Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species (original) (raw)
Abstract
Mammalian preimplantation embryonic development (PED) is thought to be governed by highly conserved processes. While it had been suggested that some plasticity of conserved signaling networks exists among different mammalian species, it was not known to what extent modulation of the genomes and the regulatory proteins could “rewire” the gene regulatory networks (GRN) that control PED. We therefore generated global transcriptional profiles from three mammalian species (human, mouse, and bovine) at representative stages of PED, including: zygote, two-cell, four-cell, eight-cell, 16-cell, morula and blastocyst. Coexpression network analysis suggested that 40.2% orthologous gene triplets exhibited different expression patterns among these species. Combining the expression data with genomic sequences and the ChIP-seq data of 16 transcription regulators, we observed two classes of genomic changes that contributed to interspecies expression difference, including single nucleotide mutations leading to turnover of transcription factor binding sites, and insertion of _cis_-regulatory modules (CRMs) by transposons. About 10% of transposons are estimated to carry CRMs, which may drive species-specific gene expression. The two classes of genomic changes act in concert to drive mouse-specific expression of MTF2, which links POU5F1/NANOG to NOTCH signaling. We reconstructed the transition of the GRN structures as a function of time during PED. A comparison of the GRN transition processes among the three species suggested that in the bovine system, POU5F1's interacting partner SOX2 may be replaced by HMGB1 (a TF sharing the same DNA binding domain with SOX2), resulting in rewiring of GRN by a trans change.
Much of the information on evolution of gene regulatory networks (GRN) was generated from studies in invertebrates (Erwin and Davidson 2009). Comparison of fungi species revealed evolution of GRNs at both cis and trans levels (Scannell and Wolfe 2004). Invertebrate GRNs can be “rewired” at the “_cis_” level through changes in genomic regions that regulate gene transcription, or _cis_-regulatory modules (CRM). CRM can gain or lose transcription factor (TF) binding sites (TFBSs) by single nucleotide mutations, insertions and deletions of neutral sequence or TFBSs (Erwin and Davidson 2009). It has been argued that at the cis level, the transcription regulatory apparatus is a “programmable” computing machine whose function can be modulated through placements of TFBSs in the regulatory region of any gene (Buchler et al. 2003; Gertz et al. 2009). A notable advantage of encoding combinatorial control in the regulatory region, as opposed to in the regulatory proteins, is evolvability (Ptashne and Gann 2002): Unlike regulatory proteins, each _cis_-regulatory region controls the expression of a given gene and hence can be programmed with minimal pleiotropic effects. Contrary to this argument, changes to the TF that are produced by the cell (“_trans_” changes) appear to be widely tolerated by Escherichia coli and may even confer evolutionary advantages (Isalan et al. 2008). Moreover, several yeast species implement compensating cis and trans changes to achieve a conserved regulatory logic (Tsong et al. 2006). This study attempted to add vertebrate data to address these arguments by analyzing a relatively conserved developmental process in three mammalian species.
Mammalian preimplantation embryonic development (PED) encompasses the period from fertilization to implantation of the embryo in the endometrial lining of the uterus. Just after fertilization, the transcriptome of the embryo is comprised of maternally deposited transcripts. After several cell divisions, maternal transcripts are specifically degraded and are replaced by zygotic transcripts produced by the new diploid nucleus containing both maternal and paternal genes. This transition is termed zygote genome activation (ZGA). The timing of ZGA and maternal transcript degradation varies among species, but in humans and bovines it reportedly occurs between the four- and eight-cell stages of development, while in mouse, this transition occurs between the one- and two-cell stages of development, consistent with an overall accelerated pace of murine embryonic development (Braude et al. 1988; Kanka et al. 2003; Adjaye et al. 2007; Kues et al. 2008).
Another major event during PED is the differentiation and specialization of cells. Immediately after ZGA, embryos undergo compaction, a process during which dividing embryonic cells become a tightly organized cell mass with an indistinguishable membrane. In most mammalian embryos, compaction is usually observed between the eight- and 16-cell stages. After compaction, the embryo is named the morula, which becomes the blastocyst through formation of an internal cavity and further cellular differentiation. At least two cell types can be distinguished in the blastocyst: an inner cell mass (ICM) and an outer layer of cells termed the trophectoderm. The ICM contributes to all three germ layers present in the mature adult (known as pluripotency). Embryonic stem (ES) cells were derived from ICM, and ES cells have been used as surrogates to analyze the molecular mechanisms in ICM and preimplantation embryos (Ogawa et al. 2008). The progression of PED is highly conserved among mammals: All progress through the same morphologic stages. Perhaps the most marked difference is the amount of time spent at each stage. The other notable interspecies differences appear after the blastocyst stage, including that bovine blastocysts initiate the gastrulation process before implantation, and the formation and functions of the placenta and yolk sac (Gossler 1992). In this study, we chose the highly conserved PED stages, i.e., from zygote to blastocyst stages for cross-species GRN comparison.
Results
Dynamic gene expression landscapes of preimplantation development in three species
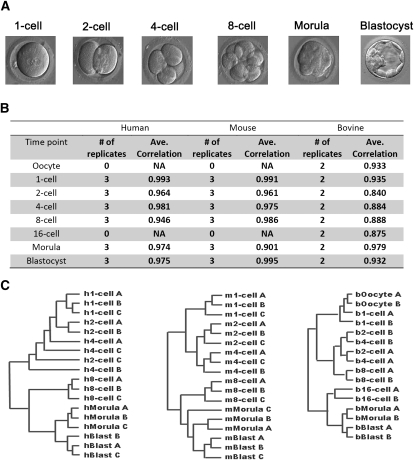
Human, mouse, and bovine embryos were collected at representative stages of PED, including oocyte (bovine only), one-, two-, four-, eight-, 16-cell (bovine only), morula, and blastocyst. Developmental stage was determined by direct microscopic visualization of each collected embryo (Fig. 1A). RNA from each stage was collected and analyzed with species–specific Affymetrix GeneChips. Biological replicates of the same developmental stage exhibited strong correlations, which were comparable to the sample correlations in previous bovine and mouse studies (Fig. 1B; Hamatani et al. 2004; Wang et al. 2004; Kues et al. 2008). Hierarchical clustering analyses revealed global transcriptional similarities among the biological replicates of the same developmental stages together, with a few exceptions of the samples at neighboring stages (Fig. 1C). The hierarchical order of all the samples almost perfectly reflected the relative time differences of all the developmental stages. The largest changes of expression were observed between four- and eight-cell stages in human and mouse and between eight- and 16-cell stages in the bovine system, in keeping with the previously reported timing of ZGA in these species.
Figure 1.
