Mammalian microRNAs: experimental evaluation of novel and previously annotated genes (original) (raw)
Abstract
MicroRNAs (miRNAs) are small regulatory RNAs that derive from distinctive hairpin transcripts. To learn more about the miRNAs of mammals, we sequenced 60 million small RNAs from mouse brain, ovary, testes, embryonic stem cells, three embryonic stages, and whole newborns. Analysis of these sequences confirmed 398 annotated miRNA genes and identified 108 novel miRNA genes. More than 150 previously annotated miRNAs and hundreds of candidates failed to yield sequenced RNAs with miRNA-like features. Ectopically expressing these previously proposed miRNA hairpins also did not yield small RNAs, whereas ectopically expressing the confirmed and newly identified hairpins usually did yield small RNAs with the classical miRNA features, including dependence on the Drosha endonuclease for processing. These experiments, which suggest that previous estimates of conserved mammalian miRNAs were inflated, provide a substantially revised list of confidently identified murine miRNAs from which to infer the general features of mammalian miRNAs. Our analyses also revealed new aspects of miRNA biogenesis and modification, including tissue-specific strand preferences, sequential Dicer cleavage of a metazoan precursor miRNA (pre-miRNA), consequential 5′ heterogeneity, newly identified instances of miRNA editing, and evidence for widespread pre-miRNA uridylation reminiscent of miRNA regulation by Lin28.
Keywords: MicroRNA, miRNA biogenesis, noncoding RNA genes, high-throughput sequencing
MicroRNAs (miRNAs) are endogenous ∼22-nucleotide (nt) RNAs that post-transcriptionally regulate gene expression (Bartel 2004). miRNAs mature through three intermediates: a primary miRNA transcript (pri-miRNA), a precursor miRNA (pre-miRNA), and a miRNA:miRNA* duplex. RNA Polymerase II transcribes the pri-miRNA, which contains one or more segments that each fold into an imperfect hairpin. For canonical metazoan miRNAs, the RNase III enzyme Drosha together with its partner, the RNA-binding protein DGCR8, recognize the hairpin, and Drosha cleaves both strands ∼11 base pairs (bp) from the base of the stem (Han et al. 2006). The cut leaves a 5′ phosphate and 2-nt 3′ overhang (Lee et al. 2003). The liberated pre-miRNA hairpin is then exported to the cytoplasm by Exportin-5 (Yi et al. 2003; Lund et al. 2004). There, the RNase III enzyme Dicer cleaves off the loop of the pre-miRNA, ∼22 nt from the Drosha cut (Lee et al. 2003), again leaving a 5′ monophosphate and 2-nt 3′ overhang. The resulting miRNA:miRNA* duplex, comprised of ∼22-nt strands from each arm of the original hairpin, then associates with an Argonaute protein such that the miRNA strand is usually the one that becomes stably incorporated, while the miRNA* strand dissociates and is degraded.
In addition to canonical miRNAs, some miRNAs mature through pathways that bypass Drosha/DGCR8 recognition and cleavage. Members of the mirtron subclass of pre-miRNAs are excised as intron lariats from the pri-miRNA by the spliceosome and, following debranching, fold into Dicer substrates (Okamura et al. 2007; Ruby et al. 2007a). For some mirtrons, known as tailed mirtrons, a longer intron is excised such that only one end of the pre-miRNA is generated by the spliceosome, whereas the other end of the pre-miRNA matures through the Drosha-independent trimming of a 5′ or 3′ tail (Ruby et al. 2007a; Babiarz et al. 2008). Members of another subclass of pre-miRNAs, called endogenous shRNAs, are suitable Dicer substrates without preprocessing by either Drosha or the spliceosome (Babiarz et al. 2008). Other small silencing RNAs are generated from the sequential processing of long hairpins or long bimolecular duplexes. These small RNAs are classified as endogenous siRNAs rather than miRNAs because they derive from extended duplexes that produce many different small RNA species, whereas miRNAs derive from distinctive hairpins that produce one or two dominant species (Bartel 2004).
The first indication of the abundance of miRNA genes came from sequencing small RNAs from mammals, flies, and worms (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Hundreds of mammalian miRNAs have been identified by Sanger sequencing of cloned small RNA-derived cDNAs (Lagos-Quintana et al. 2001, 2002, 2003; Houbaviy et al. 2003; Berezikov et al. 2006b; Landgraf et al. 2007). Some miRNAs, however, are expressed only in a limited number of cells or through a limited portion of development, and their rarity makes them difficult to detect. Computational methods have been used to identify mammalian miRNAs initially missed by sequencing, and some of these predicted miRNAs have been evaluated experimentally—e.g., by rapid amplification of cDNA ends (RACE) (Lim et al. 2003; Xie et al. 2005), hybridization to RNA blots (Berezikov et al. 2005), microarrays (Bentwich et al. 2005), and RNA-primed array-based Klenow extension (RAKE) (Berezikov et al. 2006b). Each of these experimental methods, however, can yield false positives. Indeed, recent work in invertebrates and plants (Rajagopalan et al. 2006; Ruby et al. 2006, 2007b) has shown that the fraction of erroneously annotated miRNAs can be quite high, depending on the quality of the initial computational predictions. Even when miRNA genes are predicted correctly, the resolution of the prediction is often insufficient to confidently determine the precise 5′ end of the mature miRNA. Because miRNAs repress target mRNAs by pairing to the seed sequence, which is defined relative to the position of the miRNA 5′ end, single-nucleotide resolution of 5′-end annotations is required for useful downstream analysis of their physiological consequences (Bartel 2009).
Another approach for finding miRNAs and other small RNAs missed in the early discovery efforts is high-throughput sequencing (Lu et al. 2005). In mammals, high-throughput sequencing methods that have contributed to miRNA discovery efforts have included massively parallel signature sequencing (MPSS) (Mineno et al. 2006), miRNA serial analysis of gene expression (miRAGE) (Cummins et al. 2006), 454 pyrosequencing (Berezikov et al. 2006a, 2007; Calabrese et al. 2007), and Illumina sequencing (Babiarz et al. 2008; Kuchenbauer et al. 2008).
Here we use the Illumina sequencing-by-synthesis platform (Seo et al. 2004) for miRNA discovery in mice. Analyses of these reads, combined with experimental evaluation of newly identified miRNAs as well as previous annotations, led us to substantially revise the set of confidently identified murine miRNAs, thereby providing a more accurate picture of the general features of mammalian miRNAs and their abundance in the genome. In addition, our results revealed new aspects of miRNA biogenesis and modification, including tissue-specific strand preferences, sequential Dicer cleavage of a metazoan pre-miRNA, cases of consequential 5′ heterogeneity, newly identified instances of miRNA editing, and widespread pre-miRNA uridylation reminiscent of Lin28-like miRNA regulation.
Results
We sequenced small-RNA libraries from three mouse tissues—brain, ovary, and testes—as well as embryonic day 7.5 (E7.5), E9.5, E12.5, and newborn. Combining these data with data collected similarly from mouse embryonic stem (ES) cells (Babiarz et al. 2008) yielded 28.7 million reads between 16 nt and 27 nt in length that perfectly matched the mouse genome assembly (Supplemental Table 1). Of these reads, 79.3% mapped to miRNA hairpins, and 7.1% mapped to other annotated noncoding RNA genes (Supplemental Table 2). Because the sequencing protocol was selective for RNAs with 5′ monophosphate and 3′ hydroxyl groups, this dominance of miRNA species was expected (Lau et al. 2001).
miRNA gene discovery
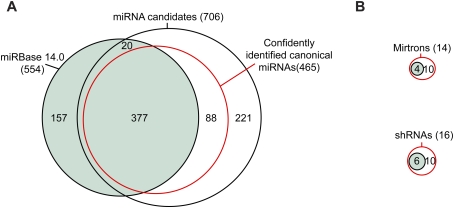
As when analyzing high-throughput data from invertebrates (Ruby et al. 2006, 2007b; Grimson et al. 2008), we identified miRNA genes in mice by applying the following criteria: (1) expression of the candidate miRNA, with a relatively uniform 5′ terminus; (2) pairing characteristics of the predicted hairpin; (3) absence of annotation suggesting non-miRNA biogenesis; (4) absence of proximal reads suggesting that the candidate is a degradation intermediate; and (5) presence of reads corresponding to a miRNA* species with potential to pair to the miRNA candidate with ∼2-nt 3′ overhangs. Using a low-stringency genomic search strategy that considered the first four criteria, 736 miRNA candidates were identified from the total data set of mouse reads. Manual inspection of these candidates, focusing on all five criteria, narrowed the list to 465 canonical miRNA genes, 377 of which were already annotated in miRBase version 14.0 (Griffiths-Jones 2004) and 88 of which were novel (Fig. 1A; Supplemental Fig. S1; Supplemental Table 3). We also found 14 mirtrons (including 10 tailed mirtrons), four of which were already annotated, and 16 endogenous shRNAs, six of which were annotated previously (Fig. 1B). When added to the 88 novel canonical miRNA genes, the newly identified mirtons and shRNAs raised the total number of novel genes to 108.
Figure 1.
Mouse miRNAs and candidates initially identified by high-throughput sequencing. (A) Overlap between previously annotated miRNA hairpins (miRBase version 14.0; green), miRNA candidates identified in the current study, and the subset of these candidates that met our criteria for classification as confidently identified canonical miRNAs (red). Additional considerations increased the number of confidently identified canonical miRNAs to 475. (B) Overlap between previously annotated mirtrons and shRNAs and the mirtrons and shRNAs supported by our study, colored as in A.
