Genome-wide association study of systemic sclerosis identifies CD247 as a novel susceptibility locus (original) (raw)
. Author manuscript; available in PMC: 2010 Nov 1.
Published in final edited form as: Nat Genet. 2010 Apr 11;42(5):426–429. doi: 10.1038/ng.565
Abstract
Systemic sclerosis (SSc) is an autoimmune disease characterized by fibrosis of the skin and internal organs that leads to profound disability and premature death. To identify novel SSc susceptibility loci we conducted the first genome wide association study (GWAS) in a population of Caucasian ancestry including a total of 2296 SSc patients and 5171 controls. Analysis of 279,621 autosomal single nucleotide polymorphisms (SNPs) followed by replication testing in an independent case-control set of European ancestry (2,753 SSc patients / 4,569 controls) identified a new susceptibility locus for systemic sclerosis at CD247 (1q22-23; rs2056626, P = 2.09 × 10−7 in the discovery samples, P = 3.39 × 10−9 in the combined analysis). Additionally, we confirm and firmly establish the role of MHC (2.31 × 10−18), IRF5 (P =1.86 × 10−13) and STAT4 (P =3.37 × 10−9) gene regions as SSc genetic risk factors.
Systemic sclerosis (SSc) is a profoundly disabling autoimmune disease characterized by vascular damage, altered immune responses and abnormal fibrosis of skin and internal organs leading to premature death in affected individuals 1. SSc etiology is complex and poorly understood, but similar to most autoimmune conditions it is widely accepted that the involvement of environmental and a multiplicity of genetic factors leads to disease. Data from familial, twin and ethnicity studies support the relevance of the genetic component in SSc etiology 2. Previous studies aimed at dissecting the genetic factors underlying SSc genetic susceptibility so far have used the candidate gene association study approach 3. In spite of the several years of research this strategy yielded a very limited characterization of SSc genetic risk factors. Except for the major histocompatibility complex (MHC) genes, that are relevant genetic markers for SSc across populations, few other loci outside the HLA region demonstrated strong and reproducible associations with SSc susceptibility 3,4. Only very recently, large case-control association studies have identified STAT4 and IRF5 genes as novel genetic factors contributing to SSc susceptibility 5–8. Similar to other complex genetic disorders it is expected that several genetic markers contribute to SSc predisposition with modest effects, and large sample sizes are required to detect novel disease associated loci 9.
Therefore, we aimed more comprehensively to identify novel SSc susceptibility loci and thus conducted the first genome wide association study (GWAS) in SSc including a total of 2296 SSc patients and 5171 healthy controls from four case-control series of Caucasian ancestry (USA, Spain, Germany and The Netherlands) (Supplementary Table 1). Genotyping of SSc case sets and Spanish controls was performed using the Illumina Bead-Array platform with chips of different single nucleotide polymorphism (SNP) densities (Supplementary Table 1). The genotypes of North American controls were obtained from the Cancer Genetic Markers of Susceptibility (CGEMS) studies and Illumina iControlDB database (www.illumina.com/iControlDB, Illumina, San Diego, CA), German and Dutch control groups were extracted from previous studies or public databases 10–13.
After rigorous genotyping quality control filters, a total of 279,621 SNPs shared between the four case-control series were extracted for analysis (Supplementary Table 1).
Genomic inflation factor (λ) was estimated for the complete data set showing evidence of a modest inflation of test statistics (λ = 1.069). When the HLA region was excluded, the inflation of test statistics somewhat decreased (λ = 1.066) (Supplementary Figure 1). To adjust for potential population stratification we applied a genomic control correction to the test statistics. The potential effect of population substructure was tested by deriving principal components on a population-specific basis. We observed that case and control individuals in each population were not significantly different by principal components and were therefore well genetically matched. We also performed an inverse variance based meta-analysis, adjusting the odds ratios for the first five country-specific principal components. This analysis showed little variation from genomic control corrected P values (Table 1).
Table 1.