Summary of preimplantation embryonic development. (A) Photomicrographs of human embryos from representative stages of preimplantation development. (B) Table of all the transcriptional arrays performed with pertinent collection parameters. The average Pearson correlations are listed for every experimental group. (C) Hierarchical clustering of gene expression in human, mouse, and bovine embryos during representative stages of PED. This clustering is based on 8479 (human), 7093 (mouse), and 9474 (bovine) informative probe sets. Adjacent stages tend to exhibit similar transcriptional profiles, except for those stages near the time of zygote genome activation. Later PED stages, characterized by expression of genes important for cellular differentiation, segregate into clusters distinct from earlier stages in all three species.
In order to explore the conservations and changes of gene expression patterns across species, we developed a Bayesian clustering method to simultaneously cluster the timecourse expression data and automatically determine the cluster number (Methods). Nine clusters were generated for 5941 orthologous gene triplets (including 1036 TF triplets) (Supplemental Fig. S1). Of these orthologous triplets, 2122 (35.7%, 383 TF triplets) fell into clusters with consistent expression patterns in all the three species, and the remaining 3290 orthologous triplets (55.4%, 444 TF triplets) showed at least two different expression patterns among these species. The large differences in the TF expression patterns suggested that there might be a substantial interspecies difference of GRN structures. To test if this result is sensitive to the input genes, we repeated this analysis with the 927 orthologous gene triplets (including 371 TF triplets) that exhibited significantly variable expression (ANOVA _P_-value < 0.05) during PED in at least two of the three species. Among them, 383 (40.2%) orthologous triplets (149 TF triplets) showed different expression patterns among these species, consistent to the estimated large-scale GRN changes.
The three mammals share more maternally deposited than ZGA-activated RNAs
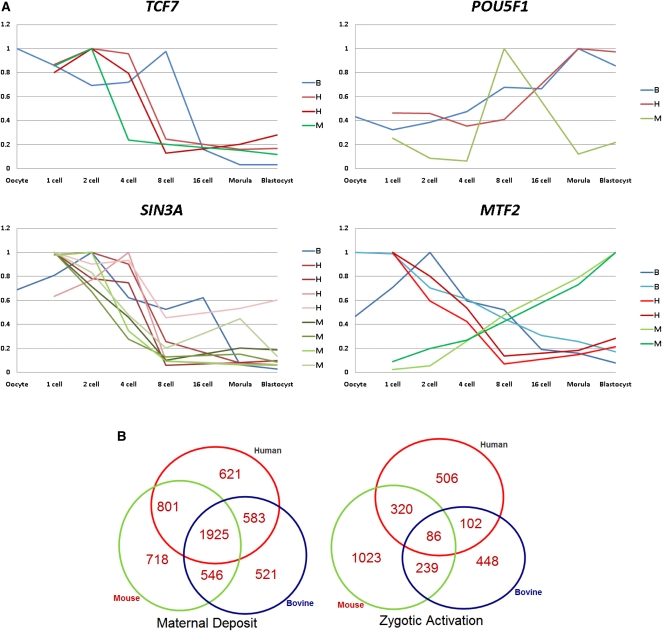
The transition from predominance of maternal transcripts to zygotic transcripts is a critical aspect of PED. We identified maternally deposited (MT) and ZGA-activated transcripts (ZGAT) in each species based on trends in transcript levels (Fig. 2B). Comparing the conservation levels of the human MTs vs. human ZGAT, we found 69.4% human MT and 40.0% human ZGAT were shared in mouse (ratio = 69.4/40.0 = 1.74, _P_-value < 10−10); 63.8% hMT and 18.5% human ZGAT were shared in bovine (ratio = 3.45, _P_-value < 10−20); 49.0% human MT and 8.5% human ZGAT were shared in both mouse and bovine (ratio = 5.76, _P_-value < 10−20). These data showed that human MT is more conserved than human ZGAT in mammals (Fig. 2A,B). Comparisons benchmarked against mouse or bovine MT and ZGAT generated similar results. Changing the algorithmic thresholds for detection of MT and ZGAT did not affect this conclusion (Supplemental Fig. S2A).
Figure 2.
Cross-species comparison of gene expression. (A) Pictorial description of trends in transcription of a subset of genes with distinct roles during PED. Human, mouse, and bovine probe sets are depicted by red, green, and blue curves, respectively. When a gene is targeted by multiple probe sets, all probe sets are shown. The maternally deposited transcript for TCF7 is degraded at two-, four-, and eight-cell stages in mouse, human, and bovine embryos, respectively. SIN3A is highly abundant in zygotes, and exhibits consistent decreasing expression patterns in all three species. POU5F1 exhibits a consistent up-regulation in all three species, however its expression peaked at the eight-cell stage in mouse and at the morula stage in human and bovine. MTF2 transcript exhibits a reversed expression pattern in mouse and in the other two species, as reproduced by two probe sets in each species. (B) Conserved and species-specific MT and ZGAT. A percentage of MT, 33.7%, is shared in all three species, whereas only 3.2% of the ZGAT is shared in all three species. MT has a 10.7-fold larger chance of being shared across species than ZGAT (_P_-value < 10−30).
Rewiring of ZGA genes by TFBS usage
We hypothesized that the large-scale changes of ZGAT across species is associated with changes of TFBSs of TFs that activate gene expression in morulas and ICM. To test this hypothesis, we intersected human and mouse ZGA genes with the genes that are bound by POU5F1, SOX2, and NANOG in human ES cells (Lister et al. 2009) and mouse ES cells (Chen et al. 2008) (Methods). POU5F1, SOX2, and NANOG are hereafter referred to as ZGA TFs. Among the ZGA genes that are conserved, human-specific and mouse-specific, 58, 64, and 335 of them are bound by ZGA TFs in a conserved, human-specific and mouse-specific manner (Supplemental Fig. S2B). Surprisingly, the TFBSs of ZGA TFs are not enriched in the conserved ZGA genes (χ2 test with continuity correction _P_-value > 0.31). Instead, mouse-specific ZGA genes are enriched with mouse-specific binding of ZGA TFs (_P_-value < 2 × 10−6). The large changes of ZGATs and the TFBSs of ZGA TFs across species may suggest a large scale of GRN rewring events during mammalian PED. We note that there can be other ZGA TFs besides POU5F1, SOX2, and NANOG, and therefore this analysis is simply a primitive attempt to estimate the extent of rewiring events associated with ZGA.
Transposons contribute to species-specific gene expression
There is a significant overlap between transposon-associated elements and TFBS-rich regions in mammalian genomes. One notable example includes association between the TP53 binding sites and transposons in the human genome (Wang et al. 2007; Bourque et al. 2008), and one of the TP53 binding transposons was stabilized in humans (Wang et al. 2007); however, it is yet to be determined to what extent the transcriptome is modulated by transposons. Therefore, we attempted to assess the overall impact of murine-specific transposons on the PED transcriptome.