Of these 108 genes, 36 appeared to be close paralogs of previously annotated miRNA genes (most of which were paralogs of mir-466, mir-467, or mir-669), producing miRNA reads that were identical to the previously annotated miRNAs, creating ambiguity as to which loci contributed to the sequenced reads. Most of these close paralogs (35 of 36), as well as 14 other novel loci, were clustered with annotated miRNAs. The 72 novel genes with reads distinguishable from those of previously identified genes were expressed at a lower level than the previously annotated genes (median read counts 27 and 8206, respectively), and, compared with previously annotated miRNAs, a higher fraction of these novel miRNAs were located within introns of annotated RefSeq (Pruitt et al. 2005) mRNAs (47% and 26%, respectively).
Experimental evaluation of unconfirmed miRNAs
Of 564 miRBase-annotated miRNA genes (including four confirmed mirtons and six confirmed shRNAs) that map to mm8 genome assembly, 157 annotated miRNAs did not pass the filters for miRNA candidates (Fig. 1A,B; Supplemental Fig. S1; Supplemental Table 4). Of these 157, 26 mapped to annotated rRNA and tRNA loci, 52 had no reads mapping to them, and another 72 had some reads but in numbers deemed insufficient for confident annotation. The remaining seven either had reads with very heterogeneous 5′ ends, which suggested nonspecific degradation of a non-pri-miRNA transcript (mir-464, mir-1937a, and mir-1937b); had many reads that mapped well into the loop of the putative hairpin, which were inconsistent with Dicer processing (mir-451, mir-469, and mir-805); or did not give a predicted fold with the requisite pairing involving the candidate and predicted miRNA* (mir-484) (Supplemental Fig. S2). For five of these seven, we have no reason to suspect that they might be authentic miRNA genes. Among the remaining two, mir-484 might be regarded as a miRNA candidate because manual refolding was able to generate a hairpin with the requisite pairing, but, even so, this candidate lacked reads for the predicted miRNA*. miR-451 is a noncanonical miRNA generated from an unusual hairpin without production of a miRNA:miRNA* duplex (S Cheloufi and G Hannon, pers comm.). We do not suspect that any other annotated miRNA genes failed to pass our filters for the same reason as mir-451.
An additional 20 annotated miRNA hairpins were in our set of candidates but failed the manual inspection because they lacked predicted miRNA* reads even after allowing for alternate hairpin structures. Hundreds of candidates from other miRNA discovery efforts (Xie et al. 2005; Berezikov et al. 2006b) also failed to pass the filters, usually because no reads mapped to them.
One of the annotated miRNA genes missing from our data sets was mir-220, which had been predicted computationally using MiRscan as a miRNA gene candidate conserved in humans, mice, and fish, and was supported experimentally using RACE analysis of zebrafish small RNAs (Lim et al. 2003). In contrast, the other 37 miRNAs newly annotated by Lim et al. (2003) were among our confirmed miRNAs. The absence of mir-220 in our data sets might have reflected either very low expression in the sequenced samples or inaccuracy of its annotation. Similarly, mir-207, annotated in a contemporaneous study that cloned novel miRNAs from mouse tissues, was missing from our data set, but another 27 miRNAs annotated from that study were confirmed (Lagos-Quintana et al. 2003).
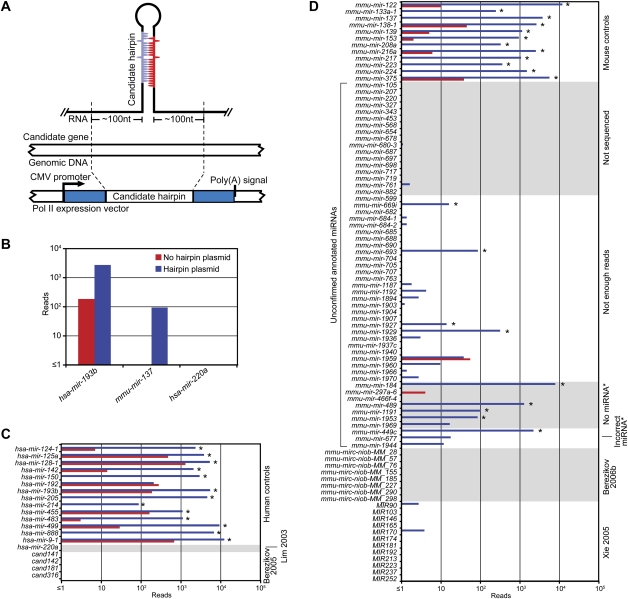
To evaluate whether the missing annotated miRNAs and candidates represented authentic miRNAs, we developed a moderate-throughput assay to examine if their respective hairpins could be processed as miRNAs in cultured cells (Fig. 2A). If these putative miRNAs were missing from our data sets because they were not expressed in the sequenced tissues or stages, we reasoned that they would probably be detected in cells ectopically expressing their respective hairpins, because most authentic miRNAs are processed correctly from heterologous transcripts that include the full hairpin flanked by ∼100 nt of genomic sequence on each side of the hairpin (Chen et al. 2004; Voorhoeve et al. 2006). Alternatively, if these putative miRNAs were missing because they were not authentic miRNAs and therefore lacked the features needed for Drosha and Dicer processing, they would not be sequenced from cells ectopically expressing their hairpins. To evaluate many hairpins simultaneously, we transfected pools of hairpin-expressing constructs into HEK293T cells and isolated small RNAs for high-throughput sequencing.
Figure 2.
Experimental evaluation of annotated miRNAs and previously proposed candidates. (A) Schematic of the expression vector transfected into HEK293T cells. (B) Examples of the standard ectopic expression assay, transfecting plasmids indicated in the key. Reads from the control transfection (no hairpin plasmid) were from endogenous expression in HEK293T cells. (C) Assay results for annotated human miRNAs and published candidates. Bars are colored as in B; asterisks indicate detectable overexpression (≥1 read from both the anticipated miRNA and miRNA*, with miRNA and miRNA* combined expressed more than threefold over endogenous levels). (D) Assay results for unconfirmed annotated mouse miRNAs and published candidates. Mouse controls were selected from miRNAs that were sequenced from our mouse samples. Bars are colored as in B; detectable overexpression is indicated (asterisks). Shown are the results compiled from two experiments (Supplemental Figs. S3, S4).
The performance of 26 positive controls, chosen from canonical human/mouse miRNAs confirmed by our sequencing from mice, illustrated the value of the assay. For all but one of these controls, miRNA and miRNA* reads were more abundant in the cells ectopically expressing the hairpin than in the cells without the hairpin constructs (Fig. 2B–D; Supplemental Figs. S3, S4). For example, both hsa-miR-193b and mmu-miR-137 (from humans and mice, respectively) were >10 fold overexpressed (Fig. 2B). The positive controls included genes of tissue-specific miRNAs, including mir-122 (liver), mir-133 (muscle), mir-223 (neutrophil), and several neuron-specific miRNAs, with the idea that hairpins of tissue-specific miRNAs might require tissue-specific factors for their processing, and therefore might be sensitive to the potential absence of such factors in HEK293T cells. Differences were observed, ranging from ∼100 to 10,000 reads above the control transfection (Fig. 2C, hsa-mir-214 and hsa-mir-9-1, respectively), consistent with the idea that factors absent in HEK293T cells might play a role in processing of some miRNAs. Alternatively, some miRNA hairpins might be processed less efficiently in all cell types, perhaps because our vectors might not present the hairpins in an optimal context for processing. Perhaps hsa-mir-192, the control gene that did not overexpress in our assay, lacked crucial processing determinants needed in all cells. In either scenario, the very high sensitivity of high-throughput sequencing enabled miRNAs to be observed from most of the less efficiently processed hairpins.
From the 52 annotated mouse miRNAs that our study did not sequence, 17 miRNAs, including mir-220 and mir-207, were tested in the ectopic expression assay. One, mir-698, generated a single read corresponding to the annotated miRNA, and the rest failed to generate any reads representing the annotated miRNA (Fig. 2D). From the 72 annotated miRNAs that we could not identify due to insufficient number of reads, 28 were tested, and only four of these were found to be overexpressed (Fig. 2D). The difficulty in overexpressing a canonical control miRNA (hsa-miR-192) illustrates that our ectopic expression assay cannot be used to prove conclusively that a particular hairpin does not represent an authentic miRNA gene. However, the inability to overexpress each of the 17 unsequenced miRNAs, as well as most of the 28 insufficiently sequenced miRNAs, strongly indicated that, overall, these annotations have been faulty, and that our failure to detect previously annotated miRNAs in mouse samples was not merely due to inadequate sequencing coverage.
We also tested 10 of the 20 annotated miRNA genes that we identified as candidates but did not confidently classify as miRNA genes because the predicted miRNA* species was not sequenced. Four of seven genes without a miRNA* read and one of three genes with substantially offset miRNA* reads produced the predicted miRNA* species in our ectopic expression assay (Fig. 2D). mir-184 and mir-489, both of which tested positive in this assay, are conserved. mir-184 is conserved throughout mammals, and mir-489 is conserved to chicken, although the miRNA seed, which is highly conserved in mammals and chickens, differs in mice and rats. Thus, these two genes, as well as mir-875, which is a broadly conserved gene without a miRNA* read, were added to our set of confidently identified miRNA genes. Also added were mir-290, mir-291a, mir-291b, mir-292, mir-293, mir-294, and mir-295, which were missing in the genome assembly (mm8) used in our analysis because they fall in the region of the genome that is difficult to assemble. Including these 10 genes, plus mir-451, brings the total number of confidently identified miRNA genes to 506, which includes 475 canonical genes.
