Viral apoptosis is induced by IRF-3-mediated activation of Bax (original) (raw)
Abstract
Upon infection with many RNA viruses, the cytoplasmic retinoic acid inducible gene-I (RIG-I) pathway activates the latent transcription factor IRF-3, causing its nuclear translocation and the induction of many antiviral genes, including those encoding interferons. Here, we report a novel and distinct activity of IRF-3, in virus-infected cells, that induces apoptosis. Using genetically defective mouse and human cell lines, we demonstrated that, although both pathways required the presence of RIG-I, IPS1, TRAF3 and TBK1, only the apoptotic pathway required the presence of TRAF2 and TRAF6 in addition. More importantly, transcriptionally inactive IRF-3 mutants, such as the one missing its DNA-binding domain, could efficiently mediate apoptosis. Apoptosis was triggered by the direct interaction of IRF-3, through a newly identified BH3 domain, with the pro-apoptotic protein Bax, their co-translocation to the mitochondria and the resulting activation of the mitochondrial apoptotic pathway. Thus, IRF-3 is a dual-action cytoplasmic protein that, upon activation, translocates to the nucleus or to the mitochondrion and triggers two complementary antiviral responses of the infected cell.
Keywords: apoptosis, Bax, IRF-3, RIG-1, Sendai virus
Introduction
Viruses are obligatory intracellular pathogens that are a threat to all eukaryotes. Mammals have developed a plethora of anti-viral mechanisms, including the triggering of innate immune response that limits viral replication and spread (Kawai and Akira, 2006). There are several innate antiviral pathways activated by double-stranded RNA (dsRNA) (Peters et al, 2002), a common byproduct of virus replication. Extracellular dsRNA, produced by dead infected cells, is endocytosed and recognized by Toll-like receptor (TLR) 3 (Alexopoulou et al, 2001), which is primarily located on the endosomal membrane. TLR3 uses the adaptor protein TRIF (Yamamoto et al, 2003) to engage the protein kinase IKK to activate the transcription factor NF-κB and the protein kinases TBK1/IKKɛ (Fitzgerald et al, 2003) to activate the transcription factor IRF-3 (Doyle et al, 2002). In contrast, intracellular dsRNA, produced by replicating viruses that multiply in the cytoplasm, is recognized by cytoplasmic sensors, such as retinoic acid inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (Mda-5) (Yoneyama et al, 2004; Kato et al, 2006), which are RNA helicases, collectively called RIG-I-like helicases (RLH). They use the mitochondrial membrane-bound protein, IPS-1 (Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005) (also known as VISA, Cardif or MAVS), as the adaptor and recruits several members of the TRAF family proteins, which, in turn, activate the same protein kinases and transcription factors as TLR3. These transcription factors drive transcription of the interferon genes and many interferon-stimulated genes (ISG), which are essential for both innate and adaptive antiviral defenses (Kawai and Akira, 2006). Another common cellular antiviral response is apoptosis (Barber, 2001); premature cell death limits virus replication and spreading. We have reported that IRF-3, the transcription factor that induces transcription of the antiviral genes, also has a central role in mediating apoptosis of paramyxovirus-infected cells, as manifested by the fact that infected cells are not killed in its absence (Peters et al, 2008). Moreover, if IRF-3 is activated by virus infection, neither interferon signalling nor NF-κB activation is needed to trigger apoptosis. In the absence of IRF-3 activation, the cells become persistently infected and continuously produce infectious virions (Peters et al, 2008). Our observations made an interesting connection between innate immune response and the apoptotic response of a virus-infected cell through the common requirement of IRF-3 for both processes. Here, we report that the pro-apoptotic action of IRF-3 is distinct and independent of its transcriptional activity. Activation of RLH signalling led to activation of both transcriptional and apoptotic activities of IRF-3 by two distinct signalling pathways, which required shared proteins as well as pathway-specific proteins. Our investigation revealed the presence of a BH3 domain in IRF-3, which enabled activated IRF-3 to interact with the pro-apoptotic protein Bax; this was followed by their translocation to the mitochondria and activation of the intrinsic apoptotic pathway.
Results
IRF-3 activation by cytoplasmic dsRNA signalling causes apoptosis
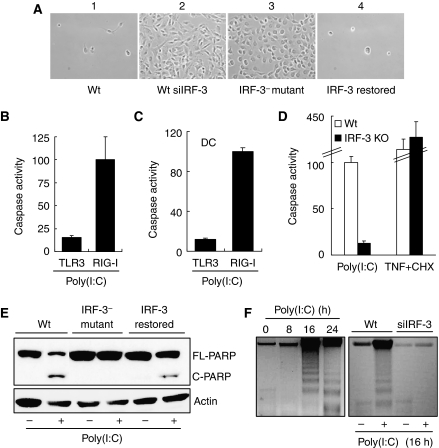
We used wild type and genetically altered human and mouse cell lines to investigate the mechanism of apoptosis caused by infection with Sendai virus (SeV), a paramyxovirus, or by transfected dsRNA, both of which activate the cytoplasmic helicases (Kato et al, 2006). Apoptosis was monitored by several complementary criteria, such as cell killing, Trypan Blue exclusion, induction of caspase 3/7 activity (Marques et al, 2005), TUNEL assay, DNA fragmentation and cleavage of PARP (Peters et al, 2008). Apoptosis caused by SeV required the presence of IRF-3 (Figure 1A); human HT1080 cells were completely killed upon SeV infection, but not when IRF-3 expression was ablated by siRNA (panels 1 and 2). As expected, P2.1, an IRF-3-deficient mutant cell line (Peters et al, 2002), was refractory to apoptosis, but when IRF-3 expression was restored in these cells they became susceptible to viral killing (panels 3 and 4). Similar results were obtained when cells were transfected with dsRNA, which activates RLH; in contrast, activation of the TLR3 pathway by dsRNA added to the culture medium (Sarkar et al, 2004) did not cause apoptosis (Figure 1B), although signalling by both TLR3 and RLH caused gene induction (Supplementary Figure S1A) and IRF-3 phosphorylation (Supplementary Figure S1B; Sarkar et al, 2004). The RLH pathway, but not the TLR3 pathway, triggered caspase activation in primary mouse dendritic cells (Figure 1C) and peritoneal macrophages (Supplementary Figure S1C), although both pathways induced genes in cells of both lineages (data not shown). Similar to IRF-3-deficient human cells, IRF-3−/− mouse fibroblasts were resistant to dsRNA-mediated apoptosis, although they were as susceptible as Wt cells to apoptosis caused by TNF (Figure 1D). IRF-3 and RLH signalling-dependent apoptotic cell death was further demonstrated by additional complementary assays such as Trypan Blue exclusion (Supplementary Figure S1D), PARP cleavage (Figure 1E), DNA fragmentation (Figure 1F) and TUNEL assay (Supplementary Figure S1E). The individual cell-based assays, such as TUNEL assay or vital dye exclusion assay, often showed that not all cells, but only a fraction, were undergoing apoptosis. This was, almost exclusively, due to inefficient transfection of the activator, dsRNA, especially into mouse primary cells. For dissecting the mechanism underlying IRF-3-mediated apoptosis, we decided to use MEF as our primary test cells because many genetically defective MEFs are easily available.
