Small molecule BDNF mimetics activate TrkB signaling and prevent neuronal degeneration in rodents (original) (raw)
Abstract
Brain-derived neurotrophic factor (BDNF) activates the receptor tropomyosin-related kinase B (TrkB) with high potency and specificity, promoting neuronal survival, differentiation, and synaptic function. Correlations between altered BDNF expression and/or function and mechanism(s) underlying numerous neurodegenerative conditions, including Alzheimer disease and traumatic brain injury, suggest that TrkB agonists might have therapeutic potential. Using in silico screening with a BDNF loop–domain pharmacophore, followed by low-throughput in vitro screening in mouse fetal hippocampal neurons, we have efficiently identified small molecules with nanomolar neurotrophic activity specific to TrkB versus other Trk family members. Neurotrophic activity was dependent on TrkB and its downstream targets, although compound-induced signaling activation kinetics differed from those triggered by BDNF. A selected prototype compound demonstrated binding specificity to the extracellular domain of TrkB. In in vitro models of neurodegenerative disease, it prevented neuronal degeneration with efficacy equal to that of BDNF, and when administered in vivo, it caused hippocampal and striatal TrkB activation in mice and improved motor learning after traumatic brain injury in rats. These studies demonstrate the utility of loop modeling in drug discovery and reveal what we believe to be the first reported small molecules derived from a targeted BDNF domain that specifically activate TrkB.We propose that these compounds constitute a novel group of tools for the study of TrkB signaling and may provide leads for developing new therapeutic agents for neurodegenerative diseases.
Introduction
Nerve growth factor (NGF), brain-derived neurotrophin factor (BDNF), and neurotrophin-3 (NT-3) are members of the neurotrophin protein family and act through their cognate tropomyosin-related kinase (Trk) receptors (NGF/TrkA, BDNF/TrkB, NT-3/TrkC) and the common neurotrophin receptor p75 (p75NTR). Trks are activated by binding of mature neurotrophin dimers and multimerization (though the precise mechanisms remain unclear), leading to phosphorylation and signaling adaptor recruitment (1). Trk activation promotes neuronal survival, differentiation, and synaptic function (1–3). p75NTR signaling is complex, involving coreceptor (e.g., sortilin) interactions, proteolytic processing, and endocytosis, with coupling to both survival and apoptosis-inducing mechanisms (e.g., refs. 4–6). Engagement of p75NTR by neurotrophins also modulates Trk activity (7, 8). BDNF and TrkB are of particular therapeutic interest. Correlations between alterations in BDNF expression and/or function and mechanism(s) occurring in Alzheimer disease (9), Huntington disease (10), Parkinson disease (11), Rett syndrome (12), traumatic brain injury (TBI), (13, 14), and aging (15) point to the therapeutic potential of TrkB agonists. The findings that TrkB is important for long-term survival, differentiation, and function of newborn neurons in the adult hippocampus (16–18), and that neurogenesis plays a fundamental role in depression, suggest that discovery of TrkB ligands might open new treatment avenues for this disorder (19). Finally, modulation of TrkB signaling could have a therapeutic role in obesity or anorexia (20).
A number of properties limit the therapeutic application of BDNF. Its plasma half-life in rats is less than 1 minute, and it has poor blood brain barrier penetration (21) and poor brain intraparenchymal penetration (22). In addition, BDNF interaction with p75NTR might contribute to its ability to promote pain (23) and other undesired effects. Thus, a long-sought goal has been the development of non-peptide, small molecule ligands capable of activating TrkB signaling with high potency and specificity (24). In the present study, we applied in silico screening with a pharmacophore modeled on a BDNF loop domain likely interacting with TrkB, coupled with low-throughput in vitro neurotrophic assays to identify the first small molecule TrkB ligands mimicking a BDNF active site.
Results
Computational modeling, pharmacophore generation, and virtual screening.
There was no available structure, for any neurotrophin-Trk pairing, of the binding of the β-loops of interest to the receptor, and no known BDNF-derived oligopeptides with TrkB-activating activity with which to constrain the model. Therefore, we began with previous studies of receptor binding and biologic activity of chimeric mutant proteins generated by successive substitution of homologous BDNF regions into an NGF backbone, which identified regions of BDNF involved in TrkB activation and Trk specificity (25, 26). Examination of those results suggested loop II subregion b (SKGQL) (Figure 1), a region sufficiently restricted to produce a small molecule model, which was associated with a substantial portion of BDNF-like activity and not apparently critical for TrkB binding, as a candidate for pharmacophore modeling. This region was modeled and a chemical feature hypothesis (shown in Figure 1A) generated, as described in Methods. Using this pharmacophore, an average of 35 conformers of each of more than 1,000,000 available compounds (including libraries from Asinex, Comgenex [now AMRI], Interbioscreen, Sigma-Aldrich [Rare Chemical Library], Timtec, and Chemstar) were screened, yielding 1,785 candidate compounds that fit with a calculated internal energy of less than 10 kcal/mol. This number was further reduced to 14 by visual inspection and other criteria, as follows: (a) since the pharmacophore used did not provide for excluded spaces, a rough (visual, qualitative) filter was applied that disfavored compounds that, in their lowest energy conformation best fitting the pharmacophore, had portions that would interfere with the interaction of pharmacophore-determined compound chemical features with a putative shallow-pocket receptor site (this criterion was the most robust in excluding compounds); (b) compounds were favored in which pharmacophore-correspondent features were connected by several rotatable bonds, providing theoretical flexibility of relative positioning; (c) compounds of lower molecular weight were favored, but those greater than 500 Da, up to 650 Da, were not automatically excluded — otherwise, Lipinski’s rule of 5 criteria was applied; and (d) compounds marginal on any of the preceding criteria, and also barely fitting the pharmacophore, could be excluded. Of these 14 compounds, 7 were obtained from commercial sources for in vitro analysis.
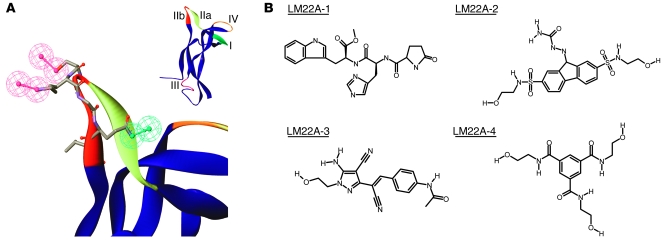
Figure 1. Pharmacophore and LM22A compound structures.
(A) Inset: Variable loop regions of a human BDNF monomer extracted from the crystallographically determined structure of a BDNF-NT3 heterodimer (56). The larger image shows the loop IIb (sequence SKGQL) pharmacophore hypothesis, as described in the text. Red, hydrogen bond donor; green, hydrogen bond acceptor. (B) Structures of the 4 LM22A compounds chosen for these studies. Compounds are abbreviated as #1–#4 in subsequent figures.
Identification of BDNF loop II mimetics with neurotrophic activity.
Compounds were screened for neurotrophic activity using E16 hippocampal neurons under conditions in which survival is, in part, dependent upon the addition of exogenous BDNF. In preliminary assays, 5 compounds emulating the loop II region of BDNF were considered to have activity significantly above baseline as defined as a percentage of BDNF maximal efficacy. Four compounds with the highest activity (LM22A-1 to -4) were selected for further characterization. Each of these 4 compounds exhibits a distinct chemotype; LM22A-1 is a tripeptide (Pro-His-Trp), and compounds LM22A-2, LM22A-3, and LM22A-4 contain no peptide bonds (Figure 1B).
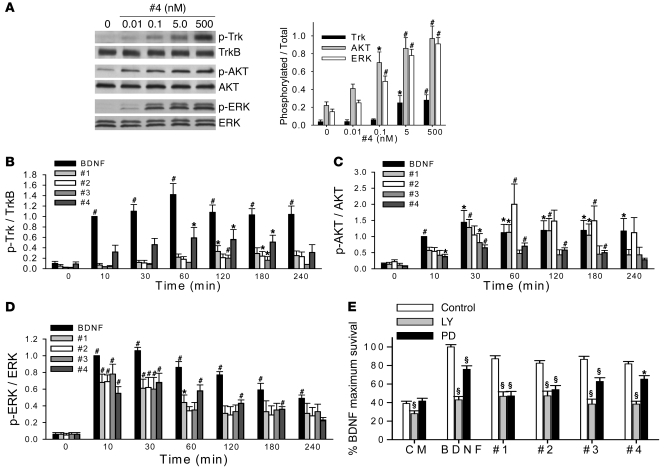
Neurotrophic activity dose-response studies with the 4 compounds demonstrated maximum levels of activity of 80%–89% of that of BDNF and EC50 values of 200–500 pM (Figure 2, A–F). TUNEL/DAPI staining confirmed the compounds’ ability to prevent neuronal death with an efficacy similar to that of BDNF (Figure 2G). Since the plateau of survival for LM22A-1 and -4 was not well established until approximately 100 nM, to ensure maximum effects, we chose 500 nM as a standard concentration for many of the subsequent mechanistic studies.