Loci showing the strongest association signal with SSc susceptibility outside the MHC region.
| Chr. | Gene | SNP | Location | BP | Minorallele | MAFCase/Control | GC correctedP value | PC correctedP value | OR 95 % CI |
|---|---|---|---|---|---|---|---|---|---|
| rs10488631 | Downstream | 128.381.419 | C | 0.145/1.102 | 1.86×10−13 | 3.84×10−14 | 1.50 (1.35–1.67) | ||
| 7q32 | TNPO3/IRF5 | rs12537284 | Intergenic | 128.505.142 | A | 0.162/0.129 | 2.74×10−7 | 1.49×10−7 | 1.30 (1.18–1.44) |
| rs4728142 | Upstream | 128.361.203 | A | 0.494/0.445 | 5.21×10−7 | 1.81×10−7 | 1.21 (1.12–1.29) | ||
| 2q32 | STAT4 | rs3821236 | Intronic | 191.611.003 | A | 0.247/0.202 | 3.37×10−9 | 3.93×10−9 | 1.30 (1.19–1.41) |
| 1q22-23 | CD247 | rs2056626 | Intronic | 165.687.049 | G | 0.370/0.421 | 2.09×10−7 | 3.27×10−7 | 0.82 (0.76–0.88) |
| 18q22 | CDH7 | rs10515998 | Intergenic | 61.521.202 | G | 0.062/0.040 | 2.25×10−7 | 1.01×10−7 | 1.53 (1.31–1.76) |
| 6p25 | EXOC2/IRF4 | rs4959270 | Intronic | 402.748 | A | 0.445/0.494 | 1.23×10−7 | 9.06×10−8 | 0.82 (0.77–0.88) |
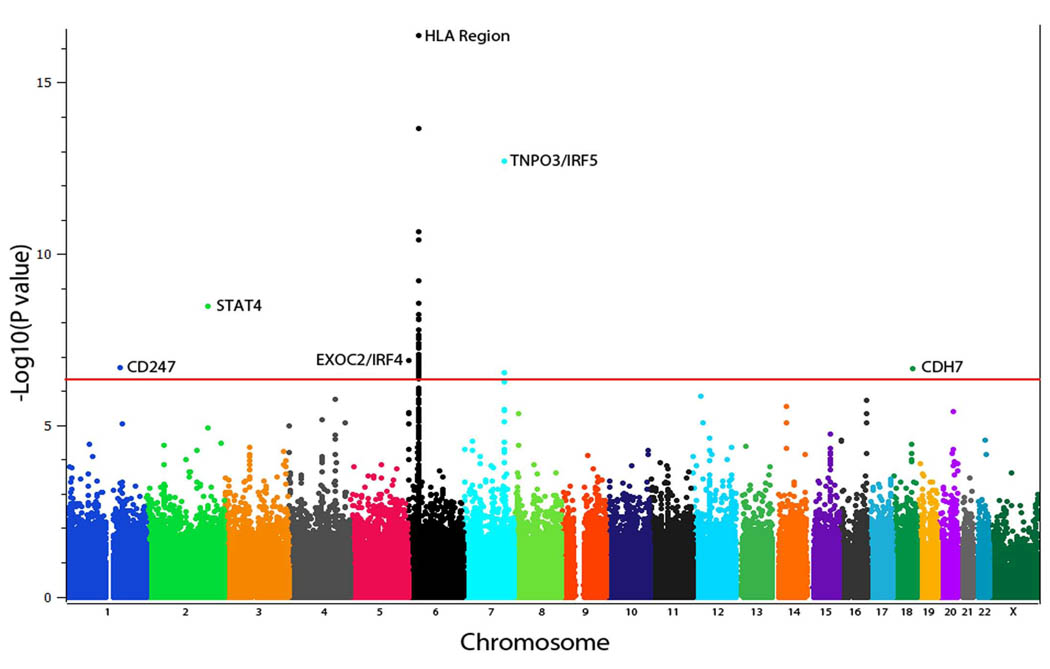
The Mantel-Haenszel test under an allelic model revealed several SNPs reaching P values at genome-wide significance after genomic control correction (P ≤ 5×10−7) (Figure 1). The strongest association signal was observed for a cluster of SNPs in an extended region at 6p21 locus within the MHC region, where the rs6457617 SNP located in the HLA*DQB1 gene region gave the highest P value (P GC corrected = 2.31 × 10−18) (Figure 1 & Supplementary Table 2). Outside the MHC region, five loci showed association at P < 10−7 namely TNPO3/IRF5 region in 7q32, STAT4 in 2q32, CD247 in 1q22-23, CDH7 in 18q22 and EXOC2/IRF4 near 6p25. The trend observed for all these loci were consistent across the different study populations (Supplementary table 3). Furthermore, the TNPO3/IRF5 locus obtained genome wide significance in the single US cohort and was further corroborated in the European cohorts (Supplementary table 3). SNPs mapping to the region of TNPO3/IRF5 and STAT4 achieved the strongest association observed for non-HLA genes (rs10488631 P =1.86 × 10−13 OR 1.50 95 % CI 1.35–1.67 and rs3821236 P =3.37 × 10−9 OR 1.30 95 % CI 1.18–1.44, respectively) (Table 1 & Supplementary table 3). Therefore, these results confirm the previously reported role of MHC, STAT4 and IRF5 genes as genetic risk factors for systemic sclerosis and identified three new candidate loci 3–8.
