Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker (original) (raw)
. 2010 Mar 20;3(4):328–337.
Abstract
Piwil2, a member of AGO/PIWI family of proteins, has been reported to be expressed in precancerous stem cells (pCSCs), tumor cell lines and various types of human cancers. However, the significance of piwil2 expression in breast cancer has not been investigated. In this study, archival formalin-fixed, paraffin-embedded breast cancer specimens at various developmental stages were prepared as tissue microarrays (TMAs) and examined for the expressions of piwil2, estrogen receptor (ER), progesterone receptor (PR) and a cell proliferation marker Ki67 by immu-nohistochemical (IHC) staining and human epidermal growth factor receptor 2 (HER2) by fluorescence in situ hybridization (FISH). The correlation of piwil2 expression with ER, PR and Ki67 were analyzed statistically. The piwil2 was detected in all of breast cancer TMA cores. In contrast, ER, PR, HER2, and Ki67 were detected only in 66.1%, 54.5%, 36.0%, and 47% of the TMA cores, respectively. Piwil2 was expressed in cytoplasm (Cyt), nucleus (N) or both cytoplasm and nucleus (C-N). The N pattern was less observed in breast precancers, whereas all three patterns were observed in invasive and metastatic cancers. While the Cyt pattern was significantly correlated with ER expression (p = 0.002); N pattern was significantly correlated with Ki67 expression (p =0.001). ER and Ki67 expressions were reduced and increased, respectively, with the expression patterns being shifted from Cyt → C-N → N. In conclusion, piwil2 is expressed in various stages of breast cancers and has the potential to be used a novel biomarker.
Keywords: Piwil2, breast cancer, precancer, estrogen receptor, progesterone receptor, HER2, and Ki67, field cancerization
Introduction
Breast cancer is one of leading causes of women death in the Untied Sates and occurs in about 13% (1/8) of women [1]. Generally, cancer development may undergo the stages of benign proliferation (hyperplasia and low grade dysplasia), precancer (high grade dysplasia and carcinoma in situ) and cancer (invasive and metastatic carcinoma) [2,3]. Breast cancer if detected early is curable by prevention or intervention of progression of precancer to invasive and metastatic types [4,5]. Thus, specific biomarker (s) that are expressed in breast precancerous lesions [atypical hyperplasia and carcinoma in situ (CIS)] are required for the early detection of breast cancers. A challenge for the early detection is the current lack of specific biomarkers expressed at the early stages of cancer development. Despite the identification of a number of oncogenes (ONGs) and tumor suppressor genes (TSGs) in breast cancers, most of them are not appropriate to be used as breast cancer biomarkers, because they are redundantly expressed in normal mammary glands and required as well for the control of normal cell cycle, cell growth and/or cell survival [6,7]. Therefore, a critical issue for early detection of breast cancer is to explore biomarkers that are specifically expressed at precancerous or both precancerous and cancerous stages of breast tumors.
The PIWIL2 gene (alias mili in mouse or hili in humans) is a member of the P-element-induced wimpy testis/Argonaute (PIWI/AGO) gene subfamily, which is essential for germ-cell development [8-12]. PIWI/AGO genes contain Piwi and PAZ domain (PPD), playing important roles for stem cell self-renewal in drosophila [13], game-togenesis [9], small RNA-mediated gene silencing [14,15] and/or chromatin remodeling [16,17]. Recently, piwil2 was found to bind a novel class of RNA called piwi-interacting RNA (piRNA) or repeat-associated small interfering RNAs (rasiRNAs), in mammal testis [18-23]. It may silence the selfish genetic elements, such as retrotransposons, in the germline stem cells (GSCs) of testis [19,23,24]. Dysregulated piwi protein expression appears to be associated with tumorigenesis [25-27]. Thus, piwil2 might play an important role in tumor development [2,27].
Recently, piwil2 was reported to be expressed in tumor cell lines and various types of human cancer [26]. We have also found that piwil2 transcripts were ectopically expressed in murine precancerous stem cells (pCSCs) [2,3,27,28], suggesting that piwil2 might be a biomarker for cancer development. Therefore, we examined piwil2 expression in various stages of human breast cancer and its association with other known breast cancer biomarkers, such as estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2/Neu or ErbB-2), and cell proliferation marker Ki67 (also known as MKI67), a cellular marker for proliferation [29]. The results indicate that piwil2 is expressed in various stages of breast cancer. Especially, it can be detected in breast precancers and its expression pattern is associated with ER and Ki67 expression. The findings suggest a potential for piwil2 to be developed into a novel biomarker of breast cancer.