Figure 1.
IRF-3 activation by cytoplasmic dsRNA signalling triggers apoptosis. (A) HT1080-derived cell lines were infected with Sendai virus and 3 days after infection culture fields were photographed. 1, U4C cells (Wt); 2, IRF-3-knocked-down HT1080 cells (Wt-siIRF-3); 3, P2.1 cells (IRF-3− mutant); 4, IRF-3-restored P2.1 cells. (B) HT1080 cells were treated (TLR3) or transfected (RIG-I) with poly(I:C) for 16 h when caspase activation was measured. (C) Primary bone-marrow-derived dendritic cells (DCs), isolated from Wt C57Bl/6 mice, were treated (TLR3) or transfected (RIG-I) with poly(I:C); caspase activity was measured 16 h after treatment. (D) Wt and IRF-3 KO MEFs were transfected with 8 μg/ml of poly(I:C) or treated with 100 ng/ml of TNF and 1 μg/ml of cycloheximide (CHX) for 16 h, when caspase activation was measured; fold induction of caspase activity of Wt MEFs was considered as 100 and all other values were normalized to this. (E) Wt, IRF-3− mutant and IRF-3-restored cells (as described in (A)) were transfected with poly(I:C) and PARP cleavage was analysed 16 h after transfection by western blot. (F) Wt and Wt-siIRF-3 cells (as described in (A)) were transfected with poly(I:C) for the indicated time, when cellular DNA was isolated and analysed by running on 1.5% agarose gel. The left panel shows kinetics of DNA fragmentation for Wt cells. For the above experiments, 2 μg/ml of poly(I:C) was transfected (B, C, E, F) or 100 μg/ml of poly(I:C) was added into the culture medium in (B, C); fold induction of caspase activity by RIG-I signalling in response to transfected poly(I:C) was considered as 100 and other values were normalized to this (B, C).
Shared need of dsRNA-signalling proteins for both gene induction and apoptotic actions of IRF-3
To investigate the obvious possibility that IRF-3-mediated transcriptional induction of pro-apoptotic genes causes the observed apoptosis, we compared the signalling proteins that are required for the two effects. Several proteins that mediate transcriptional signalling by virus or dsRNA-activated pathways have been identified. In the next series of experiments, we investigated whether these proteins are also required for the apoptotic pathway. For this purpose, we used knock-out MEFs or other methods of selective functional disruption of the test genes. A human cell line expressing a dominant-negative mutant of RIG-I (Yoneyama et al, 2004; Kato et al, 2006) was completely resistant to apoptosis caused by SeV infection (Figure 2A), confirming that, for SeV, RIG-I was the relevant receptor. RIG-I and Mda-5 are the two major cytoplasmic RNA helicases that recognize viral or transfected RNA to trigger transcriptional signalling. Between these two, Mda-5 was not required for apoptosis, but caspase 3/7 activation and TUNEL positivity were greatly reduced in RIG-I−/− MEF (Figure 2B and C). The residual activation observed in the RIG-I−/− MEF could be mediated by Mda-5, which can substitute for RIG-I in its absence. It is important to note that the dsRNA stimulator used in these experiments was a mixture of long and short RNAs; for gene induction, the former is recognized by Mda-5, whereas the latter is recognized by RIG-I (Kato et al, 2008). The mitochondrial adaptor protein, IPS-1, was also required for apoptosis: mouse IPS-1−/− cells were resistant to dsRNA-induced apoptosis (Figure 2B and C). Expression of the HCV protease, NS3.4A, that cleaved IPS-1 (Meylan et al, 2005) blocked apoptosis by transfected dsRNA (Figure 2D) or SeV infection (Supplementary Figure S2A), and gene induction (Figure 2E) by SeV infection of human cells. Requirement of TRAF3, an essential component of RIG-I signalling, was also tested by using TRAF3−/− MEF cells. The results indicate that this protein was also essential for both dsRNA-induced apoptosis (Figure 2B) and gene induction (Supplementary Figure S2B). For mediating transcriptional signalling, IPS-1 recruits the protein kinase, TBK1, which phosphorylates and activates IRF-3. We, therefore, tested the requirement of TBK1 in causing apoptosis by using TBK1−/− MEF and human cells expressing a dominant-negative mutant of TBK1. In both cell types, dsRNA-mediated caspase activation and cell killing were severely inhibited (Figure 2B and D and Supplementary Figure S2C). As expected, the induction of the IRF-3-dependent ISG56 gene was blocked in the HT1080 cells expressing TBK1-DN (Figure 2F). These results showed that RIG-I, IPS-1, TRAF3 and TBK1 were all needed for mediating both gene induction and the apoptotic effects of IRF-3.
Figure 2.
Components of RIG-I signalling are required for apoptosis. (A) HT1080 cells (Wt) and HT1080 cells expressing a dominant-negative mutant of RIG-I (RIG-Ic) were infected with SeV; 3 days after infection culture fields were photographed. (B) Wt and KO MEF cells (as indicated) were transfected with poly(I:C), caspase activity was measured after 16 h; fold induction of caspase activity of Wt MEFs was considered as 100 and all other values were normalized to this. (C) Wt and knockout MEF cells (as indicated), grown on coverslips, were transfected with poly(I:C) for 16 h, when the cells were stained with FITC-conjugated TUNEL; TUNEL-positive and DAPI-stained cells were counted under the microscope and percent TUNEL-positive cells are presented. (D) Human cells expressing Hepatitis C virus protease NS3.4A or HT1080 cells expressing a dominant-negative mutant of TBK1 (TBK1-DN) were used for this experiment. The cells were transfected with poly(I:C), caspase activity was measured 16 h after transfection. (E) Cell lysates from NS3.4A-expressing cells (as described in (C)) were analysed for induction of P56 by SeV infection (6 h after infection) by western blot. (F) TBK1-DN (DN)-expressing cells (as described in (C)) were analysed for induction of P56 by western blot, 6 h after poly(I:C) transfection.