Our sets of confirmed and novel murine miRNAs also provided the opportunity to evaluate results of more recent computational efforts to find miRNAs conserved among mammals. One set of studies predicted miRNAs based on phylogenetic conservation, and then tested these and additional murine-specific hairpins using RAKE and cloning (Berezikov et al. 2005, 2006b). Among the 322 candidates supported by these experiments, 11 were in our sets of miRNAs (two in our confirmed set, and nine in our novel set), and another nine did not satisfy our annotation criteria but had at least one read consistent with the predictions. Another study started with MiRscan predictions conserved in four mammals, and filtered these predictions for potential seed pairing to conserved motifs in 3′ untranslated regions (UTRs) (Xie et al. 2005). Of their 144 final candidates, 45 were paralogs of miRNAs already published at the time of prediction. Of the remaining 99 candidates, 27 were in our sets of miRNAs (26 in our confirmed set and one in our novel set), and one did not satisfy our annotation criteria but had three reads consistent with the miRNA* of the predicted miRNA. However, only four of the 27 confirmed miRNA genes (4% of the 99 novel predictions) gave rise to the mature miRNA with the predicted seed, suggesting that filtering MiRscan predictions for potential seed pairing provided little, if any, added benefit. This conclusion concurs with a recent analysis of miRNA targeting: miRNAs that are not conserved beyond mammals do not have enough preferentially conserved sites to place these sites as among the most conserved UTR motifs (Friedman et al. 2009). Therefore, it stands to reason that preferentially conserved UTR motifs would provide little value for predicting such miRNAs.
To investigate whether the computational candidates might have been missed because of low expression in tissues and stages from which we sequenced, we included representatives from each study in our ectopic expression assay. We randomly selected 12 Xie et al. (2005) candidates and eight Berezikov et al. (2006b) candidates that our study did not sequence, as well as four human candidates from the Berezikov et al. (2005) set whose mouse orthologs were not sequenced. None generated reads representing the candidate miRNAs (Fig. 2C,D). Taken together, our results raise new questions regarding the authenticity of these candidates, and suggest that previous extrapolation from these candidates, which had suggested that mammals have a surprisingly high number of conserved miRNA genes (as many as 1000) (Berezikov et al. 2005), should be revised accordingly.
Experimental evaluation of novel miRNAs and new candidates
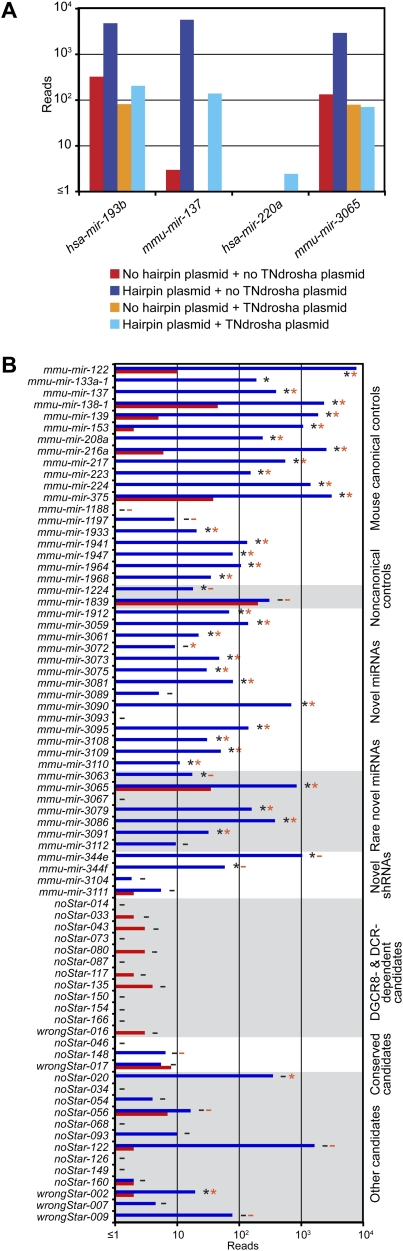
We also used the ectopic expression assay to evaluate novel miRNAs identified from our sequencing. Of the 25 evaluated hairpins, 18 (72%) generated a significant number of miRNA-like reads in HEK293T cells, indicating that most, although perhaps not all, of our 108 novel annotations represented authentic miRNAs (Fig. 3; Supplemental Figs. S5, S6). These 25 hairpins were selected arbitrarily for evaluation, except for a preference for rare miRNAs; i.e., those that had <10 mature miRNA reads. The rare miRNAs and the higher-abundance miRNAs performed similarly (five of seven and 11 of 14 positives, respectively).
Figure 3.
Experimental evaluation of novel miRNAs and candidates. (A) Examples of assays evaluating Drosha dependence, transfecting plasmids indicated in the key. (B) Assay results for control miRNAs, novel miRNAs, and miRNA candidates. Bars are colored as in A; detectable overexpression (black asterisks), overexpression attempted but not detected (black minus sign), detectable Drosha dependence (orange asterisks), and Drosha dependence assayed but not detected (orange minus sign) are all indicated. Shown are the results compiled from three experiments (Supplemental Figs. S5–S7).
To evaluate Drosha and Dicer dependence of the overexpressed hairpins, the experiment was repeated with and without a plasmid encoding a dominant-negative allele of either Drosha or Dicer (Fig. 3A; Han et al. 2009). All but two canonical miRNA controls and most of the novel canonical miRNAs (16 of 17) responded to TNdrosha coexpression (Fig. 3B; Supplemental Fig. S7). Fewer responded to TNdicer, suggesting that this construct was less disruptive of normal miRNA processing (Supplemental Fig. S7).
The tested hairpins included several noncanonical miRNA precursors. The level of mmu-miR-1224, an annotated mirtronic miRNA (Berezikov et al. 2007), increased in the presence of TNdrosha, as expected if this pre-miRNA had more access to Exportin-5 and Dicer when the canonical pre-miRNAs were reduced (Grimm et al. 2006). Although mmu-miR-1839, an annotated shRNA (Babiarz et al. 2008), did not overexpress, mmu-miR-344e and mmu-miR-344f, novel shRNAs, did overexpress from our vector, and, as expected for shRNAs, their biogenesis was Drosha-independent (Fig. 3B; Supplemental Figs. S5–S7). Repeating the ectopic expression assay in Dicer knockout and control cells confirmed that mmu-miR-344e biogenesis was Dicer-dependent (data not shown).
We also evaluated our candidates that had not satisfied our criteria for confident annotation as miRNAs, usually because they lacked reads representing the predicted miRNA*. We tested three sets of these candidates. One set represented our candidates that lacked predicted miRNA* reads, yet, based on small RNA sequencing results from wild-type and mutant ES cells (Babiarz et al. 2008), appeared DGCR8- and Dicer-dependent. Another set represented candidates that appeared conserved in syntenic regions of other mammalian genomes, and the third set was selected at random from among the remaining candidates. All but one of the 28 tested candidates failed to generate miRNA-like reads, and the processing of the candidate that did generate miRNA-like reads in HEK293T cells was not dependent on Dicer, based on its presence in Dicer knockout ES cells (Babiarz et al. 2008).
The results evaluating the novel miRNAs and candidates illustrated the importance of requiring a convincing miRNA* read as a criterion for confident miRNA annotation. Five previously annotated miRNAs that were initially rejected due to lack of a convincing miRNA* read had tested positive in our overexpression assay (Fig. 2D), which indicated that this criterion was too stringent for some of the previously annotated genes. However, the results for the newly identified miRNAs and candidates showed that the presence of a convincing miRNA* read was the primary criterion that distinguished the novel canonical miRNAs (most of which tested positive) from the remaining candidates (nearly all of which tested negative). By requiring a convincing miRNA* read in addition to the other four annotation criteria, our approach accurately distinguished miRNA reads from the millions of other small RNA reads generated by high-throughput sequencing, with relatively few false positives among the novel annotations and few false negatives among the rejected candidates.
miRNA expression profiles
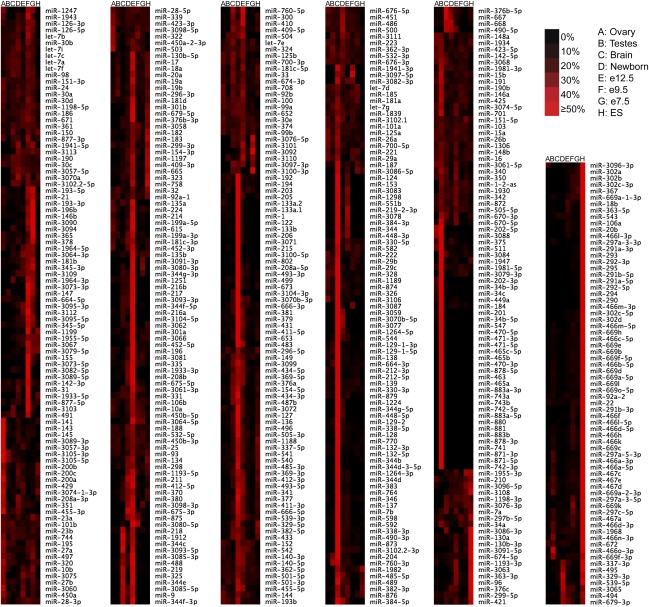
To compare expression levels of each miRNA in different sequenced samples, we constructed relative miRNA expression profiles (Fig. 4; Supplemental Table 5), and to compare the relative expression of various miRNAs with each other, we generated a table of overall miRNA abundance (Supplemental Table 5). Most miRNAs had substantially stronger expression in some tissues or stages than in others, in agreement with previous observations (Wienholds et al. 2005). We expect that strong tissue- or stage-specific expression preferences inferred from our limited sample set will be revised as more tissues and stages are surveyed.
