Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double strand break repair (original) (raw)
. Author manuscript; available in PMC: 2010 Dec 1.
Abstract
As single agents, chemical inhibitors of poly(ADP-ribose) polymerase (PARP) are non-toxic and have clinical efficacy against BRCA1 and BRCA2 deficient tumours. PARP inhibitors also enhance the cytotoxicity of ionizing radiation (IR) and alkylating agents, but will only improve clinical outcomes if tumour sensitization exceeds effects on normal tissues. It is also unclear how tumour DNA repair proficiency affects the degree of sensitization. We have previously shown that the radiosensitizing effect of PARP inhibition requires DNA replication and will therefore affect rapidly proliferating tumours more than normal tissues. Since many tumours exhibit defective DNA repair, we investigated the impact of double strand break (DSB) repair integrity on the sensitizing effects of the PARP inhibitor olaparib. Sensitization to IR and the alkylating agent methylmethane sulfonate (MMS) was enhanced in DSB repair deficient cells. In _Artemis_−/− and _ATM_−/− mouse embryo fibroblasts (MEFs), sensitization was replication-dependent and associated with defective repair of replication-associated damage. Radiosensitization of _Ligase IV_−/− MEFs was independent of DNA replication and is explained by inhibition of alternative end-joining. After MMS treatment, PARP inhibition promoted replication-independent accumulation of DSB, repair of which required Ligase IV. Our findings predict that the sensitizing effects of PARP inhibitors will be more pronounced in rapidly dividing and/or DNA repair defective tumours than normal tissues and demonstrate their potential to enhance the therapeutic ratio achieved by conventional DNA-damaging agents.
Keywords: Poly(ADP-ribose) polymerase, non-homologous end-joining, radiation sensitivity, olaparib, alkylating agents
INTRODUCTION
The cytotoxic effects of ionizing radiation (IR) and alkylating chemotherapeutic drugs are mediated via DNA damage. IR induces single and double stranded DNA breaks in an approximate ratio of 25:1 (1), while alkylating agents induce base damage that gives rise to single strand breaks (SSB) and stalled replication forks. SSB induced by IR or alkylating agents are predominately repaired by base excision repair (BER), whereas double strand breaks (DSB) are repaired mainly by the non-homologous end-joining (NHEJ) pathway (2). DSB associated with DNA replication or occurring during G2 phase are also repaired by homologous recombination (HR) (2). Radiation-induced DSB are frequently associated with base or sugar damage, repair of which requires contributions from BER proteins such as polynucleotide kinase (PNK) (3). Furthermore, DSB can arise from SSB occurring in close proximity, or as a consequence of DNA replication.
Since the cytotoxicity of DNA damaging agents correlates with the number of unrepaired DSB (4), inhibition of DNA repair represents a mechanism by which the therapeutic effects of these agents might by enhanced. Such enhancement must be tumour specific if outcomes are to be improved. Whereas cancer cells are typically characterized by aberrant cell cycle checkpoint control, defective DNA repair pathways and accelerated proliferation rates, normal tissue cells have intact cell cycle checkpoints and DNA repair pathways (5). In addition, some critical normal tissues are composed almost entirely of non-replicating cells. A sensitizing agent that was effective only in cells with high replication rates and/or DNA repair defects would therefore have great clinical potential. Poly(ADP-ribose) polymerase-1 (PARP-1) is a DNA damage sensing protein that binds to SSB (reviewed in (6)), a process that activates its catalytic function and facilitates DNA repair. Inhibition of PARP activity reduces the rate of SSB repair and increases cellular sensitivity to IR and alkylating agents such as methylmethane sulfonate (MMS) (7-10). We have previously demonstrated that the radiosensitizing effect of PARP inhibition requires DNA replication, and that enhanced conversion of unrepaired SSB to DSB during S phase is the likely mechanism (11). Another study showed that PARP inhibition exacerbates the cytotoxicity of the alkylating agent temozolomide by enhancing conversion of SSB to DSB during S phase (12).