Figure 1.
Manhattan plot of the Genome wide association study of the discovery cohort comprising 2346 SSc patients and 5193 healthy controls. The –log10 of the Mantel-Haenszel test P value of 279.621 SNPs after correction by λ is plotted against its physical chromosomal position. Chromosomes are showed in alternate colors. SNPs above the red line represent those with a _P_-value of 5 < 10−7. Plot corresponds to the combined analysis of the study cohorts.
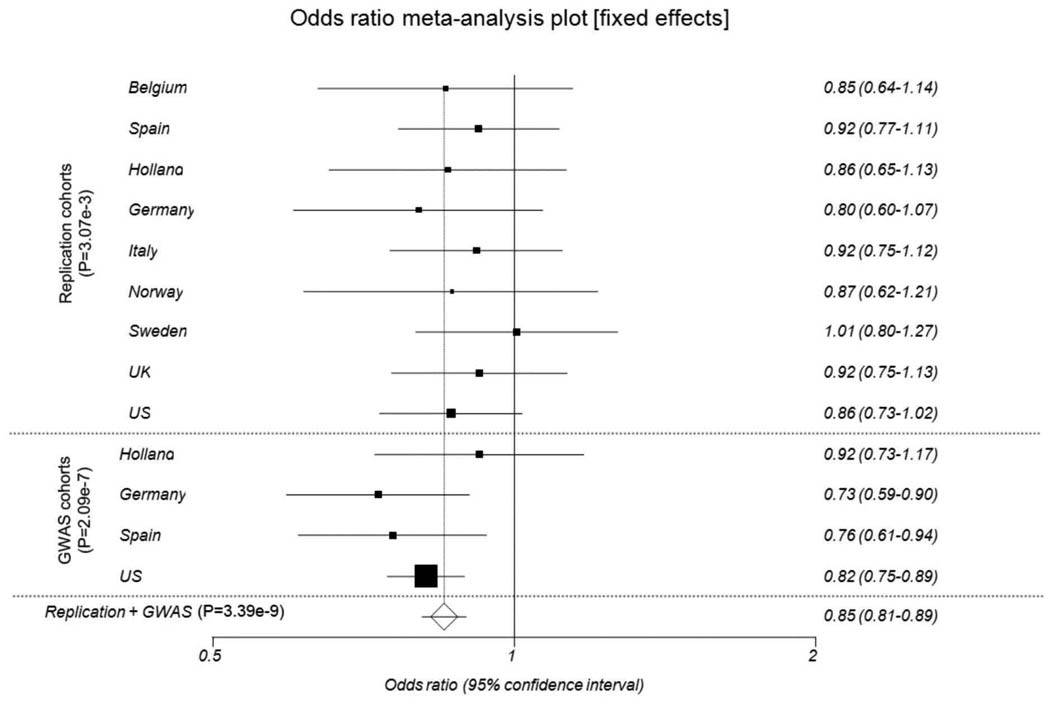
We then aimed to confirm the association of the CD247, CDH7 and EXOC2/IRF4 loci with SSc susceptibility using a large independent replication case-control set comprising 2753 SSc patients and 4569 controls of Caucasian ancestry (Supplementary table 4). The SNPs showing the strongest GWAS association on each region (rs2056626 for CD247, rs10515998 for CDH7 and rs4959270 for EXOC2/IRF4) were genotyped in the replication cohorts using TaqMan 5′ allelic discrimination assay technology. The association analysis by Mantel-Haenszel test revealed a significant association of the rs2056626 genetic variant in the CD247 region (_P_=3.07×10−3 OR 0.89 95 % CI 0.83–0.96) (Table 2 & figure 2). The combined analysis of the GWAS and replication cohort for this SNP revealed highly significant association (P =3.39×10−9 OR 0.86 95 % CI 0.81–0.90). The association of the SNPs in the CDH7 and EXOC2/IRF4 regions was not confirmed in this replication cohort (Table 2 and Supplementary figure 2). Considering that the frequency observed for the CDH7 rs10515998 genetic variant, is quite low (around 5%) the population size of the replication cohort reached only 13% of statistical power to detect an association at a significance level similar to that observed in the replication analysis (OR 1.05). Therefore, the possible implication of the CDH7 locus in SSc genetic predisposition should be still further investigated. In contrast, due to the high MAF frequency of the rs4959270 polymorphism in the EXOC2/IRF4 region, great heterogeneity of the association was observed in the replication cohorts (supplementary figure 1). These findings are concordant with previous GWAS studies in which great population allelic heterogeneity has been reported for