Figure 4.
miRNA relative expression profiles. Profiles of mature miRNAs were constructed as described (Ruby et al. 2007b). The relative contribution of each miRNA from each sample and the sum of the normalized reads of all samples are provided (Supplemental Table 5).
General features of mammalian miRNAs
Our analyses of high-throughput sequencing data and subsequent experimental evaluation reshaped the set of known murine miRNAs, setting aside 173 questionable annotations and adding 108 novel miRNA genes to bring the total number of confidently identified murine genes to 506. A majority (60%) of the 506 genes appeared conserved in other mammals (Supplemental Fig. S1; Supplemental Table 6). However, only 15 of the 108 novel miRNA genes were conserved in other mammals, suggesting that the number of nonconserved miRNA genes will soon surpass that of conserved ones as high-throughput sequencing is applied more deeply and more broadly.
Five novel miRNAs (mir-3065, mir-3071, mir-3074-1, mir-3074-2, and mir-3111) mapped to the antisense strand of previously annotated miRNAs (mir-338, mir-136, mir-24-1, mir-24-2, and mir-374, respectively), which, when added to the previously identified mir-1-2/mir-1-2-as pair, brings the total number of sense/antisense miRNA pairs to six. In addition, the mir-486 hairpin has a palindromic sequence, which resulted in the same reads mapping to both the sense (mir-486) and antisense (mir-3107) hairpins. Analysis of the antisense loci of all 498 miRNA genes identified six additional loci that gave rise to some antisense reads resembling miRNAs (antisense loci of mir-21, mir-126, mir-150, mir-337, mir-434, and mir-3073). As more high-throughput data is acquired, these as well as other antisense loci are likely to be annotated as miRNA genes. However, <0.00002 of our miRNA reads corresponded to miRNAs from antisense loci (excluding the reads mapping ambiguously to mir-486/mir-3107), raising the possibility that none of the murine antisense miRNAs have a function comparable with that of miR-iab-as in flies (Bender 2008; Stark et al. 2008; Tyler et al. 2008).
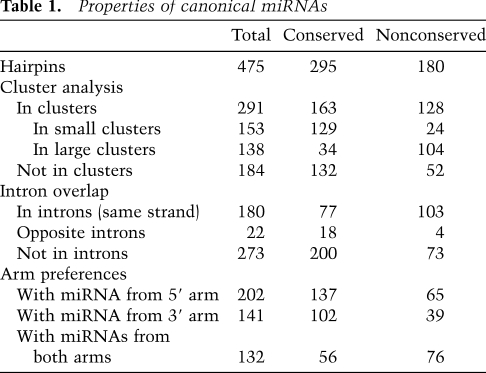
Our substantially revised set of miRNA genes provided the opportunity to speak to the general features of 475 canonical miRNAs in mice, with the properties of the 295 conserved genes applying also to the conserved genes of humans and other mammals (Table 1). Most canonical miRNA genes (61%) were clustered in the genome, falling within 50 kb of another miRNA gene, on the same genomic strand. Even when excluding the four known megaclusters (Calabrese et al. 2007), which are on chromosomes 2, 12 (two clusters), and X (with 69, 35, 16, and 18 genes, respectively), a sizable fraction of the remaining genes (153 of 337) were in clusters of two to seven genes. As observed in humans (Baskerville and Bartel 2005), miRNAs from these loci within 50 kb of each other tended to have correlated expression, consistent with their processing from polycistronic pri-miRNA transcripts (Supplemental Fig. S8). In a scenario of one transcript per cluster, the 475 canonical miRNA genes would derive from 245 transcription units. In addition, many miRNA hairpins mapped to introns. Just over a third (38%) of the hairpins fell within introns of annotated mRNAs. Several lines of evidence—including coexpression correlations, chromatin marks, and directed experiments—indicate that miRNAs can be processed from introns (Baskerville and Bartel 2005; Kim and Kim 2007; Marson et al. 2008). In this scenario, as many as 107 (44%) of the 245 transcription units could double as pre-mRNAs. Other hairpins were found within transcripts that lacked other annotated functions, falling either within introns or exons, or in transcripts without evidence of splicing.
Table 1.
Properties of canonical miRNAs
miRNA hairpins are generally thought to each give rise to a single dominant mature guide RNA. This was usually the case for the murine miRNAs, although, as in other species, this result relied on grouping together as a single functional species all the isoforms that share the same 5′ terminus. This grouping is justified based on the current understanding of miRNA target recognition, which stipulates that heterogeneity often observed at miRNA 3′ termini should have no effect on miRNA target recognition (Bartel 2009). Most mature miRNA reads (97%) were 20–24 nt in length, with 20mer, 21mer, 22mer, 23mer, and 24mer comprising 5%, 19%, 47%, 21%, and 4% of the reads, respectively (Supplemental Fig. S9). Although a single dominant mature species appears to be the most frequent outcome of miRNA biogenesis, some miRNA hairpins give rise to two or more species that each could function to target different sets of mRNAs. This expanded targeting potential arises from multiple mechanisms, including utilization of both strands of the miRNA:miRNA* duplex with similar frequency, 5′ heterogeneity, sequential Dicer cleavage, and RNA editing. Addition of untemplated nucleotides to the 3′ termini of the miRNAs can also occur, and although not thought to change targeting specificity, these changes could indicate post-transcriptional regulation of miRNA stability. Occurrence of each of these phenomena is described below.
miRNAs from both arms, with occasional tissue-specific differences in the preferred arm
Most canonical miRNA genes produced one dominant mature miRNA species, from either the 5′ or 3′ arm of the pre-miRNA hairpin, with an overall tendency to derive from the 5′ arm (Table 1), as reported for previously annotated human miRNAs (Hu et al. 2009). Some, however, yielded a similar number of reads from both arms, suggesting that the two species enter the silencing complex with similar frequencies. For these genes, mature species from the 5′ and 3′ arms were annotated using the -5p and -3p suffixes, as is conventional in such cases (Griffiths-Jones 2004). Discrimination favoring one arm over the other was less pronounced for both the nonconserved miRNAs and the less highly expressed miRNAs (Fig. 5A), although for the miRNAs with very few reads this trend was likely enhanced by our requirement for a miRNA* read. Overall, the discrimination was high, with the species from the less dominant arm comprising 4.1% of the reads that map to a miRNA or miRNA*. For the 10 most abundant miRNAs (sampling just the most abundant member in cases of repetitive miRNAs), discrimination was even higher, with the less dominant arm comprising only 1.3% of the reads. Nevertheless, the miRNA* species of these more highly expressed miRNAs were sequenced at a median frequency 13-fold greater than that of the median nonconserved miRNA, suggesting that a search for biological function for these miRNA* species might be at least as fruitful as that for the poorly expressed nonconserved miRNAs.
Figure 5.
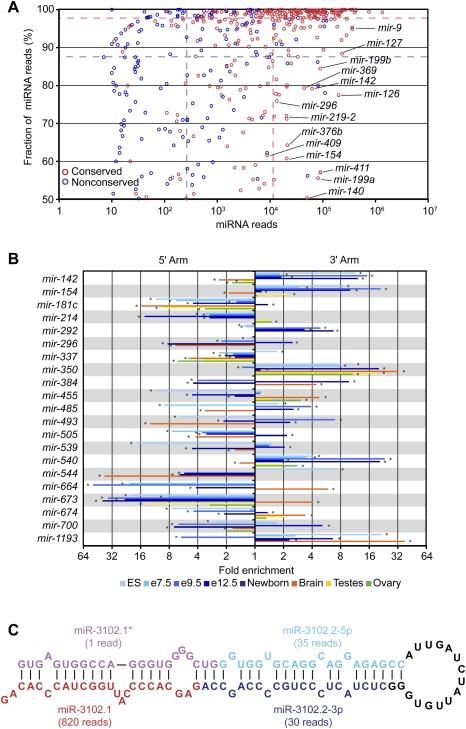
Reads from both arms of a hairpin, and sequential reads from the same arm. (A) Fraction and abundance of miRNA reads from each miRNA hairpin. To calculate the fraction, the miRNA reads were divided by the total number of miRNA and miRNA* reads, considering on each arm only the major 5′ terminus. The dashed lines indicate the median fraction of miRNA reads and the median number of miRNA reads for conserved (red) and nonconserved (blue) miRNAs. (B) Switching of the dominant arm in different samples. For each sample, the fold enrichment of miRNA reads produced from the 5′ arm over those produced from the 3′ arm and vice versa was calculated. Shown are results for nonrepetitive miRNAs that switch dominant arms, with at least a fivefold differential between two samples. The samples are color-coded (key), and an asterisk indicates samples with statistically significant enrichment of miRNAs produced from one arm over the other (P < 0.05, χ2 test). (C) Sequential Dicer cleavage. Predicted secondary structure of mmu-mir-3102 pre-miRNA (Hofacker et al. 1994).