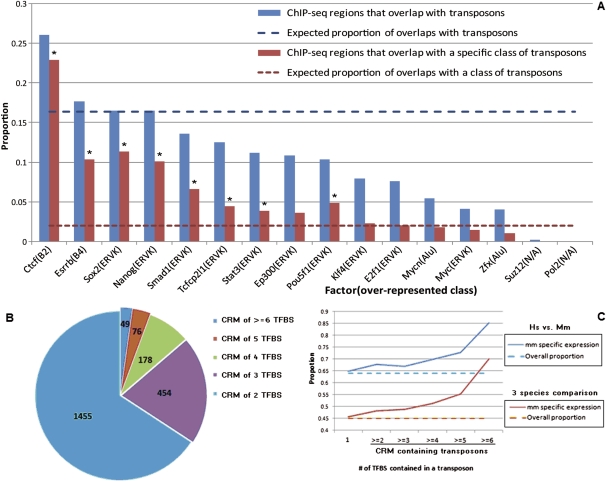
Previously reported chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) mapping of mouse ES cells identified binding sites of 16 transcriptional regulators to the mouse genome (Chen et al. 2008). Based on this information, we identified the binding regions that resided in murine-specific transposable regions (Fig. 3A). CTCF-binding regions exhibited the largest number of overlap with known transposons (10,313 overlapping regions, representing 26.0% of total CTCF-binding regions), among which 9074 (22.9%) overlapped with the B2 family of transposons. In addition to the CTCF-binding regions, the binding regions for the following factors exhibited nontrivial overlaps (>10% of the binding regions of a factor) with transposons: ESRRB, SOX2, NANOG, SMAD1, TCFCP2l1, STAT3, EP300, and POU5F1. Transposon families B2, ERVK, and B4 were most strongly associated with the binding regions of these transcription regulators. This suggests that these families of transposable elements contain the DNA binding motifs of the listed TFs. On the contrary, 10 (0.2%) and 1 (0%) of Suz12 and RNA polymerase II (Pol II) binding regions overlapped with transposons, far below the expected amount of overlaps (16.3%), suggesting that the insertion of transposable elements to the binding regions may negatively affect the function of these factors.
Figure 3.
CRM-containing transposons. (A) Proportions of mouse ChIP-seq detected binding regions that overlap with transposons (blue) and a specific class of transposons (red). An asterisk (*) denotes a specific class of transposons that is significantly overrepresented in the ChIP-seq detected regions. (B) Numbers of CRM-containing transposons detected by ChIP-seq. (C) The percentage of transposon-flanked genes with mouse-specific expression (solid lines) and the overall percentage of genes with mouse-specific expression (dashed lines).
A number of the TFs included in the original ChIP-seq analysis (Chen et al. 2008), especially NANOG and POU5F1, exhibited increase in expression after ZGA. In addition, these TFs are known to specify ICM expression of their target genes. Therefore, if the binding of these TFs to the murine-specific transposons manifests functional consequences to the transcriptome, we would expect to see mouse-specific expression of the genes near these transposons. Among the 7450 orthologous gene triplets in the PED data set, 3369 (45.2%) exhibited mouse-specific expression patterns. Within the 3066 gene triplets whose mouse gene had a nearby transcription-factor-bound transposon, 1403 (45.8%) exhibited mouse-specific expression patterns. The genes near TF-bound transposons did not exhibit a significantly larger percentage of mouse specifically expressed genes, suggesting that the majority of binding events between the transcription regulators to the transposons do not manifest functional consequences. Therefore, the majority of mutants generated by the insertion of TFBS-carrying transposons are likely to be selectively neutral (Kimura 1991).
We hypothesized that the transposons containing binding regions of multiple TFs (CRM-containing transposons) were more likely to affect the expression of neighboring genes. We tested this hypothesis by first classifying the transposons by the number of transcription regulators bound to them and then analyzing the effect on gene expression of each class of transposons (Fig. 3B). A total of 20,784 murine transcription-regulator bound transposons were identified, among which 2212 (10.2%) transposons were bound with at least two transcription regulators. For each class of transposons, we identified the subset whose nearest genes exhibited mouse-specific expression patterns (Methods). First, we compared mouse PED to human PED, ignoring the bovine expression data. For the transposons bound by one transcription regulator, 65% of the nearest mouse genes exhibited mouse-specific expression changes, which is comparable to the total percentage of genes that exhibit mouse-specific expression changes (64%). For the transposons bound by at least two (and up to six) factors, the percentages of the nearest genes with mouse-specific expression changes increased consistently from 67.8% to 85.2% (Fig. 3C). Second, we compared mouse PED to both human PED and bovine PED. From the single-factor-bound transposons to the six-factor-bound transposons, the percentages of the nearby genes that exhibited mouse-specific expression increased consistently from 45.1% to 70%. These data suggest that species-specific transposons bound by a single TF generally do not change gene expression, whereas CRM-carrying transposons bound with multiple TFs are statistically associated with species–specific expression. About 10% of the transposons in our analyses carry CRMs.
Gain of functional TFBSs and CRM by species–specific transposons
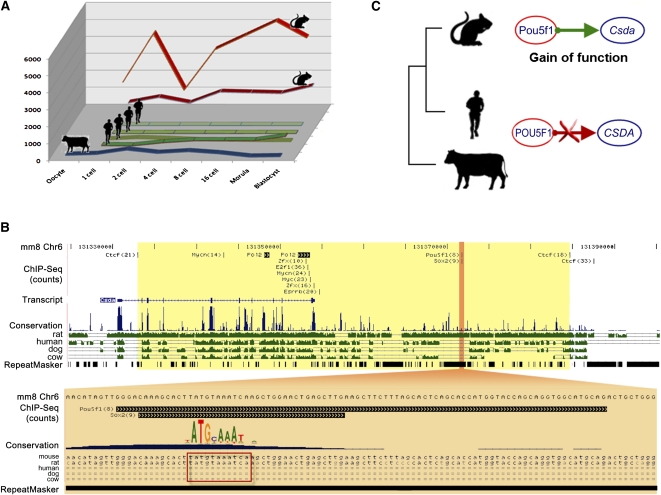
To confirm the functional importance of the transposons to gene regulation, we further characterized two murine-specific transposons that carry functional TFBSs and CRM and, in turn, regulate mouse-specific gene expression in early development. One such transposon is Lx8, a member of non-LTR retrotransposons, which we estimated to be inserted into the murine genome after the divergence of murine and primate species, ∼60 million years ago (Supplemental Fig. S3). A copy of Lx8 is present upstream of the cold shock domain–containing protein A (Csda) gene in the mouse genome. Csda is a transcription repressor, highly conserved among eukaryotes from yeast to human. In the mouse, Csda is known to be critical for several processes in reproduction and development, including spermatogenesis and in utero fetal development. The function of Csda in other mammals has not been tested. Transcription of Csda in mouse PED starts at the time of ZGA (two-cell stage), with an apparent pause during the four-cell stage, followed by continued increase in transcription from the eight-cell stage to the morula stage (Fig. 4). The increase in Csda transcription during mouse PED was correlated with Pou5f1 expression (R = 0.41, permutation _P_-value = 0.046). Like Pou5f1, Csda transcripts were restricted to the ICM at the blastocyst stage (Yoshikawa et al. 2006); however, ZGA of CSDA was not conserved in human or bovine, suggesting that the mouse Csda is associated with species-specific regulatory sequences. The insertion of Lx8 upstream of the mouse Csda gene produces a consensus POU5F1 binding site ([T/A]ATG[C/T]AAAT). This POU5F1 TFBS was functional in mouse ES cells: Analysis of published ChIP-seq data (Chen et al. 2008) revealed that the POU5F1–SOX2 protein complex was attached to this transposon-associated TFBS, which was at the center of the 36 base pair (bp) minimal overlap of the ChIP-seq reads (Fig. 4). Analysis of the sequence flanking this site revealed no other POU5F1–TFBS-like sequence within 300 bp in each direction. It appears unlikely that other regions of the mouse genome regulate the expression of Csda, as two CTCF-bound “insulator” TFBSs flank the POU5F1–SOX2-bound regions together with the Pol II-bound Csda promoter. ChIP-chip analyses of human ES cells did not identify any POU5F1, SOX2, or NANOG binding sites upstream of the human CSDA gene. Furthermore, motif scans based on the POU5F1 and NANOG motifs did not detect any high-affinity TFBS within 20 kilobases (kb) upstream or downstream of the human CSDA transcription start site (TSS). The data produced by gene expression, DNA motif, and TF binding analyses collectively suggest that the unique, transposon-mediated change in the regulatory sequence of the mouse Csda gene is associated with a gain of function.