EXOC2/IRF4 genetic variants leading to not true disease associations, as possibly occurred in our screening phase 14. Interestingly, the novel identified SSc susceptibility locus, CD247, participates in the regulation of the immune response and thus could have a role in SSc pathogenesis. The CD247 gene encodes the T-cell receptor zeta (CD3ζ) subunit, a component of the T cell receptor (TCR)/CD3 complex 15. This chain plays an important role in the assembly and transport of the TCR/CD3 complex to the cell surface and is crucial to receptor signaling function. It has been observed that the expression of the CD3ζ chain is altered in chronic autoimmune/inflammatory disorders and that its low expression results in impaired immune response 16–18. Notably, the CD247 gene has been associated with susceptibility to systemic lupus erythematosus (SLE), another systemic autoimmune disease 19,20. Moreover, genetic variants in the 3′ untranslated region of this gene have shown functional implication leading to a reduced expression of this molecule that could be manifested by systemic autoimmunity 19. Therefore, further studies aiming to dissect the exact role of this molecule in SSc will be of interest.
Table 2.
Association results for three loci genotyped in the replication samples.
| CHR | Gene | SNP | Position | MinorAllele | Stage | N(case/control) | MAF(case/control) | P value | OR 95 % CI |
|---|---|---|---|---|---|---|---|---|---|
| 1q22–23 | CD247 | rs2056626 | 165,687,049 | G | GWAS | 2296/5014 | 0.370/0.421 | 2.09×10−7 | 0.82(1.76–0.88) |
| Replication | 2566/4387 | 0.366/0.394 | 3.07×10−3 | 0.89(0.83–0.96) | |||||
| Combined | 4867/9401 | 0.368/0.409 | 3.39×10−9 | 0.86(0.81–0.90) | |||||
| 18q22 | CDH7 | rs10515998 | 61,521,202 | G | GWAS | 2296/5014 | 0.062/0.040 | 2.25×10−7 | 1.53(1.31–1.79) |
| Replication | 2594/4414 | 0.058/0.056 | 4.98×10−1 | 1.05(0.91–1.22) | |||||
| Combined | 4895/9428 | 0.060/0.048 | 3.99×10−5 | 1.25(1.13–1.40) | |||||
| 6p25 | EXOC2/IRF4 | rs4959270 | 402,748 | A | GWAS | 2296/5171 | 0.445/0.494 | 1.23×10−7 | 0.82(0.77–0.88) |
| Replication | 2361/4372 | 0.466/0.469 | 6.34×10−1 | 0.98(0.91–1.05) | |||||
| Combined | 4662/9554 | 0.456/0.483 | 2.16×10−5 | 0.90(0.85–0.94) |
Figure 2.