The cellular consequences of PARP inhibition are dictated by DNA repair proficiency. This is illustrated by the extreme sensitivity to PARP inhibitors of tumour cells deficient in BRCA1 or BRCA2 (13, 14). This phenomenon is caused by accumulation of endogenously arising DNA damage that would normally be repaired by PARP-dependent processes. In replicating cells this damage triggers replication fork collapse, repair of which requires HR and is therefore dependent on BRCA1 and BRCA2. The impact of DNA repair defects on the sensitising effects of PARP inhibitors in combination with DNA damaging agents remains unclear, however. Various studies have indicated that PARP inhibition exacerbates radiosensitivity (15) or chromosomal instability associated with NHEJ deficiency (16), but the evidence is inconsistent (17). Although it has been reported that PARP-1 binds to DSB and interacts with NHEJ proteins (18), there is no evidence that PARP activity is required for resolution of DSB in NHEJ proficient cells. However, an alternative repair pathway has been postulated to execute end-joining of DSB in cells that are deficient in NHEJ (19). This process has been termed ‘backup’ or ‘alternative’ end-joining and there is evidence that it is compromised by inhibition of PARP (20). The aim of this study was to investigate the relationship between NHEJ integrity and the sensitising effects of PARP inhibition.
NHEJ comprises core components (Ku70/80, DNA-PKcs, DNA Ligase IV, XRCC4 and XLF) which are responsible and sufficient for repair of the majority of radiation-induced DSB (reviewed in (21)). Loss of function of these proteins is associated with marked radiosensitivity and immunodeficiency, the latter as a consequence of impaired V(D)J recombination. A subset of radiation-induced DSB (approximately 15%) requires more extensive processing. Repair of these breaks occurs with slow kinetics and requires additional factors including Artemis and Ataxia Telangiectasia Mutated (ATM). Despite the relatively small size of this subset, defects in Artemis function are associated with significant radiosensitivity (22, 23).
To elucidate the relative impact of defects in core NHEJ and the Artemis/ATM pathway on the sensitizing properties of PARP inhibition, we evaluated the effects of olaparib (AZD2281, previously KU-005946) on radiation and MMS responses in repair defective cell lines. Olaparib is a potent and specific inhibitor of PARP-1 and PARP-2 and is currently undergoing phase II clinical trials as a single agent (24). Our data indicate that the sensitizing effects of olaparib are enhanced in cells with defective DSB repair and support the hypothesis that the sensitising effects of PARP inhibitors will be more pronounced in tumours than in normal tissues.
MATERIALS AND METHODS
Cell culture
CJ176 primary and hTERT (immortalized derivative) are Artemis-deficient human fibroblast cell lines. AT5-BIVA are ATM-deficient, SV40 immortalized human fibroblasts. HSC62 (kindly provided by Dr. M. Digweed) are primary fibroblasts from a patient with a homozygous mutation (IVS19-1 G to A) in BRCA2. 48BR primary and 1BR hTERT cells are WT controls. WT, _Artemis_−/−, _ATM_−/− and _LigaseIV_−/−_p53_−/− MEFs were kind gifts from Dr. F Alt (_Artemis_−/−), Dr. T Mak (WT, _ATM_−/−) and Dr. P McKinnon (_LigaseIV_−/−_p53_−/−). Human hTERT cell lines and MEFs were cultured in minimal essential medium (MEM) or α-MEM respectively, supplemented with 10% foetal calf serum, 2 mM L-glutamate, penicillin (100 units/ml) and streptomycin (0.1 mg/ml). Primary cell lines and HeLa cells (obtained from ECACC) were cultured in MEM with 15% foetal calf serum. All cell culture reagents from Gibco, other reagents from Sigma unless otherwise stated.