Material and methods
Specimens and reagents
This study was approved by the Institutional Review Board (IRB #2007EO686) at the Ohio State University (OSU). One hundred and twenty six breast cancer specimens at various developmental stages were obtained from the Tissue Procurement Shared Resource (TPSR), Comprehensive Cancer Center, Ohio State University (Table 1). Tumor specimens were fixed in 10% formalin and embedded in paraffin for pathological and immunohistochemical analysis. Tissue microarrays (TMAs) with 2 mm cores were built by the Histological Core Facility, Department of Pathology. Normal mammary specimens (n=14) from the patients without breast cancer were used as negative controls for piwil2 expression.
Table 1.
Clinicopathological diagnosis and piwil2 expression patterns
| Pathological Diagnosis | Cyt | N | C-N | No. |
|---|---|---|---|---|
| DH | + | ND | ND | 1 |
| ADH | ND | ND | + | 1 |
| DCIS | + | - | + | 9 |
| IDC | + | + | + | 76 |
| ILC | + | + | + | 7 |
| MET | + | + | + | 7 |
| METSqCC | ND | ND | + | 1 |
| MEDCA | ND | ND | + | 1 |
| MICCA | + | ND | ND | 1 |
| SRCC | + | ND | ND | 1 |
| TC | + | ND | ND | 1 |
| IDC+DCIS | + | - | + | 14 |
| Benign | + | +/- | + | 6 |
| Normal | - | - | - | 14 |
Polyclonal rabbit anti-mili antibody was generated and purified as previously described [9]. mAbs to ER (1D5), PR (PgR636) and Ki67 (MIB-1) were purchased from Dako (Carpinteria, California, USA).
Histological and immunohistochemical (IHC) analysis
IHC analysis was performed as previously described [27,28,30]. TMAs were examined for piwil2, ER, PR and Ki67 by immunohistochemistry (IHC). HER2 expression was determined by fluorescence in situ hybridization (FISH). Sections (4 ∼ 5 μm thick) were stained by H. & E. for pathological analysis, or immunostained with a primary antibody to mili (piwil2), ER, PR, or Ki67 followed by a horseradish peroxidase (HRP)-conjugated secondary antibody. The immunostained sections were counterstained with hematoxylin. The specimens were analyzed based on the staining score and staining patterns (Cyt, N and N-Cyt). The staining score (0 ∼ 3) was determined blindly by two pathologists: 0 (-) or negative, 0 ∼ <5% piwil2+ cells; 1 (+) or weak, 5 ∼ 25% piwil2+ cells; 2 (++) or medium, 25% ∼ 50% piwil2+ cells; 3 (+++) or strong, > 50% piwil2+ cells. ER+, PR+, and Ki67+ cells were also scored as for piwil2+ cells. HER2 is based on clinical positive or negative diagnosis.
Statistical analysis
McNemar Test was used to compare the expression in percentage between piwil2 and ER, PR, HER2 or Ki67 in breast cancer. This test is used to compare two paired measurements from the same subject. When the sample size is large, the McNemar test follows the same χ2 distribution but uses a slightly different formula [31]. Fisher's Exact Test was used to compare difference of piwil2 expression patterns between various types of breast lesions. The correlation of piwil2 expression with ER, PR or Ki67 expression, the correlation between ER, PR and Ki67, and the correlation between each piwil2 expression pattern and ER, PR or Ki67 expression were analyzed by a least-squares linear regression using SAS statistical software (SAS Institute Inc, NC, USA). A value of p ≤ 0.05 was considered significant. Data are represented as mean ±SD. *, p≤0.05; **, p≤0.01.
Results
Archival formalin-fixed, paraffin-embedded breast cancer tissues (n=126) were prepared as tissue microarrays (TMA), and analyzed for piwil2 expression by IHC staining (Table 1). Although these specimens were all procured from the patients with breast cancers, the TMA cores might not be consistent with the clinical diagnosis based on regular histological sections because of multiple primary tumors or field can-cerization [32], which might cause technical error. Therefore, the TMA cores were double-blindly re-examined by two pathologists. While six cores from the patients with invasive ductal carcinoma exhibited histologically normal appearance, and others (n=120) exhibited consistent lesions with clinicopathological diagnosis, including ductal hyperplasia (DH), atypical ductal dysplasia (ADH), ductal carcinoma in situ (DCIS), invasive ductal or lobular carcinoma (IDC or ILC), and metastatic carcinoma (Met-ca) (Table 1).