Forest plot showing the odds ratios and confidence intervals of the CD247 association in the various populations studied both in the discovery and replication cohorts.
This work represents the first large GWAS study conducted to date in SSc. Of note, the results obtained confirm and firmly establish the role of HLA, STAT4 and IRF5 in the genetic predisposition to SSc, genes that are also known to be risk factors for several other autoimmune conditions. In addition, a new susceptibility locus not previously considered as susceptibility factors for SSc has been identified. All these findings support the strong autoimmune component underlying SSc pathogenesis and highlight that the development of SSc seems to be determined by shared common genetic and pathogenic mechanisms with other autoimmune diseases and specific disease pathways that should be further characterized.
METHODS
Subjects
Because SSc is a relatively rare autoimmune disorder (estimated prevalence in Caucasian populations ~ 0.01 %) large sets of SSc patients can best be recruited through international collaboration. Consequently, to reach the total of 2296 SSc patients and 5171 healthy control individuals analyzed in the present study, four series with Caucasian European-ancestry were included: USA, Spain, Germany and The Netherlands. The North American cases (initial n=1,678; after applying quality control criteria, n=1486; 179 men, 1307 women; mean age=54.5 (median, 55.0); SD=12.9) were obtained from May, 2001 to December, 2008 from three U.S. sources: University of Texas (UT) Health Science Center-Houston, The Johns Hopkins University Medical Center and Fred Hutchinson Cancer Center, each enrolling patients from a US-wide catchment area. Whole genome genotyping data from USA control individuals (initial n=5,520) were obtained from the following 3 publicly available databases: (1) breast cancer controls from the Cancer Genetic Markers of Susceptibility (CGEMS) studies; (2) prostate cancer controls from CGEMS; and (3) controls from Illumina iControlDB. After sex-matching and applying quality control criteria, 419 men and 3058 women controls were analyzed.
The initial European SSc cases series came from previously established collections with nationally representative recruitment of in total 380 Spanish, 288 German and 190 Dutch SSc patients. Main demographical and clinical data of European SSc patients have been described previously 5,21. As control population, healthy unrelated individuals of Spanish (initial n=414), German (initial n=678) and Dutch (initial n=643) origin were included in the study. Whole genome genotyping data from German controls were from the Popgen Biobank and from a previous study in the case of Dutch 12,13.
To further confirm associations found on GWAS stage we collected a large independent replication cohort of individuals with Caucasian ancestry from Belgium, Spain, Holland, Germany, Italy, Norway, Sweden, UK and USA. A total of 2753 SSc patients and 4569 healthy controls where recruited for this second stage (supplementary table 4).
All cases either met the American College of Rheumatology Preliminary criteria for the classification of SSc or had at least 3 of the 5 CREST (Calcinosis, Raynaud’s phenomenon, Esophageal dysmotility, Sclerodactyly, Telangiectasias) features22. Main clinical features of SSc patients are included in supplementary table 5.
Collection of blood samples and clinical information from case and control subjects was undertaken with informed consent and relevant ethical review board approval from each contributing center in accordance with the tenets of the Declaration of Helsinki.
Genotyping
The GWAS genotyping of the Spanish SSc and controls together with Dutch and German SSc patients was performed at the Department of Medical Genetics of the University Medical Center Utrecht (The Netherlands) using the commercial release Illumina human CNV370K BeadChip that contains 300.000 standard SNPs with an additional 52,167 markers designed to specifically target nearly 14,000 copy number variant (CNV) regions of the genome, for a total of over 370,000 markers. This system delivers high genomic coverage of the SNPs from Phase I and II of the HapMap Project (www.hapmap.org), capturing 81% of the HapMap variation at r2 > 0.8 in Caucasian populations. Genotype data for Dutch and German controls was obtained from the Illumina Human 550K BeadChip available from a previous study 12,13. The North American SSc patient group was genotyped at Boas Center for Genomics and Human Genetics, Feinstein Institute for Medical Research, North Shore LIJ Health System, using the Illumina Human610-Quad BeadChip capturing 89% of the HapMap CEU variation at r2 > 0.8. CGEMS and Illumina iControlDB controls were genotyped on Illumina Hap550K-BeadChip. For the replication phase, SNPs reaching GWAS significance located in novel potential SSc susceptibility loci (rs2056626 for CD247, rs10515998 for CDH7 and rs4959270 for EXOC2/IRF4) were genotyped in the replication cohorts using Applied Biosystems’ TaqMan SNP genotyping Assays on an Abiprism 7900 HT real-time thermocycler. Markers with call rates of 95% or less were excluded, as were markers whose allele distributions deviated strongly from Hardy-Weinberg equilibrium in controls (P <10−5). Only markers with minor allele frequencies of ≥1% in both cases and controls were included in the analyses.