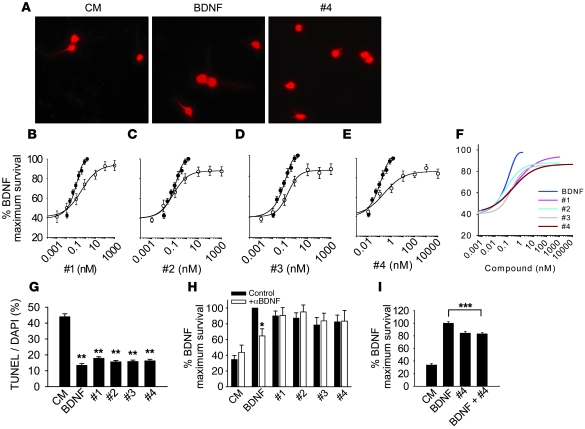
Figure 2. Neurotrophic activities of LM22A compounds.
(A) Fluorescence photomicrographs (original magnification, ×40) of GAP43-immunostained E16 mouse hippocampal neuronal cultures treated with culture medium (CM), BDNF, or LM22A-4 (#4) for 48 hours. (B–E) Neuron survival dose-response curves for BDNF and LM22A compounds. Counts were normalized to survival achieved with 20 ng/ml (~0.7 nM) BDNF. For BDNF, survival values for each concentration were derived from n = 29–32 wells obtained from 12 separate experiments; for LM22A-1, -2, and -3, values for each concentration were derived from n = 11–19 wells obtained from 6 separate experiments; for LM22A-4, values for each concentration were derived from n = 13–21 wells obtained from 8 separate experiments. White circles, compound responses; black circles, BDNF responses (F) Comparison of survival curves from all compounds and BDNF. (G) Quantitation of the fraction of TUNEL-positive/DAPI-staining cells demonstrates that LM22A compounds decrease the number of TUNEL-positive cells to a degree similar to that of BDNF. A total of n = 93–95 fields per condition derived from 3 separate assays were counted. (H) Survival analysis of E16 hippocampal neurons treated with nonimmune serum (Control) or antibody to BDNF (Ab; 1:400) and BDNF (0.7 nM) or LM22A compounds (500 nM). Counts made in a total of n = 10–12 wells were derived from 5–6 experiments. (I) Survival of E16 hippocampal neurons treated with BDNF alone (0.7 nM), LM22A-4 alone (500 nM), or BDNF plus LM22A-4. n = 24–72 wells for each condition derived from 3 experiments were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001.
In beginning to examine the mechanism of action of these compounds, we considered the possibility that they might act as secretagogues, increasing BDNF levels. Anti-BDNF antibody treatment of hippocampal cultures resulted in a decrease in BDNF neurotrophic activity but had no effect on the activity of the 4 test compounds (Figure 2H), supporting the view that these compounds are not BDNF secretagogues.
LM22A compounds interact with and act through TrkB.
The hypothesis that the LM22A compounds act through TrkB was examined through multiple approaches. The finding that compounds had maximal activities at 80%–89% of that of BDNF suggested that if they functioned as a ligand, or by otherwise co-opting TrkB or its downstream signaling pathways, they might inhibit BDNF function by approximately 10%–20%. In contrast, additive activity would indicate a parallel mechanism. In hippocampal cultures, addition of 500 nM LM22A-4 with 20 ng/ml (~0.7 nM) BDNF, concentrations of each which induce a maximal response, produced no additive effect on survival (Figure 2I). Similar results were obtained with LM22A-3 (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI41356DS1). Also of note, addition of these compounds resulted in an approximately 15% reduction in the BDNF neurotrophic response, consistent with the profile of a TrkB partial agonist. Finally, the co-addition of LM22A-3 and -4 did not result in additive effects (Supplemental Figure 1B).
If these compounds work through activation of TrkB, they should be blocked by inhibition of receptor activation. Treatment of hippocampal neuron cultures with the Trk inhibitor K252a led to a reduction in the neurotrophic activity of BDNF and all 4 test compounds (Figure 3A). K252a has been reported to also block mixed-lineage kinases, leading to inhibition of JNK activation, and to activate AKT and ERK through the Src pathway (27), which lead to improved survival; however, we saw no improvement in survival with K252a alone in our cellular systems. In addition, as an alternative approach, we examined the effects of an antibody directed against the extracellular domain (ECD) of TrkB (anti-TrkBECD) that is known to inhibit BDNF function (28). This resulted in a reduction in cell survival (Figure 3B) and increased numbers of TUNEL-positive cells (Figure 3C) in the presence of BDNF, LM22A-3, and LM22A-4.
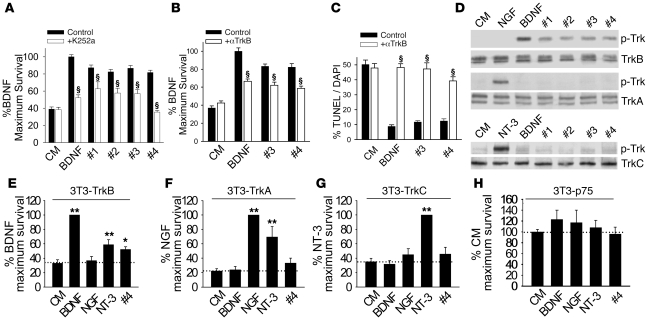
Figure 3. LM22A compounds function through TrkB.
(A) Survival analysis of hippocampal neurons treated with BDNF (0.7 nM) or LM22A compounds (500 nM) with or without the Trk inhibitor K252a (200 nM). For BDNF, n = 37 wells derived from 7 experiments; for LM22A-1, -2, and -3, n = 17–21 wells derived from 6 experiments; and for LM22A-4, n = 28–57 wells derived from 4 experiments were assessed. (B) Survival analysis of hippocampal neurons treated with CM, BDNF (0.7 nM), or LM22A compounds (500 nM) with or without TrkBECD antibody. For each condition, n = 37–42 wells derived from 5 experiments were assessed. (C) TUNEL/DAPI analysis of E16 hippocampal neurons treated with CM, BDNF (0.7 nM), or LM22A compounds (500 nM) with or without TrkBECD antibody. For each condition, n = 17–31 fields derived from 3 experiments were assessed. (D) LM22A compounds or neurotrophic factors were incubated for 60 minutes with NIH-3T3 cells stably expressing specific Trk receptors. Western blot analysis (anti–pan-phospho-TrkY490 [p-Trk]) demonstrated that LM22A compounds activated TrkB (upper panels) but not TrkA (middle panels) or TrkC (lower panels), while all 3 Trks were activated by their cognate ligands. Activation patterns of 3 additional independent assays were identical. (E–H) Trk- and p75NTR-specific NIH-3T3 cells were incubated in serum-free medium in the presence of the designated ligands for 72–96 hours, and survival was measured using the EnzyLight assay. n = 18–28 wells derived from 11–14 experiments. *P < 0.05, **P < 0.01, §P < 0.001.
LM22A compounds specifically activate TrkB.
To examine the effects of LM22A compounds on different Trk family members, we determined TrkY490 phosphorylation in NIH-3T3 cells expressing single Trks. In 3T3-TrkB cultures, addition of BDNF or LM22A compounds resulted in TrkB activation, while NGF was inactive (Figure 3D). However, in 3T3-TrkA and 3T3-TrkC cultures, the neurotrophins produced the expected results, while the LM22A compounds induced no Trk activation (Figure 3D). Thus, the LM22A compounds selectively activate TrkB.
LM22A-4 was selected as a prototype compound to further examine Trk interactions, as it appeared the simplest and most flexible structurally, and most amenable to chemical modifications that might improve its medicinal properties. To extend the examination of specificity to receptor function, we investigated whether the ability of LM22A-4 to prevent death is specific for TrkB relative to TrkA and TrkC. BDNF, NGF, and NT-3 supported survival in serum-free medium of 3T3-TrkB (Figure 3E), 3T3-TrkA (Figure 3F), and 3T3-TrkC (Figure 3G) cells, respectively; and, survival-promoting activity of NT-3 in 3T3-TrkA and 3T3-TrkB cells was consistent with its known receptor cross-activity (1). In 3T3-TrkB cells (Figure 3E), LM22A-4 increased survival by 56%, while no significant increase in survival was detected in 3T3-TrkA (Figure 3F), 3T3-TrkC (Figure 3G), or 3T3-p75NTR (Figure 3H) cells. These results are consistent with LM22A-4 acting selectively through TrkB.
LM22A-4 binds specifically to TrkB.
