De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia (original) (raw)
Abstract
Schizophrenia likely results from poorly understood genetic and environmental factors. We studied the gene encoding the synaptic protein SHANK3 in 285 controls and 185 schizophrenia patients with unaffected parents. Two de novo mutations (R1117X and R536W) were identified in two families, one being found in three affected brothers, suggesting germline mosaicism. Zebrafish and rat hippocampal neuron assays revealed behavior and differentiation defects resulting from the R1117X mutant. As mutations in SHANK3 were previously reported in autism, the occurrence of SHANK3 mutations in subjects with a schizophrenia phenotype suggests a molecular genetic link between these two neurodevelopmental disorders.
Schizophrenia (SCZ) is a chronic psychiatric disorder characterized by a profound disruption in cognition, behavior, and emotion which begins in adolescence or early adulthood. There is significant clinical variability among SCZ patients, suggesting that it is etiologically heterogeneous. There are several hypotheses to explain genetic factors underlying SCZ, such as polygenic inheritance (1) or, in a fraction of cases, variably penetrant de novo mutations. The de novo hypothesis is based on several observations. One is that relatives of an individual with SCZ have a higher risk of being affected (parents 6%, offspring 13%, and siblings 9% compared with 1% for the general population) (2). The greater frequency in offspring than in parents may occur if new mutations account for a fraction of SCZ cases. Also, there is a significantly increased risk of SCZ with increasing paternal age (3), which could result from the age-related increase in paternal de novo mutations. Furthermore, despite reduced reproductive fitness (4) and extremely variable environmental factors, the incidence of SCZ is maintained at ∼1% worldwide. Interestingly, recent studies reported de novo copy-number variants in SCZ, providing further support for the de novo mutation hypothesis (5, 6).
As part of the Synapse to Disease (S2D) project aimed at exploring the de novo mutation hypothesis in brain diseases, we are sequencing synaptic genes in individuals with SCZ and autism spectrum disorder (ASD), two neurodevelopmental disorders. Recently, mutations in the SHANK3 (SH3 and multiple ankyrin repeat domains 3) gene, encoding a scaffolding protein abundant in the postsynaptic density of excitatory synapses on dendritic spines, were found in patients with ASD (7–9). Considering that ASD and SCZ share some features, we decided to screen the SHANK3 gene in our cohort of SCZ probands. Given our hypothesis that a significant fraction of SCZ cases are the result of new mutations, we selected SCZ cases with unaffected parents and screened for de novo mutations.
Results
We screened the coding region of the SHANK3 gene in 185 unrelated SCZ probands (Table S1) and identified two unrelated SCZ patients who were heterozygous for de novo mutations (R1117X and R536W; Fig. 1). Parentage of proband/parent trios and DNA authenticity in all family members were confirmed using a panel of microsatellite markers (Table S2).
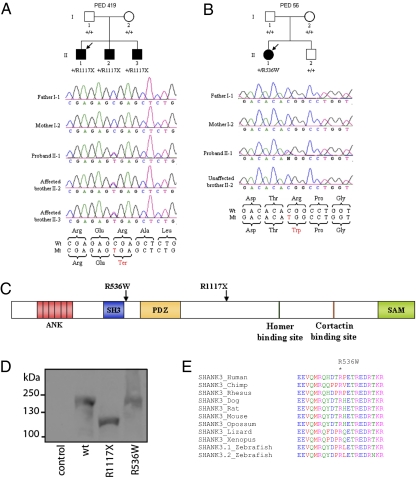
Fig. 1.
Families with de novo mutations in the SHANK3 gene. (A) Segregation of the R1117X nonsense mutation in three affected brothers of family PED 419. The proband is indicated by the arrow. (B) Segregation of the R536W missense mutation in the proband but not her unaffected brother in PED 56. (C) Localization on the linear protein structure of SHANK3 of the de novo mutations found in families with schizophrenia. ANK, ankyrin repeats; SH3, Src homology 3 domain; PDZ, postsynaptic density protein (PSD95), Drosophila disk large tumor suppressor (DlgA), and zonula occludens-1 protein (z _o_-1) domain; SAM, sterile α motif domain. (D) Western blot analysis using HA antibody of HEK293T cell lysate transfected with empty vector (control), HA-Shank3 (WT), R1117X, and R536W. Shank3 WT and R536W have a similar size (200 kDa), whereas the nonsense R1117X results in a truncated protein (123 kDa). (E) Alignment of SHANK3 orthologous peptide sequences near the R536W missense (indicated by the asterisk showing amino acid conservation of the R536 residues in 10 species). GenBank accession numbers: chimp, AC145340; Rhesus monkey, AANU01101358; dog, XP_848271; rat, P_067708; opossum, XP_001366729; lizard, AAWZ01039630; Xenopus, CX494866; zebrafish: zs3.1, CAI20675 and zs3.2, XR_028926.