Statistical analysis
Statistical analyses were undertaken using R (v2.6), STATA (v8; State College, Texas, US) and PLINK (v1.06) software (http://pngu.mgh.harvard.edu/purcell/plink/)23. All reported P values are two-sided. Using PLINK we identified and excluded pairs of genetically related subjects or duplicates and excluded pair members with lower call rate. To identify individuals who might have non-Western European ancestry, we merged our case and control data with the data from the HapMap Project (60 western European (CEU), 60 Nigerian (YRI), 90 Japanese (JPT) and 90 Han Chinese (CHB) samples). We used principal component analysis as implemented in HelixTree, plotting the first two principal components for each individual. All individuals who were not clustering with the main CEU cluster (deviating more than 4 SD from cluster centroids) were excluded from subsequent analyses. The principal components derived on the resulting sample look typical for populations of European origin (Supplementary Figure 3)24. Additionally, we excluded individuals with low call rate (11 on US, 24 on Spanish, 1 on German and 1 on Dutch), relatedness (50 on US, 2 on Spanish, 1 on German and 1 on Dutch), non-Caucasian ancestry (42 on US, 5 on Spanish, 6 on German and 4 on Dutch) and inconsistent gender (83 on US, 2 on Spanish, 2 on German and 2 on Dutch). Then we filtered for SNP quality, removing SNPs with less than 98% genotyping success call rate and those showing minor allele frequency below 1%. Deviation of the genotype frequencies in the controls from those expected under Hardy-Weinberg equilibrium (HWE) was assessed by a χ2 test or Fisher's exact test when an expected cell count was <5. SNPs strongly deviating from HW equilibrium (P <10−5) were eliminated from the study. For the combined analysis of the four datasets the same quality controls per individual and per SNP were re-applied except HW equilibrium. The genotyping success call rate on the merged data set after all these quality filters was 99.83% in the GWAS cohorts. In the replication cohorts, genotyping success call rate was 98.16% after quality filters. The association between each SNP and risk of scleroderma in each set of data was assessed by the Cochran-Armitage trend test. Odds ratios and associated 95% CIs were calculated by unconditional logistic regression.
To determine if genome wide significant associated SNPs belonged to extensive linkage disequilibrium (LD) blocks we investigated the LD pattern (using r2 parameter) on a 1 Mb length region surrounding significant SNPs (Supplementary figures 4A–E). No strong LD (r2 > 0.8) was observed among investigated SNPs and other variants on the region, except rs12537284 which was on LD with rs10488631 (r2 = 0.82) in TNPO3/IRF5 region, both found to be genome wide significantly associated with SSc (table 1). The Meta-analysis of the four study series was conducted using standard methods based on Cochran-Mantel-Haenszel test. Breslow-Day test was performed for all SNPs to assess heterogeneity of the effect in different populations. We tested for the population structure and possibility of differential genotyping of cases and controls using quantile-quantile plots of test statistics and calculated inflation factor λ, by dividing the median of the test statistic s by the expected median from a χ2 distribution with one d.f. There was evidence of modest inflation of the test statistics (λ = 1.069, or 1.066 after excluding the HLA region), indicating a potential effect of the population substructure on the results. We therefore applied a genomic control correction to our results. Alternatively, we also derived principal components on a population-specific basis using HelixTree software, and applied an adjustment for the five first principal components as well as gender, separately for each country using logistic regression, after which the effects for each SNP were combined by meta-analysis, using inverse variance method (corresponding _P_-values are presented in Table 1 and Supplementary table 2). The results from this analysis were consistent with the results from the GC corrected Mantel-Haenszel meta-analysis. We then proceeded analyzing three new SNPs association found on the GWAS screen on the replication cohorts. Data was filtered according to same proceedings as the GWAS stage. Analysis was carried out by Mantel-Haenszel meta-analysis of all the independent replication cohorts to control for differences between groups. We then did meta-analysis of all the replication and GWAS cohorts for these SNPs with the same Mantel-Haenszel statistical procedure. Results are shown in table 2.