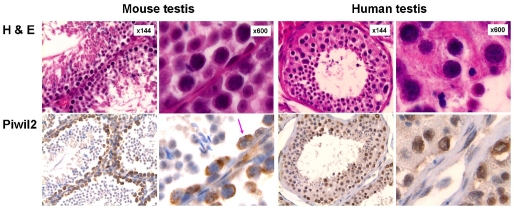
Polyclonal rabbit anti-mili antibody, which was generated as previously described [9] was used to detect piwil2, because it detected not only mili in the GSCs of murine testis but also hili in the GSCs of human testis (Figure 1). The piwil2 was detected essentially in the cytoplasm (Cyt) of murine GSCs with fine granules in some nuclei (N) but mainly in the nuclei or both cytoplasm and nucleus (C-N) of human GSCs. In the human testis, piwil2 was also detected in hyper-plastic testicular cells (Figure 1). Similarly, piwil2 expression patterns of Cyt, N and N-C were also observed in breast cancers, but not in normal mammary tissues (Figure 2).
Figure 1.
Expression pattern of piwil2 in the testis. The murine testes were derived from C57BL/6 mice showing normal histology; and the human testes were derived from TPSR, Ohio State University, exhibiting hyperplasia. The piwil2 was detected essentially in the cytoplasm of murine germline stem cells with fine granular in nucleus, and in both cytoplasm and nucleus of human germline stem cells as well as in hyperplastic cells.
Figure 2.
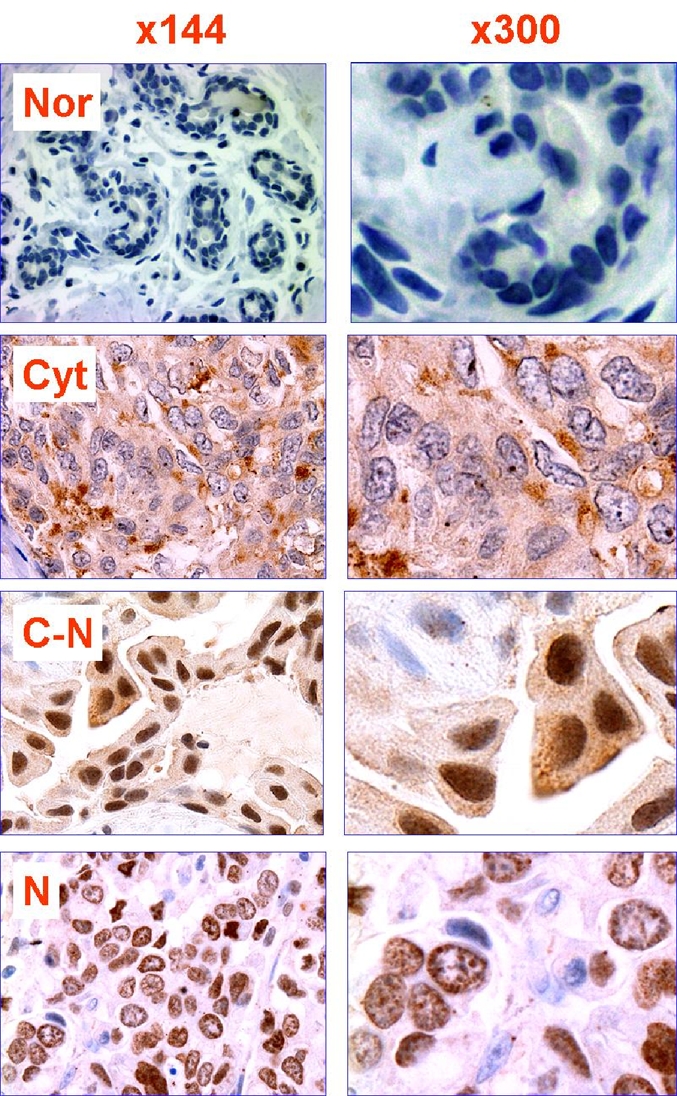
Expression pattern of Piwil2 in human breast cancer. Typical staining patterns of Piwil2 in human breast cancer include cytoplasmic (Cyto), nuclear (N) and nuclear and cytoplasmic (C-N). The examples of staining patterns are derived from the tissues of IDC, which were stained with rabbit anti-mili IgG. Nor: normal mammary tissue.