If the mature miRNA accumulated preferentially from one arm of the pre-miRNA hairpin, the preferred arm generally remained consistent across the various libraries. For a few miRNAs, however, the preferred arms switched between samples (Fig. 5B), as reported previously using PCR-based miRNA quantification (Ro et al. 2007). For example, miR-142-5p was sequenced more frequently in ovary, testes, and brain, and miR-142-3p was sequenced more frequently in embryonic and newborn samples. These results imply a developmental switch in targeting preferences. A similar arm-switching phenomena has been reported for a sponge miRNA (Grimson et al. 2008), and was observed for 20 other nonrepetitive mouse miRNA genes (Fig. 5B).
Sequential Dicer cleavage of a mirtron hairpin
In plants, a few pri-miRNA hairpins with long, continuous RNA duplexes are cleaved sequentially by Dicer to generate two adjacent miRNA:miRNA* duplexes (Kurihara and Watanabe 2004; Rajagopalan et al. 2006). Those precursors bear little resemblance to the shorter, imperfectly base-paired hairpins of metazoan miRNA genes. In mice, similar precursors are found in the form of hairpin siRNA (hp-siRNA) precursors, but their expression appears to be limited to germline tissues and totipotent ES cells, which lack a robust interferon response to intracellular dsRNA (Babiarz et al. 2008; Tam et al. 2008; Watanabe et al. 2008). However, we detected two miRNA:miRNA* duplexes deriving from the mmu-mir-3102 pre-miRNA hairpin, an apparent mirtron as evidenced by reads mapping to both boundaries of an intron (Fig. 5C; Supplemental Table 3). After splicing and debranching, the excised intron was predicted to fold into a 104-nt pre-miRNA hairpin—substantially longer than the average pre-miRNA length of 61 nt (calculated from the set of confirmed miRNAs). Reads from this locus suggested that Dicer cleaved this pre-miRNA twice, with the first cut generating the outer miRNA:miRNA* duplex and the second cut generating the inner miRNA:miRNA* duplex (Fig. 5C). The inner miRNA (miR-3102.2-3p) was among a set of proposed miRNA candidates (Berezikov et al. 2006b), but the most frequently sequenced species from this hairpin was the outer miRNA (miR-3102.1) (Fig. 5C). Of the 16 genomes examined, the extended mir-3102 hairpin with both the inner and outer miRNAs appeared conserved only in rats, although the orthologous loci in cows, dogs, and humans also could fold into shorter hairpins, with miR-3102.1 potentially conserved in cows.
We suspect that it is more than a coincidence that the single metazoan example of a sequentially diced miRNA is initially processed by the spliceosome rather than by Drosha. One way to explain this observation is that DGCR8/Drosha interacts directly with the loop of pri-miRNA stem–loops when recognizing its substrates (Zeng et al. 2005), and that the lack of sequentially diced Drosha-dependent miRNA hairpins in animals reflects the limited reach of this complex.
5′ Heterogeneity
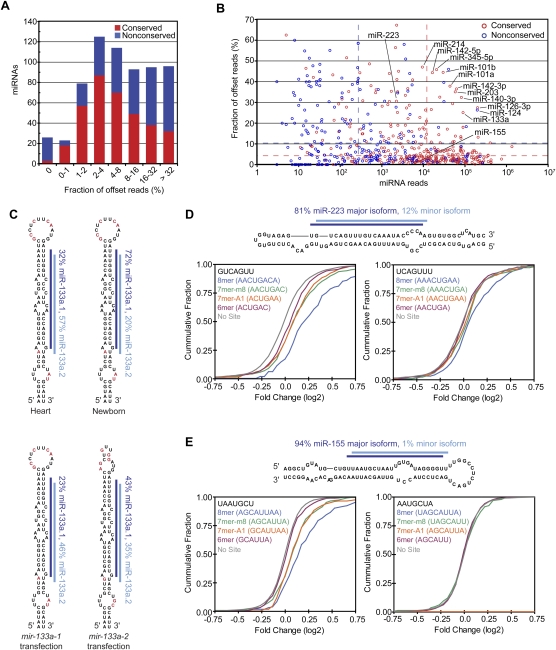
Most conserved miRNAs had very precise 5′ processing, with alternative 5′ isoforms comprising only 8% of all miRNA reads (Fig. 6A,B). These results, analogous to those observed in worms and flies (Ruby et al. 2006, 2007b), are consistent with the idea that selective pressure to avoid off-targeting acts to optimize precision of the cleavage event that produces the 5′ terminus of the dominant species so as to prevent a consequential number of molecules with seed sequences in the wrong register. Moreover, 5′ termini of conserved miRNAs were more precise than those of miRNA* reads (4% and 12% offset reads, respectively, excluding those that produce comparable numbers of small RNAs from each arm). For cases in which Dicer produced the 5′ terminus of the miRNA, the Dicer cut appeared somewhat more precise than the Drosha cut (5% offset reads for miRNAs on the 3′ arm, compared with 7% offset reads for miRNA* on the 5′ arm), hinting that features of the pre-miRNA structure may supplement the distance from the Drosha cut as determinants of Dicer cleavage specificity (Ruby et al. 2006, 2007b).
Figure 6.
miRNAs with 5′ heterogeneity. (A) The distribution of conserved (red) and nonconserved (blue) miRNAs with reads ≤5 nt offset at their 5′ terminus. (B) The fraction of offset reads and abundance of reads for each miRNA hairpin, colored as in A. The dashed lines indicate the median level of reads for conserved (red) and nonconserved (blue) miRNAs. (C) 5′ Heterogeneity of miR-133a. Data from mouse heart (Rao et al. 2009) and newborn are mapped to the mir-133a-1 hairpin (top), and data from the ectopic expression assay are mapped to the indicated transfected hairpin (bottom). The lines indicate miR-133a.1 (dark blue) and miR-133a.2 (light blue), and red nucleotides indicate those that differ between mir-133a-1 and mir-133a-2. (D) Effect of losing miR-223 on messages with 3′ UTR sites for miR-223 major and minor isoforms. (Top) Small RNA sequencing data from mouse neutrophils (Baek et al. 2008) were mapped to the mir-223 hairpin as in C. For each set of messages with the indicated 3′ UTR site for miR-233 (major isoform sites, bottom left; minor isoform sites, bottom right), the fraction that changed at least to the degree indicated following loss of miR-223 is plotted, using data published for neutrophils differentiated in vivo (Baek et al. 2008). (E) Effect of losing miR-155 on messages with 3′ UTR sites for miR-155 major and minor isoforms, plotted as in D using published data from T cells (Rodriguez et al. 2007). (Top) Sequencing data from our study are mapped to the mir-155 hairpin as in C. The mRNAs with 8mer and 7mer-A1 sites for the minor isoform were excluded from the analysis because these sites overlapped with 7mer-m8 sites for the major isoform.
A few miRNAs had less uniform 5′ termini (Fig. 6A,B). For some miRNAs, 5′ heterogeneity has been documented previously (Ruby et al. 2007b; Stark et al. 2007; Azuma-Mukai et al. 2008; Wu et al. 2009), the most prominent example being hsa-miR-124, a conserved neuronal miRNA for which the 5′-shifted isoform was initially annotated as the miRNA and eventually replaced by the more prominent isoform following more extensive sequencing (Lagos-Quintana et al. 2002; Landgraf et al. 2007). Another prominent miRNA with unusually diverse 5′ termini was miR-133a. This conserved miRNA, which is highly expressed in heart and muscle, had a second dominant isoform (miR-133a.2) that was shifted 1 nt downstream from the annotated miRNA (miR-133a.1) (Fig. 6C; Supplemental Table 3). To test whether this heterogeneity might be explained by differential processing of the two mir-133a paralogous hairpins, as observed for the two Drosophila mir-2 hairpins (Ruby et al. 2007b), we tested the two mir-133a hairpins in our ectopic expression assay. Although mir-133a-1 was somewhat more prone to produce the miR-133a.2 isoform, both hairpins produced a substantial amount of both isoforms (Fig. 6C).
To investigate the functional consequences of miRNA 5′ heterogeneity, we examined published array data showing the responses of mRNAs after deleting either mir-223, a miRNA with substantial heterogeneity, or mir-155, a miRNA with little heterogeneity. miR-223 is highly expressed in neutrophils, and analysis of small RNA sequences from isolated neutrophils (Baek et al. 2008) was consistent with our sequencing results (Supplemental Table 3) in showing 5′ heterogeneity, with 81% of the reads mapping to the 5′ end of the major isoform miRNA and 12% mapping to the 5′ end of a second isoform that was shifted by 1 nt in the 3′ direction (Fig. 6D). As expected, mRNAs with canonical 7–8mer sites (Bartel 2009) matching the seed of the major isoform were significantly derepressed in the mir-223 deletion mutant (P < 10−12, Kolmogorov–Smirnov [K–S] test, compared with no site distribution). mRNAs with canonical sites matching the minor isoform also showed a significant tendency to be derepressed, albeit to a lesser degree (P = 0.0022 × 10−7, 0.013 × 10−7, and 1.7 × 10−7, for 8mer, 7mer-m8, and 7–8mers combined, respectively) (Fig. 6D). This result could not be attributed to the overlap between sites matching the major and minor isoforms because all mRNAs with a 6mer seed match to the major isoform (ACUGAC) were excluded, and additional analyses ruled out participation of the “shifted 6mer” match (Friedman et al. 2009) to the major isoform (AACUGA) (Supplemental Fig. S10A). Analogous analysis of miR-155 yielded strong evidence for function of the major isoform (Rodriguez et al. 2007) but no sign of function for the minor isoform, which comprised very few (1%) of our miR-155 reads (Fig. 6E; Supplemental Table 3).