We detected a nonsense de novo R1117X mutation, in a proband and his two affected brothers (Fig. 1_A_), which appeared to be inherited from the paternal strand based on haplotype analysis (Table S2) and is likely due to germline mosaicism. The proband (II-1) is of European ancestry and has a diagnosis of schizoaffective disorder (age of onset 19 years). This patient was considered suffering from borderline mental retardation (MR) since childhood but went through school and graduated from high school in a special educational program for children with intellectual difficulties. He had a Wechsler Adult Intelligence Scale verbal intelligence quotient (IQ) of 73, performance of 73, and full-scale of 72. He presents no evidence of any autistic features. Brother II-2 was diagnosed with SCZ at age 21, presented symptoms of hyperactivity disorder (medicated with Ritalin) in childhood, and had one seizure episode at age 10. He was described as having mild MR (no IQ available). Brother II-3 was diagnosed with SCZ and atypical chronic psychosis at age 16. His medical record revealed moderate MR with an IQ of 36. He attended education institutions for children with intellectual deficits. None of the three affected brothers had evidence of dysmorphic features and no psychiatric illness was known to be present in the extended family on either side, although the mother was diagnosed with major depression during her lifetime. The R1117X mutation results in a truncated protein, as confirmed by expression analysis, lacking the Homer- and Cortactin-binding sites and the sterile α motif (SAM) domain (Fig. 1 C and D).
We also identified a de novo missense mutation, R536W, in a 23-year-old woman, also of European ancestry, who was diagnosed as schizoaffective (age of onset 11 years). This patient had normal growth, no dysmorphic features, speech impairment, poor academic and social performance, and an IQ of 73. To exclude a possible diagnosis of autism, the Autism Screening Questionnaire (10) was completed and her score was 1 (score > 15 = autism). The R536W mutation was absent from her healthy brother and unaffected parents’ DNA (Fig. 1_B_). It was not possible to determine whether the de novo R536W mutation had occurred in the paternal or the maternal germlines, or in the conceptus (Table S2).
To evaluate whether SHANK3 supports deleterious mutations in healthy individuals, we resequenced its entire coding and splice junction regions in 285 unaffected controls (CTL) with no history of psychiatric diseases. Neither of the two de novo mutations, nor any new protein-truncating mutations, were detected in this control group. From both cohorts, we identified a total of 31 coding variants, including the following variants that were detected in either the SCZ or CTL cohorts but not both: 8 nonsynonymous (1 nonsense, 7 missenses) and 14 synonymous variants (Table S3). Among the 6 nonsynonymous variants found only in the SCZ cohort, only the R1117X and R536W were de novo, whereas the other 4 (H494Q, S952T, G1011V, and P1134H) were transmitted from an unaffected parent. Therefore, these four transmitted nonsynonymous variants can be excluded from a direct role as dominant mutations in SCZ. We observed a slight increase in the mutational load (nonsynonymous) in potentially “damaging” mutations between cases and controls (P = 0.0369; odds ratio: 4.76). Due to unavailability of parents’ DNA, we could not determine the inheritance status of the two nonsynonymous mutations (I1546V and R1298K) that were found only in the CTL cohort. These mutations were predicted not to affect protein function by three commonly used prediction programs (PolyPhen, SIFT, and SNAP) (11–13). In addition, one of these missenses (I1546V) has also been observed elsewhere (8), suggesting that it may be a common polymorphism.
To evaluate whether SHANK3 is accumulating deleterious mutations in human populations, we performed a standard population genetic test (14) which asks whether there is an excess of potentially disruptive amino acid mutations accumulating at this locus more so than expected by chance (Table S4). We did not detect a significant excess of amino acid mutations segregating at this locus relative to a neutral expectation (two-tailed, P = 0.6796). Furthermore, over the entire coding locus (5,196 nucleotides), we observed only 3 fixed amino acid differences between humans and chimps, relative to 16 silent substitutions, which are similarly constrained compared with other brain-expressed genes (15). This suggests that SHANK3 is not accumulating deleterious mutations or mutating at an unusual rate in the population at large and that the observations in the SCZ cohort are an exception.