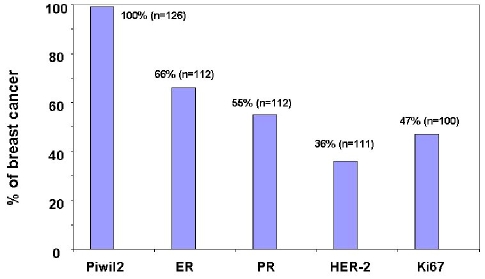
Piwil2 was detected in 126/126 TMA cores of breast cancers, regardless of their developmental stages (Figure 3 and Table 1). Among them, six cores exhibited normal histology. Since these specimens were all procured from the patients with breast tumors, the sensitivity for detection of piwil2 in breast tumors was 100% (Figure 3). However, the level of piwil2 expression in the breast lesions was highly variable with individuals rather than with the types of lesion (from about 5% to >50% of cancer cells) (Figure 4). To further exclude the false positivity of piwil2 expressed in some histologically normal cores, we further examined piwil2 expression in the normal breast tissues without hyperplasia (n=14); none of them was detected with piwil2 (Figure 2 and not shown). The results suggest that piwil2 may express in various types and various stages of breast lesions but unlikely in normal mammary tissue.
Figure 3.
Expression of piwil2 in breast cancer. The breast cancer TMAs were stained with antibodies to piwil2, ER, PR, and Ki67. The TMA cores with >5% positive cells were considered as positive one for each marker. The data of HER2 were derived from clinicopathological records of the patients with breast cancer and revealed by FISH. The percentage of piwil2 expression is significantly higher than that of ER, PR, HER2 or Ki67, as determined by McNemar Test. The p-value of all comparisons (piwil2:ER, PR, HER2 or Ki67) are < 0.0001.
Figure 4.
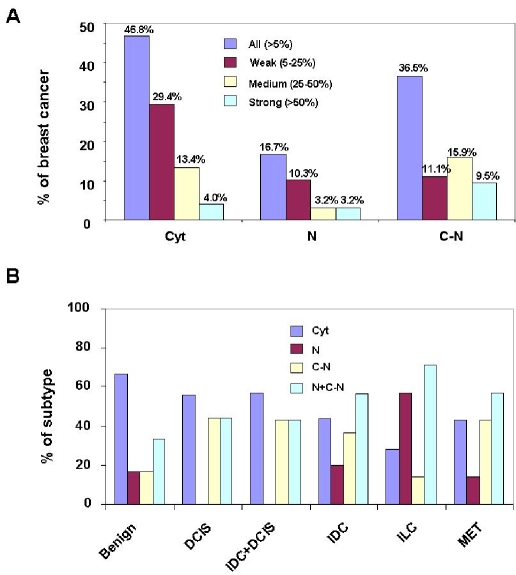
Piwil2 expression pattern in various stages of breast cancer. Piwil2 can be detected in cytoplasm (Cyt), nucleus (N) or both cytoplasm and nucleus (C-N), as demonstrated in Figure 2. Each pattern was scored as weak (5-25%), medium (25-50%), and strong (>50%), based on the percentage of piwil2-expressiong cells. A, proportion of each piwil2 expression pattern in breast cancer (n=126). B, distribution of piwil2 expression pattern in various types of breast cancer: Benign (histological normal tissue within breast cancer): n=6; DCIS: n=9; DCIS + IDC: n=14; IDC: n=76; ILC: n=7; MET (metastatic carcinoma) n=7. N + C-N: the summation of N and C-N pattern. The percentage between groups is not significantly different as tested by Fisher's Exact Test.
To compare sensitivity of piwil2 as a breast biomarker with other known breast cancer markers, such as ER, PR, HER2 and ki67, breast cancer TMAs were immunohistochemically stained with anti-ER, PR or Ki67 antibody, and the data of HER2 detected by FISH was procured from clinicopathological records. The ER, PR, and Ki6 expression in the cores were consistent with clinicopathological data. In striking contrast to piwil2, ER, PR, HER2, and Ki67 were detected only in 66.1 (74/112), 54.5% (61/112), 36.0% (40/111), and 47% (47/100) of breast cancer, respectively. The differences between groups were statistically significant (p < 0.0001) (Figure 3). The total case reduction for each marker in the Figure 3 was due to TMA cores that dropped off slides during staining. These data suggest that piwil2 as a breast cancer biomarker is broader in expression than HER-2, ER, PR and Ki67. Because ER, PR, HER2 and Ki67 are required as well for normal cell development but piwil2 is silenced in adult tissues except in testis [8,10,26], piwil2 as a breast cancer biomarker is obviously more tumor-specific than HER2, ER, PR, and Ki67.