Figure 4.
Murine-specific expression of the transcription factor Csda is induced by a murine-specific transposome carrying POU5F1 binding site. (A) CSDA is expressed during mouse PED, but not in human and bovine PED. CSDA is analyzed by two mouse probe sets (red), four human probe sets (green), and one bovine probe set (blue). (B) ChIP-seq data in mouse ES cells. The region of the mouse genome upstream of the Csda gene contains a functional POU5F1 binding site, which would appear to be competent to bind the POU5F1 protein. Two CTCF-associated TFBSs attract the CTCF insulator and define the boundaries of a genomic regulatory region (yellow) between them. The functional POU5F1 TFBS is carried by a murine-specific transposon Lx8, which is conserved in the mouse and the rat, but not in other mammals. (C) Expression and genome sequence data jointly suggest that the murine lineage gained a regulatory relationship between Pou5f1 and Csda.
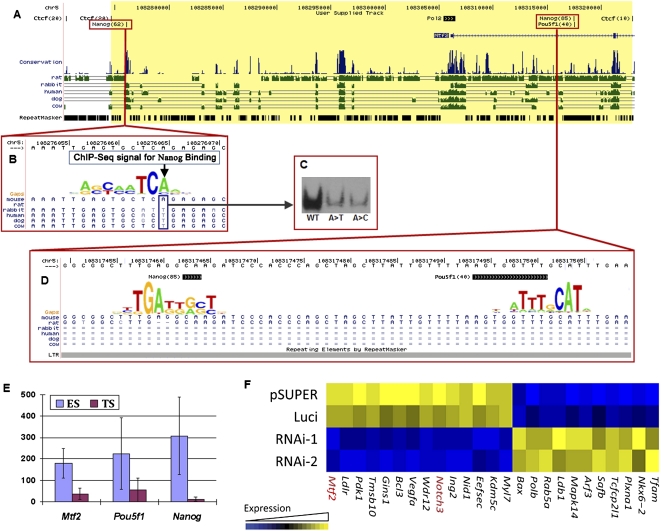
A second transposon in the mouse genome is associated with species-specific gene expression during PED. This transposon, Orr1b1-int, a member of the ERV3 family of retroviruses, is inserted upstream of the murine Mtf2 gene (Fig. 5A,D). Orr1b1-int is a “younger” transposon than Lx8 (Supplemental Fig. S3), but may have more potent gene regulatory effects than Lx8. Orr1b1-int contains a CRM comprised of two TFBSs (POU5F1 and a NANOG), in contrast with the single POU5F1 site in Lx8. ChIP-seq data revealed strong binding of POU5F1 and NANOG to their corresponding TFBSs within this Orr1b1-int, as reflected by large sequence counts: (the NANOG site ranked 434 among 10,345 detected NANOG sites, and the POU5F1 site ranked 66 among 3736 detected POU5F1 sites) (Chen et al. 2008). The POU5F1 binding on this Orr1b1-int region was five times stronger than on the Lx8 Csda enhancer. For both NANOG and POU5F1, the minimal overlaps of the ChIP-seq reads pinpointed to sites that matched their known DNA binding motifs (Fig. 5D), further confirming that the TF–DNA binding was a result of the presence of TFBSs within the Orr1bi-int sequence. These data, together with the gain of another NANOG TFBS by mutation (see below), offer an explanation for the dramatic difference in MTF2 expression among the three mammals analyzed in this study (Fig. 2A). Although it is not known why MTF2 transcripts are present in only human and bovine oocytes, the dramatic, mouse-specific induction of the Mtf2 gene during PED is consistent with the gain of murine-specific regulatory sequences.
Figure 5.
Mouse-specific expression of Mtf2 is due to the creation of transcription factor binding sites produced through the combined effects of a single nucleotide mutation and transposon-introduced sequences. (A) Binding sites for POU5F1, SOX2, NANOG, and RNA Polymerase II lie in close proximity to the mouse Mtf2 gene, between two CTCF binding sites (yellow region). (B) A single nucleotide mutation generates a NANOG binding site in the mouse, but not in human or cow. ChIP-seq data accumulated over 62 sequence reads, overlapping on a single nucleotide in the mouse genome. (C) Electrophoretic mobility shift assay results confirm that mutation of the mouse binding site back to the version in other mammals (A-to-T) eliminates NANOG binding in vitro. (A>T) A-to-T mutation; (A>C) A-to-C mutation. (D) A mouse-specific transposable element inserts a CRM for POU5F1 and NANOG into the region upstream of Mtf2: This CRM is bound by POU5F1 and NANOG in mouse ES cells. (E) Expression levels of Mtf2, Pou5f1, and Nanog in mouse ES and trophoblastic stem (TS) cells. (F) Mtf2 knockdown in mouse ES cells affects the expression of other transcription factors and signaling proteins, including the down-regulation of another ICM-specific gene, Notch3. pSUPER and Luci are control vectors that carry an empty vector (pSUPER) and shRNA against a luciferase gene (Luci). RNAi-1 and RNAi-2 are knockdowns mediated by two different shRNAs.
Turnover of TFBSs caused by single nucleotide mutations
Besides transposons, we postulated that single nucleotide changes may cause TFBS turnover and thus be responsible for interspecies differences in mammalian PED. Associating TFBSs with single nucleotide mutation with the genes with species-specific expression, we found three single nucleotide mutations that appear to influence gene expression by modulating the binding affinities of the TFBSs. Two of the three mutations locate within the same TFBS upstream to the FRAT2 gene, and the third mutation locates upstream to the MTF2 gene.