Taken together, our results show that some miRNAs have alternative 5′ miRNA isoforms that are expressed at levels sufficient to direct the repression of a distinct set of endogenous targets and thereby broaden the regulatory impact of the miRNA genes. Therefore, we suggest that, rather than choosing one isoform over the other for annotation as the authentic miRNA, more of these alternative isoforms should be annotated, with the expectation that, for some highly expressed miRNAs, more than one 5′ isoform contributes to miRNA function.
RNA editing
RNA editing in which adenosine is deaminated and thereby converted to inosine (I) has been reported for some miRNA precursors (Blow et al. 2006; Landgraf et al. 2007; Kawahara et al. 2008). Because I pairs with C, such edits could change miRNA target recognition. Reasoning that the mammalian adenosine deaminases (ADARs) responsible for A-to-I editing are expressed primarily in the brain, we searched for sequencing reads from the brain that did not match the genome and had as their closest match a mature miRNA or miRNA*. After filtering for mismatches occurring >2 nt from the 3′ end, a step taken to avoid considering instances of untemplated 3′-terminal addition, only 4% of the reads had single mismatches to the genome (Supplemental Fig. S11A). Moreover, the fraction of sequences with A-to-G changes (indicative of A-to-I editing) was only 0.61%, a fraction resembling that of other mismatches (Supplemental Fig. S11A). This fraction was also similar to that of the A-to-G changes in our synthetic internal standards used for preparing the sequencing libraries. These results indicate that mature edited miRNAs are very rare and difficult to distinguish above the background level of sequencing errors. The low frequency of editing in mature miRNAs was consistent with the findings that edited processed miRNAs are more than fourfold less common in mice relative to humans (Landgraf et al. 2007), and are less common than edited miRNA precursors (Kawahara et al. 2008). The latter observation might be due to rapid degradation or impaired processing, which has been shown for miR-142 (Yang et al. 2006) and miR-151 (Kawahara et al. 2007a).
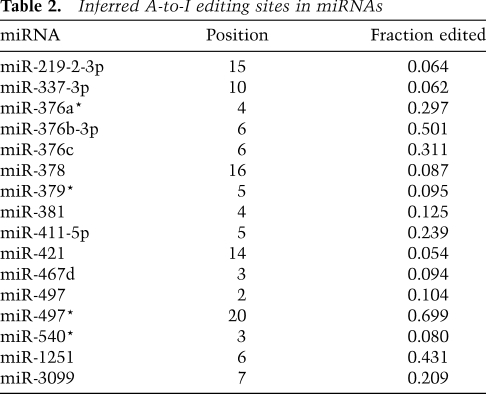
Although editing did not appear to be a widespread phenomenon among all mature miRNAs, editing at specific sites might still be important for a few individual miRNAs. To investigate this possibility, mismatch fractions were calculated as the fraction of reads bearing a particular mismatch over all reads covering that genomic position. For each library, a change was considered significant if the fraction exceeded 5% and at least 10 reads contained the mismatch. Additional filters designed to remove sequencing errors, alignment artifacts, and instances of untemplated nucleotide addition preferentially retained A-to-G changes while removing nearly all other events (Supplemental Fig. S11B). Sixteen A-to-G events passed the filters and subsequent manual examination, all of which occurred only in the brain library (Table 2). Five of these inferred editing sites were also observed in a low-throughput sequencing effort in human brain samples (Kawahara et al. 2008), indicating that editing of some miRNAs is conserved between mammals. Consistent with that study, eight of 16 editing sites occurred in a UAG motif. A separate examination of read alignments with up to three mismatches showed that the vast majority of edited reads were edited at one position, suggesting that either editing of multiple sites in the same RNA molecule is rare, or multiply edited RNAs are degraded more rapidly.
Table 2.
Inferred A-to-I editing sites in miRNAs
A-to-I editing of a seed nucleotide would dramatically affect targeting. In addition to editing in the miR-376 cluster described previously (Kawahara et al. 2007b, 2008), we found another eight miRNAs that are edited within the seed of either the miRNA or the miRNA*. A-to-I editing could also affect miRNA loading, and thereby indirectly affect targeting. Indeed, the editing of miR-540 might help explain why the 5′ arm is more abundant in the brain than in other tissues, although editing is too infrequent to fully explain the switch in strand bias. Altering Drosha and Dicer processing could also indirectly affect targeting. Analysis of 5′ ends showed that seven of 16 instances of editing were associated with a statistically significant (P < 0.05) shift in the 5′ nucleotide, presumably due to changes in the Drosha and Dicer cleavage site (Supplemental Fig. S11D).
Untemplated nucleotide addition
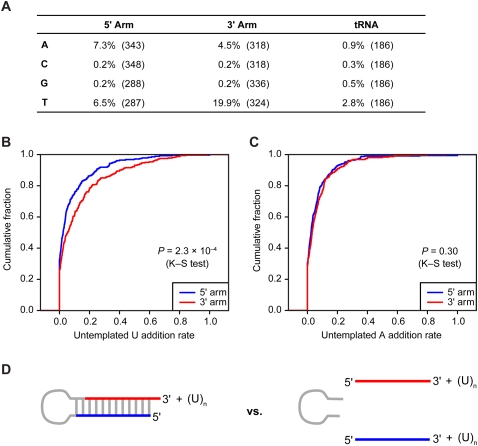
Much more prevalent than editing of internal nucleotides was addition of untemplated nucleotides to miRNA 3′ termini. As reported previously for miRNAs in mammals (Landgraf et al. 2007), and also observed for those of worms and flies (Ruby et al. 2006, 2007b), nucleotides most frequently added to murine miRNAs were U and A (Fig. 7A). Addition of C or G was no higher than background, as estimated by monitoring apparent addition to tRNA fragments (Fig. 7A). Possible sources of the background rate could be sequencing error, transcription error, or a low level of biological nucleotide addition. Some miRNAs were much more frequently extended than others (Supplemental Table 7). One very frequently extended miRNA was miR-143, for which the extended reads outnumbered the nonextended ones (196,565 compared with 114,980 reads, respectively).
Figure 7.
Untemplated nucleotide addition. (A) Untemplated nucleotide addition rate for miRNA and miRNA* reads from the indicated arm. Rates for each miRNA are provided (Supplemental Table 6). As a control, tRNA degradation fragments were analyzed similarly. Numbers of genes analyzed are indicated in parentheses. (B) Distribution of rates for untemplated U addition to RNAs from the 5′ arm (blue) and from the 3′ arm (red). (C) Distribution of rates for untemplated A addition to RNAs from the 5′ arm (blue) and from the 3′ arm (red). (D) Schematic of the biogenesis stage in which U could be added to the RNA of only one arm (pre-miRNA, left), and the stage in which U could be added to the RNA of either arm (mature miRNA and miRNA*, right).
For extension by U, RNAs from the pre-miRNA 3′ arm were three times more frequently extended than were those from the 5′ arm (Fig. 7A,B, P = 2.3 × 10−4, K–S test). This preference, not observed for the A extension (Fig. 7A,C), suggests that much of the U extension occurs to the pre-miRNA, prior to Dicer cleavage—a state in which the 3′ arm but not the 5′ arm would be available for extension (Fig. 7D). TUT4-catalyzed poly(U) addition to the let-7 pre-miRNA, which is specified by Lin28, plays an important role in post-transcriptional repression of let-7 expression (Heo et al. 2008, 2009; Hagan et al. 2009). Our analyses indicating untemplated U extension to many other pre-miRNAs hint that this type of regulation may not be limited to let-7, but that analogous pathways, presumably using mediators other than Lin28, act to regulate the expression of other murine miRNAs.
Discussion
The status of miRNA gene discovery in mammals
Our current study sets aside nearly a third (173 of 564) of the miRBase version 14.0 gene annotations for lack of convincing evidence that these produce authentic miRNAs. It also adds another 108 novel miRNA loci, raising the question of how many more authentic loci remain undiscovered. This question is difficult to answer. Ever since the recognition that the poorly conserved miRNAs are also the ones expressed at lower levels in mammals, and thus are the most difficult to detect by both computational and experimental methods, we have known that it is impossible to provide a meaningful estimate of the number of mammalian miRNA genes remaining to be discovered (Bartel 2004). The broadly conserved miRNAs are another matter. Only three of the 88 novel canonical miRNAs had recognizable orthologs sequenced in chickens, lizards, frogs, or fish, and these three were antisense to previously annotated broadly conserved miRNA genes. Therefore, apart from miRNAs expressed at very low levels from the antisense strand of known genes, we suspect that the list of broadly conserved miRNA gene families is nearing completion. The current set of murine miRNA genes includes 192 genes that fall into 89 broadly conserved miRNA gene families (Supplemental Table 6).