As mentioned above, three distinct expression patterns (Cyt, N and Cyt-N) of piwil2 in breast cancer were identified by IHC staining (Figure 2 & 4). The expression patterns were highly variable between individuals, because not all three patterns were observed in the same core. About 46.8%, 16.7% and 36.5% of the TMA cores was expressed with the pattern of Cyt, N and C-N, respectively (Figure 4A). Interestingly, the expression patterns of piwil2 appeared to be associated with the developmental stages of breast cancer (Figure 4B). In the histologically normal cores, Cyt pattern was dominant compared to breast precancer and invasive/metastatic cancer. The N pattern was almost absent in ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) +DCIS (both IDC and DCIS were detected in the same cores), and all three patterns were detected in IDC, invasive lobular carcinoma (ILC) and metastatic cancers (MET) (Figure 4B & Table 1). In addition, piwil2 was also detected in ductal hyperplasia (DH), atypical ductal hyperplasia (ADH) and other histologi-cal types of breast cancer, although the sample size is very small (Table 1). Although the percentage of the N + C-N pattern was lower in pre-cancers (benign lesions: 33.4%, DCIS: 44.44% and IDC + DCIS: 42.86%) than in invasive/ metastatic cancers (IDC: 56.58%; ILC: 71.39%; and MET: 57.15%), the difference was not statistically significant, as tested by Fisher's Exact Test (Figure 4B).
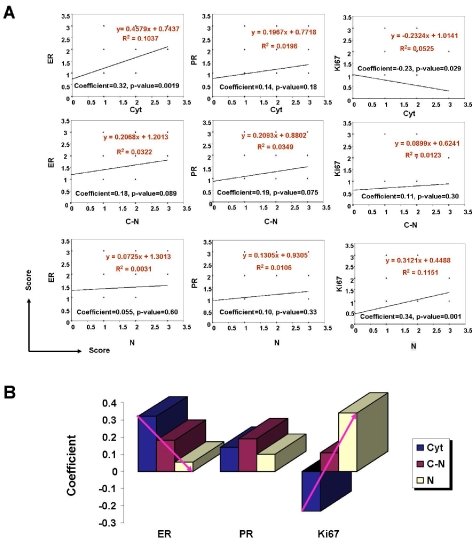
To determine the significance of piwil2 expression patterns in breast cancer development, we statistically analyzed the correlation of piwil2 with the prognostic markers of breast cancer including ER, PR or Ki67. As shown in Figure 5, Cyt pattern of piwil2 expression was positively correlated with ER and PR expressions, especially significantly correlated with ER expression (p = 0.002), but inversely correlated with Ki67 expression (p = 0.03) (Figure 5A). In contrast, N pattern of piwil2 expression was positively correlated with Ki67 expression (p = 0.001) and had no correlation with ER and PR expression (Figure 5A). C-N pattern of piwil2 expression appeared not to be correlated significantly with ER, PR and Ki67 expression. Interestingly, the coefficient of correlation was decreased between piwil2 and ER but increased between piwil2 and Ki67, with the expression pattern being shifted from Cyt → C-N → N, suggesting an important role of piwil2 in regulation of ER and Ki67 expression in breast cancer (Figure 5B).
Figure 5.
Correlation of piwil2 expression pattern with the expressions of ER, PR and Ki67. Breast cancer TMAs cores (n=126) were immunohistochemically stained with antibody to mili, ER, PR, or Ki67. The intensity of staining for each maker was scored as negative (0); weak (1), moderate (2), and strong (3). The correlation of piwil2 expression patterns with ER, PR and Ki67 was determined by least-squares linear regression. A, correlation of each piwil2 expression pattern with ER, PR, or Ki67; B, Correlation coefficient between piwil2 expression patterns and ER, PR or Ki67. Cyt: cytoplasm; N: nucleus; C-N: both cytoplasm and nucleus. The red arrows indicate the trends of correlation coefficient between piwil2 expression and ER or Ki67 expression in breast cancer.
Discussion
Identification of biomarkers for detection of breast cancer is a critical issue for the treatment and cure of breast cancer [4,5]. To reach the goal, it is a necessary step to identify a biomarker widely expressed in various developmental stages of breast cancer, especially at the initial stage of cancer development [4,33]. Recently, it has been demonstrated that piwil2 is widely expressed in various types of human cancers [26], but not in normal tissues [2,8,26]. The significance of piwil2 expression in breast cancer, however, has not been defined.