Identification of pathway-specific signalling proteins
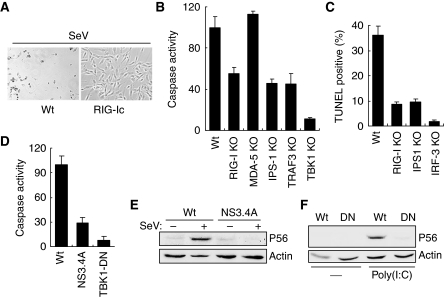
In addition to the signalling proteins discussed above, several other adaptor proteins, including TRAF2 and TRAF6, have been implicated in the dsRNA-signalling pathways. As mentioned above, TRAF3 was required for both transcriptional and apoptotic pathways (Figure 2B and Supplementary Figure S2B); we tested the roles of the other TRAF proteins for these pathways. It has been shown that TRAF2 and TRAF6 are recruited by mitochondrial adaptor protein IPS-1 upon RIG-I activation (Xu et al, 2005). We investigated their requirement by testing genetically deficient MEF cells and observed that both TRAF2 and TRAF6 proteins were essential for the IRF-3-mediated apoptotic pathway; caspase activation (Figure 3A) and TUNEL staining (Figure 3B) in response to dsRNA signalling were significantly inhibited in the deficient cells. However, neither of these two adaptor proteins was needed for IRF-3-mediated gene induction (Figure 3C and D). Taken together, the above analyses of the needs for specific signalling proteins strongly suggest that dsRNA-elicited activation of IRF-3 for gene induction and apoptosis uses partially overlapping but distinct pathways.
Figure 3.
Additional components are required for apoptotic pathway. (A) Wt, TRAF2 KO and TRAF6 KO MEFs were transfected with poly(I:C); caspase activity was measured 16 h after transfection. (B) Wt and KO MEF cells (as indicated) were transfected with poly(I:C); 16 h after transfection the cells were stained with FITC-conjugated TUNEL; TUNEL-positive and DAPI-stained cells were counted under the microscope and the percentage of positive cells are shown here. (C, D) Wt and KO MEF cells (as indicated) were transfected with poly(I:C); induction of P54 was analysed 12 h after transfection, by western blot.
dsRNA-mediated apoptosis is independent of IRF-3-driven gene induction
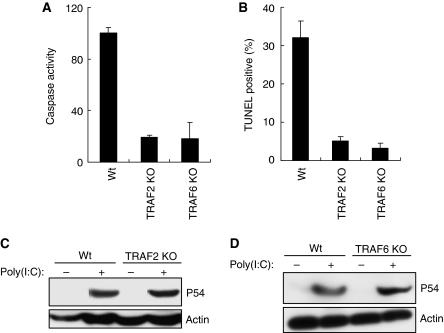
The above observation of the pathway-specific requirement of signalling components raised the intriguing possibility that the apoptotic pathway, although IRF-3 dependent, does not require gene induction. Indeed, when new gene expression was blocked by the transcriptional inhibitor, actinomycin D, or the translational inhibitor, cycloheximide (CHX), dsRNA-induced PARP cleavage remained unabated, although, as expected, gene induction was completely blocked (Figure 4A). The unexpected observation that apoptosis needed IRF-3, but not new gene expression, raised the possibility that IRF-3 might have a separate function that is distinct and separable from its role as a transcription factor. As shown in Figure 4B, the amino-terminal region of IRF-3 contains the DNA-binding domain (DBD), the nuclear localization signal (NLS) and the nuclear export signal, whereas the carboxyl-terminal domain contains the critical serine residues (S385, S386, S396 and S398), of which signal-dependent phosphorylation and the resultant IRF-3 dimerization and nuclear translocation are required for activating IRF-3 as a transcription factor (Yoneyama et al, 2004). To determine whether the same changes in the properties of IRF-3 are required for mediating apoptosis, we used mutants of IRF-3 that are deficient in driving transcription. For this purpose, we used IRF-3-deficient human cells, in which IRF-3 expression had been ablated by siRNAs that are targeted to the untranslated regions of IRF-3 mRNA. As expected, restoring Wt IRF-3 expression (by transfecting a cDNA that encoded only the translated region of human IRF-3 mRNA) in IRF-3-ablated HT1080 cells restored apoptosis and gene induction in response to dsRNA (Figure 4C and Supplementary Figures S3A and S3B) or SeV (Supplementary Figure S3C). We used this system to examine whether apoptosis was dependent on and mediated by IRF-3-induced genes and tested the apoptotic properties of IRF-3 mutants that are incapable of supporting gene transcription. One such mutant of IRF-3, 396AA, is incapable of inducing gene transcription (Supplementary Figure S3A) because it lacks two critical serine residues, 396 and 398, that are targets of signal-induced phosphorylation (Lin et al, 1998). However, this mutant was perfectly functional in the apoptosis assay (Figure 4C and Supplementary Figure S3C). Two other mutants, missing Ser385 or Ser386, were similarly effective in inducing apoptosis (Figure 4C), but not gene expression (Supplementary Figure S3B). A more drastic mutant of IRF-3, IRF-3ΔDBD (Figure 4B), which has a deletion of the entire DBD and the NLS, could also mediate apoptosis as assayed by PARP cleavage (Figure 4D) and caspase activation (Supplementary Figure S3D); it obviously could not induce gene transcription because of the defects in nuclear translocation and promoter-binding ability (Figure 4E). These results clearly showed that the transcriptional and the apoptotic functions of IRF-3 are distinct, and that the apoptotic effect is not mediated by the products of IRF-3-inducible genes.
Figure 4.
dsRNA-signalling-mediated apoptosis is independent of IRF-3-driven gene induction. (A) HT1080 cells were transfected with poly(I:C) in the presence or absence of the protein synthesis inhibitor, cycloheximide, or the RNA synthesis inhibitor, actinomycin D, for 12 h, when the cell extracts were analysed for cleaved PARP and P56 induction by western blot. (B) A schematic representation of IRF-3; DNA-binding domain (DBD), nuclear localization signal (NLS), nuclear export signal (NES) and critical phosphoacceptor sites on the C-terminus are indicated. (C) HT1080-siIRF-3 cells expressing Wt IRF-3 (Wt), IRF-3 396/98AA mutant (396AA), IRF-3 385/86AA mutant (385AA) or IRF-3 386A (a stable pool of cells) were transfected with poly(I:C); caspase activity was measured after 16 h and the cell lysates were analysed for the expression of IRF-3 mutant proteins. (D, E) HT1080-siIRF-3 cells expressing Wt or DBD-deleted IRF-3 mutant (ΔDBD, IRF-3 residues 1–112 were deleted, as in (A)) were transfected with poly(I:C) and the cell extracts were analysed for cleaved PARP (16 h after transfection) or P56 induction (6 h after transfection) by western blot.