FRAT2 belongs to the GSK-3-binding protein family and is a positive regulator of the WNT signaling pathway. Expression of the human and bovine FRAT2 genes is strongly induced in late PED, whereas the mouse Frat2 gene appears to be transcriptionally silent throughout PED. (Supplemental Fig. S4A). The absence of expression in mouse is attributable to an A-to-C mutation at the center position of the POU5F1 binding site (Supplemental Fig. S4C,D), which eliminated the POU5F1 binding, as indicated by a chromatin immunoprecipitation with a microarray hybridization (ChIP-chip) peak on the human POU5F1 site (Boyer et al. 2005), but no ChIP-seq signal in the genomic neighborhood of the mouse Frat2 gene (Chen et al. 2008) (Supplemental Fig. S4B). This mutation at the center of the DNA binding motif caused a loss of the TFBS. Furthermore, the human FRAT2 exhibits larger induction of expression in response to the increased concentration of POU5F1 during PED, as compared with the bovine system. This delicate difference is attributable to a primate-specific T-to-A mutation at the second position (a peripheral position) of the same POU5F1 TFBS (Supplemental Fig. S4C,D). This mutation made the human TFBS identical to the consensus sequence of the POU5F1 motif, thus generating a stronger TFBS than the bovine version (Matys et al. 2003). In summary, in the case of an orthologous group of POU5F1 TFBSs, two mutations are present in DNA motif. The central mutation was consistent with a dramatic change on gene expression, and the peripheral mutation was reflected by a delicate change of expression.
A NANOG TFBS resides upstream of the Mtf2 gene in the mouse, but not in the human or the cow. This gain is associated with mouse-specific regulation of Mtf2. Sixty-two NANOG ChIP-seq reads overlapped a single nucleotide upstream of mouse Mtf2, at a location corresponding to a NANOG binding site (Fig. 5A). The position in question is occupied by an A in the mouse and a T in all other mammals: This suggests that the murine lineage contains a distinct T-to-A mutation (Fig. 5B). We sought to assess whether the putative “ancestral” sequence (not containing the T-to-A mutation) might harbor a NANOG TFBS. To this end, we mutated the functional binding site from A (mouse) back to T (ancestral). This mutation abrogated NANOG binding to the site in vitro (Fig. 5C). An A-to-C mutation had the same effect. These data suggest that the mouse TFBS was “born” from this mutation. Flanking CTCF insulator TFBSs grouped this newborn mouse NANOG TFBS together with the Orr1b1-int transposon reported above and the Pol II bound Mtf2 promoter, creating an autonomous _cis_-regulatory region within (Fig. 5A). This autonomous _cis_-regulatory region contained two murine-specific enhancers, which were generated by different evolutionary mechanisms, but synergistically contributed to the dramatic change of Mtf2 expression (Fig. 2A).
What is the consequence of the _MTF2_-associated gain of regulatory sequences on GRN?
We suspect that the altered regulatory sequence is responsible for the murine-specific expression of Mtf2. We sought to further explore the impact of this gain-of-function on the GRN of mouse blastocyst embryos. To this end, mouse ES cells and trophoblast stem (TS) cells, derived from the ICM and the trophectoderm of blastocysts, respectively, were used to analyze tissue-specific gene regulation. Mtf2 exhibited a 4.22-fold increase of expression level in ES cells than in TS cells (Fig. 5E), consistent with ICM-specific expression of Pou5f1 and Nanog in the mouse (Yoshikawa et al. 2006). The up-regulation of Mtf2 in ES cells is consistent with a previous computational result, which predicted Mtf2 should bear functional importance in mouse ES cells (Zhou et al. 2007). To test this, we knocked down Mtf2 in mouse ES cells with shRNA. Upon two days of transfection, two shRNA constructs both reduced Mtf2 expression to about 33% of the expression levels in the ES cells transfected with control plasmids. These cells were subjected to expression microarray analysis. Mtf2 knockdown resulted in the repression of expression of 14 genes that are involved in signaling and transcription (false discovery rate = 0.06), notably Notch3 (ranked ninth among 28,064 genes, Fig. 5F). Notch3 is one of the receptors in the Notch signaling pathway, which modulates specification of germ layers and differentiation of ES cells. These data are consistent with the notion that the gain of regulatory sequences detailed here may implement a Pou5f1/Nanog → Mtf2 → Notch3 pathway in the ICM of mouse blastocyst embryos. This model predicted that the expression of Notch3 should be restricted to the ICM. This prediction was confirmed by whole-mount, in situ hybridization (Yoshikawa et al. 2006).
Dynamic changes in GRNs during PED
The static GRNs active in human ES cells were recently described (Muller et al. 2008), although the dynamic processes that are capable of generating pluripotent GRNs remain elusive (Mikkelsen et al. 2008). Since many varieties of ES cells are procured from the inner cell mass of blastocyst-stage embryos, a more comprehensive understanding of the molecular events that drive PED could illuminate the mechanisms by which pluripotency-associated GRNs active in ES cells are constructed. Species-specific differences in the dynamic development of these GRNs may also help us better understand the different behaviors of ES cells from different species.
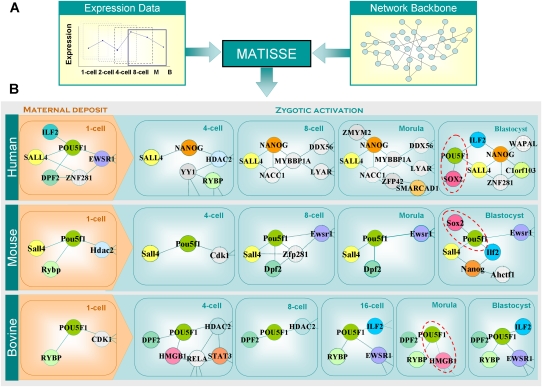
We therefore reconstructed the dynamic transitions of GRNs during PED in the human, murine, and bovine systems. For each stage of PED, we identified the interacting modules of genes (in each species) by jointly modeling the expression data and a reference protein interaction network with the MATISSE program as described in Methods (Fig. 6A; Ulitsky and Shamir 2007). The expression data from a given developmental stage were used in conjunction with expression data from adjoining stages: e.g., one-, two-, and four-cell expression data were used for reconstructing the interacting modules at the two-cell stage. The reference protein interaction network was generated by integrating the data from the Human Protein Reference Database (Mishra et al. 2006) with high throughput yeast two-hybrid screens (Ulitsky and Shamir 2007) and mass spectrometry analyses (Wang et al. 2006; Ewing et al. 2007). After constructing the state transition of the GRNs (http://biocomp.bioen.uiuc.edu/ped), we focused our analyses on the modules that contained the key regulatory genes that were induced in ES cells, as opposed to differentiated cells (Fig. 6B). Although expression data from the blastocyst-stage embryos were generated from a mixture of both ICM and trophectodermal cells, the reconstructed GRN in human blastocysts resembles the GRN of human ES cells (Muller et al. 2008). This similarity is highlighted by the interaction of POU5F1, SOX2, NANOG, SALL4, and C1orf103. The resemblance of the two GRNs lends credibility to the GRN reconstruction procedure and also suggests the genes involved in the POU5F1 interaction module exhibited a strong coexpression pattern during the morula-to-blastocyst transition.