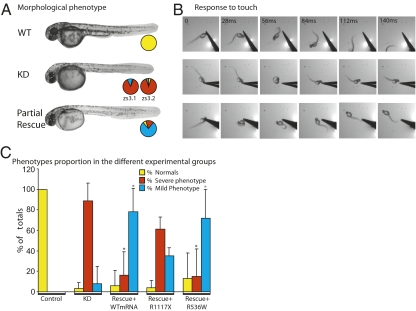
To determine whether the R1117X and R536W mutations affect SHANK3 function in vivo, we tested their ability to rescue a Shank3 knockdown phenotype in the zebrafish embryo by monitoring swimming activity that is due to a well-integrated synaptic drive (16). We knocked down the expression of zebrafish zshank3 orthologous genes (zs3.1 and zs3.2) by injecting selective antisense morpholino oligonucleotides (AMOs) into blastocysts and observed a dose-dependent effect which was most reproducible at 0.75 mM. Knockdown of either gene (n = 99 for zs3.1, n = 92 for zs3.2) resulted in a reduction in size of the head, eyes, and trunk (Fig. 2_A_) and embryos that were unable to swim in response to touch (Fig. 2_B_; n = 191). A small proportion (8%) showed milder deficits and could slowly swim in response to touch (Fig. 2 B and C). Because human SHANK3 cDNA was not readily available, we tested the ability of wild-type (WT) rat Shank3 mRNA to rescue the knockdown phenotype and observed a dose-dependent rescue with a reproducible partial rescue upon injection of 100 ng (Fig. 2_C_; n = 107), resulting in a significantly increased proportion of mild (78%) compared with severe (16%) phenotypes (P < 0.001). In contrast, coinjection of AMO with rat Shank3 mRNA bearing the equivalent R1117X mutation (n = 43) failed to rescue the phenotype, whereas mRNA bearing the equivalent R536W mutation (n = 39) partially rescued the phenotype (Fig. 2 B and C). Overexpression of WT (n = 76) or either of the mutations (n = 24 for R1117X, n = 39 for R536W) was without effect (P = 0.2).
Fig. 2.
Validation of Shank3 mutations in zebrafish. Knockdown (KD) of either of the zebrafish Shank3 genes (zs3.1, zs3.2) using selective AMOs resulted in severe morphological (A) and behavioral deficits (B) compared with wild type (WT), as illustrated using representative images taken from high-speed video films. Partial rescue was observed with coinjection of AMO and rat WT Shank3 mRNA. The pie charts depict the proportion (% of totals) of normal (control-like; yellow), severely affected (no swimming; red), and mildly affected (slow swimming; blue) embryos in each group. The results with coinjection of AMO and rat WT Shank3 or mutated (R1117X or R536W) mRNA are summarized in bar graphs (C). The asterisk denotes significant (P < 0.001) differences from the KD group.
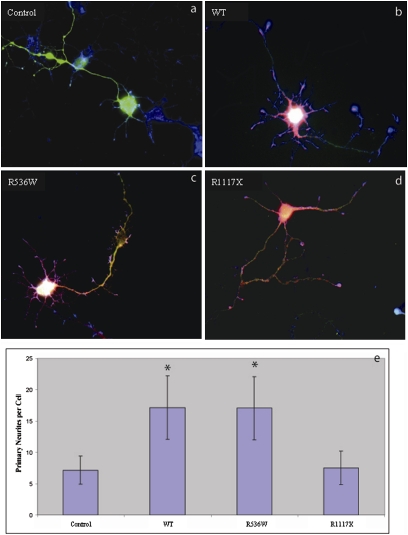
We also examined the consequences of these mutations on the overexpression of Shank3 in transfected rat hippocampal neurons. Expression of WT (Fig. 3_B_) or R536W (Fig. 3_C_) Shank3 resulted in increased somatic sprouting of neurites compared with control neurons (Fig. 3_A_; summarized in Fig. 3_E_), whereas the equivalent truncation mutation R1117X failed to promote sprouting (Fig. 3_D_). These results suggest that the R1117X mutation has a dramatic loss-of-function and not a gain-of-function effect in vivo. Although the R536W mutation had no obvious effect in either of our assays, this may simply be a reflection of the limitations of the assays used, as our genetic findings suggest that it is likely to exert a pathogenic effect in humans in the form of SCZ. The R536 residue is located in close proximity to the SH3 domain, is perfectly conserved from mammals to fish (Fig. 1_D_), and is predicted to have a damaging effect on SHANK3 function [according to PolyPhen, SIFT, and SNAP (12, 17, 18)].
Fig. 3.
Effect of Shank3 mutants on differentiation of hippocampal neurons. Transfected hippocampal neurons were identified by GFP expression (A). Overexpression of WT Shank3 in neurons leads to an increase in primary neurite outgrowth from somata (B). Overexpression of R536W (C) similarly stimulated neurite outgrowth. In contrast, expression of the R1117X truncating mutation (D) failed to do so. In E, the data are quantified in a bar histogram along with standard deviations for each bar. Neurite outgrowth significantly different from control levels (P < 0.001) is indicated with an asterisk.
Discussion
We identified two de novo mutations in the gene encoding the SHANK3 synaptic scaffolding protein in patients ascertained for SCZ and validated in our biological models a pathogenic consequence for at least one of these. The most likely explanation for the identical de novo mutation in three affected brothers (PED 419 family) is germline mosaicism, as described previously for a de novo 300-kb deletion of the NRXN1 gene in two autistic sisters (19), for a de novo 1-Mb duplication at 17p12 in two affected autistic sibs (19), and for a de novo 4.36-Mb deletion involving the SHANK3 gene in two autistic sibs (9). Our PED 419 pedigree represents a partial explanation of why conventional genetic approaches have mostly failed to identify SCZ risk factors, as even a low frequency of de novo mutations could lead to significant genetic heterogeneity and render linkage and association studies problematic.