dsRNA-induced apoptosis is accompanied by mitochondrial translocation of IRF-3
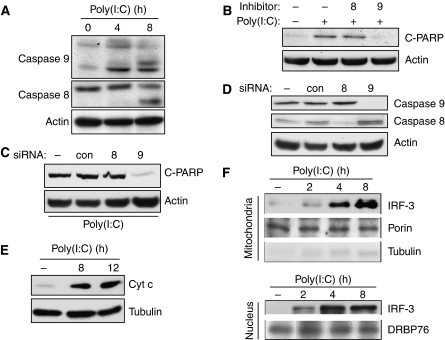
The two major branches of apoptosis, the intrinsic and the extrinsic pathways, are triggered, respectively, by the activation of caspase 9 and caspase 8 (Galluzzi et al, 2008); however, the two pathways interact, and often primary activation of one causes secondary activation of the other as well. To understand the nature of the apoptosis caused by RLH signalling, we investigated the involved pathway. Both apoptotic pathways were activated by RLH signalling, as indicated by the cleavage of both caspase 9 and caspase 8 (Figure 5A). However, an inhibitor of caspase 9, but not of caspase 8, blocked apoptosis (Figure 5B), suggesting that the IRF-3-mediated apoptosis might be mediated by the intrinsic mitochondrial pathway. Moreover, ablation of caspase 9 by the corresponding siRNA blocked the dsRNA-induced apoptosis; however, an siRNA-mediated ablation of caspase 8 did not affect the PARP cleavage induced by dsRNA (Figure 5C and D). As expected from activation of the mitochondrial apoptotic pathway, cytochrome c (Cyt c) was released from the mitochondria to the cytoplasm when RLH signalling was triggered (Figure 5E). To delineate the mechanism of IRF-3-mediated activation of the mitochondrial apoptotic pathway, we inquired whether IRF-3 itself was translocated to the mitochondria. Indeed, signal-dependent translocation of IRF-3 to the mitochondria could be demonstrated by subcellular fractionation followed by western blotting; much more IRF-3 was present in the mitochondrial and the nuclear fractions after dsRNA treatment of cells (Figure 5F). These results demonstrated that IRF-3-mediated apoptosis is accompanied by mitochondrial translocation of IRF-3 and concomitant activation of the mitochondrial apoptotic pathway.
Figure 5.
dsRNA-induced apoptosis is accompanied by mitochondrial translocation of IRF-3. (A) HT1080 cells were transfected with poly(I:C) for the indicated time, when the cell extracts were analysed for the activation of caspase 8 and caspase 9 by western blot. (B) HT1080 cells were transfected with poly(I:C) in the presence or absence of caspase inhibitors (caspase 8 inhibitor, z-IETD-FMK, and caspase 9 inhibitor, z-LEHD-FMK, as indicated); cell extracts were analysed for cleaved PARP by western blot. (C, D) HT1080 cells were transfected with siRNAs specific for caspase 8, caspase 9 or a control siRNA against cyclophilin A (Dharmacon); 48 h after transfection, the cells were transfected with poly(I:C) and analysed for PARP cleavage or, as indicated, by western blot. (E) Wt IRF-3-expressing cells were transfected with poly(I:C) for the indicated time, when the cytosolic fractions were analysed for Cyt c release by western blot. (F) Wt IRF-3-expressing cells (doubly tagged with N-terminal V5 and C-terminal Flag) were transfected with poly(I:C) for the indicated time, when the mitochondrial and nuclear fractions were isolated and analysed for the presence of IRF-3 and other proteins, as indicated, by western blot.
Bax interacts with IRF-3 and is translocated to the mitochondria
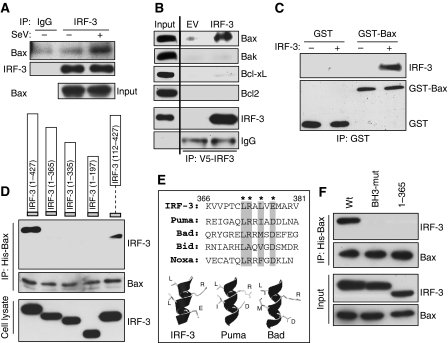
To further analyse the mechanism of IRF-3-mediated activation of the mitochondrial apoptotic pathway, we wondered whether IRF-3 interacts with any pro-apoptotic protein of the Bcl-2 family that is known to trigger this process. Surprisingly, there was a strong interaction between IRF-3 and the pro-apoptotic protein Bax, in SeV-infected cells, as revealed by the co-immunoprecipitation of the two proteins (Figure 6A). A similar interaction was observed in dsRNA-transfected cells (Supplementary Figure S4A). For these co-immunoprecipitation experiments, cells were extracted in a buffer containing the detergent Triton X-100, which sometimes promotes artifactual interactions between proteins. To eliminate this possibility we repeated the co-immunoprecipitation experiment in a buffer containing a different detergent, CHAPS, which does not create similar problems, and confirmed IRF-3/Bax interaction (Supplementary Figure S4B). The observed interaction was Bax-specific; several other members of the Bcl-2 family, including Bak, did not interact with IRF-3 (Figure 6B). To establish that the IRF-3/Bax interaction was direct, and not mediated by an intermediate protein, we purified activated IRF-3 from dsRNA-stimulated cells and used it for binding to recombinant GST-Bax. IRF-3 was pulled down by GST-Bax, but not by GST (Figure 6C). To identify the structural element in IRF-3 that mediates its interaction with Bax, several deletion mutants of IRF-3 were tested (see Figure 4B). Deletion of even a short region (residues 366–427) from the carboxyl terminal of IRF-3 eliminated the interaction (Figure 6D). In contrast, as expected (see Figure 4D), the amino-terminal region was dispensable for interaction; a mutant missing residues 1–112, encompassing the DBD still interacted with Bax. Examination of the sequence of the essential region of IRF-3 revealed the presence of a putative BH3 domain with a strong homology to the corresponding domains of a number of pro-apoptotic proteins. Molecular modelling confirmed that this domain could assume the helical structure of a BH3 domain with charged residues on one side and hydrophobic residues on the other (Figure 6E). When we introduced point mutations in IRF-3, which were expected to functionally disrupt the BH3 domain, IRF-3/Bax interaction was eliminated (Figure 6F). These results clearly showed that IRF-3 contains a BH3 domain, near its carboxyl terminus, which enables it to interact with Bax.
Figure 6.