Supplementary Material
Supp Materials
ACKNOWLEGDMENTS
This work was supported by the following grants: T.R.D.J.R. was funded by the VIDI laureate from the Dutch association of research (NWO) and Dutch arthritis foundation (National Reumafonds). GEN-FER from the Spanish Society of Rheumatology, SAF2009-11110 from the Spanish Ministry of Science, CTS-4977 from Junta de Andalucía, Spain and in part by RETICS Program, RD08/0075 (RIER) from Instituto de Salud Carlos III (ISCIII), Spain (J.M.). R.B. is supported by the I3P CSIC program funded by the “Fondo Social Europeo”. BZA is supported by the Netherlands Organization for Health Research and Development (ZonMW grant 016.096.121). B.K. is supported by the Dutch Diabetes Research Foundation (grant 2008.40.001) and the Dutch arthritis foundation (reumafonds, grant NR 09-1-408). Genotyping of the Dutch control samples was sponsored by NIMH funding, R01 MH078075 (R.O.A.). The German controls were the PopGen biobank [to B.K.]. The PopGen project received infrastructure support through the German Research Foundation excellence cluster “Inflammation at Interfaces”. The US analyses were supported by the NIH/NIAMS R01 AR055258, Two-Stage Genome Wide Association Study in Systemic Sclerosis, (M.D.M.) and by the NIH/NIAMS Center of Research Translation (CORT) in SSc (P50AR054144) (F.C.A.), the NIH/NIAMS SSc Family Registry and DNA Repository (N01-AR-0-2251) (M.D.M.), UTHSC-H Center for Clinical and Translational Sciences (Houston CTSA Program) (NIH/NCRR 3UL1RR024148) (F.C.A.), NIH/NIAMS K08 Award (K08AR054404) (S.K.A.), SSc Foundation New Investigator Award (S.K.A.)
Footnotes
AUTHOR CONTRIBUTIONS
Study Design: T.R., O.G., B.R., J.E.M., B.P.C.K., F.C.A., J.M., M.D.M.
Collection of data: T.R., M.J.C., M.C.V., A.V., A.S., J.B., B.A.L., A.M.H.V., R.A.O., G.R., N.H., C.P.S., N.O.C., M.A.G.G., M.F.G.E., P.A., J.v.L., A.H., J.W., R.H., V.S., F.d.K., F.H., M.M.C., R.M., P.S., R.W., A.K., H.K., E.d.B., T.W., L.P., L.K., L.B., R.S., J.V., M.H., P.G., J.L.N., F.M.W., L.H.
Interpretation and analysis of results: T.R., O.G., B.R., J.E.M., B.A., R.P.M., J.Y., Y.H., S.F.W., R.S., P.G., A.T.L., J.Y., Y.H., S.F., C.I.A., S.K.A., B.P.C.K., J.M., M.D.M.
Critical reading of manuscript: T.R., O.G., B.R., J.E.M., B.A., J.Y., M.J.C., M.C.V., A.V., A.S., J.B., P.L.C.M.R., R.S., B.A.L., A.M.H.V., G.R., N.H., C.P.S., N.O.C., M.A.G.G., M.F.G.E., P.A., J.v.L., A.H., J.W., R.H., V.S., F.d.K., F.H., M.M.C., R.M., P.S., R.W., A.K., H.K., E.d.B., T.W., L.P., L.B., R.S., J.V., M.H., P.G., C.I.A., J.L.N., F.M.W., L.H., S.K.A., P.G., F.K.T., B.P.C.K., F.C.A., J.M. M.D.M.