In this study, Archival formalin-fixed, paraffin-embedded breast cancer tissues were prepared for tissue microarray (TMA), and analyzed for piwil2, ER, PR and Ki67 expression by IHC staining as well as HER2 by FISH. The correlation of piwil2 expression with ER, PR, and Ki67 was also analyzed statistically. Our results indicate that the piwil2 was not expressed in normal mammary tissues but in all breast cancer TMA cores (100%), including some histologically normal cores derived from the tissues adjacent to carcinoma, precancerous cores, and invasive/ metastatic cancers. In contrast, ER, PR, HER2 and Ki67 were detected only in 66.1 (74/112), 54.5% (61/112), 36.0% (40/111), and 47% (47/100) of breast cancers, respectively. While ER expression was significantly correlated with PR expression (p < 0.001), there was no significant expression correlation between piwil2 and ER, PR, or HER2 (p > 0.05); however, the expression pattern of piwil2 was significantly correlated with ER and Ki67 expressions. Three expression patterns of piwil2 were observed in breast cancer cells, including cytoplasmic (Cyt), nuclear (N) and cytoplasmic and nuclear expression (C-N). The Cyt pattern of piwil2 was positively correlated with ER expression (p = 0.02); whereas the N pattern was positively correlated with Ki67 expression (p = 0.001). Interestingly, the expression pattern of piwil2 from Cyt → C-N → N was associated with the decreased expression of ER but with the increased expression of Ki67. N pattern of piwil2 expression was less observed in breast precancers. The results suggest an important role of piwil2 for breast cancer development. Because piwil2 can be expressed at the initiation stages of breast tumors, it is likely that piwil2 could be further developed into a biomarker for diagnosis and/or prognosis of breast cancers when complemented with other biomarkers and a therapeutic target for cure of breast cancers. In addition, piwil2 might be an ideal target for breast cancer therapy, because it is expressed in all stages of breast tumors.
Generally, a cancer may undergo the developmental stages of benign proliferation (initiation), precancer and malignant cancer [2,3,34-36], which is supposed corresponding to pathological changes of hyperplasia and atypical hyperplasia (benign proliferations), dysplasia and carcinoma in situ (precancer), and invasive / metastatic carcinoma (malignant cancer) [2,3,36,37]; and is mediated by tumor stem cells (TSCs), including tumor initiating stem cells (TISCs), pCSCs, and cancer stem cells (CSCs) [2,3,27,28]. Tumor initiation stage and precancer stage are highly reversible and thus are the ideal targets for caner prevention and therapy [2,3,34,38]. In this study, piwil2 was detected in breast benign proliferations [DH and histological normal tissues within cancer (benign)], precan-cers (ADH and DCIS), and malignant cancers (IDC, ILD and MET), suggesting that PIWIL2 can be activated as early at the initial stage (benign proliferation) of breast cancer. This feature is of importance for piwil2 to be developed into a biomarker for detection, prevention and therapy of breast cancer. One potential concern is that detection of piwil2 in the histologically normal tissues within malignant breast cancers might lead to false-positivity in practice. This issue needs to be further addressed with a large cohort of patients, despite unlikely to happen.
Detection of piwil2 in the histologically normal tissues adjacent to cancers might be related to field cancerization [39,40] or “field effects” of cancer, which may reflect the precancerous epigenetic alteration of normal epithelial cells or mammary stem cells [41,42], or the seeding of breast tumor cells such as pCSCs and CSCs in the distant areas [3,43]. In fact, the detection of piwil2 in the histologically normal tissues further confirms the “field effect” of epigenetic and oncogenetic alterations in breast cancer [41,42]. Because piwil2 transcripts are constitu-tively expressed in pCSCs [27], it is likely that the piwil2-expressing “normal” tissues surrounding or within malignant breast cancer reflect that the precancerous lesions occur at molecular levels rather than at cellular and/or histological levels [3,38]. The specificity of piwil2 for early breast lesions was further supported by the failure to detect piwil2 in the normal mammary tissues without hyperplastic lesions, which were derived from the patients with lumpectomy. Because the case number for normal mammary tissues was relatively small, further experiments are warranted to validate the sensitivity and specificity of piwil2 for detection of breast cancers.