IRF-3 interacts with Bax using a BH3-like domain on its C-terminus. (A) HT1080-siIRF-3 cells expressing Wt IRF-3 were infected with SeV; 2 h after infection IRF-3 was immunoprecipitated and analysed for Bax by western blot. (B) HT1080-siIRF-3 cells expressing Wt IRF-3 (or empty vector, EV) were transfected with poly(I:C) for 2 h and the cell lysates were immunoprecipitated with IRF-3 antibody and the immunoprecipitates were analysed for the indicated proteins by western blot. (C) IRF-3 was purified from human cells as described in Materials and methods and used for in vitro interaction assay with recombinant GST-conjugated human Bax (ABNOVA) and the reaction mixture was pulled down with GST beads and analysed for IRF-3 by western blot. (D) HT1080-siIRF-3 cells were transiently co-transfected with N-terminally V5-tagged Wt or deletion mutants of IRF-3 (as indicated) and N-terminally His-tagged Bax; 16 h after transfection cells were transfected with dsRNA for 2 h, when the cell lysates were treated with Ni-NTA and bound proteins were analysed for IRF-3 by western blot. (E) Human IRF-3 residues (366–381) were aligned with the BH3 domains of pro-apoptotic proteins (as indicated) and a molecular modelling of this domain is shown in comparison with the BH3 domains of Puma and Bad. (F) HT1080-siIRF-3 cells were co-transfected with Wt or mutants of IRF-3 (BH-3 domain mutant (BH3-mut)), wherein the critical residues (as shown by * in (E)) were mutated to Ala, (1–365) mutant, IRF-3 mutant consisting of residues 1–365, and human Bax (as in (D)); the cells were transfected with poly(I:C) for 2 h, when the cell lysates were treated with Ni-NTA and bound proteins were analysed for IRF-3 by western blot.
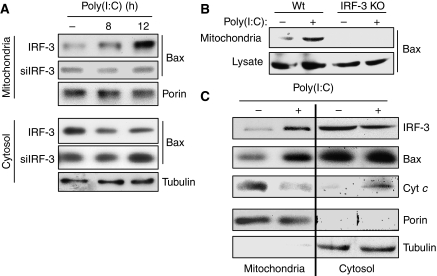
As we had seen that activated IRF-3 could translocate to the mitochondria, we wondered whether it brought along Bax to the mitochondria. Indeed, mitochondrial translocation of Bax was observed after dsRNA stimulation (Figure 7A, upper block, top panel); this was accompanied by a concomitant decrease in the cytoplasmic level of Bax (Figure 7A, lower block, top panel). Bax translocation did not occur in cells in which IRF-3 expression had been knocked down (Figure 7A, upper block, middle panel) or knocked out (Figure 7B). The cytoplasm-to-mitochondria translocation of IRF-3 and Bax and the concomitant cytoplasmic release of Cyt c in stimulated cells were confirmed by simultaneously analysing the cytoplasmic and mitochondrial fractions of stimulated and unstimulated cells. The presence and absence of porin, a mitochondrial protein, and tubulin, a cytoplasmic protein, were used as quality controls of the preparations. The abundance of IRF-3 and Bax was higher in the mitochondria, whereas that of Cyt c was higher in the cytoplasm after dsRNA treatment of the cells (Figure 7C). These experiments demonstrated that dsRNA signalling induced the interaction of the BH3 domain of IRF-3 with Bax, followed by their co-translocation to the mitochondria.
Figure 7.
IRF-3 interaction translocates Bax to the mitochondria. (A) HT1080-siIRF-3 (siIRF-3) or IRF-3-restored cells (IRF-3) were transfected with poly(I:C); the mitochondrial and the cytosolic fractions were analysed by western blot for the indicated proteins. (B) Wt and IRF-3 KO MEFs were transfected with poly(I:C); the mitochondrial fractions and the cell lysates were analysed by western blot. (C) HT1080-siRIF-3 cells expressing IRF-3 were transfected with poly(I:C); the mitochondrial and cytosolic fractions were isolated and analysed as indicated.
IRF-3 interaction activates Bax and causes Cyt c release from the mitochondria
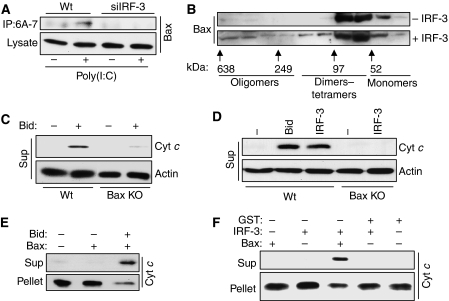
For acting as a pro-apoptotic protein, Bax needs to be activated by a conformational change, often triggered by a binding partner. We inquired whether Bax is activated in dsRNA-transfected cells by using an antibody that recognizes activated Bax (Ohtsuka et al, 2004). Indeed, dsRNA signalling caused increased activation of Bax and, more importantly, no activated Bax could be detected in cells devoid of IRF-3 (Figure 8A). Activation of Bax also causes its oligomerization in the mitochondria. We used purified Bax, mouse liver mitochondria and activated IRF-3 to measure Bax oligomerization in the presence of mitochondria in vitro. Gel filtration analysis confirmed the presence of Bax oligomers only in the presence of IRF-3 (Figure 8B). IRF-3-induced mitochondrial leakage of Cyt c was further investigated using in vitro assays. For this purpose, the mitochondria were purified from mouse liver, incubated with different sources of IRF-3 and Bax, and Cyt c release was monitored. In the first experiment, the assay was standardized by using purified activated Bid as a positive control and by providing Bax from the cytoplasmic fraction. Cyt c was released to the cytoplasm only when activated Bid was added and the cytoplasmic extract was from Wt cells, but not from Bax−/− cells (Figure 8C). Cyt c release was also observed when IRF-3 purified from stimulated cells was used instead of Bid; again the cytoplasmic extract had to come from Wt cells, but not from Bax−/− cells (Figure 8D). To develop the right conditions for these experiments, different amounts of extracts and pure proteins were titrated into the reactions (Supplementary Figure S5). Finally, similar assays were done, in the absence of cell extracts, using purified activated IRF-3 and purified recombinant GST-Bax, which were added to freshly isolated mitochondria. Cyt c was released by activated Bid (Figure 8E) and by activated IRF-3 (Figure 8F). These results showed that activated IRF-3 can trigger mitochondrial apoptosis by directly interacting with Bax, activating it and translocating it to the mitochondria.
Figure 8.
IRF-3 interaction activates Bax and causes Cyt c release from the mitochondria. (A) HT1080 (Wt) and HT1080-siIRF-3 (siIRF-3) cells were transfected with poly(I:C) for 6 h, after which the cell lysates were immunoprecipitated with Bax 6A-7 antibody and the immunoprecipitates were analysed by western blot using polyclonal Bax antibody. (B) Purified human IRF-3 was used for Bax oligomerization assay as described in Materials and methods; at the end of the reaction, the mitochondrial pellet was extracted in HE buffer containing 1% CHAPS and the extract was applied to Superdex 200 PG XK16/60 column; the fractions were analysed by western blot for Bax; column calibration was performed using the molecular weight standards kit as indicated. (C) Caspase-8-cleaved Bid (Bid, R&D Systems, 50 nM) was used for mitochondrial activation assay using the mitochondria-free cytosolic extract from Wt or Bax KO MEF cells as described in Materials and methods; at the end of the reaction, the mitochondrial pellet and the supernatant (sup) were analysed for released Cyt c, as indicated. (D) Purified activated human IRF-3 (500 nM) was used in mitochondrial activation assay using the mitochondria-free cytosolic extract from Wt or Bax KO MEF cells and analysed as described in (C). (E) Bid (50 nM) was used for in vitro Cyt c release assay using recombinant human Bax (120 nM) as described in Materials and methods; at the end of the reaction, the supernatant (sup) and the pellet were analysed for Cyt c by western blot. (F) Purified activated IRF-3 (500 nM) was used for in vitro Cyt c release assay using recombinant human Bax (120 nM) and analysed as described in (E).