Figure 6.
Comparison of the state transition of GRN across species. For each time point in each species, the active state of a GRN module is drawn. The nodes represent the genes with detectable transcripts; the edges represent the predicted interactions of the connecting nodes. Interactions were predicted by the MATISSE program, using correlation of expression changes in a window of three time points centered at the current time point, and protein–protein interaction data.
We found a few shared protein interactions among the three species. For example, HDAC2 is predicted to interact with POU5F1 (mouse, bovine) or NANOG (human) in early PED stages (four-cell human and bovine, one-cell mouse). HDAC2 is one of the histone deacetylase (HDAC) family proteins and is responsible for transcriptional repression. The fact that HDAC2 exhibited conserved interacting partners may indicate that its role of regulating cleavage of mouse preimplantation embryos (Ito et al. 2000) is conserved across species as well. An interaction module involving POU5F1, SOX2, NANOG, SALL4, and ILF2 is shared between human and mouse blastocyst embryos. In particular, the predicted POU5F1–ILF2 interaction at the blastocyst stage is shared in all three species. ILF2, also known as NF45, is a TF required for T-cell expression of the interleukin 2 gene. Although its role in non-T cells is largely unexplored, Ilf2 does physically interact with NANOG and POU5F1 in mouse ES cells (Wang et al. 2006). The conserved POU5F1–ILF2 interaction might suggest a novel transcription regulatory module responsible for regulating blastocyst embryos and ES cells.
A notable interspecies difference is that SOX2 is recruited to the POU5F1 module in human and mouse morula and blastocysts, but not in bovine morula or blastocysts. Instead, POU5F1 is predicted to interact with HMGB1 in bovine morulas (Fig. 6, dashed circles). Analyses of the probe sequences for the HMGB1 and SOX2 genes excluded cross-hybridization as a confounding factor to this prediction. HMGB1 (high-mobility group box 1) is a DNA-binding protein containing the HMG-box DNA binding domain. SOX2 is characterized by the same HMG-box protein domain, which is the only protein domain identified thus far in SOX2 (Supplemental Fig. S5A). A reanalysis of gene expression levels using real-time PCR confirmed the increased expression level of HMGB1 in bovine morulae (Supplemental Fig. S5B). The expression and protein domain data suggest that the POU5F1–SOX2 interaction may have been replaced by the POU5F1–HMGB1 interaction without changing target CRMs in bovine preimplantation embryos. Further experiments will be required to test this hypothesis.
Discussion
An evolutionary model for TFBS insertion by transposons
It has been recently observed that species-specific transposable elements carried a substantial fraction of TFBSs that are associated with TFs in vivo (Wang et al. 2007; Bourque et al. 2008). The influence of these transposable elements on the transcriptome has not been extensively explored. Our analyses suggest that although tens of thousands of transposons bind with TFs in vivo, the majority of such TF-bound transposons do not affect the expression of the surrounding genes. We estimate that 10% of murine-specific transposons, generally those that carry CRMs, may affect gene expression. These data suggest a model for the evolution of cis regulatory sequences by insertion of transposons. The TFBSs associated with transposons are inserted into the host genome and bind with corresponding TFs with similar stoichiometric coefficients as the parent TFBSs. The majority of these inserted TFBSs do not affect the expression of the surrounding genes and are evolutionarily neutral. A small fraction of the transposons, especially those that carry CRMs and attract a module of TFs, can change the expression of surrounding genes, and may be subject to positive or negative selections.
ES cells better represent ICM than EpiSCs
Pluripotent stem cells have been derived from several embryonic source tissues in mouse: ES cells are derived from the inner cell mass of blastocyst stage embryos, and epiblast stem cells are derived from the late epiblast layer of post-implantation mouse embryos (mEpiSCs) (Brons et al. 2007). mEpiSCs and mES cells exhibit nonidentical TF binding patterns and gene expression patterns. In terms of both enhancer usage and gene expression patterns, mES cells resemble ICM, whereas mEpiSCs are different. Based on these data, it was suggested that mES cells and mEpiSCs are two distinct pluripotent states representing cells of the preimplantation embryo and later epiblast cells (Brons et al. 2007), which is in line with our approach of using TF-binding data in ES cells to study TF-binding in PED.
Pluripotency is a conserved phenotype that can be sustained by evolvable GRNs
Human and mouse ES (hES and mES) cells are similar in the sense that they are both pluripotent cell populations that are capable of self-renewal that are derived from the ICM of blastocyst embryos. Even so, the growth factor requirements for hES and mES maintenance are different: mES cells require the presence of Lif, whereas hES cells do not. The differences in media conditions are thought to reflect differences in critical signaling networks responsible for maintenance of pluripotency and self-renewal capability between species. For example, TF Foxd3 is required for mES cell self-renewal, but its expression appears to be nonessential for hES (Ginis et al. 2004). Similarly, STAT3, a TF downstream to LIF signaling, is also required for self-renewal and the maintenance of pluripotency of mES cells, but it seems to be dispensable in hES cells (Schuringa et al. 2002). Compared to mES cells, mEpiSCs are functionally more similar to hES cells in several ways, including their responses to BMB4 and Activin/Nodal signaling (Brons et al. 2007). These observations led to the hypothesis that pluripotent cell identity can be established and maintained through more than one GRN. These GRNs may share core components universally indispensable for pluripotency. Peripheral components, though critical for cell fate, can be implemented using alternative designs. Because ES cells are derived from the ICM of blastocyst embryos, the natural processes of building pluripotent GRN are reflected by the transitions of the maternal cellular environment to ZGA-activated GRN. The comparison of GRN across multiple stages of PED suggested novel factors, such as MTF2 and HMGB1 for building a pluripotent GRN. These factors serve as candidates of dispensable reprogramming factors, which may be tested for potential enhancement of reprogramming efficiency. Since these factors may already be expressed in some tissues, they may contribute to customizing the combination of reprogramming factors for each tissue.
Methods
IVF embryos
The IVF conditions were optimized for each species. To minimize the influence of IVF conditions to cross-species comparison, we subsequently based the comparisons on either relative expression changes or coexpression patterns (Stuart et al. 2003; Gilad et al. 2006; Tirosh et al. 2007).
Human embryos
Supernumerary human embryos were obtained from patients at the Boston IVF Clinic through informed consent with the approval of the Internal Review Board of Harvard University. Embryos thus obtained were cultured in a two-step culture system (Sage/Biopharma) in microdrops at 37°C and 5% CO2 under oil (Sage) as described previously (Kaeberlein and Powers 2007). Embryos from each stage were segregated into three groups. Each group was considered to be a biologic replicate: Embryos within each replicate were pooled for further analysis. The blastocyst embryos were graded according to the Gardner scale (Brook and Gardner 1997; Van Royen et al. 1999).