Different lines of evidence strongly suggest that mutations in the SHANK3 gene are involved in the SCZ phenotype. First, de novo heterozygous mutations in SHANK3 have been reported in ASD, another neurodevelopmental disorder, but not in control individuals. Moreover, a 22q13.3-qter duplication including the SHANK3 gene has been described in a girl diagnosed with borderline intellectual functioning and SCZ of the disorganized subtype [according to the Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV-TR] without any familial neuropsychiatric disease (20). Previously, it was shown that SHANK3 is sensitive to gene dosage and that duplications and deletions are both implicated in ASD (7). Second, both mutations detected herein are predicted to affect protein function and, in the case of R1117X, were functionally validated to cause loss of SHANK3 function. Third, numerous studies have shown evidence of linkage in SCZ families at 22q11-13 locus, which includes SHANK3 (21–27). Finally, comparative genomic methods estimate that in any single conceptus there are ∼1–3 new deleterious mutations that lead to an altered amino acid per genome, which is on average 1 new mutation in 10,000 genes/zygote (28). Assuming that mutation rates are Poisson-distributed, the rate of mutation (μ) in the SCZ cohort at SHANK3 is μ = 1.9 × 10−6 [95% confidence intervals (CI): 4.65 × 10−7, 1.52449 × 10−5] and is significantly higher than published estimates (28) of human mutation rates (μ = 3 × 10−8; CI: 0.18 × 10−8, 8.7 × 10−8) (P < 0.002). The simple statistics presented in this paper further support our hypothesis that rare de novo mutations occur more frequently in SCZ patients than in non-SCZ individuals in a cross-sectional population survey, particularly in those with a severe type of illness accompanied by variable levels of MR. The empirical de novo rate in controls at this locus remains to be tested given the unavailability of the parents in this cohort. These rare SCZ-associated variants will only exist in the gene pool transiently and will only happen sporadically in an individual genome. Therefore, we cannot expect them to be common, nor expect such variants to be concentrated in one or a few genes of the genome. It is most likely that SCZ will result from many rare mutations in dozens or more likely hundreds of genes. The two deleterious mutations found in three affected siblings and one sporadic patient reported in this paper were not found in 285 non-SCZ ethnically matched controls, nor reported in any database of human genomic variation with or without disease phenotype. Given that the population genetic evidence suggests that SHANK3 is not accumulating deleterious mutations in the population at large, these de novo deleterious mutations at SHANK3 should be associated with diseases characterized by reduced reproductive fitness, such as SCZ and ASD. The variant burden test was performed on a modest sample size, and we acknowledge that the real test will be to replicate these results (find new de novo mutations) in a larger sample. Finally, a recent observation showing that heterozygous Shank3 knockout mice display deficits in hippocampal long-term potentiation supports the role of Shank3 in synaptic transmission and plasticity, and further suggests that SHANK3 haploinsufficiency could similarly cause related synaptic dysfunctions in humans (29). Likewise, heterozygous knockout of the closely related gene Shank1 resulted in mice with smaller dendritic spines and synapses but without gross brain abnormalities (30). This is consistent with neuropathological observations seen in SCZ patients reporting aberrant neurodevelopment affecting neuronal migration and connectivity (31).
Deleterious SHANK3 mutations have previously been reported in ASD (7, 8, 32), suggesting that the phenotypic spectrum of such mutations is rather broad. It is well-established that individuals suffering from SCZ perform cognitively below normal comparison (33), and a linear relationship has been found between low premorbid IQ and the risk of SCZ (34). Whereas the common mean IQ found in the literature is around 94 ± 15 (33, 35), the range of values is wide (45–145) and premorbid IQs < 75 were associated with the highest odds ratio for SCZ (2.49; CI: 2.09–2.97) (35). In addition to lower premorbid IQ, the onset of the disease is also associated with an additional cognitive decline (33). Altogether, the literature supports that impaired intellectual functioning and SCZ might share etiological factors. SCZ is not a unifactorial disorder, and other factors (genetic and/or environmental) could influence the final phenotype and the severity of MR. The interplay between SCZ and premorbid intellectual deficit is highlighted in the patients described in this study. Indeed, the probands included in the present study were ascertained as part of ongoing SCZ genetic studies. However, these were atypical cases in that they were at one end of the severity spectrum and had variable premorbid degrees of mental retardation. At least one family (PED 419) carrying the SHANK3 mutation seems to display an atypical form of SCZ and might represent some syndrome with intellectual deficit and SCZ. Although determining psychotic symptoms in the setting of low IQ can be difficult, experienced psychiatrists are trained to search for reliable symptoms such as bizarre thinking (and not just stereotyped ideas) and persistent and prominent delusions and/or hallucinations, especially auditivo-verbal hallucinations. The standardized procedures used in the present study (Materials and Methods), together with consensual diagnosis by two or three experienced psychiatrists, review of medical records, and additional information from family members, are likely to have minimized the risk of misdiagnosis. Screening larger SCZ cohorts with or without premorbid MR would be needed to clarify the role of SHANK3 in SCZ. The identification of genetic anomalies that could play a role in the etiology of SCZ and MR will probably clarify this issue of the nature of the overlap between cognitive dysfunction in these diseases. The presence of MR in patients presenting deleterious SHANK3 mutations is consistent with the role of SHANK3 as a regulator of dendritic spines (7, 29).