High sensitivity and high specificity are essential criteria for a biomarker to be used for detection of cancer. Currently, none of the known breast cancer biomarkers such as ER, PR, HER2, BRST2 [anti-gross cystic disease fluid protein (GCDFP15)] and Ki67 meets the criteria. HER2, ER, PR, BRST2 or Ki67 have been used in clinic as complementary markers for prognosis of breast cancer [4,44,45]. However, these markers are not appropriate for diagnosis of breast cancer. In this study, piwil2 was detected in all TMA cores of breast cancers with various developmental stages but not in normal mammary tissues, suggesting that piwil2 as a biomarker is more sensitive and specific than other known breast cancer biomarkers. Thus, piwil2 has the potential of developing into a novel biomarker for detection of various stages of breast cancers.
Based on the expression of HER2, ER and PR, breast cancer can be categorized by IHC staining into the following subtypes: luminal A: ER+ or PR+ and HER2-; luminal B: ER+ or PR+ and HER2+ (triple positive); HER2: ER-and PR-and HER-2+; and basal: ER-and PR-and HER2-(triple negative) [46,47]. Although the classification is useful for determining therapeutic mode for breast cancer patients, it can not be used to precisely predict progression of a breast cancer. Piwil2 might also have the potential to be used as a novel marker for breast cancer classification when complemented with other biomarkers. In this study, we demonstrate that the frequency and patterns of piwil2-expressing cells are individually variable in breast cancer. This might reflect differential developmental status of breast cancer between individuals. The association of piwil2 expression with ER and Ki67 expression further suggests that piwil2 expression pattern might be an indicator of tumor progression. For example, the shift of piwil2 expression pattern from Cyt → C-N → N may lead to an increase of Ki67-expressing cells in breast cancer; as a consequence, tumor growth is accelerated. Interestingly, N pattern was not detected in the TMA cores of DCIS or DCIS + IDC. Thus, the N pattern might be a demarcation to distinguish true IDC from false IDC. The hypothesis needs to be verified in a larger cohort of breast cancer patients.
Our studies have demonstrated that piwil2 is ubiquitously and uniquely expressed in various stages of breast cancers and its expression patterns are associated with ER and Ki67 as well as cancer development, suggesting that piwil2 plays an important role in breast cancer development. Therefore, piwil2 can be targeted for the study of the mechanisms underlying breast cancer development and has the potential to be developed into a novel biomarker for detection, prognosis and therapy of breast cancers.
Acknowledgments
This work is supported from Department of Pathology, OSU, Strategy Initiative (SIG), 2006/2007, Immunology Program Award 2008 (OSUCCC), American Cancer society IRG-112367 (JXG), and National Cancer Institute CA109527 (RKG).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Gao JX. Cancer stem cells: The lessons from precancerous stem cells. J Cell Mol Med. 2008;12:67–96. doi: 10.1111/j.1582-4934.2007.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao JX, Zhou Q. Epigenetic progenitors in tumor initiation and development. Drug Discovery Today: Disease Models. In Press, Corrected Proof. [Google Scholar]
- 4.Levenson VV. Biomarkers for early detection of breast cancer: What, when, and where? Biochim Biophys Acta. 2007;1770:847–856. doi: 10.1016/j.bbagen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 5.Esserman LJ, Shieh Y, Park JW, Ozanne EM. A role for biomarkers in the screening and diagnosis of breast cancer in younger women. Expert Rev Mol Diagn. 2007;7:533–544. doi: 10.1586/14737159.7.5.533. [DOI] [PubMed] [Google Scholar]
- 6.Osborne C, Wilson P, Tripathy D. Oncogenes and tumor suppressor genes in breast cancer: Potential diagnostic and therapeutic applications. Oncologist. 2004;9:361–377. doi: 10.1634/theoncologist.9-4-361. [DOI] [PubMed] [Google Scholar]
- 7.Sporn MB. Dichotomies in cancer research: Some suggestions for a new synthesis. Nat Clin Pract Oncol. 2006;3:364–373. doi: 10.1038/ncponc0536. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 9.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Engel W, Nayernia K. Stem cell protein piwil2 modulates expression of murine sper-matogonial stem cell expressed genes. Mol Re-prod Dev. 2006;73:173–179. doi: 10.1002/mrd.20391. [DOI] [PubMed] [Google Scholar]