Mouse embryos
Mouse embryos were obtained as described previously (Wang et al. 2004). Briefly, female mice were impregnated, and embryos were flushed from the oviducts. Individual embryos were morphologically staged by light microscopy.
Bovine embryos
Ovaries were washed, and 2–6 mm follicles were aspirated. Only oocytes surrounded by several cell layers of dense cumulus were selected for culture. Oocytes were washed three times in TL-HEPES (0.22 mM sodium pyruvate, 3 mg/mL BSA at pH 7.4), and we placed maturation media that had been equilibrated in 5% carbon dioxide and air (39°C, high humidity). The maturation medium consisted of TC199 supplemented with 3 mL of bovine LH and FSH (Sioux Biochemical), 0.25 mM sodium pyruvate, and 10% fetal calf serum. After 20 h of incubation in maturation media, oocytes were removed, washed in TL-HEPES and placed in groups of 10 oocytes each into 44-μL microdrops of IVF-Talp (Biowhittaker) supplemented with 0.22 mM sodium pyruvate, and 6 mg/mL essentially fatty acid-free BSA. Live sperm were separated by centrifugation in a percoll gradient (Sigma, P1985). Eggs were fertilized by adding one million/mL of sperm, 2.0 μg of heparin (Sigma), pencillamine, hypotaurine, and epinephrine (Sigma) to each drop of fertilization media. Oocytes were removed from the fertilization media after 18–22 h, washed in TL-HEPES, placed in a 1.5-mL tube, and vortexed for 2 min to remove cumulus cells. Twenty-five cumulus-free oocytes were placed in each 50-μL drop of SOF culture medium, supplemented with 8 mg/mL of fatty acid–free BSA, nonessential and essential amino acids, and pyruvate.
Transcription profiling
RNA was isolated from embryos using the PicoPure Isolation system (Arcturus) according to the manufacturer's instructions. Two rounds of linear amplification were performed for each sample to generate cDNA probes from total RNA as previously described (Baugh et al. 2001) using the PicoAmp system (Arcturus). The cDNA produced by this system was converted to aRNA and labeled using a T7-based Invitro-Transcription Labeling system (Enzo Lifesciences). Labeled aRNA was hybridized to the Affymetrix U133 2.0 plus, MOE430A 2.0 and Bovine Genome Genechips array (Affymetrix) according to the manufacturer's instructions. These expression data were submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE18290. The raw data for each species were normalized and modeled with dChip software.
Clustering analysis
We developed a Bayesian clustering method to cluster the timecourse expression data (W Huang, X Cao, G Lin, D Xie, and S Zhong, in prep.; see Supplemental material). The cluster generating process is modeled with a Dirichlet Process (Rasmussen 2000) and the time course profile of each cluster is modeled with a mixed-effects model with splines (Luan and Li 2003). The advantages of this method are: (1) it explicitly utilizes the timecourse data structure to minimize the adverse effects of noise in the data; (2) it automatically identifies the proper cluster number; (3) it is based on a fully described probabilistic model and therefore it minimizes the use of heuristic algorithmic steps. The difference in developmental time had little contribution to the detected difference of expression patterns. For example, maternal deposited RNA for TCF7, the canonical TF for the WNT signaling pathway, started degradation at four-, two-, and eight-cell stages in human, mouse, and bovine, respectively (Fig. 2A), but TCF7 was grouped into clusters of consistent expression patterns.
Identifying genes with mouse-specific expression changes
For the mouse vs. human comparison, we defined mouse-specific expression change as a gene exhibiting a non-flat expression trajectory in mouse PED, but its orthologous human gene exhibiting a flat expression trajectory in human PED, ignoring the bovine expression data. For the mouse vs. human and bovine comparison, we defined mouse-specific expression change as a gene exhibiting a non-flat expression trajectory in mouse PED, but its orthologous genes exhibiting flat expression trajectories in both human PED and bovine PED. To detect if a gene has a flat expression trajectory, we compared the expression levels between the first time point and every later time point, using _t_-tests. If any of the _t_-tests showed a significant difference (_P_-value < 0.05), then this gene was considered to have a non-flat trajectory. If none of the _t_-tests showed a significant difference, this gene was considered to not have any expression changes.
Detection of maternal and ZGA transcripts
The MT genes were required to have presence call in all the replicates at one-cell stage (oocyte and one-cell stages for bovine). The ZGA genes were identified with the following criteria: There is a detectable increase in expression levels during PED. To detect an increase of expression, we compared every time point to its previous time point. A detectable increase was defined as: fold change > 2, difference of dChip gene expression index > 100 and two-sample comparison _P_-value < 0.05.
ChIP-seq data
ChIP-seq data in human and mouse ES cells were obtained from GSE17917, GSE18292 (Lister et al. 2009), and GSE11431 (Chen et al. 2008). ChIP-seq identified TFBSs were associated with a gene if it is within a 50,000-bp flanking region (25,000 upstream and downstream) of the transcription start site of the gene. A ZGA gene is called bound by a ZGA TF, if it is bound by any of the three TFs: POU5F1, SOX2, and NANOG.
Estimation of transposon ages
All the Lx8 and Orr1b1-int transposons were downloaded from the RepeatMasker track on the UCSC Genome Browser (http://genome.ucsc.edu). The ages of the repeats were estimated by age = divergence/substitution rate (Bourque et al. 2008). The substitution rate of the mouse genome used in this analysis was 4.5 × 10−9 (Mouse Genome Sequencing Consortium 2002).
Detection of TFBS-carrying transposons
Coordinates of ChIP-seq detected binding regions of 16 transcription regulators in mouse ES cells were downloaded from GEO (Chen et al. 2008). Coordinates of transposable elements were downloaded from the UCSC Genome Browser, and the murine specific transposons were identified by the annotation of RepBase (Jurka et al. 2005). We took the intersection of these two sets of sequences and obtained a list of TFBS-carrying transposons.