Differences in the mutation type (for example, point mutation vs. large deletions) may be one possible explanation for the phenotypic variability of SHANK3 mutations. Another possibility is that differences in genetic background may contribute to the phenotypic variability of SHANK3 mutations. Although this observation is intriguing, multiple phenotypic effects from a single gene have already been described (36) and are supported by recent studies reporting CNVs in both ASD and SCZ for the same locus [for example, the 16p11.2 region and _NRXN1_ genes (37)]. The fact that the two zebrafish Shank3 genes yielded similar dose-dependent phenotypes when knocked down indicates that partial suppression of zShank3 expression was sufficient to yield a phenotype, as expected for a clinically relevant haploinsufficiency. Additional work is needed to determine the extent of the variable phenotype due to SHANK3 mutations and which other genetic and/or environmental factors may affect the expressivity of SHANK3 mutations.
Our findings highlight three very important points in the field of SCZ genetics with serious implications for research strategies. First, it is important in SCZ genetic studies to examine subgroups of families based on family history (familial vs. sporadic) in addition to whether parents are affected or not, and available parents should be carefully phenotyped as well. Second, for nontransmitted sporadic SCZ, deep resequencing in individual genomes will probably be the only way to find the genetic causes. Finally, screening SHANK3 and its interacting partners could provide additional information on the pathogenic pathways of SCZ.
SHANK3 is a scaffolding protein that promotes the formation and maturation of dendritic spines (38, 39). It is also expressed in growth cones, where it appears to regulate process outgrowth by binding to Densin to inhibit outgrowth or Abi-1 to facilitate outgrowth (38). Our Shank3 neuronal overexpression assay shows that the Shank3 mutant R1117X is not promoting neurite outgrowth, indicating that the C-terminal domain is important for this function. In fact, Roussignol et al. demonstrated that the C-terminal domain (including Homer, Cortactin, and SAM domains) is important for spine induction (38). The fact that the two zebrafish Shank3 genes yielded similar phenotypes when knocked down separately and that the morpholinos and mRNA effects were dose-dependent indicate that partial suppression of expression was sufficient to yield a phenotype, as expected for a clinically relevant haploinsufficiency. Although the R536W missense mutation requires additional study, our zebrafish and hippocampal experiments clearly show that the R1117X mutation results in a loss of SHANK3 function. Accordingly, individuals harboring a mutated SHANK3 allele may be expected to display immature or abnormal dendritic structures, especially because the C-terminal region (deleted in the R1117X mutation) of the protein is important for spine induction (38). Indeed, abnormalities in dendritic spine structures have been previously observed in the hippocampus and neocortex in some SCZ patients (40). This would also be compatible with current hypotheses suggesting that synaptic dysfunction occurs in a significant fraction of SCZ cases. Finally, we propose that the de novo mutation mechanism described here for SCZ is plausible for other brain diseases which have so far resisted conventional genetic approaches.
Materials and Methods
SCZ and Control Cohorts.
The SCZ samples included in this study were selected from over 1,000 SCZ families, and 3,000 DNA samples were collected from several large SCZ clinical genetic research centers around the world. These include: (i) Lynn E. DeLisi cohort of cases from USA and Europe (47 cases): L.E.D. and her collaborators have identified and collected over 500 families with schizophrenia or schizoaffective disorder in at least two siblings over the last two decades. Diagnoses were made by using DSM-III-R criteria on the basis of structured interviews, review of medical records from all hospitalizations or other relevant treatment, and structured information obtained from at least one reliable family member about each individual. Two independent diagnoses (one made by L.E.D.) were made for each individual in the study. In cases of disagreement between the diagnosing clinicians, a third clinician was consulted, and final diagnoses were made by consensus after discussion. The sibling with earlier age of onset and more definite SCZ diagnosis was selected for the initial screening. (ii) Ridha Joober cohort (103 cases): R.J. has collected over 300 SCZ families in Montréal in the past 10 years. The same clinical assessment procedures have been followed as in L.E.D.’s study. In addition, extensive pharmacological data have been collected in this cohort. (iii) Judith L. Rapoport cohort of childhood SCZ in USA (33 cases): Cases with childhood-onset schizophrenia were recruited nationwide and assessed as previously described. Individuals in this cohort known to carry the VCFS deletion on chromosome 22q11 were excluded. To summarize briefly, all patients met DSM-IIIR/DSM-IV criteria for schizophrenia or psychosis not otherwise specified, had premorbid full-scale IQ scores of 70 or above, and onset of psychotic symptoms by age 12 years. (iv) Marie-Odile Krebs cohort in France (2 cases): All subjects were examined according to a standardized interview, the Diagnostic Interview for Genetic Studies (DIGS 3.0) (translated into French by M.