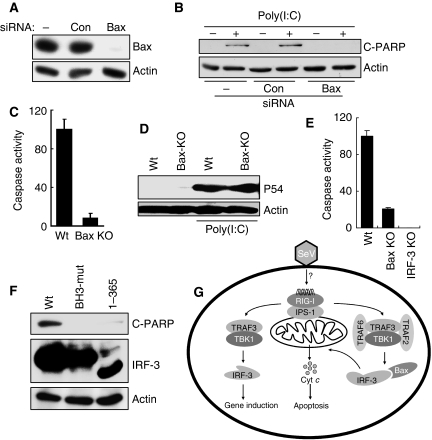
Bax is essential for dsRNA-induced apoptosis
The functional importance of Bax activation and mitochondrial translocation was tested in genetically altered cells. In human cells, in which Bax expression had been ablated by expressing the corresponding siRNA (Figure 9A), there was no dsRNA-triggered apoptosis (Figure 9B). The same was true for Bax−/− MEFs (Figure 9C), although IRF-3-mediated gene induction was normal in the absence of Bax (Figure 9D). Similarly, in mouse primary dendritic cells, apoptosis induction by SeV infection needed both IRF-3 and Bax (Figure 9E). As anticipated, IRF-3 mutants that did not interact with Bax could not mediate apoptosis (Figure 9F). These results showed that Bax and its interaction with IRF-3 were essential for mediating the apoptotic effect of IRF-3.
Figure 9.
Bax is essential for dsRNA-induced apoptosis. (A) HT1080 cells were transfected with 100 nmol/ml of Bax siRNA or a control siRNA (against cyclophilin A) (Dharmacon) using DharmaFECT 4 reagent; cell extracts were subjected to western blot analysis after 48 h. (B) RNAi-mediated knockdown of Bax was carried out by transfection of HT1080 cells as in (A); 48 h later, these cells were transfected with poly(I:C) and the cell extracts were analysed for cleaved PARP by western blot. (C) Wt and Bax KO MEFs were transfected with poly(I:C); caspase activation was measured after 16 h. (D) Wt and Bax KO MEF cells were transfected with poly(I:C) and cell extracts were analysed for P54 induction by western blot. (E) Primary bone-marrow-derived DCs were isolated from Wt, IRF-3 KO and Bax KO mice and infected with SeV (at MOI: 10) for 72 h, when the cell lysates were analysed for caspase activity. (F) HT1080-siIRF-3 cells expressing Wt or mutants of IRF-3 (as in Figure 6D; a stable pool was generated) were transfected with poly(I:C) for 16 h, when the cell lysates were analysed for PARP cleavage. (G) A model showing the transcriptional and apoptotic pathways mediated by dual actions of IRF-3.
Discussion
The results presented above led us to propose a model for the dual actions of IRF-3 in infected cells (Figure 9G). It is known that IRF-3 activation by virus infection causes apoptosis. As IRF-3 induces the transcription of several pro-apoptotic genes, such as TRAIL, Noxa, etc. (Kirshner et al, 2005; Goubau et al, 2009), it was assumed that the observed apoptosis was mediated exclusively by the products of these genes. This notion was reinforced by the observation that ectopic expression of a constitutively active mutant of IRF-3 caused apoptosis as well (Heylbroeck et al, 2000; Weaver et al, 2001); however, the action of the mutant probably does not reflect the physiological situation, because signal-dependent activation and action of IRF-3 is transient in nature and of short duration, whereas the constitutively active mutant transcribes the target genes continuously (Lin et al, 1999). It is more likely, as demonstrated here, that at least one pro-apoptotic effect of IRF-3 is distinct from its transcriptional effect, but for both activities of IRF-3, its activation by the RLH-signalling pathway is required. Added support for the two-branch model comes from the observation that IRF-3 activation by the alternative dsRNA receptor, TLR3, did not cause apoptosis, although very similar set of genes are induced by the two receptors (Elco et al, 2005). Several common proteins were required for activating IRF-3 in both transcriptional and apoptotic pathways. For causing apoptosis, the relevant signalling receptor is, one of the RLH proteins, because the absence of another dsRNA sensor, PKR (Der et al, 1997), did not diminish the apoptotic activity (data not shown); on the other hand, blocking RIG-I signalling protected the infected cells from apoptosis. IPS-1, TRAF3 and TBK1 are common components of both apoptotic and transcriptional pathways, but the apoptotic pathway requires TRAF2 and TRAF6 in addition.
For the apoptotic pathway, the exact biochemical nature of activation of IRF-3 remains to be delineated. However, it is clear that apoptotic activation does not require its DNA-binding activity, its ability to translocate to the nucleus and its phosphorylation at serine residues 385, 386, 396 and 398, properties all of which are necessary for its transcriptional functions. The observed need of TBK1 suggests that induced phosphorylation of different Ser/Thr residues causes the apoptotic conformational change of IRF-3, allows its binding to Bax and targets it to the mitochondria. Identification of such residues is technically difficult because IRF-3 is phosphorylated at many sites, even in unstimulated cells. We speculate that the activation process causes a conformational change of IRF-3 that makes the BH3 domain accessible to Bax. The BH3 domain of IRF-3, identified in this study, was necessary for the interaction with Bax, a Bcl-2 family member. However, activated IRF-3 did not bind to several other Bcl2-family proteins; moreover, such member-specific binding of BH3-only proteins is not unprecedented. The apoptotic effect of IRF-3 was mediated by its binding and activation of Bax and co-translocation of the two proteins to the mitochondria. Translocation of activated Bax to the mitochondria triggered, as expected, the mitochondrial apoptotic pathway by activating caspase 9 and releasing Cyt c to the cytoplasm. In the absence of IRF-3, or its BH3 domain, the mitochondrial translocation of Bax and apoptosis were blocked. As expected, in the absence of Bax, apoptosis did not happen. Using in vitro assays, we established that IRF-3 directly bound to Bax and the two proteins were both necessary and sufficient to cause the release of Cyt c from the mitochondria. These observations provide strong genetic and biochemical evidence for the proposed model of IRF-3-mediated apoptosis.
Our findings help explain the observation that virus-infected IRF-3-deficient mice have significantly decreased survival, despite IFN induction being largely normal (Honda et al, 2005). Thus, in virus-infected cells, IRF-3 is essential for triggering a direct apoptotic response in addition to inducing the synthesis of many antiviral proteins. It remains to be seen whether other members of the IRF family share the dual-function paradigm of IRF-3. In this respect, it is interesting to note that although IRF-7 is a major regulator of IFN production (Honda et al, 2005), we observed that IRF-7 expression could not compensate for the absence of IRF-3 in the apoptotic response (data not shown).