Another 107 miRNA gene families appeared conserved in other mammals (Supplemental Table 6). These were represented by 120 murine genes, including 14 novel genes. Of these novel genes, 11 were founding members of novel conserved gene families. Some of these were identified with only 11 reads, indicating that additional pan-mammalian gene families remain to be found, although we have no evidence supporting the idea that the number of conserved gene families will rise to the very high levels suggested by some earlier computational studies (Berezikov et al. 2005, 2006b; Xie et al. 2005). For now, we can say that mammals have at least 196 conserved miRNA gene families represented in mice by at least 312 pre-miRNA hairpins (303 canonical and nine noncanonical hairpins) produced from at least 194 unique transcription units.
Because a single miRNA hairpin can produce multiple functional isoforms, generated by either 5′ processing heterogeneity or utilization of both arms of the miRNA duplex, a single conserved hairpin can produce more than one conserved miRNA isoform. Because the different isoforms have different seed sequences, they fall into different families of mature miRNAs. Thus, the number of conserved families of miRNAs (i.e., mature guide RNAs) will exceed the number of conserved families of genes (i.e., hairpins). Perhaps the best known example of a hairpin with two broadly conserved isoforms is mir-9, for which conserved miRNAs from both arms of the hairpin are readily detected by using in situ hybridization in both zebrafish and marine annelids (Wienholds et al. 2005; Christodoulou et al. 2010). Numerous conserved genes produce more than one miRNA isoform (Figs. 5A, 6A), but for most of these we do not yet know whether production of the alternative isoform is conserved in other species. High-throughput sequencing from other species will help identify many additional conserved isoforms. We anticipate that the discovery of multiple conserved isoforms will contribute much more to the future growth in the list of broadly conserved miRNA families than will the discovery of new conserved genes.
As expected, the conserved miRNAs tended to be expressed at much higher levels than were the nonconserved ones, with the median read frequency of conserved miRNAs 44-fold greater than that of the nonconserved miRNAs (Figs. 5A, 6B). Therefore, even if many nonconserved miRNA genes remained to be found, these would add little to the number of annotated miRNA molecules in a given cell or tissue, and presumably even less to the impact of miRNAs on gene expression (Bartel 2009). Indeed, even more pressing than the question of how many poorly conserved miRNAs remain undetected is the question of whether any of the known poorly conserved miRNAs have any consequential function in the animal.
Most of these poorly conserved miRNAs could have derived from transcripts that fortuitously acquired hairpin regions with features needed for some Drosha/Dicer processing. In this scenario, most of these newly emergent miRNAs will be lost during the course of evolution before ever acquiring the expression levels needed to have a targeting function sufficient for their selective retention in the genome. Consistent with the hypothesis that most of these miRNAs play inconsequential regulatory roles, these miRNAs generally accumulated to much lower levels in our ectopic expression assay, (Fig. 3B, median read frequencies of 58 and 844 for nonconserved and conserved miRNAs, respectively), and they displayed weaker specificity for one arm of the hairpin (Fig. 5A), as would be expected if there was no advantage for the cell to efficiently use their respective hairpins. Nonetheless, some were processed efficiently, and at least a few poorly conserved miRNAs probably have acquired consequential species-specific functions. Although none have known functions, such hairpins are worthy of annotation as miRNA loci (just as protein-coding genes can be annotated before the protein is known to be functional), and as a class these newly emergent miRNAs could provide an important evolutionary substrate for the emergence of new regulatory activities.
The major challenge for miRNA gene discovery stems from the difficulty in proving that a nonconserved, poorly expressed candidate is an authentic miRNA, combined with the even greater difficulty in proving that a questionable candidate is not an authentic miRNA. This challenge has become all the more acute as miRNA discovery has reached the point to which nearly all of the novel candidates are both nonconserved and poorly expressed. Our approach of testing pools of candidates in an ectopic expression assay provides useful data for evaluating miRNA authenticity. However, our approach cannot provide conclusive proof for or against the authenticity of a proposed candidate, leaving open the possibility that some of the nonconserved, poorly expressed candidates that we classify as “confidently identified miRNAs” are false positives. When considering the limitations of the current tools for miRNA gene identification, this possibility cannot be avoided. Therefore, if any nonconserved, poorly expressed miRNAs are annotated as miRNAs, the resulting list of miRNAs will have to be somewhat fuzzy, with an expectation that some of the annotated genes will not be authentic miRNAs. This expectation should not be viewed as advocating the indiscriminant annotation of all candidates as miRNAs. Our proposal is that miRNA gene discovery efforts should annotate as miRNAs only those novel candidates that both are found in high-thoughput sequencing libraries and pass a set of criteria that is sufficiently stringent such that a majority of the novel canonical miRNAs are cleanly processed in a Drosha-dependent manner when using the ectopic expression assay. Although implementing this proposal would not prevent all false positives from entering the databases, it would preserve a higher quality set of miRNAs while eliminating few authentic annotations. Those wanting to take additional measures to avoid false positives could focus on only the subset of miRNAs that both meet these criteria and are conserved in other species.
Unknown features required for Drosha/Dicer processing
Before learning the results of our experiments, we wondered whether any ectopically overexpressed hairpin of suitable length would be processed as if it were a miRNA, a result that would have rendered our assay too permissive to be of value. In this scenario, most of the specificity that distinguished authentic miRNA genes from other regions of the genome with the potential to produce transcripts that fold into seemingly miRNA-like hairpins would have been a function of whether or not the regions were transcribed. This scenario was not realized, however, and our assay turned out to be informative, which illustrates how much of Drosha/Dicer substrate recognition still remains unknown. Many of the previously proposed miRNA hairpins that had no reads in our mouse samples were indistinguishable from authentic miRNA hairpins with regard to the known determinants for Drosha/Dicer recognition, yet none of these unconfirmed hairpins produced miRNA and miRNA* molecules in our very sensitive assay (Fig. 2C,D). These results showing that major processing specificity determinants still remain undiscovered point to the importance of finding these determinants—efforts that, if successful, will mark the next substantive advance in accurately predicting and annotating metazoan miRNAs.
Materials and methods
Library preparation
Total RNA samples from mouse ovary, testes, and brain were purchased from Ambion, and total RNA from mouse E7.5, E9.5, E12.5, and newborn were obtained from the Chess laboratory. The small RNA cDNA libraries were made as described (Grimson et al. 2008), except for the 3′ adaptor ligation, which was 5′ adenylated pTCGTATGCCGTCTTCTGCTTGidT. For a detailed protocol, see http://web.wi.mit.edu/bartel/pub/protocols.html.
miRNA discovery
The reads with inserts of 16–27 nt were processed as described (Babiarz et al. 2008). The miRNA candidates were identified using reads matching genomic regions that were not very highly repetitive (reads with <500 genomic matches). Reads from all data sets were combined and grouped by their 5′-terminal loci, requiring that each candidate 5′ locus pass five criteria listed in the text. (1) To pass the expression criterion, a candidate required ≥10 normalized reads. (2) To address the hairpin requirement, the secondary structure of the candidate was evaluated by selecting for each 5′-terminal locus the most abundant sequence and extending its 5′ end by 2 nt to define the range of the potential miRNA/miRNA* duplex. Three genomic windows were extracted with the 5′ end extended an additional 10 nt and the 3′ end extended either 50 nt, 100 nt, or 150 nt. Three more windows were extracted extending the 3′ end by 10 nt and the 5′ end another 50 nt, 100 nt, or 150 nt. The secondary structure of each of the six windows was predicted using RNAfold (Hofacker et al. 1994), and the number of hairpin base pairs (denoted using bracket notation) involving the 5′-extended miRNA candidate was calculated as the absolute value of ([number of 5′-facing brackets] − [number of 3′-facing brackets]). A candidate with a minimum of 16 bp using at least one of the six genomic windows satisfied the hairpin criteria. (3) The candidates with non-miRNA biogenesis were found by mapping to annotated noncoding RNA loci (rRNA, tRNA, snRNA, and srpRNA). (4) The candidates likely produced by degradation were defined as those failing the 5′ homogeneity requirement. A candidate satisfied the 5′ homogeneity requirement if at least half of the reads within 30 nt of the candidate 5′ end were present within 2 nt of the candidate 5′ end and if the candidate 5′ end comprised at least half of the reads within 2 nt of the candidate 5′ end, or if there was only one other 5′ end within 30 nt of the candidate 5′ end that had more than half of the reads mapping to the candidate 5′ end. (5) Manual inspection of reads mapped to predicted secondary structures identified candidates accompanied by potential miRNA* reads. For 10 previously annotated miRNAs and seven novel miRNAs, a suitable miRNA* read was found only after considering alternative hairpin folds predicted to be suboptimal using mfold (Mathews et al. 1999; Zuker 2003).
For the analysis of mir-290, mir-291a, mir-291b, mir-292, mir293, mir-294, and mir-295, which are not present in mm8 genome assembly, we mapped all reads to mm9 genome assembly corresponding to the region [chr7(+): 3,218,627–3,220,842].