-O.K. and colleagues). Family histories of psychiatric disorders were also collected using the Family Interview for Genetic Studies (FIGS). All DIGS and FIGS have been reviewed by two or more psychiatrists for a final consensus diagnosis based on DSM-IIIR or DSM-IV at each center. Exclusion criteria for all subjects included neurologic hard signs, a history of head trauma, and substance abuse or dependence. We obtained an SCZ cohort of 185 trios from the following population ancestries: 135 European, 35 non-European Caucasians, 5 African, and 10 mixed origins. This included 8 probands with second-degree relatives affected with SCZ, 45 probands having one or more affected sibs, 126 probands with no history of SCZ (sporadic), and 6 of unknown familial history of SCZ. IQ information was available for 47 of the patients, and it ranged from 49 to 130. All parents and siblings from all SCZ cohorts were evaluated with clinical and structured psychiatric interviews. The 285 controls (225 European, 58 non-European Caucasians, 1 African, and 1 Asian) were recruited by advertisements in local newspapers; the responding volunteers were interviewed using DIGS and clinical examinations. Only individuals without any neuropsychiatric symptoms or family history of neuropsychiatric problems, including any psychotic symptoms, were included as negative controls. The ethnicity was determined by self-reported ethnic origin of four grandparents. A total of 95 samples were recruited by M.-O.K. and 190 samples by R.J. All samples were collected through informed consent following approval of each of the studies by the respective institutional ethics review committees. Genomic DNA was extracted from blood using a Puregene Extraction Kit (Gentra Systems). In all cases, rare mutations were confirmed using blood-derived DNA. Unfortunately, parents were not available for the schizophrenia-negative controls.
Paternity Assays.
Paternity, maternity, and unique genetic identification of each individual of all families were confirmed using at least five highly informative unlinked microsatellite markers. Families for which genotyping data were previously available (used in other genotyping projects or published data) were genotyped using a panel of five highly informative microsatellite markers (D3S1754, D4S3351, D6S1043, D8S1179, and D15S659). Families not previously studied were genotyped using a panel of additional markers (D1S533, D2S1327, D9S1118, D10S677, D11S1984, D12S1294, D14S587, D16S748, and D17S2196). PCR fragments were labeled by incorporation of radiolabeled [35S]dATP; the fragments were separated on 5% denaturing polyacrylamide gels, and the gels were exposed to Hyperfilm MP film (Amersham). All fragments were amplified using a common PCR amplification protocol: 50 ng of DNA template was used with Taq polymerase (Qiagen); PCR initiation at 94 °C for 5 min, denaturation at 94 °C for 30 s, 35 cycles at 55 °C for 30 s, elongation at 72 °C for 30 s, and final elongation at 72 °C for 10 min. Allele sizes were determined by comparison with an M13mp18 sequence ladder and numbered according to the Fondation Jean Dausset CEPH database (http://www.cephb.fr). Microsatellite primer pair sequences were obtained from the Human Genome Database web site (http://www.gdb.org). Primer pairs for markers used to determine the parental origin of the de novo mutations are described in Table S5. PCR conditions were as described above, although a cycling temperature of 55 °C was used. Parentage was confirmed using the CERVUS (version 3.0) program1.
Gene Screening, Variation Analysis, and Bioinformatics.
Coding exons and flanking splice junctions of the SHANK3 gene were sequenced on a 3730xl DNA Analyzer System (Applied Biosystems). Primers were designed using the ExonPrimer program (Table S6). Exon numbering (32) and cDNA sequences (9) are as previously described. PolyPhred (version 6.11) and Mutation Surveyor (version 3.10; Soft Genetics) were used for mutation detection analysis. All variations were confirmed by reamplifying the fragment and resequencing the proband and both parents using forward and reverse primers.
Test for Excess of Deleterious Mutations at SHANK3 Locus.
The McDonald-Kreitman test2 compares the ratio of synonymous and nonsynonymous polymorphisms within a species with the ratio of synonymous and nonsynonymous fixations between species (in comparison with an outgroup species) within a single gene region; the expectation is that this ratio is the same under neutrality (14) (Table S4). A Fisher's exact test was performed. Even though the ratio of polymorphism is two times larger than that of substitutions, the two-tailed P value = 0.22 suggests that there is no significant excess of deleterious mutations.
Molecular Cloning.
Full-length (HA) rat Shank3 in pRK5 was a gift from Paul F. Worley, Baltimore, MD. The HA-rat Shank3-Δ3′UTR was cloned into pcGlobin2 (41) to synthesize the mRNA by using the mMESSAGE mMACHINE T7 Kit (Applied Biosystems). The human R536W and R1117X mutations, corresponding to R535W and R1119X of the rat Shank3 protein, respectively, were generated by site-directed mutagenesis (Stratagene) and confirmed by sequencing. The human and rat Shank3 peptide sequences share 94.7% identity.
HEK293 Shank3 Expression.