LM22A-4 was incubated with chimeric TrkB-Fc and ephrin-A5-Fc (as a negative control) and found to bind to the TrkBECD-Fc but not ephrin-A5ECD-Fc protein, and addition of BDNF blocked the binding of LM22A-4 to TrkBECD-Fc (Figure 4A). To further examine LM22A-4-TrkB interactions, the displacement of BDNF from TrkBECD-Fc-Cy3B by the compound was determined by measuring changes in fluorescence anisotropy of the receptor with increasing LM22A-4; this was compared to the effects of the compound on labeled TrkB in the presence of a nonbinding neurotrophin, NGF (Figure 4B). Measured anisotropy (), proportional to TrkB bound, fell from approximately 0.31 to approximately 0.17 between 0 and 75 nM LM22A-4, with an IC50 of 47 nM or less. In the presence of NGF, without LM22A-4, there was no increase in fluorescence anisotropy above baseline, and the further addition of the compound had no effect. To further address specificity of receptor binding and binding in a cellular context, we performed competition studies with 3T3 cells expressing each of the neurotrophin receptors. LM22A-4 inhibited binding of BDNF to TrkB-expressing cells but had no detectable effect on cells expressing TrkA, TrkC, or p75NTR (Figure 4, C–F). In addition, a screen of 57 receptors performed by Cerep Inc. detected no significant binding (Supplemental Figure 2). Within this screen, inhibition of binding by a test ligand of greater than 50% is considered to demonstrate significant binding by the test compound. At the standard screening concentration of 10 μM, LM22A-4 demonstrated no significant binding within the panel. Together, these studies demonstrate that LM22A-4 binds to TrkB directly, can relatively potently displace BDNF from TrkB, and demonstrates binding selectivity for TrkB.
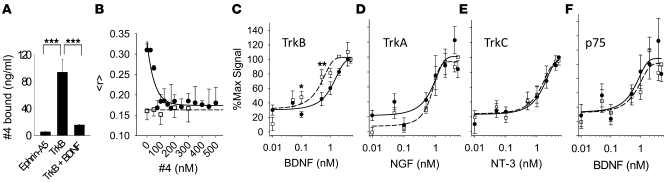
Figure 4. LM22A-4 binds selectively to TrkB.
(A) LM22A-4 was incubated with ephrin A5-Fc receptor as a negative control or with TrkBECD-Fc in the absence or presence of BDNF (600 pmol). After rinsing of receptor complexes and subsequent ligand elution, LC/MS-MS demonstrated readily detectable LM22A-4 binding to TrkBECD-Fc that was blocked by BDNF. Values were derived from n = 3 independent binding studies. (B) BDNF (100 nM, black circles) or NGF (100 nM, white squares) was incubated with TrkBECD-Fc-Cy3B (100 nM) and increasing concentrations of LM22A-4; fluorescence anisotropy () was measured n = 6–9 times in single samples at each concentration. (C–F) NIH-3T3 cells expressing TrkB, TrkA, TrkC, or p75NTR as indicated were incubated with increasing concentrations of the indicated neurotrophin in the absence (white squares, dashed line) or presence (black circles, solid line) of 100 nM LM22A-4. Symbols represent average fluorescent signal after background (i.e., binding to cells lacking expressed receptors) subtraction, as described in Methods. n = 14–18 wells from 7–9 experiments at each concentration. Significant rightward shifting of the binding curve by the compound was observed only with TrkB expressing cells. *P < 0.05, **P < 0.01, ***P < 0.001.
LM22A-induced TrkB and downstream signaling activation kinetics: differences and similarities with BDNF.
“Prosurvival” signaling triggered by TrkB activation includes activation of the PI3K/AKT and MAPK/ERK pathways mediated by SHC binding to TrkB phospho-Y490 (1, 3). LM22A-4 induced the activation of TrkB, AKT, and ERK at 60 minutes over a concentration range of 0.01–500 nM, a range encompassing the onset and plateau of its promotion of survival (Figure 5A). Of note, TrkB activation was not significantly increased at an LM22A-4 concentration of 0.1 nM, though ERK and AKT were detectably activated at this level. In examining kinetics, BDNF caused substantial TrkB Y490 phosphorylation within 10 minutes of application to hippocampal neurons in culture, and these levels were maintained for up to 240 minutes (Figure 5B). In contrast, the response to 500 nM LM22A-1, -2, and -3 was nominal for up to 30 minutes, with increased but relatively low levels of activation at later time points. LM22A-4 showed intermediate levels of TrkB activation over a time course similar to that of BDNF. Interestingly, treatment of hippocampal neurons with BDNF and LM22A compounds led to robust increases in AKT and ERK phosphorylation within 30 minutes, with profiles similar to BDNF thereafter (Figure 5, C and D). This early discrepancy between TrkB phosphorylation and AKT/ERK activation is reminiscent of that seen in the LM22A-4 concentration series (Figure 5A) and suggested the possibilities that in these cells, downstream mechanisms are highly sensitive to small changes in TrkB phosphorylation state or, less likely, that the compounds stimulate coordinated signaling to AKT and ERK through another site on TrkB or act through some other mechanism. To test whether LM22A compound signaling is dependent on the presence and activation of TrkB, we examined the effects Trk inhibition on compound signaling in cultured hippocampal neurons and in cells expressing one of TrkA, TrkB, or TrkC. We found that anti-TrkB antibodies substantially inhibited TrkB, AKT, and ERK activation induced by LM22A-3 and -4 in hippocampal neurons (Supplemental Figure 3, A–D). In addition, LM22A-1 to -4 each induced AKT and ERK phosphorylation in TrkB-expressing fibroblasts but not in TrkA- or TrkC-expressing cells (Supplemental Figure 4, A and B). Finally, we found that inhibiting Trk phosphorylation in 3T3-TrkB cells with K252a was associated with inhibition of compound-induced AKT and ERK activation (Supplemental Figure 5, A–D). Of note, K252a alone had no effect on AKT or ERK activation in these cells. These findings suggest that signaling induced by the LM22A compounds specifically requires TrkB and is associated with TrkB activation/phosphorylation. The question of the differences in TrkB activation kinetics between BDNF and the compounds remains for further study.
Figure 5. Comparison of BDNF’s and LM22A compound’s signaling activation kinetics and dependence of survival on downstream signaling.
(A) Left: Representative Western blot showing activation of Trk, AKT, and ERK in cultured E16 hippocampal neurons incubated with LM22A-4 at the indicated concentrations for 60 minutes. Right: Western blot analyses were quantitated for Trk, AKT, and ERK activation (ratio of p-TrkY490, using a pan–phospho-Trk antibody, to total TrkB; p-AKT to total AKT; and p-ERK to total ERK signal). n = 15–18 Western analyses from 4–9 independent protein preparations. (B–D) Cultured E16 hippocampal neurons were incubated with BDNF (0.7 nM) or LM22A compounds (500 nM) for the indicated times, and quantitative analysis for Trk, AKT, and ERK activation was performed as in A. n = 8–11 Western blot analyses from 4–5 independent protein preparations. For each Western blot, the activation ratio induced by BDNF at the 10-minute time point was normalized to 1.0 and the other values adjusted accordingly. (E) E16 hippocampal neurons were incubated with BDNF (0.7 nM) or LM22A compounds (500 nM) for 48 hours in the presence or absence of the PI3K inhibitor LY294002 (LY, 10 μM) or the MAPK kinase inhibitor PD98059 (PD, 50 μM). For BDNF, n = 33–37 wells derived from 7 experiments were assessed. For LM22A-1, -2, and -3, n = 17–21 wells derived from 6 experiments were analyzed. For LM22A-4, n = 28–57 wells derived from 4 experiments were assessed. *P < 0.05, #P < 0.01, §P < 0.001.
To further establish the pathways involved in LM22A activity, we investigated whether the compound’s neurotrophic activity was dependent on AKT and ERK activation. LY294002, a specific inhibitor of PI3K (an upstream activator of AKT), and the MAPK kinase (an upstream activator of ERK) inhibitor PD98059 reduced the neurotrophic activity of BDNF and the LM22A compounds (Figure 5E). Together, these findings suggest that LM22A compounds induce survival of hippocampal neurons in culture through interactions with TrkB and downstream activation of PI3K/AKT and MAPK/ERK pathways.
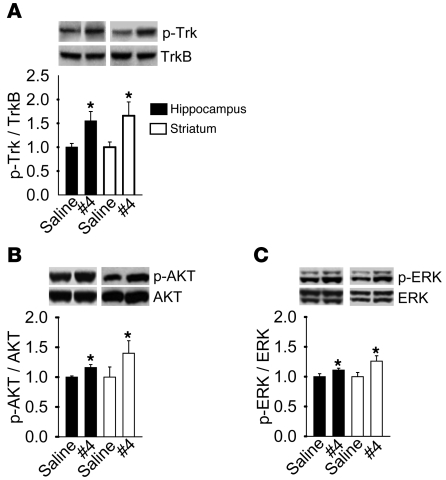
LM22A-4 activates TrkB, AKT, and ERK in vivo.