Detection of species-specific expression
The map of orthologous human, mouse, and bovine probe sets was downloaded from the Affymetrix website (http://www.affymetrix.com/Auth/analysis/downloads/na28/ivt/Bovine.na28.ortholog.csv.zip). To resolve ambiguous mapping, for each gene we chose the probe set with the largest mean signal as the representative probe set for that gene. A gene was identified as exhibiting species-specific expression when either of the following two criteria was satisfied: (1) It was called “absent” in all time points in one species, but it had “presence” calls in other species. (2) When it was present in all species, its fold change of early (one- to eight-cell) and late (morula and blastocyst) embryos were computed. The fold change was more than twice as large in one species as compared to another.
shRNA-mediated RNA knockdown
Feeder-free E14 mouse ES cells were cultured at 37°C with 5% CO2. All cells were maintained on gelatin-coated dishes in DMEM (GIBCO), supplemented with 15% heat-inactivated FBS (GIBCO), 0.055 mM β-mercaptoethanol (GIBCO), 2 mM l-glutamine, 0.1 mM MEM nonessential amino acid, 5000 units/mL penicillin–streptomycin, and 1000 units/mL LIF (Chemicon), as described previously. Transfection of shRNA constructs was performed using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Briefly, 1.5 μg of plasmid DNA was transfected into ES cells on 60-mm plates for RNA extraction. Puromycin (Sigma) selection was introduced 1 d after transfection at 1.0 μg/mL and maintained for 2 and 4 d before harvesting.
shRNA targeting specific genes was designed as previously described (Reynolds et al. 2004; Ui-Tei et al. 2004). The 19-nucleotide (nt) hairpin-type shRNAs with a 9-nt loop were cloned into pSUPER.puro (BglII and HindIII sites, Oligoengine). Two shRNA, targeting different regions of respective transcripts, were designed for MTF2 to ensure specificity. These short nucleotides are: GGAGACTGGTATTTAAAGA, GCACACCTATGCCTTTACA. pSuperpuro constructs expressing shRNA against luciferase (Firefly) were used as controls.
EMSA
Electrophoretic mobility shift assay (EMSA) was used to analyze the binding affinities of putative NANOG TFBSs. The mouse wild-type “AGTGCTCA” was compared to two site-directed mutagenesis sites “AGTGCTCt” and “AGTGCTCc”, denoted as “A > T” and “A > C,” respectively. A recombinant NANOG protein (GST-tagged) was used for EMSA. The full-length NANOG protein was cloned into the pET42b (Novagen) vector, expressed and purified with GSH-sepharose beads (Amersham). DNA oligonucleotides (Proligo) labeled with biotin at the 5′ end of the sense strands were annealed with the antisense strands and were purified with agarose gel DNA extraction kit (Qiagen). A LightShift Chemiluminescent EMSA kit (Pierce Biotechnologies) was used. One-hundred nanograms of protein were added to a 5-μL reaction mixture and incubated for 20 min at room temperature. The binding mixture was resolved on 6% native polyacrylamide gels. The gels were transferred to Biodyne B nylon membranes (Pierce Biotechnologies) using Western blot techniques and detected using chemiluminescence.
Microarray data for Mtf2 knockdown ES cells and trophoblast stem (TS) cells
Two Mtf2 knockdown ES cell samples and two samples with control vectors were analyzed with Agilent Whole Mouse Genome Oligo Microarray Kit, representing over 41,000 mouse genes and transcripts. These expression data were submitted to GEO under accession no. GSE18319. Affymetrix MU74Av2 microarray data for ES and trophoblast stem (TS) cells were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/, accession no. GSE3766).
Acknowledgments
We thank the Douglas Melton lab for initiating this collaboration by supplying human IVF embryos, all duly obtained by full review of an ESCRO committee and IRB with certified consent forms. We thank Carole Wilson and Mark Band for microarray experiments. We thank Tetsuya Tanaka, Yanen Li, and Wei Huang for useful discussions. This work is partially supported by NSF DBI 08-45823 (S.Z.) and USDA Agricultural Research Service AG 58-1265-7-317 (H.A.L.).
Footnotes
References
- Adjaye J, Herwig R, Brink TC, Herrmann D, Greber B, Sudheer S, Groth D, Carnwath JW, Lehrach H, Niemann H 2007. Conserved molecular portraits of bovine and human blastocysts as a consequence of the transition from maternal to embryonic control of gene expression. Physiol Genomics 31: 315–327 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 29: e29 http://nar.oxfordjournals.org/cgi/content/full/29/5/e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S 1988. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332: 459–461 [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Brook FA, Gardner RL 1997. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci 94: 5709–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet 10: 141–148 [DOI] [PubMed] [Google Scholar]
- Ewing RMP, Chu F, Elisma H, Li P, Taylor S, Climie L, McBroom-Cerajewski MD, Robinson L, O'Connor M, Li R, et al. 2007. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3: 89 doi: 10.1038/msb4100134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245 [DOI] [PubMed] [Google Scholar]
- Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, et al. 2004. Differences between human and mouse embryonic stem cells. Dev Biol 269: 360–380 [DOI] [PubMed] [Google Scholar]
- Gossler A 1992. Early embryonic development of animals Springer, Berlin [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6: 117–131 [DOI] [PubMed] [Google Scholar]
- Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L 2008. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452: 840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Sakai S, Nagata M, Aoki F 2000. Effect of histone deacetylase inhibitors on early preimplantation development in mouse embryo. J Mammal Ova Res 17: 90–95 [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW III 2007. Sir2 and calorie restriction in yeast: A skeptical perspective. Ageing Res Rev 6: 128–140 [DOI] [PubMed] [Google Scholar]
- Kanka J, Bryova A, Duranthon V, Oudin JF, Peynot N, Renard JP 2003. Identification of differentially expressed mRNAs in bovine preimplantation embryos. Zygote 11: 43–52 [DOI] [PubMed] [Google Scholar]
- Kimura M 1991. The neutral theory of molecular evolution: A review of recent evidence. Jpn J Genet 66: 367–386 [DOI] [PubMed] [Google Scholar]
- Kues WA, Sudheer S, Herrmann D, Carnwath JW, Havlicek V, Besenfelder U, Lehrach H, Adjaye J, Niemann H 2008. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc Natl Acad Sci 105: 19768–19773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y, Li H 2003. Clustering of time-course gene expression data using a mixed-effects model with B-splines. Bioinformatics 19: 474–482 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. 2006. Human protein reference database—2006 update. Nucleic Acids Res 34: D411–D414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. 2008. Regulatory networks define phenotypic classes of human stem cell lines. Nature 455: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A 2002. Genes and signals Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rasmussen C 2000. The infinite Gaussian mixture model. Adv Neural Inf Process Syst 12: 554–560 [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A 2004. Rational siRNA design for RNA interference. Nat Biotechnol 22: 326–330 [DOI] [PubMed] [Google Scholar]
- Scannell DR, Wolfe K 2004. Rewiring the transcriptional regulatory circuits of cells. Genome Biol 5: 206 doi: 10.1186/gb-2004-5-2-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuringa JJ, van der Schaaf S, Vellenga E, Eggen BJ, Kruijer W 2002. LIF-induced STAT3 signaling in murine versus human embryonal carcinoma (EC) cells. Exp Cell Res 274: 119–129 [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Bilu Y, Barkai N 2007. Comparative biology: Beyond sequence analysis. Curr Opin Biotechnol 18: 371–377 [DOI] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443: 415–420 [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shamir R 2007. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol 1: 8 doi: 10.1186/1752-0509-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Royen E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermans W, Gerris J 1999. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod 14: 2345–2349 [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M 2004. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6: 133–144 [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci 104: 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Piao Y, Zhong J, Matoba R, Carter MG, Wang Y, Goldberg I, Ko MS 2006. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr Patterns 6: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH 2007. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci 104: 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]