HEK293 cells were transfected with a control plasmid or with plasmids encoding the HA-rat Shank3 WT, R1117X, and R536W using Lipofectamine 2000 (Invitrogen). After cell lysis, the total protein extracts were subjected to SDS/PAGE, transferred to Hybond-P PVDF membrane (GE Healthcare), and immunoblotted with an anti-HA antibody (HA.C5; Abcam).
Validation of Shank3 Mutations in Zebrafish.
Two antisense morpholino oligonucleotides (AMO) were used: 5′-AGATCCTCCATAGGTTCGGAGCCAC-3′ and 5′-CTCCTCGCAAGACAAAGCCGAATCC-3′. The amino acid identity for the two zebrafish genes is 71% between one another and 63% (zs3.1) and 65% (zs3.2) compared with the human sequence. The morpholinos were selective for either zebrafish homolog. In the absence of a zebrafish antibody (no expression studies have been reported), we could not quantify the knockdown (KD). The latter, however, was dose-dependent, as expected. Further, the similarity of the phenotypes caused by KD of either gene (zs3.1 or zs3.2; data combined) and the partial rescue by rat Shank3 in both cases are consistent with a significant and specific KD. We agree that the KD is likely partial rather than complete and that this would also be consistent with the heterozygous disease phenotype. AMOs (Gene Tools) were used to knock down both zebrafish Shank3 orthologous genes (zs3.1 or zs3.2). The AMO was pressure-injected into one- to four-cell-stage blastulae. After establishing the phenotype (0.75 mM AMO), rescue experiments were performed in which AMO and rat Shank3 wild-type mRNA or mutated mRNA (100 ng) were coinjected. The response to touch was documented at high speed (250 frames/sec). Because no human SHANK3 cDNA was available for our analysis, and the human and rat SHANK3 protein sequences are 95% identical, we considered the rat gene to be a reasonable substitute. The rat cDNA is what has been used primarily in functional studies of Shank3.
Neuronal Culture and Transfection.
Hippocampal neuronal cultures were prepared from embryonic rats (E18) as described (42). Briefly, dissociated neurons were cotransfected with GFP and wild-type or mutant forms of Shank3 by electroporation using a Nucleofector Kit (Amaxa). Shank3 expression was detected in GFP-transfected cells with monoclonal antibodies (NeuroMabs) and a secondary Alexa Fluor 555-labeled donkey anti-mouse IgG1 antibody (Molecular Probes). F-actin was visualized by coumarin-labeled phalloidin (Sigma-Aldrich). To quantify neurite numbers, neurites extending from the cell bodies (primary neurites) were quantified with Northern Eclipse version 7.0 image analysis software (Empix Imaging).
Supplementary Material
Supporting Information
Acknowledgments
We thank all the families involved in this study. We would like to thank Dr. Chawki Benkelfat for several helpful discussions. We also acknowledge the efforts of the members of the Génome Québec Innovation Centre Sequencing (Pierre Lepage, Sébastien Brunet, and Hao Fan Yam) and Bioinformatic (Louis Létourneau and Louis Dumond Joseph) groups. This work was supported by Genome Canada and Génome Québec, and received cofunding from Université de Montréal for the Synapse to Disease (S2D) project as well as funding from the Canadian Foundation for Innovation. G.A.R. holds the Canada Research Chair in Genetics of the Nervous System; P.D. holds the Canada Research Chair in Neuroscience. A.P.H. holds the Canada Research Chair in Drosophila Neurobiology. P.A. holds a career award from the Fonds de la recherche en Santé du Québec. A portion of the schizophrenia cohort was collected through the Collaborative Network for Family Study in Psychiatry (Réseau d’étude Familiale en Psychiatry), supported by the Fondation Pierre Deniker.
Footnotes
The S2D team is composed of the following additional members: Kathleen Daignault, Ousmane Diallo, Joannie Duguay, Marina Drits, Edouard Henrion, Philippe Jolivet, Frédéric Kuku, Karine Lachapelle, Guy Laliberté, Sandra Laurent, Meijiang Liao, Carlos Marino, Huashan Peng, Amélie Piton, Annie Raymond, Annie Reynolds, Daniel Rochefort, Judith St-Onge, Pascale Thibodeau, Kazuya Tsurudome, Yan Yang, Sophie Leroy, Katia Ossian, Mélanie Chayet, and David Gourion.