To determine whether a TrkB small molecule, non-peptide ligand would be capable of activating TrkB in the adult mammalian brain, LM22A-4 was selected for in vivo studies. Preliminary pharmacokinetic studies in mice indicated that LM22A-4 exhibited very low blood brain barrier penetration (brain concentrations for 10 mg/kg LM22A-4 given i.p. for 1 hour, 62 ± 7 nM; 3 hours, below detectable limit; by liquid chromatography/tandem mass spectrometry [LC-MS/MS], Absorption Systems); therefore, the compound was applied to adult mice by intranasal administration. After once-daily administration of vehicle or LM22A-4 for 7 days, brains were harvested 2.5 hours after the last dose, and hippocampal and striatal tissue were assessed for Trk, AKT, and ERK activation. LM22A-4 induced an increase in activation of Trk, AKT, and ERK in hippocampus and striatum (Figure 6, A–C). Given that a small portion of the cell population within each sampled tissue expressed TrkB, the apparent increases in TrkB, AKT, and ERK activation in whole tissue extracts point to particularly robust signaling effects in these brain regions.
Figure 6. LM22A-4 activates Trk, AKT, and ERK signaling in vivo.
LM22A-4 or saline control was administered intranasally (0.22 mg/kg/d) to adult mice for 7 days. Protein extracts of hippocampus and striatum were assessed by Western blot analysis for (A) Trk, (B) AKT, or (C) ERK phosphorylation. n = 11 mice for each assay. *P < 0.05.
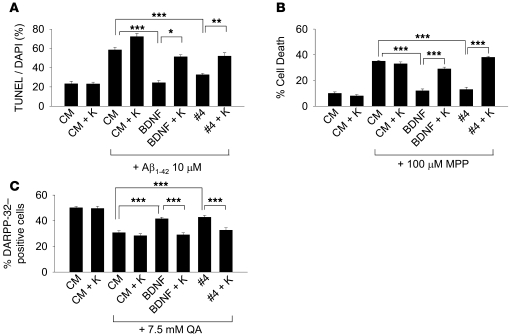
LM22A-4 matches BDNF efficacy in preventing neuronal death in in vitro models of neurodegenerative diseases.
Since the LM22A compounds upregulated fundamental survival/stress resistance mechanisms, we hypothesized that they would mitigate neuronal death across a variety of mechanisms and disease models and examined the effects of LM22A-4 in models of Alzheimer, Huntington, and Parkinson diseases. Accumulation of toxic species of Aβ is likely to play a critical role in the pathogenesis of Alzheimer disease, and the neurodegenerative effect of Aβ in neuronal cultures serves as an in vitro model (29). Previous studies have demonstrated that BDNF protects neurons from Aβ-induced neuronal death (30). In 6–7 days in vitro (DIV) hippocampal neurons treated with oligomeric Aβ, BDNF and LM22A-4 each led to a similar substantial reduction in Aβ-induced death, and this protective activity was inhibited by the Trk inhibitor K252a, consistent with a Trk-dependent mechanism (Figure 7A). Application of MPP+ (metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine [MPTP]), a mitochondrial toxin, to SH-SY5Y human tumor line cells (31) is a widely used Parkinson disease model. Here, BDNF and LM22A-4 blocked MPP+-induced cell death to a similar extent, and this protective activity was inhibited by K252a (Figure 7B). Finally, a model for Huntington disease involves the administration of the excitotoxin quinolinic acid (QA), an _N_-methyl-d-aspartate receptor agonist (32, 33). In preliminary studies, QA caused increasing cell death in cultures of striatal neurons, from 1.25 to 50 mM; 7.5 mM was selected in order to ensure a clear toxic effect. BDNF and LM22A-4 led to a similar substantial reduction in QA-induced death, and this protective activity was again inhibited by K252a (Figure 7C). Together, these studies indicate the potential broad application of TrkB agonists to neurodegenerative processes.
Figure 7. LM22A-4 inhibits neuronal death in in vitro neurodegenerative disease models.
(A) Six- to 7-DIV hippocampal neurons from E16 mice were treated in the absence of Aβ with CM alone or CM + K252a (K) or in the presence of oligomeric Aβ with CM alone, CM + K252a, BDNF (0.7 nM), BDNF + K252a, LM22A-4 (500 nM), LM22A-4 + K252a. K252a was used at 200 nM. After 72 hours, cultures were assessed by TUNEL/DAPI staining. n = 28–29 fields for each condition derived from a total of 3 experiments. (B) SH-SY5Y human neuroblastoma cells were pretreated for 3 days in the following conditions: CM alone, CM + K252a, BDNF (0.7 nM), BDNF + K252a, LM22A-4 (500 nM), LM22A-4 + K252a. MPP+ (100 μM) was added, and 48 hours later cell survival was assessed by MTT/cell count assay. (C) Six- to 7-DIV striatal neurons from E16 mice were pretreated for 2 hours in the following conditions: CM, CM + K252a, BDNF (0.7 nM), BDNF + K252a, LM22A-4 (500 nM), LM22A-4 + K252a. QA (7.5 mM) was added, and after 24 hours numbers of DARPP-32–positive/total surviving cells was determined. n = 50–100 fields for each condition derived from a total 5 experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
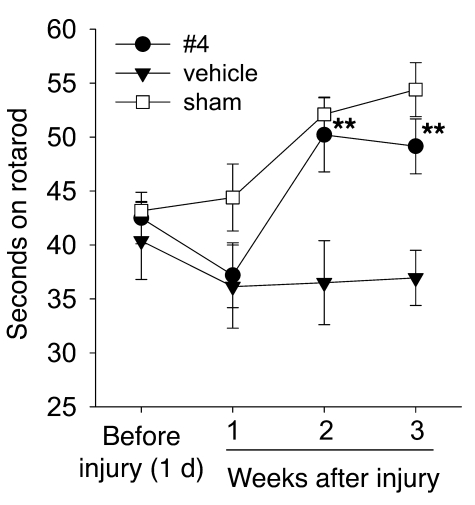
LM22A-4 improves motor learning after TBI.
Motor learning and plasticity have been associated with TrkB and/or BDNF expression in the cortex (34, 35) and cerebellum (34), and BDNF has been reported to improve motor recovery and increase measures of neuronal remodeling after cortical ischemia (36, 37) and cerebellar pedunculotomy (38). Though BDNF infusion has been reported to have no effect on spatial learning after TBI (39, 40), motor learning was not assessed in those studies. We therefore examined the effects of LM22A-4 on changes in rotarod performance over repeated trials, a measure of motor learning (41), after TBI. Rats subjected to parietal controlled cortical impact injury showed a trend toward decreased rotarod performance relative to sham-operated animals at 1 week after TBI, and this was similar with or without LM22A-4 treatment (Figure 8). With repetition at 2 and 3 weeks after injury, LM22A-4–treated animals showed significant improvements in performance, indistinguishable from sham-operated controls, whereas animals subjected to TBI and treated with vehicle showed no improvements (Figure 8). In addition, at 21 days after injury, there was no difference between the lesion volumes of compound-treated and vehicle alone–treated groups (Supplemental Figure 6). This suggests that LM22A-4 may reverse deficits in motor task learning caused by TBI, though it has no effect on gross total injury.
Figure 8. LM22A-4 restores motor learning after TBI.
Adult male Sprague-Dawley rats were trained on an accelerating rotarod task and subjected to parietal controlled cortical impact injury (filled symbols) or sham surgery (open symbols). Injured animals were treated intranasally with vehicle alone or containing LM22A-4 beginning immediately after injury then daily for 2 weeks, as indicated. Symbols indicate the mean ± SEM of the dwell time on the rod from the average of 2 trials per animal/week; n = 5 animals for vehicle and sham, and n = 6 for LM22A-4; significance was determined by repeated-measures ANOVA and post-hoc Holm-Sidak analysis. **P < 0.01.
Discussion
These studies describe what we believe to be the first reported small molecules mimicking a specific neurotrophin domain, capable of functioning as ligands to selectively and potently activate the TrkB receptor.