Materials and methods
Cells and reagents
HT1080, U4C, P2.1, P2.1.17, HT1080/RIG-Ic and HT1080/siIRF-3 cells have been described before (Peters et al, 2002, 2008; Matikainen et al, 2006). RIG-I−/−, Mda-5−/−, IPS-1−/−, TRAF2−/−, TRAF6−/−, TBK1−/−, IRF-3−/− MEFs and their corresponding Wt cells were kind gifts from Michael Gale (University of Washington, Seattle, WA), Michael Diamond and Marco Colonna (Washington University School of Medicine, St Louis, MO), Xiaxioa Li, Nywana Sizemore, George Stark and Robert Silverman(Cleveland Clinic). These cells were grown in DMEM supplemented with 10% Fetal Bovine Serum (FBS). NS3.4A expressing cell line was a kind gift from Michael Gale (University of Washington). Primary peritoneal macrophages and bone-marrow-derived dendritic cells were isolated from Wt and Bax−/− mice using the protocol described elsewhere (Hamilton and Major, 1996; Fensterl et al, 2008). Antibody against human IRF-3 was a gift from Michael David (University of California, San Diego, CA); phospho-IRF-3 S396 was a gift from John Hiscott (Lady Davis Institute for Medical Research, Canada); SeV C protein was a gift from Atsushi Kato (National Institute of Infectious Diseases, Tokyo, Japan); human Bax plasmid was a gift from Dr Shigemi Matsuyama (CWRU, Cleveland); P54 and P56 were raised in our laboratory; PARP was from Cell Signaling; Bax and Cyt c were from Santa Cruz; Bax-6A-7 was from BD Pharmingen; porin and tubulin were from Calbiochem; β-actin was from Sigma; and GAPDH was from Chemicon International. Poly(I:C) and CHX were purchased from GE Healthcare and Sigma Aldrich, respectively. FuGENE 6 and Lipofectamine were purchased from Roche Diagnostics (Indianapolis, IN) and Invitrogen, respectively. Inhibitors of caspase 8 and caspase 9 were obtained from Calbiochem. Recombinant full-length human Bax protein was obtained from Abnova, recombinant human Bid (caspase 8 cleaved) from R&D Systems and TUNEL staining reagent from Promega.
Poly(I:C) transfections
dsRNA stocks were prepared by re-suspending poly(I):poly(C) in PBS and shearing RNA by passing through a 26-gauge needle. Poly (I:C) was transfected using FuGENE 6 reagent in all experiments according to the protocols provided by the manufacturer. Briefly, 2 μg of poly(I:C) per 3 μl of FuGENE was incubated in 100 μl of serum-free DMEM for 15–30 min before being added to the supernatant of cells containing 10% of FBS.
Virus infections
SeV (Cantell strain) was obtained from Charles Rivers SPAFAS (Preston, CT). For infections, cells were washed two times with virus infection media (DMEM supplemented with 2% FBS) and then placed in a minimal amount of virus infection media. SeV was added at a concentration of 80 HAU/ml. Cells were incubated with virus for 1 h with gentle agitation every 10 min. The virus was removed, and cells were washed twice with complete media. The cells were placed in complete media until they were harvested (Peters et al, 2008).
Retrovirus transduction
Phoenix cells were transfected with pBabePuro-IRF3 plasmids using Lipofectamine Plus reagent (Invitrogen). The cell supernatant was collected after 24 h, filtered through a 0.22-μM filter and combined with polybrene (4 μg/ml) (Sigma), and used to infect the target cells. After three rounds of infections, the target cells were grown in a medium containing an antibiotic for selection.
Caspase 3/7 activity
Cells were grown in black-walled 96-well plates with a transparent bottom and treated as indicated. All treatments were performed in biological triplicates and the results shown represent the average and s.d. of the three independent wells. Caspase activity was measured directly from the 96-well plate using the Apo-ONE Homogeneous caspase 3/7 assay, according to the protocols provided by the manufacturer (Promega, Madison, WI). Fold induction of caspase activity was obtained by dividing the caspase activity in treated samples by the value obtained from untreated cells. Induction of caspase activity for Wt cells was considered as 100 and all other values were normalized to this. Results are representative of at least two independent experiments. It is noteworthy that we and others have found a good correlation between the levels of caspase 3 activity and other methods used to measure apoptosis, such as DNA fragmentation, TUNEL assay and PARP cleavage (Figures 1D–F, 2C and 3B; Marques et al, 2005). We therefore chose to use caspase 3/7 activity as the standard method to quantitatively measure apoptosis. For measuring the caspase activation of the primary bone-marrow-derived DCs from IRF-3 and Bax KO mice, the cells were infected by SeV, and the cell lysates were used for testing the caspase activity using the manufacturer's instructions and the data represented as indicated in the figure legend (Figure 9E).
Cell fractionation
Mitochondrial and cytosolic fractions were isolated using the Mitochondria Isolation Kit (Pierce Biotechnology) following the manufacturer's instructions. Isolated mitochondrial fractions were washed with PBS and extracted in lysis buffer for analysis. The mitochondrial fractions were isolated from livers of Wt C57Bl/6 mice using the above method and were washed twice with HE buffer (10 mM HEPES, pH 7.4, 1 mM EDTA) before being used in the reactions. Nuclear fractions were isolated from the unstimulated and the dsRNA-transfected cells by procedures described earlier (Sarkar et al, 2004).
siRNA experiments
HT1080-siIRF-3 cells were generated as described previously (Peters et al, 2008) and were routinely cleaned by transfecting poly(I:C) for any revertant cells. The siRNAs against caspase 8, caspase 9, Bax and cyclophillin A (100 nmol/ml, Dharmacon) were transfected using DharmaFECT 4 reagent for 48 h when the cells were transfected with poly(I:C) as indicated.
Western blot and immunoprecipitation
Western blots were performed as described elsewhere (Marques et al, 2005; Peters et al, 2008). Briefly, cells were lysed in 50 mM Tris buffer, pH 7.4 containing 150 mM of NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 10 mM of NaF, 10 mM of β-glycerophosphate, 5 mM sodium pyrophosphate and protease/phosphatase inhibitors; the total protein extracts were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by western blot. For immunoprecipitation, cell lysates were pre-cleared using protein A–G agarose (Santa Cruz) beads and then reacted with V5-agarose beads (Sigma) for 16 h. The beads were washed in lysis buffer and analysed by SDS–PAGE and western blot. For the reverse IP reaction using Bax antibody, an ExactaCruz system (Santa Cruz) was used using the manufacturer's instructions. For immunoprecipitation in CHAPS-containing lysis buffer, the cells were lysed in 10 mM Hepes pH 7.4, 150 mM NaCl and 1% CHAPS-containing buffer, and immunoprecipitated using Bax 6A-7 monoclonal antibody (BD Pharmingen) or IRF-3 antibody and protein A–G agarose. The beads were then washed with lysis buffer and analysed by SDS–PAGE and western blot using a polyclonal Bax antibody.