For conservation analysis, a candidate was considered broadly conserved if the hairpin structure and the seed sequence were conserved to chickens, fish, frogs, or lizards (galGal3, danRer5, xenTro2, and anoCar1, respectively) in the University of California at Santa Cruz whole-genome alignments (Kuhn et al. 2009). To identify a candidate conserved in mammals, we looked at 12 additional genomes (bosTau3, canFam2, cavPor2, equCab1, hg18, loxAfr1, monDom4, ornAna1, panTro2, ponAbe2, rheMac2, and rn4) and calculated the branch length score from a phylogenetic tree trained on mouse 3′ UTR data (Friedman et al. 2009), using the cutoff score of 0.7. A gene was considered to be in a conserved miRNA gene family if the hairpin produced a miRNA with a seed matching that of a conserved miRNA (Supplemental Table 6).
Ectopic overexpression assays
To generate expression constructs, pre-miRNA hairpins and the surrounding regions were amplified from human genomic DNA (NCI-BL2126) or from mouse BL6 genomic DNA using Pfu Ultra II polymerase (Stratagene) and primers with Gateway (Invitrogen)-compatible ends designed to anneal ∼100 nt upstream of and downstream from the miRNA hairpins. PCR products were inserted into Gateway vector pDONR221 and subsequently into pcDNA3.2/V5-DEST, and the resulting plasmids were transformed into DH5-α cells. Positive clones were selected by colony PCR and were sequenced. Clones that did not have a mutation within pre-miRNA hairpins were selected. Plasmid DNA from the confirmed expression clones was purified for transfection using the Plasmid Mini Kit (Qiagen). For each standard assay, plasmids for up to 10 hairpin expression constructs were mixed in equal amounts to create seven or eight pools of ∼1.4 μg of DNA each, with each pool including one to three positive control hairpins.
HEK293T cells were cultured in DMEM supplemented with 10% FBS, and were plated in 12-well plates ∼24 h prior to transfection to reach ∼80%–90% confluency. Each well of cells was transfected with one pool of DNA using Lipofectamine 2000 (Invitrogen). For the standard assays, 145–200 ng of pMaxGFP (Amaxa) was cotransfected with each pool to enable transfection efficiency to be confirmed by GFP expression. Control wells (no hairpin plasmid) were transfected only with 145 ng of pMaxGFP. For the Drosha/Dicer dependency assays, seven to eight hairpin constructs were combined to create six pools of ∼400 ng each. Each pool was mixed with 1.2 μg of the pCK-Drosha-Flag(TN) (TNdrosha), pCK-Flag-Dicer(TN) (TNdicer), or pCK-dsRed.T4 (control vector, constructed by replacing the Drosha-coding sequence of TNdrosha with dsRed-coding sequence) and used to transfect one well of HEK293T cells as above. Control wells were transfected with 1.2 μg of either TNdrosha, TNdicer, or control vector. For the dependency assays, each transfection was performed in duplicate wells. Cells from all assays were harvested 39–48 h after transfection. Cells from each treatment were combined, total RNA was extracted using TriReagent (Ambion), and small RNA libraries were prepared for Illumina sequencing.
The reads were processed as above, and RNA species were matched to the transfected hairpins. In the standard assay, reads were normalized by the median of the 30 most frequently sequenced endogenous miRNAs. For assays testing Drosha/Dicer dependency, reads were normalized based on the number of reads corresponding to an 18-nt internal standard that had been spiked into equivalent amounts of total RNA prior to beginning library preparation. Reads matching the transfected hairpins were grouped by their 5′ termini (5′-terminal locus). The locus with the largest number of reads was considered the 5′-terminal locus of the mature miRNA produced by the hairpin, and similarly, the most dominant 5′ locus on the opposite arm was considered the miRNA*. The normalized miRNA and miRNA* read numbers were summed to calculate the expression level.
If an overexpressed hairpin generated mature miRNA with the dominant 5′-terminal locus corresponding to the expected locus and at least one read corresponding to the miRNA* with an ∼2-nt 3′ overhang, it was considered expressed. A hairpin was classified as overexpressed if there were at least threefold more reads in the hairpin transfection than in the control transfection, after adding psuedocounts of five to both. A hairpin was classified as Drosha- or Dicer-dependent if the knockdown was at least threefold.
Identification of arm-switching miRNAs
To determine the read numbers from the 5′ and 3′ arms, reads from each sample were grouped based on their 5′ termini, and the read numbers were tallied for those corresponding to the miRNA or miRNA* 5′ terminus. Only samples with five or more reads on either arm were considered. The fold enrichment was calculated as the ratio of 5′ and 3′ arm reads after adding pseudocounts of one.
RNA editing analysis
Sequencing libraries from individual tissues were combined and mapped to the genome using the Bowtie alignment tool (Langmead et al. 2009). The alignments were filtered for sequences that uniquely aligned to the genome, contained at most one mismatch to the genome, and had 5′ ends that mapped to within 1 nt of an annotated miRNA or miRNA* 5′ end. The 12 possible mismatch types were then quantified at each position covered by the filtered reads. For example, to screen for A-to-G mismatches indicative of A-to-I editing sites, the editing fraction was calculated as the number of reads containing an A-to-G mismatch at a particular position, divided by the number of filtered reads covering that position. Sites were considered editing candidates if the editing fraction was >5%, had at least 10 A-to-G mismatch reads, and did not occur in the last 2 nt of the corresponding miRNA or miRNA*. Candidate editing sites were then manually examined and discarded if an alternative explanation was more parsimonious. For example, the only nonbrain editing candidate mapped to let-7c-1, but was most likely due to a handful of let-7b reads containing untemplated nucleotide additions that fortuitously matched the let-7c-1 locus. Consistent with this explanation, the putatively edited reads were unusually long and at unusually low abundance. Candidate editing sites were also checked in the Perlegen SNP database (Frazer et al. 2007) and dbSNP; no editing candidates corresponded to known SNPs.
Untemplated nucleotide analysis
To examine untemplated nucleotide addition, non-genome-mapping reads were filtered for those that matched miRNA or miRNA* sequences but also included a nongenomic poly(N) at the 3′ end. The untemplated nucleotide addition rate was calculated as the ratio of reads with the untemplated nucleotide to the sum of the reads with and without the untemplated nucleotide. After excluding miRNAs that map to multiple loci, and any miRNAs or miRNA*s with a genomic T at the position immediately 3′ of the annotated sequence, there were 343 miRNA/miRNA* species with untemplated U on the 5′ arm and 318 on the 3′ arm. Similarly, there were 287 5′ arm species with untemplated A on the 5′ arm and 324 on the 3′ arm. The background tRNA untemplated U addition rate was calculated similarly. A two-sided K–S test was used to assess significant differences in distributions.
Accession numbers
All small RNA reads are available at the GEO database with accession number GSE20384.
Acknowledgments
We thank N. Lau and A. Chess for embryonic and newborn total RNA, R. Friedman for calculating branch length scores for the analysis of conservation, A. Marson and N. Hannet for technical advice, and V.N. Kim for TNdrosha and TNdicer plasmids. This work was supported by a grant from the NIH (GM067031) to D.B.
Footnotes
References
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang YM, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W 2008. MicroRNAs in the Drosophila bithorax complex. Genes & Dev 22: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RHA, Cuppen E 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24 [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RHA 2006a. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 38: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. 2006b. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res 16: 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC 2007. Mammalian mirtron genes. Mol Cell 28: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR 2006. RNA editing of human microRNAs. Genome Biol 7: R27 doi: 10.1186/gb-2006-7-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA 2007. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci 104: 18097–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Z, Li L, Lodish HF, Bartel DP 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D 2010. Ancient animal microRNAs and the evolution of tissue identity. Nature 463: 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He YP, Leary RJ, Pagliarini R, Diaz LA, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. 2006. The colorectal microRNAome. Proc Natl Acad Sci 103: 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KKH, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S 2004. The microRNA registry. Nucleic Acids Res 32: D109–D111 doi: 10.1093/nar/gkh023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541 [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JJ, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho YJ, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim Y-K, Yeom K-H, Yang W-Y, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han JJ, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P 1994. Fast folding and comparison of rna secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Hu H, Yan Z, Xu Y, Hu H, Menzel C, Zhou Y, Chen W, Khaitovich P 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 10: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K 2007a. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K 2007b. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, Kim VN 2007. Processing of intronic microRNAs. EMBO J 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. 2008. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res 18: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 doi: 10.1093/nar/gkn875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y 2004. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T 2003. New microRNAs from mouse and human. Rna 9: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han JJ, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP 2003. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ 2005. Elucidation of the small RNA component of the transcriptome. Science 309: 1567–1569 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. 2006. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res 34: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2005. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–D504 doi: 10.1093/nar/gki025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, et al. 2009. Loss of cardiac microRNA-mediated regulation leads to dilated xardiomyopathy and heart failure. Circ Res 105: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W 2007. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007a. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007b. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo TS, Bai XP, Ruparel H, Li ZM, Turro NJ, Ju JY 2004. Photocleavable fluorescent nucleotides for DNA sequencing on a chip constructed by site-specific coupling chemistry. Proc Natl Acad Sci 101: 5488–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M 2007. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 17: 1865–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes & Dev 22: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung W-J, Hagen JW, Berezikov E, Hannon GJ, Lai EC 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes & Dev 22: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJM, Stoop H, Nagel R, Liu Y-P, van Duijse J, Drost J, Griekspoor A, et al. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N 2009. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One 4: e7566 doi: 10.1371/journal.pone.0007566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XH, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]