We initially theorized that using loop subdomain pharmacophore modeling and virtual screening, the a priori likelihood of identifying compounds functioning through TrkB would be substantially greater than that for compounds identified in conventional high-throughput screening for neurotrophic activity (which would identify compounds functioning through any mechanism). As in our previous studies on p75NTR (42), we found that this method provides a manageable group of compounds for biologic testing, with a high rate of recovery (5 activated of 7 tested) of the desired activity relative to that which would be obtained using high-throughput screening (<1%, refs. 43, 44), and similar to that of other virtual screens beginning with more structural information (45). Multiple lines of experimental evidence suggest that these small molecules function as TrkB ligands, including: specific activation of TrkB and two primary downstream signaling intermediates; inhibition of activity by the kinase inhibitor K252a and a monoclonal antibody directed to the ECD of TrkB; blocking of compound binding to the ECD of TrkB by BDNF; compound effects on TrkB-expressing, but not TrkA- or TrkC-expressing, NIH-3T3 cells; and inhibition of BDNF binding to TrkB, with no inhibition of neurotrophin binding to TrkA, TrkC, or p75NTR, in addition to a negative Cerep screen. While the present studies failed to detect interaction of LM22A-4 with p75NTR or prevention of cell death through p75NTR, future studies will address whether such ligands might nevertheless affect its complex intracellular adaptor/signaling milieu. The lack of effect of BDNF antibodies, compound inhibition of BDNF maximal activity, and LM22A-4 induction of greater neurite outgrowth than maximal BDNF suggest that the LM22A compounds do not act as BDNF secretagogues. Thus, we suggest that the identification, within this small group of tested compounds, of chemically diverse compounds sharing the common mechanism of TrkB activation supports the validity of the model employed in these studies.
Recent studies show that small molecules may bind with relatively high affinity to protein surfaces through “hot spots,” small groups of residues that contribute significant energy to the protein-protein interaction of interest. Small molecule binding can disrupt protein-protein interactions and inhibit the functions they mediate (46–50). Moreover, the small molecule may induce conformational changes in the binding domain that differ from those associated with binding of the natural ligand (48). Given the findings of small molecule–mediated TrkB agonism presented here, and our previous findings with p75NTR ligands (42), we suggest that hot-spot binding and induction of conformational changes in the receptor may allow these small molecules to act as activating ligands, though there may be differences from the natural ligand with respect to the coupling and kinetics of the induced signaling. Further structural studies of compound-receptor complexes will be needed to confirm this hypothesis.
Of note, other natural ligands of TrkB may also induce signaling quantitatively and qualitatively different from that induced by BDNF. Mutation of the SHC-binding site of TrkB in mice caused loss of NT-4–dependent, but not BDNF-dependent, neurons in vivo and decreased survival of sensory neurons in culture and ERK activation in response to NT-4 relative to BDNF (51). In another study, c-fos promoter activity in cortical neurons was found to be more responsive to exogenous NT-4 than BDNF (52). Such differential activation of downstream signaling by ligands may reflect and/or play an active role in partial agonism and may therefore be significant in the effects of LM22A compounds on endogenous BDNF actions. Moreover, the effects of differential activation by small molecule ligands may vary with respect to other native TrkB ligands (NT-4, NT-3) and lead to unexpected inhibition or synergy. Further studies comparing signaling activation kinetics and pathways between the compounds and protein ligands, including coexposure, will be needed to evaluate these possibilities.
Additionally, it would be reasonable to expect that the methodology presented here would also yield pure BDNF/TrkB antagonists, though we have not specifically assayed for these as of yet. Unfortunately, the small numbers of compounds and lack of available close structural analogs make detailed structure-activity relationship analysis infeasible at this point; this will be addressed in future studies employing larger libraries and/or synthesis of directed analogs.
The lack of rotational symmetry of LM22A-1 and -3 and the small size of the compounds suggest that they are functionally monovalent (i.e., do not interact with more than one receptor monomer). This apparent monovalency raises interesting questions regarding mechanisms of TrkB activation by the compounds. Previous studies have characterized synthetic peptides modeled on loop regions of BDNF, with the goal of using active peptides to derive active non-peptide small molecules that might function as BDNF mimetics (24, 53, 54). In each of these studies, peptides with neurotrophic activity were found; however, the ability to activate TrkB was either not described (24, 53) or not detected (54). Hence whether such peptides can function as ligands that activate TrkB remains to be determined. Unlike TrkA and TrkC, for which there is evidence that “monovalent” cyclic peptidomimetics can exhibit small degrees of agonist activity (55, 56), for TrkB only “dimeric” peptides have as yet been found to demonstrate trophic activity. This finding is consistent with the classical model in which the BDNF dimer is thought to induce dimerization leading to activation (autophosphorylation) of TrkB. If the compounds act in a monomeric fashion, one possible mechanism by which this activation might occur is that the compound binds to a TrkB monomer, inducing a conformational change that promotes dimerization. Also, if, as has been proposed for TrkA, the unliganded receptor monomer is in equilibrium with dimeric or higher multimeric states (57), then the compounds might stabilize and/or promote phosphorylation of complexes, without inducing dimerization. It is unlikely that each of the 4 compounds characterized here is capable of forming high-affinity, bivalent dimers in solution, which then function as dimeric ligands. Co-crystallization and other future kinetic and structural studies will address the questions of how the mechanisms of activation and binding of the small molecules to TrkB compare with those of BDNF.
The ability of the prototype compound LM22A-4 to trigger activation of TrkB and its downstream signaling intermediates in the hippocampus and striatum is consistent with the observed effects on motor learning after TBI, though the anatomic substrates and mechanisms of that effect will require further investigation.
These findings, along with the compound’s ability to improve cell survival in several in vitro neurodegenerative disease models, open new opportunities for the further development of derivatives that may have therapeutic effects in a large number of acute and chronic neurological and neuropsychiatric disorders. The prospect that small molecule TrkB ligands could have the BDNF-like effects of promoting neurogenesis and synaptic function suggests further novel therapeutic applications of such compounds. The finding that BDNF interaction with p75NTR promotes mechanisms underlying pain (23) raises the interesting possibility that small molecules activating TrkB without mimicking BDNF-p75NTR interactions might reduce the likelihood of pain as a side effect occurring in BDNF-based therapeutics. In addition, the partial agonist profile of the compounds described here may support their use under circumstances where excessive TrkB signaling is undesirable.
The methods and compounds described here provide a basis for drug discovery in other receptor systems and for designing and characterizing novel non-peptide TrkB ligands optimized for blood brain barrier penetration and other pharmacological features.
Methods
Materials.
5-Oxo-l-prolyl-l-histidyl-l-tryptophan methyl ester (LM22A-1, catalog BAS 0380372), 2-[2,7-bis[[(2-hydroxyethyl)amino]sulfonyl]-9H-fluoren-9-ylidene]-hydrazinecarboxamide (LM22A-2, catalog BAS 0461519), and _N_-[4-[2-[5-amino-4-cyano-1-(2-hydroxyethyl)-1H-pyrazol-3-yl]-2-cyanoethenyl]phenyl]-acetamide (LM22A-3, catalog BAS 0548227) were purchased from Asinex Corp., with purity of more than 90% determined by NMR spectroscopy. N,_N_′,_N_′′-tris(2-hydroxyethyl)-1,3,5-benzenetricarboxamide (LM22A-4, catalog CHS 0233913) was purchased from ChemBridge Corp., with purity of more than 95% determined by NMR and LC-MS. The molecular weights and formulae of these 4 compounds were further confirmed in our laboratory by high-resolution mass spectrometry. Other chemicals were purchased from Sigma-Aldrich unless otherwise stated. Other sources include the following: LY294002, K252a, and PD98059, Calbiochem; mouse monoclonal anti–phospho-ERKT202/Y204, rabbit polyclonal anti-ERK42/44, mouse monoclonal anti–phospho-AKTS473, rabbit polyclonal anti-AKT, and site-specific rabbit polyclonal anti-TrkY490, Cell Signaling Technology Inc. Antibodies were obtained from the following sources: mouse monoclonal anti-BDNF, Sigma-Aldrich; rabbit polyclonal TrkA and TrkB antibodies, Upstate USA Inc.; mouse monoclonal TrkB antibody, BD Transduction Laboratories; TrkC rabbit polyclonal antibody, Santa Cruz Biotechnology Inc.; rabbit DARPP-32 polyclonal antibody, Millipore; recombinant human NGF, Invitrogen; recombinant human BDNF, Sigma-Aldrich; recombinant chimeric proteins ephrin-A5-Fc and Trk-Fc, R&D Systems.
Institutional review of animal protocols.
All animal protocols used in the present study were approved by the Institutional Animal Care and Use Committee of the University of North Carolina–Chapel Hill, Stanford University, or the San Francisco VA Medical Center.
Computational modeling and pharmacophore generation.
Beginning with a configuration derived from the crystal structure of a BDNF/NT-3 heterodimer (58), the structure of loop II was further processed by the addition of a hydration shell and 100 ps of molecular dynamics simulation (using the CFF91 force field with the Discover MD engine of InsightII, sampled every 50 fs [Accelrys Inc.]), followed by extraction of a structure averaged spatially over the simulation. The loop structure was exported to Catalyst (Accelrys Inc.) to generate the chemical feature hypothesis used for virtual screening, as illustrated in Figure 1A.