For mapping the IRF-3/Bax interaction, the cells were co-transfected with epitope-tagged IRF-3 mutants and Bax; the Bax protein was pulled down by Ni-NTA beads in the presence of 25 μM of immidazole and the immunoprecipitates were washed in the presence of 50 μM immidazole and analysed by western blot. For the molecular modelling of BH3 domains, the amino-acid sequences of IRF-3, Puma and Bad (as shown in Figure 6E) were analysed using the program Swiss PDB Viewer and the critical residues were mutated to Ala as described before (Ghosh et al, 2001).
In vitro interaction, Cyt C release and oligomerization assays
Human IRF-3 was purified from P2.1 cells expressing doubly-tagged (N-His and C-Flag) IRF-3. The cells were transfected with poly(I:C) for 12 h before purification of IRF-3. Briefly, the cell lysate was first incubated with Ni beads and bound proteins were eluted using immidazole; the resultant eluate was incubated with Flag beads and eluted in the presence of Flag peptide in HE buffer. The final preparation was used for the following experiments. For the in vitro interaction experiment, the purified IRF-3 was incubated with recombinant Bax in mitochondrial reaction buffer (HE buffer containing 200 mM mannitol, 68 mM sucrose, 10 mM HEPES-KOH pH 7.4, 100 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.1% BSA, 5 mM succinate, 2 mM ATP, 10 μM phosphocreatine, 10 μg/ml creatine kinase and protease inhibitors) at 37 °C for 60 min, when the reaction mixture was pulled down using GST beads and analysed by SDS–PAGE.
Mitochondria were freshly isolated from the liver of 6-week-old Wt C57Bl/6 mice using the techniques described above; mitochondria-free cytosolic extracts were isolated from Wt or Bax−/− MEF cells and used for the following reactions as indicated. The indicated proteins were incubated with mitochondria in the mitochondrial reaction buffer at 37 °C for 60 min. Samples were centrifuged at 5500 g for 15 min and the supernatants and pellets (washed with mitochondrial buffer) were analysed by SDS–PAGE.
For Bax oligomerization assay, isolated mitochondria were incubated with recombinant Bax and IRF3 as described above. Membranes were pelleted by centrifugation at 5500 g for 15 min and washed with the mitochondrial buffer. Proteins were extracted from the mitochondrial pellet in HE buffer containing 1% CHAPS. Extracts were centrifuged at 18 000 g for 30 min and the soluble material was applied to a Superdex 200 PG XK16/60 column (GE Life Sciences) equilibrated in low-salt buffer (20 mM HEPES-KOH pH 7.4, 250 mM KCl, 0.25 mM EDTA, 0.25 mM DTTand 10% glycerol) and eluted isocratically; 3.6-ml fractions were collected, precipitated with trichloroacetic acid, and analysed by western blot for Bax. Column calibration was performed in the same buffer using the molecular weight standards kit for gel filtration, in the 12 000–200 000 range (Sigma).
Supplementary Material
Supplementary Figures S1–S5
Review Process File
Acknowledgments
We thank Shizuo Akira, Michael Diamond, Michael Knudson, Marco Colonna, Michael Gale, Xiaxioa Li, Nywana Sizemore, George Stark and Robert Silverman for providing knock-out cells and John Hiscott, Michael David and Shigemi Matsuyama for providing the important reagents. We also thank Christine White for assistance with DC isolation. This work was supported by National Institutes of Health grants AI073303 (GCS) and CA062220 (GCS and BRGW).
Author contributions: SC and JTM planned and performed the experiments and participated in writing the paper; KLP, MY, KS and AD performed the experiments; BRGW designed the experiments and edited the paper; GCS planned the experiments, interpreted the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738 [DOI] [PubMed] [Google Scholar]
- Barber GN (2001) Host defense, viruses and apoptosis. Cell Death Differ 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Der SD, Yang YL, Weissmann C, Williams BR (1997) A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA 94: 3279–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G (2002) IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251–263 [DOI] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Williams BR, Sen GC (2005) Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol 79: 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, White CL, Yamashita M, Sen GC (2008) Novel characteristics of the function and induction of murine p56 family proteins. J Virol 82: 11045–11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4: 491–496 [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G (2008) Viral control of mitochondrial apoptosis. PLoS Pathog 4: e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Sarkar SN, Rowe TM, Sen GC (2001) A specific isozyme of 2′-5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J Biol Chem 276: 25447–25455 [DOI] [PubMed] [Google Scholar]
- Goubau D, Romieu-Mourez R, Solis M, Hernandez E, Mesplede T, Lin R, Leaman D, Hiscott J (2009) Transcriptional re-programming of primary macrophages reveals distinct apoptotic and anti-tumoral functions of IRF-3 and IRF-7. Eur J Immunol 39: 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TA, Major JA (1996) Oxidized LDL potentiates LPS-induced transcription of the chemokine KC gene. J Leukoc Biol 59: 940–947 [DOI] [PubMed] [Google Scholar]
- Heylbroeck C, Balachandran S, Servant MJ, DeLuca C, Barber GN, Lin R, Hiscott J (2000) The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J Virol 74: 3781–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434: 772–777 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2006) Innate immune recognition of viral infection. Nat Immunol 7: 131–137 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Kirshner JR, Karpova AY, Kops M, Howley PM (2005) Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J Virol 79: 9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Genin P, Pitha PM, Hiscott J (1999) Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol 19: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18: 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Rebouillat D, Ramana CV, Murakami J, Hill JE, Gudkov A, Silverman RH, Stark GR, Williams BR (2005) Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J Virol 79: 11105–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matikainen S, Siren J, Tissari J, Veckman V, Pirhonen J, Severa M, Sun Q, Lin R, Meri S, Uze G, Hiscott J, Julkunen I (2006) Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J Virol 80: 3515–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW (2004) ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol 6: 121–128 [DOI] [PubMed] [Google Scholar]
- Peters K, Chattopadhyay S, Sen GC (2008) IRF-3 activation by sendai virus infection is required for cellular apoptosis and avoidance of persistence. J Virol 82: 3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KL, Smith HL, Stark GR, Sen GC (2002) IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc Natl Acad Sci USA 99: 6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SN, Peters KL, Elco CP, Sakamoto S, Pal S, Sen GC (2004) Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat Struct Mol Biol 11: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Weaver BK, Ando O, Kumar KP, Reich NC (2001) Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. Faseb J 15: 501–515 [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301: 640–643 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S5
Review Process File