [Google Scholar]
- 74.Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362(9392):1275–1281. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]