Aβ preparations.
Aβ1–42 peptide oligomeric preparations were derived as described previously, and their oligomeric state was confirmed using atomic force microscopy (29).
Primary neuronal cultures and bioassays.
Hippocampal neuron cultures were prepared from E16 CF1 mouse fetuses as described previously (59). Under the low-density conditions used here, neuronal survival is dependent, in part, on the addition of exogenous neurotrophins (60). LM22A-1, -2, and -3 were initially dissolved in 100% DMSO and diluted in culture medium to 0.005% DMSO for use. LM22A-4 was dissolved in water. Solvents were diluted in culture media in equivalent amounts for controls. Cell survival after a 48-hour exposure to BDNF or compounds was measured by morphological criteria using staining for GAP-43, a standard method that allows visualization of neurites and cell bodies (42) in combination with assessment of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) metabolism (42); or by TUNEL with DAPI staining (29, 59). Fitting of dose-response curves was performed with SigmaPlot (Systat Software Inc.). For signaling inhibitor studies, K252a, LY294002, or PD98059 were added to cultures at final concentrations of 200 nM, 10 μM, and 50 μM, respectively, concomitantly with BDNF or LM22A compounds. For antibody inhibition studies, mouse monoclonal BDNF or TrkB antibodies and control non-immune serum were used at a final dilution of 1:400 (0.25 μg/ml) and added concomitantly with BDNF or LM22A compounds.
Assays employing in vitro models for Alzheimer, Huntington, and Parkinson disease were based on previously published protocols and are described below.
NIH-3T3 cell cultures and survival assays.
Mouse NIH-3T3 cells expressing TrkA (NIH-3T3-TrkA) or p75NTR (NIH-3T3-p75NTR) were provided by William Mobley, Stanford University. NIH-3T3 cells expressing TrkB (NIH-3T3-TrkB) or TrkC (NIH-3T3-TrkC) were provided by David Kaplan, Montreal Neurological Institute, Montreal, Quebec, Canada. Cells were propagated in DMEM supplemented with 10% fetal bovine serum (Invitrogen) and 200–400 μg/ml geneticin (for Trk-expressing cells) or 400 μg/ml hygromycin (for p75NTR-expressing cells). Cells were seeded into 24-well plates (30,000 cells/well) and cultured in medium consisting of 50% PBS and 50% DMEM without supplements. After exposure to growth factors or compounds for 72–96 hours, cells were suspended in 50 μl PBS and transferred to 96-well white, opaque tissue culture plates, and survival was measured using the EnzyLight Cytotoxicity Assay Kit (BioAssay Systems).
LM22A-4 binding assays.
For direct binding measurements, protein G agarose beads (10 μl) were washed twice with PBS binding buffer (pH 8.0) and incubated with 300 pmol TrkBECD-Fc or ephrin A5ECD-Fc at room temperature overnight. After 2 PBS washes, 600 pmol of BDNF or PBS was added. After incubation at room temperature for 1 hour, 300 pmol of LM22A-4 was added, followed by a 1 hour incubation. Bead-receptor-ligand complexes were washed twice with 500 μl PBS over a filter vacuum system to remove excess ligand. After washing, filters were placed in vials, and receptor-ligand complexes were separated from beads by eluting for 15 minutes at 4°C with 1 ml of 0.1-M glycine pH 2.5, followed by incubation with 100 μl of 1-M Tris pH 8.0 neutralizing buffer. The mixture was centrifuged, and the supernatant elution mixture was submitted for determination of LM22A-4 levels using LC-MS/MS by Absorption Systems Inc. For the fluorescence anisotropy competition binding assay, TrkB ECD was labeled with Cy3B-NHS ester (Amersham) using standard protein coupling protocols recommended by the manufacturer. Samples containing varying LM22A-4 concentrations (0 nM–500 nM) with 100 nM BDNF or NGF, and 100 nM Cy3B-TrkB in binding buffer (PBS, 0.7% BSA), were prepared in 150 μl total volume and incubated overnight at 4°C. Fluorescence anisotropy was measured 6–9 times per sample using Fluorolog-3 (HORIBA) in quartz cells (Starna Cells).
In cell-based binding assays, NIH-3T3 wild-type cells and cell expressing one of the 4 neurotrophin receptors were seeded on poly-l-lysine–coated 96-well plates (Fisher Scientific). After reaching 85%–95% confluency, Trk-expressing cells were incubated with their cognate neurotrophin, and p75NTR cells were incubated with BDNF, each at multiple neurotrophin concentrations with or without LM22A-4 (100 nM) for 45 minutes at 4°C with gentle shaking. After 1 wash with ice-cold PBS, cells were fixed with 4% paraldehyde for 15 minutes, rinsed twice with PBS, and then incubated with anti-mouse monoclonal NGF antibody (1 μg/ml, Abcam), anti-mouse monoclonal BDNF antibody (1 μg/ml, Sigma-Aldrich), or anti-rabbit polyclonal NT3 antibody (1 μg/ml, Abcam) for 1 hour at room temperature. Cells were then rinsed twice with PBS and incubated with DyLight 549–conjugated secondary antibody (1:500, Jackson ImmunoResearch Laboratories Inc.) for 1 hour at room temperature, followed by 2 washes. Fluorescent signal was measured at an excitation wavelength of 549 nm and emission of 568 nm using a plate reader (Molecular Devices). Duplicate wells were measured for each neurotrophin concentration. For each condition, data were derived by subtracting background signal, which was established as the signal obtained from wells containing NIH-3T3 cells lacking neurotrophin receptors incubated with a maximal level of neurotrophin. Data were normalized to the signal representing maximal binding of each neurotrophin to cells expressing its corresponding receptor. LM22A-4 was submitted to Cerep Inc. for receptor binding screening using their standard protocols.
Protein extraction and Western blot analysis.
For assays of Trk, AKT, and ERK activation, hippocampal neurons derived from E16 mice were cultured in poly-l-lysine–coated 6-well plates (Corning Inc.) (5 × 106 cells/well) in DMEM/F12 containing 10% FBS for 2 hours, Neurobasal culture medium plus B27 for an additional 44 hours, and then serum-free DMEM/F12 for 2 hours before addition of neurotrophins or compounds. At the indicated time points, neurons were harvested and processed for Western blot analysis as described previously (61).
In vitro models for Alzheimer, Huntington, and Parkinson diseases.
In Aβ assays, E16 hippocampal neurons were plated at 80,000–100,000 cells per well and matured for 6–7 days (29). BDNF, LM22A compounds, and signaling inhibitors were added concomitantly with 10 μM Aβ, followed by 72 hours incubation. Cell death was assessed using TUNEL/DAPI staining. For QA assays, striatal neurons derived from (E16) CF1 mice were seeded on glass coverslips in 24-well plates, matured for 6–7 days, and then treated with fresh Neurobasal B27 medium containing QA at a final concentration of 7.5 mM in the presence or absence of BDNF or LM22A-4 with or without K252a (200 nM). After 24 hours, neurons were immunostained with DARPP-32 antibody, followed by Cy3-conjugated secondary antibody. DARPP-32–positive neurons were counted in systematically preselected fields. MPP+ assays were conducted as described previously using human SH-SY5Y neuroblastoma cells (31). After retinoic acid– and TPA-induced differentiation, cells were pretreated with LM22A-4 for 3 days. Fresh medium was added containing LM22A-4 (10 nM) with or without K252a (200 nM), and 30 minutes later MPP+ (100 μM) was added. After incubation for 48 hours, the number of surviving cells in each of 8 wells per condition was determined by MTT/morphology criteria.
LM22A-4–induced signaling in vivo.
Adult male C57BL/6J mice (2–4 months of age) were randomly divided into vehicle (normal saline [NS]) and treatment (LM22A-4 dissolved in NS) groups. Mice were given 10–15 μl vehicle or a 0.5-mg/ml solution of LM22A-4 into the right naris 3–5 times at 2- to 4-minute intervals (providing a dose of 0.22 mg/kg body weight), once daily for 1 week. Hippocampal and striatal brain regions were harvested 2.5 hours after the last dose and processed for Western blot analysis.
TBI, LM22A-4 treatment, and rotarod performance.