Project conception: T.R., B.P.C.K., F.C.A., J.M., M.D.M.
REFERENCES
- 1.Gabrielli A, Avvedimento EV, Krieg T. Scleroderma. N Engl J Med. 2009;360:1989–2003. doi: 10.1056/NEJMra0806188. [DOI] [PubMed] [Google Scholar]
- 2.Jimenez SA, Derk CT. Following the molecular pathways toward an understanding of the pathogenesis of systemic sclerosis. Ann Intern Med. 2004;140:37–50. [PubMed] [Google Scholar]
- 3.Agarwal SK, Tan FK, Arnett FC. Genetics and genomic studies in scleroderma (systemic sclerosis) Rheum Dis Clin North Am. 2008;34:17–40. doi: 10.1016/j.rdc.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Arnett FC, et al. Major Histocompatibility Complex (MHC) class II alleles, haplotypes, and epitopes which confer susceptibility or protection in the fibrosing autoimmune disease systemic sclerosis: analyses in 1300 Caucasian, African-American and Hispanic cases and 1000 controls. Ann Rheum Dis. 2009 doi: 10.1136/ard.2009.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rueda B, et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009;18:2071–2077. doi: 10.1093/hmg/ddp119. [DOI] [PubMed] [Google Scholar]
- 6.Tsuchiya N, et al. Association of STAT4 polymorphism with systemic sclerosis in a Japanese population. Ann Rheum Dis. 2009;68:1375–1376. doi: 10.1136/ard.2009.111310. [DOI] [PubMed] [Google Scholar]
- 7.Ito I, et al. Association of a functional polymorphism in the IRF5 region with systemic sclerosis in a Japanese population. Arthritis Rheum. 2009;60:1845–1850. doi: 10.1002/art.24600. [DOI] [PubMed] [Google Scholar]
- 8.Dieude P, et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 2009;60:225–233. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 9.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 12.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawczak M, et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 14.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Call ME, Wucherpfennig KW. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol Immunol. 2004;40:1295–1305. doi: 10.1016/j.molimm.2003.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan S, et al. Increased caspase-3 expression and activity contribute to reduced CD3zeta expression in systemic lupus erythematosus T cells. J Immunol. 2005;175:3417–3423. doi: 10.4049/jimmunol.175.5.3417. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan S, et al. Generation and biochemical analysis of human effector CD4 T cells: alterations in tyrosine phosphorylation and loss of CD3zeta expression. Blood. 2001;97:3851–3859. doi: 10.1182/blood.v97.12.3851. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan S, Warke VG, Nambiar MP, Tsokos GC, Farber DL. The FcR gamma subunit and Syk kinase replace the CD3 zeta-chain and ZAP-70 kinase in the TCR signaling complex of human effector CD4 T cells. J Immunol. 2003;170:4189–4195. doi: 10.4049/jimmunol.170.8.4189. [DOI] [PubMed] [Google Scholar]
- 19.Gorman CL, et al. Polymorphisms in the CD3Z gene influence TCRzeta expression in systemic lupus erythematosus patients and healthy controls. J Immunol. 2008;180:1060–1070. doi: 10.4049/jimmunol.180.2.1060. [DOI] [PubMed] [Google Scholar]
- 20.Warchol T, et al. The CD3Z 844 T>A polymorphism within the 3'-UTR of CD3Z confers increased risk of incidence of systemic lupus erythematosus. Tissue Antigens. 2009;74:68–72. doi: 10.1111/j.1399-0039.2009.01264.x. [DOI] [PubMed] [Google Scholar]
- 21.Gourh P, et al. Association of the PTPN22 R620W polymorphism with anti-topoisomerase I- and anticentromere antibody-positive systemic sclerosis. Arthritis Rheum. 2006;54:3945–3953. doi: 10.1002/art.22196. [DOI] [PubMed] [Google Scholar]
- 22.Preliminary criteria for the classification of systemic sclerosis (scleroderma). Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 1980;23:581–590. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian C, et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med. 2009;15:371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Materials