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Risch N. Genetic linkage and complex diseases, with special reference to psychiatric disorders. Genet Epidemiol. 1990;7:3–16. doi: 10.1002/gepi.1370070103. discussion 17−45. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman II, Erlenmeyer-Kimling L. Family and twin strategies as a head start in defining prodromes and endophenotypes for hypothetical early-interventions in schizophrenia. Schizophr Res. 2001;51:93–102. doi: 10.1016/s0920-9964(01)00245-6. [DOI] [PubMed] [Google Scholar]
- 3.Malaspina D, et al. Schizophrenia risk and paternal age: A potential role for de novo mutations in schizophrenia vulnerability genes. CNS Spectr. 2002;7:26–29. doi: 10.1017/s1092852900022239. [DOI] [PubMed] [Google Scholar]
- 4.Bassett AS, Bury A, Hodgkinson KA, Honer WG. Reproductive fitness in familial schizophrenia. Schizophr Res. 1996;21:151–160. doi: 10.1016/0920-9964(96)00018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 6.Stefansson H, et al. GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauthier J, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 9.Moessner R, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berument SK, Rutter M, Lord C, Pickles A, Bailey A. Autism screening questionnaire: Diagnostic validity. Br J Psychiatry. 1999;175:444–451. doi: 10.1192/bjp.175.5.444. [DOI] [PubMed] [Google Scholar]
- 11.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromberg Y, Rost B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila . Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 15.Osada N. Inference of expression-dependent negative selection based on polymorphism and divergence in the human genome. Mol Biol Evol. 2007;24:1622–1626. doi: 10.1093/molbev/msm080. [DOI] [PubMed] [Google Scholar]
- 16.Drapeau P, et al. Development of the locomotor network in zebrafish. Prog Neurobiol. 2002;68:85–111. doi: 10.1016/s0301-0082(02)00075-8. [DOI] [PubMed] [Google Scholar]
- 17.Sunyaev S, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 18.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szatmari P, et al. Autism Genome Project Consortium Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Failla P, et al. Schizophrenia in a patient with subtelomeric duplication of chromosome 22q. Clin Genet. 2007;71:599–601. doi: 10.1111/j.1399-0004.2007.00819.x. [DOI] [PubMed] [Google Scholar]
- 21.Condra JA, Neibergs H, Wei W, Brennan MD. Evidence for two schizophrenia susceptibility genes on chromosome 22q13. Psychiatr Genet. 2007;17:292–298. doi: 10.1097/YPG.0b013e3281ac2345. [DOI] [PubMed] [Google Scholar]
- 22.Verma R, et al. MLC1 gene is associated with schizophrenia and bipolar disorder in Southern India. Biol Psychiatry. 2005;58:16–22. doi: 10.1016/j.biopsych.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen TH, et al. Search for common haplotypes on chromosome 22q in patients with schizophrenia or bipolar disorder from the Faroe Islands. Am J Med Genet. 2002;114:245–252. doi: 10.1002/ajmg.10191. [DOI] [PubMed] [Google Scholar]
- 24.DeLisi LE, et al. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am J Psychiatry. 2002;159:803–812. doi: 10.1176/appi.ajp.159.5.803. [DOI] [PubMed] [Google Scholar]
- 25.Vallada H, et al. Chromosome 22 markers demonstrate transmission disequilibrium with schizophrenia. Psychiatr Genet. 1995;5:127–130. doi: 10.1097/00041444-199505030-00005. [DOI] [PubMed] [Google Scholar]
- 26.Gill M, et al. A combined analysis of D22S278 marker alleles in affected sib-pairs: Support for a susceptibility locus for schizophrenia at chromosome 22q12. Schizophrenia Collaborative Linkage Group (Chromosome 22) Am J Med Genet. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Mowry BJ, et al. Multicenter linkage study of schizophrenia loci on chromosome 22q. Mol Psychiatry. 2004;9:784–795. doi: 10.1038/sj.mp.4001481. [DOI] [PubMed] [Google Scholar]
- 28.Eyre-Walker A, Keightley PD. High genomic deleterious mutation rates in hominids. Nature. 1999;397:344–347. doi: 10.1038/16915. [DOI] [PubMed] [Google Scholar]
- 29.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung AY, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 32.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodberry KA, Giuliano AJ, Seidman LJ. Premorbid IQ in schizophrenia: A meta-analytic review. Am J Psychiatry. 2008;165:579–587. doi: 10.1176/appi.ajp.2008.07081242. [DOI] [PubMed] [Google Scholar]
- 34.David AS, Malmberg A, Brandt L, Allebeck P, Lewis G. IQ and risk for schizophrenia: A population-based cohort study. Psychol Med. 1997;27:1311–1323. doi: 10.1017/s0033291797005680. [DOI] [PubMed] [Google Scholar]
- 35.Urfer-Parnas A, Lykke Mortensen E, Saebye D, Parnas J. Pre-morbid IQ in mental disorders: A Danish draft-board study of 7486 psychiatric patients. Psychol Med. 2010;40:547–556. doi: 10.1017/S0033291709990754. [DOI] [PubMed] [Google Scholar]
- 36.Zoghbi HY. Postnatal neurodevelopmental disorders: Meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 37.Kirov G, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 38.Roussignol G, et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sala C, et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 40.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 41.Ro H, Soun K, Kim EJ, Rhee M. Novel vector systems optimized for injecting in vitro-synthesized mRNA into zebrafish embryos. Mol Cells. 2004;17:373–376. [PubMed] [Google Scholar]
- 42.Goslin K, Asmussen H, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing Nerve Cells. Cambridge, MA: MIT Press; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information