Male Sprague-Dawley rats (8 weeks old) were anesthetized with 80 mg/kg ketamine/10 mg/kg xylazine i.p. A rectal thermistor and heating pad were used to maintain rectal temperature at 37°C, throughout the procedure and recovery. The skull was exposed by midline incision and reflection of the scalp and temporalis muscle, and a 4-mm craniotomy was placed over the left parietal cortex midway between the lambda and bregma sutures, 2 mm from the lambda, and 1 mm from the sagittal suture. Injury was induced using a computer-controlled electromagnetic linear motor fitted with a slightly rounded 2-mm-diameter impact tip positioned perpendicularly to the meningeal surface (62, 63). Impact velocity was 1.5 m/s, depth 2.5 mm, and dwell time 120 ms, producing cortical disruption and mild damage to the underlying hippocampus. The incisions were then sutured, and the animals were allowed to recover. Sham-operated animals received a craniotomy but no impact. Injured animals received 3.4 μg LM22A-4 in a gel containing Carbopol 934P (0.7%), hydroxypropyl methylcellulose (1.3%), triethanolamine (2.5%), sodium deoxycholate (1%), sodium metabisulfite (0.01%), and methylparaben (0.05%) (to enhance retention and delivery; ref. 64) or vehicle alone, with 15 μl delivered to the left nares immediately after injury, then daily for 14 days. Motor testing was performed on an accelerating rotarod (increasing 1 rpm/5 s; SDI Rotor-Rod, San Diego Instruments Inc.) with time to fall recorded. Animals received training of 2 trials on the 2 consecutive days prior to TBI and then were tested with 2 trials weekly thereafter by an observer blinded to treatment. To evaluate compound effects on lesion size, male Sprague-Dawley rats injured and treated as above were terminally perfused with phosphate-buffered saline containing 4% paraformaldehyde (PFA) 21 days after injury, and their brains were postfixed in 4% PFA overnight, followed by soaking in 30% sucrose for 48 hours. Serial sections encompassing the injured area were cut at 40-μm intervals using a Leica CM1850 microtome. Sections were Nissl stained and every fifth section evaluated in a blinded fashion for lesion size, using Adobe Photoshop pixel counting.
Statistics.
Statistical analyses applied the Student’s t test, ANOVA with Dunnett’s or Tukey-Kramer correction, or repeated-measures ANOVA, with pairwise multiple comparisons using the Holm-Sidak method. For all graphs, mean ± SEM is shown. α = 0.05 for all tests.
Supplementary Material
Supplemental data
Acknowledgments
These studies were supported by the Institute for the Study on Aging (F.M. Longo), the Alzheimer’s Association (F.M. Longo), the Eastern Chapter of the North Carolina Alzheimer’s Association (F.M. Longo), Taube Philanthropies (F.M. Longo), the Koret Foundation (F.M. Longo), the Jean Perkins Foundation (F.M. Longo), and the Department of Defense (DOD DAMD17-03-1-0532) and the Department of Veterans Affairs (S.M. Massa).
Footnotes
Conflict of interest: F.M. Longo is a founder of PharmatrophiX, a company focused on the development of small molecule ligands for neurotrophin receptors.
Citation for this article: J Clin Invest. 2010;120(5):1774–1785. doi:10.1172/JCI41356.
References
- 1.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10(3):381–391. doi: 10.1016/S0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 3.Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4(4):299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 4.Mamidipudi V, Li X, Wooten MW. Identification of interleukin 1 receptor-associated kinase as a conserved component in the p75-neurotrophin receptor activation of nuclear factor-kappa B. J Biol Chem. 2002;277(31):28010–28018. doi: 10.1074/jbc.M109730200. [DOI] [PubMed] [Google Scholar]
- 5.Roux PP, Bhakar AL, Kennedy TE, Barker PA. The p75 neurotrophin receptor activates Akt (protein kinase B) through a phosphatidylinositol 3-kinase-dependent pathway. J Biol Chem. 2001;276(25):23097–23104. doi: 10.1074/jbc.M011520200. [DOI] [PubMed] [Google Scholar]
- 6.Salehi AH, et al. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27(2):279–288. doi: 10.1016/S0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 7.Wehrman T, He X, Raab B, Dukipatti A, Blau H, Garcia KC. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53(1):25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 8.Barker PA. High affinity not in the vicinity? Neuron. 2007;53(1):1–4. doi: 10.1016/j.neuron.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Schindowski K, Belarbi K, Buee L. Neurotrophic factors in Alzheimer’s disease: role of axonal transport. Genes Brain Behav. 2008;7(suppl 1):43–56. doi: 10.1111/j.1601-183X.2007.00378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington’s disease. Prog Neurobiol. 2007;81(5–6):294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli F, Racagni G, Riva MA. Shedding light into the role of BDNF in the pharmacotherapy of Parkinson’s disease. Pharmacogenomics J. 2006;6(2):95–104. doi: 10.1038/sj.tpj.6500360. [DOI] [PubMed] [Google Scholar]
- 12.Ogier M, Wang H, Hong E, Wang Q, Greenberg ME, Katz DM. Brain-derived neurotrophic factor expression and respiratory function improve after ampakine treatment in a mouse model of Rett syndrome. J Neurosci. 2007;27(40):10912–10917. doi: 10.1523/JNEUROSCI.1869-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griesbach GS, Sutton RL, Hovda DA, Ying Z, Gomez-Pinilla F. Controlled contusion injury alters molecular systems associated with cognitive performance. J Neurosci Res. 2009;87(3):795–805. doi: 10.1002/jnr.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griesbach GS, Hovda DA, Gomez-Pinilla F. Exercise-induced improvement in cognitive performance after traumatic brain injury in rats is dependent on BDNF activation. Brain Res. 2009;1288:105–115. doi: 10.1016/j.brainres.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7(4):278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sairanen M, Lucas G, Ernfors P, Castren M, Castren E. Brain-derived neurotrophic factor and antidepressant drugs have different but coordinated effects on neuronal turnover, proliferation, and survival in the adult dentate gyrus. J Neurosci. 2005;25(5):1089–1094. doi: 10.1523/JNEUROSCI.3741-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergami M, Rimondini R, Santi S, Blum R, Gotz M, Canossa M. Deletion of TrkB in adult progenitors alters newborn neuron integration into hippocampal circuits and increases anxiety-like behavior. Proc Natl Acad Sci U S A. 2008;105(40):15570–15575. doi: 10.1073/pnas.0803702105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. TrkB regulates hippocampal neurogenesis and governs sensitivity to antidepressive treatment. Neuron. 2008;59(3):399–412. doi: 10.1016/j.neuron.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455(7215):894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsao D, et al. TrkB agonists ameliorate obesity and associated metabolic conditions in mice. Endocrinology. 2008;149(3):1038–1048. doi: 10.1210/en.2007-1166. [DOI] [PubMed] [Google Scholar]
- 21.Poduslo JF, Curran GL. Permeability at the blood-brain and blood-nerve barriers of the neurotrophic factors: NGF, CNTF, NT-3, BDNF. Brain Res Mol Brain Res. 1996;36(2):280–286. doi: 10.1016/0169-328X(95)00250-V. [DOI] [PubMed] [Google Scholar]
- 22.Morse JK, et al. Brain-derived neurotrophic factor (BDNF) prevents the degeneration of medial septal cholinergic neurons following fimbria transection. J Neurosci. 1993;13(10):4146–4156. doi: 10.1523/JNEUROSCI.13-10-04146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Chi XX, Nicol GD. BDNF enhances the excitability of rat sensory neurons through activation of the p75 neurotrophin receptor and the sphingomyelin pathway. J Physiol. 2008;586(13):3113–3127. doi: 10.1113/jphysiol.2008.152439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher JM, Hughes RA. Novel monocyclic and bicyclic loop mimetics of brain-derived neurotrophic factor. J Pept Sci. 2006;12(8):515–524. doi: 10.1002/psc.760. [DOI] [PubMed] [Google Scholar]
- 25.Ibanez CF, Ebendal T, Persson H. Chimeric molecules with multiple neurotrophic activities reveal structural elements determining the specificities of NGF and BDNF. EMBO J. 1991;10(8):2105–2110. doi: 10.1002/j.1460-2075.1991.tb07743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibanez CF, Ilag LL, Murray-Rust J, Persson H. An extended surface of binding to Trk tyrosine kinase receptors in NGF and BDNF allows the engineering of a multifunctional pan-neurotrophin. EMBO J. 1993;12(6):2281–2293. doi: 10.1002/j.1460-2075.1993.tb05882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux PP, et al. K252a and CEP1347 are neuroprotective compounds that inhibit mixed-lineage kinase-3 and induce activation of Akt and ERK. J Biol Chem. 2002;277(51):49473–49480. doi: 10.1074/jbc.M203428200. [DOI] [PubMed] [Google Scholar]