- 11.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. Mili, a piwi-interacting rna-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. Journal of Biological Chemistry. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Current Biology. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The argonaute family: Tentacles that reach into rnai, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian rnai. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 16.Buhler M, Verdel A, Moazed D. Tethering rits to a nascent transcript initiates rnai- and heterochro-matin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Yin H, Lin H. An epigenetic activation role of piwi and a piwi-associated pirna in drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 18.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small rnas bind to mili protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 19.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated pirna clusters implicate mili in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 20.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small rnas in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grivna ST, Pyhtila B, Lin H. Miwi associates with translational machinery and piwi-interacting rnas (pirnas) in regulating spermatogenesis. Proc Natl AcadSciUSA. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the pirna complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 23.Saito K, Nishida KM, Mori T, Kawamura Y, Miyo-shi K, Nagami T, Siomi H, Siomi MC. Specific association of piwi with rasirnas derived from retrotransposon and heterochromatic regions in the drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small rna-generating loci as master regulators of transposon activity in drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 25.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overex-pression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein piwil2 is widely expressed in tumors and inhibits apoptosis through activation of stat3/bcl-xl pathway. Hum Mol Genet. 2006;15:201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan W, Wen J, Zimmerer J, Wang Y, Liu Y, Lasky LC, Heerema NA, Perrotti D, Ozato K, Kuramochi-Miyagawa S, Nakano T, Yates AJ, Carson Iii WE, Lin H, Barsky SH, Gao JX. Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS ONE. 2007;2:e293. doi: 10.1371/journal.pone.0000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen R, Ye Y, Chen L, Yan Q, Barsky SH, Gao JX. Precancerous stem cells can serve as tumor vasculogenic progenitors. PLoS ONE. 2008;3:e1652. doi: 10.1371/journal.pone.0001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas S, Johannes G. The ki-67 proteFrom the known and the unknown. Journal of Cellular Physiology. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Gao JX, Liu X, Wen J, Zhang H, Durbin J, Liu Y, Zheng P. Differentiation of monocytic cell clones into cd8alpha(+) dendritic cells (dc) suggests that monocytes can be direct precursors for both cd8alpha(+) and cd8alpha(-) dc in the mouse. J Immunol. 2003;170:5927–5935. doi: 10.4049/jimmunol.170.12.5927. [DOI] [PubMed] [Google Scholar]
- 31.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- 32.Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of slaughter's concept of field cancerization: Evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 33.Kumar S, Mohan A, Guleria R. Biomarkers in cancer screening, research and detection: Present and future: A review. Biomarkers. 2006;11:385–405. doi: 10.1080/13547500600775011. [DOI] [PubMed] [Google Scholar]
- 34.Werbowetski-Ogilvie TE, Bhatia M. Pluripotent human stem cell lines: What we can learn about cancer initiation. Trends in Molecular Medicine. 2008;14:323–332. doi: 10.1016/j.molmed.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 36.Berman JJ, Albores-Saavedra J, Bostwick D, Delellis R, Eble J, Hamilton SR, Hruban RH, Mutter GL, Page D, Rohan T, Travis W, Henson DE. Precancer: A conceptual working definition - results of a consensus conference. Cancer Detect Prev. 2006;30:387–394. doi: 10.1016/j.cdp.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Cardiff RD, Anver MR, Boivin GP, Bosenberg MW, Maronpot RR, Molinolo AA, Nikitin AY, Rehg JE, Thomas GV, Russell RG, Ward JM. Precancer in mice: Animal models used to understand, prevent, and treat human precancers. Toxicol Pathol. 2006;34:699–707. doi: 10.1080/01926230600930129. [DOI] [PubMed] [Google Scholar]
- 38.Shen R, Tao L, Xu Y, Chang S, Brocklyn JV, Gao JX. Reversibility of aberrant global DNA and estrogen receptor-a gene methylation distinguishes the colorectal precancer from cancer. Int J Clin Exp Pathol. 2009;2:21–33. [PMC free article] [PubMed] [Google Scholar]
- 39.Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danely P. Slaughter HWSWS. “Field cancerization” In oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 41.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12:6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 42.Cheng ASL, Culhane AC, Chan MWY, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BAT, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang THM. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M-Y, Oskarsson T, Acharyya S, Nguyen DX, Zhang XHF, Norton L, Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duffy MJ. Role of tumor markers in patients with solid cancers: A critical review. Eur J Intern Med. 2007;18:175–184. doi: 10.1016/j.ejim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 45.de Almeida PC, Pestana CB. Immunohistochemical markers in the identification of metastatic breast cancer. Breast Cancer Res Treat. 1992;21:201–210. doi: 10.1007/BF01975003. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen PL, Taghian AG, Katz MS, Niemierko A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL, Harris JR. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and her-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26:2373–2378. doi: 10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 47.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]