- 28.Balkowiec A, Katz DM. Activity-dependent release of endogenous brain-derived neurotrophic factor from primary sensory neurons detected by ELISA in situ. J Neurosci. 2000;20(19):7417–7423. doi: 10.1523/JNEUROSCI.20-19-07417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T, et al. Small molecule, non-peptide p75 ligands inhibit Aβ-induced neurodegeneration and synaptic impairment. PLoS ONE. 2008;3(11):e3604. doi: 10.1371/journal.pone.0003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arancibia S, et al. Protective effect of BDNF against beta-amyloid induced neurotoxicity in vitro and in vivo in rats. Neurobiol Dis. 2008;31(3):316–326. doi: 10.1016/j.nbd.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Presgraves SP, Borwege S, Millan MJ, Joyce JN. Involvement of dopamine D(2)/D(3) receptors and BDNF in the neuroprotective effects of S32504 and pramipexole against 1-methyl-4-phenylpyridinium in terminally differentiated SH-SY5Y cells. Exp Neurol. 2004;190(1):157–170. doi: 10.1016/j.expneurol.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Kells AP, Fong DM, Dragunow M, During MJ, Young D, Connor B. AAV-mediated gene delivery of BDNF or GDNF is neuroprotective in a model of Huntington disease. Mol Ther. 2004;9(5):682–688. doi: 10.1016/j.ymthe.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Bjugstad KB, Zawada WM, Goodman S, Freed CR. IGF-1 and bFGF reduce glutaric acid and 3-hydroxyglutaric acid toxicity in striatal cultures. J Inherit Metab Dis. 2001;24(6):631–647. doi: 10.1023/A:1012706908779. [DOI] [PubMed] [Google Scholar]
- 34.Klintsova AY, Dickson E, Yoshida R, Greenough WT. Altered expression of BDNF and its high-affinity receptor TrkB in response to complex motor learning and moderate exercise. Brain Res. 2004;1028(1):92–104. doi: 10.1016/j.brainres.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Ishibashi H, Hihara S, Takahashi M, Heike T, Yokota T, Iriki A. Tool-use learning induces BDNF expression in a selective portion of monkey anterior parietal cortex. Brain Res Mol Brain Res. 2002;102(1–2):110–112. doi: 10.1016/S0169-328X(02)00201-2. [DOI] [PubMed] [Google Scholar]
- 36.Schabitz WR, et al. Effect of brain-derived neurotrophic factor treatment and forced arm use on functional motor recovery after small cortical ischemia. Stroke. 2004;35(4):992–997. doi: 10.1161/01.STR.0000119754.85848.0D. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Pardridge WM. Blood-brain barrier targeting of BDNF improves motor function in rats with middle cerebral artery occlusion. Brain Res. 2006;1111(1):227–229. doi: 10.1016/j.brainres.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Willson ML, McElnea C, Mariani J, Lohof AM, Sherrard RM. BDNF increases homotypic olivocerebellar reinnervation and associated fine motor and cognitive skill. Brain. 2008;131(pt 4):1099–1112. doi: 10.1093/brain/awn024. [DOI] [PubMed] [Google Scholar]
- 39.Blaha GR, Raghupathi R, Saatman KE, McIntosh TK. Brain-derived neurotrophic factor administration after traumatic brain injury in the rat does not protect against behavioral or histological deficits. NeuroScience. 2000;99(3):483–493. doi: 10.1016/S0306-4522(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 40.Conte V, et al. TrkB gene transfer does not alter hippocampal neuronal loss and cognitive deficits following traumatic brain injury in mice. Restor Neurol Neurosci. 2008;26(1):45–56. [PMC free article] [PubMed] [Google Scholar]
- 41.Buitrago MM, Schulz JB, Dichgans J, Luft AR. Short and long-term motor skill learning in an accelerated rotarod training paradigm. Neurobiol Learn Mem. 2004;81(3):211–216. doi: 10.1016/j.nlm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Massa SM, et al. Small, nonpeptide p75NTR ligands induce survival signaling and inhibit proNGF-induced death. . J Neurosci. 2006;26(20):5288–5300. doi: 10.1523/JNEUROSCI.3547-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukuru SC, et al. Plate-based diversity selection based on empirical HTS data to enhance the number of hits and their chemical diversity. J Biomol Screen. 2009;14(6):690–699. doi: 10.1177/1087057109335678. [DOI] [PubMed] [Google Scholar]
- 45.Triballeau N, et al. High-potency olfactory receptor agonists discovered by virtual high-throughput screening: molecular probes for receptor structure and olfactory function. Neuron. 2008;60(5):767–774. doi: 10.1016/j.neuron.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonacci TM, et al. Differential targeting of Gβg-subunit signaling with small molecules. Science. 2006;312(5772):443–446. doi: 10.1126/science.1120378. [DOI] [PubMed] [Google Scholar]
- 47.Tesmer JJ. Pharmacology. Hitting the hot spots of cell signaling cascades. Science. 2006;312(5772):377–378. doi: 10.1126/science.1126903. [DOI] [PubMed] [Google Scholar]
- 48.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci U S A. 2006;103(42):15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruncko M, et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem. 2007;50(4):641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 50.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 51.Minichiello L, et al. Point mutation in trkB causes loss of NT4-dependent neurons without major effects on diverse BDNF responses. Neuron. 1998;21(2):335–345. doi: 10.1016/S0896-6273(00)80543-7. [DOI] [PubMed] [Google Scholar]
- 52.Fan G, et al. Knocking the NT4 gene into the BDNF locus rescues BDNF deficient mice and reveals distinct NT4 and BDNF activities. Nat Neurosci. 2000;3(4):350–357. doi: 10.1038/73921. [DOI] [PubMed] [Google Scholar]
- 53.O’Leary PD, Hughes RA. Design of potent peptide mimetics of brain-derived neurotrophic factor. J Biol Chem. 2003;278(28):25738–25744. doi: 10.1074/jbc.M303209200. [DOI] [PubMed] [Google Scholar]
- 54.Williams G, Williams EJ, Maison P, Pangalos MN, Walsh FS, Doherty P. Overcoming the inhibitors of myelin with a novel neurotrophin strategy. J Biol Chem. 2005;280(7):5862–5869. doi: 10.1074/jbc.M411121200. [DOI] [PubMed] [Google Scholar]
- 55.Maliartchouk S, Debeir T, Beglova N, Cuello AC, Gehring K, Saragovi HU. Genuine monovalent ligands of TrkA nerve growth factor receptors reveal a novel pharmacological mechanism of action. J Biol Chem. 2000;275(14):9946–9956. doi: 10.1074/jbc.275.14.9946. [DOI] [PubMed] [Google Scholar]
- 56.Zhang AJ, Khare S, Gokulan K, Linthicum DS, Burgess K. Dimeric β-turn peptidomimetics as ligands for the neurotrophin receptor TrkC. Bioorg Med Chem Lett. 2001;11(2):207–210. doi: 10.1016/S0960-894X(00)00618-1. [DOI] [PubMed] [Google Scholar]
- 57.Mischel PS, Umbach JA, Eskandari S, Smith SG, Gundersen CB, Zampighi GA. Nerve growth factor signals via preexisting TrkA receptor oligomers. Biophys J. 2002;83(2):968–976. doi: 10.1016/S0006-3495(02)75222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson RC, Radziejewski C, Stuart DI, Jones EY. Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer. Biochemistry. 1995;34(13):4139–4146. doi: 10.1021/bi00013a001. [DOI] [PubMed] [Google Scholar]
- 59.Yang T, Massa SM, Longo FM. LAR protein tyrosine phosphatase receptor associates with TrkB and modulates neurotrophic signaling pathways. J Neurobiol. 2006;66(13):1420–1436. doi: 10.1002/neu.20291. [DOI] [PubMed] [Google Scholar]
- 60.Lindholm D, Carroll P, Tzimagiogis G, Thoenen H. Autocrine-paracrine regulation of hippocampal neuron survival by IGF-1 and the neurotrophins BDNF, NT-3 and NT-4. Eur J Neurosci. 1996;8(7):1452–1460. doi: 10.1111/j.1460-9568.1996.tb01607.x. [DOI] [PubMed] [Google Scholar]
- 61.Yang T, et al. Leukocyte antigen-related protein tyrosine phosphatase receptor: a small ectodomain isoform functions as a homophilic ligand and promotes neurite outgrowth. J Neurosci. 2003;23(8):3353–3363. doi: 10.1523/JNEUROSCI.23-08-03353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bilgen M. A new device for experimental modeling of central nervous system injuries. Neurorehabil Neural Repair. 2005;19(3):219–226. doi: 10.1177/1545968305278635. [DOI] [PubMed] [Google Scholar]
- 63.Onyszchuk G, Al-Hafez B, He YY, Bilgen M, Berman NE, Brooks WM. A mouse model of sensorimotor controlled cortical impact: characterization using longitudinal magnetic resonance imaging, behavioral assessments and histology. J Neurosci Methods. 2007;160(2):187–196. doi: 10.1016/j.jneumeth.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Souza R, Mutalik S, Venkatesh M, Vidyasagar S, Udupa N. Insulin gel as an alternate to parenteral insulin: formulation, preclinical, and clinical studies. AAPS PharmSciTech. 2005;6(2):E184–E189. doi: 10.1208/pt060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data