Radiation and drug treatments
Cells were irradiated in medium using either a 137Cs gamma source (Gammacell 1000, dose rate 7.5 Gray (Gy)/min) or 250 kVp X-rays delivered at 12 mA (dose rate 0.5 Gy/min). Olaparib (gift of KuDOS Pharmaceuticals/AstraZeneca) was used at an end-concentration of 500 nM, a non-cytotoxic dose that abolished PARP activity in the cell lines indicated (Figure 1A). PJ34 (Calbiochem) and KU55933 (ATM inhibitor, gift of KuDOS Pharmaceuticals/AstraZeneca) were used at end-concentrations of 10 μM. To inhibit DNA replication during PARP inhibition, aphidicolin (5 μM) was administered for three hours, commencing one hour pre-radiation. For clonogenic survival assays using MMS, aphidicolin and olaparib were administered one hour pre-, during (one hour) and for one or 22 hours after treatment.
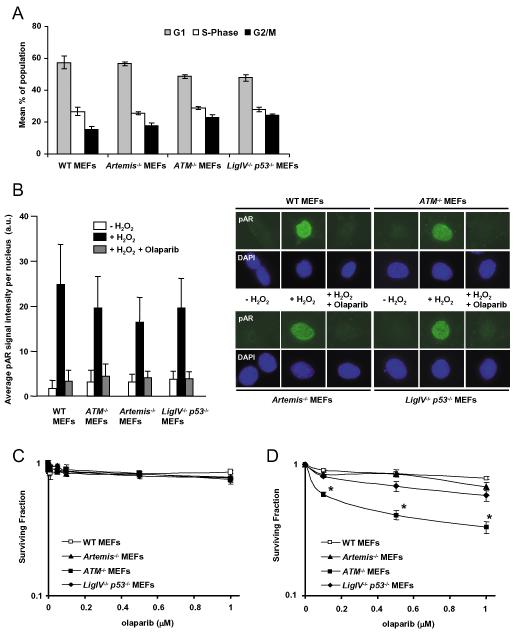
Figure 1.
A. Mean percentage of cells in G1, S phase and G2 phases of the cell cycle in exponential phase populations of MEFs. Cells fixed and stained with propidium iodide for flow cytometric analysis of DNA content. B. WT, _ATM_−/−, _Artemis_−/− and _Ligase IV_−/−_p53_−/− MEFs were pre-treated with 0.5 μM olaparib for 2 hours before treatment with 10 mM H2O2 (with or without 0.5 μM olaparib) for 10 min in the dark. Cells were stained for poly(ADP-ribose) (pAR) and counterstained with DAPI. Fluorescence intensity of pAR signal in each nucleus was quantified using NIH Image-J. Mean values of 100-200 cells +/− SD are presented. C. Clonogenic survival of exponential phase populations of WT, _ATM_−/−, _Artemis_−/− and _Ligase IV_−/−_p53_−/− MEFs exposed to olaparib at the indicated doses for 24 hours after plating. D. Clonogenic survival of WT, _ATM_−/−, _Artemis_−/− and _Ligase IV_−/−_p53_−/− MEFs continuously exposed to olaparib for the duration of the assay. To maintain levels of olaparib, medium was replaced every 48 hours. *p<0.01 (_ATM_−/− compared with WT, _Artemis_−/− and _Ligase IV_−/−_p53_−/− at each olaparib dose).
Clonogenic cell survival assay
Cells were trypsinised, seeded into 4 cm plates and allowed to adhere at 37°C before drug treatment or irradiation. For radiation or MMS assays, cells were exposed to olaparib for 24 hours, commencing two hours pre-treatment. For continuous olaparib exposure, medium was replaced every 48 hours to ensure constant levels of the drug. Plates were incubated at 37°C for 7 to 8 days then stained with methylene blue. Colonies of fifty cells or more were counted manually and survival curves derived from a minimum of three independent experiments, each performed in triplicate. Surviving fraction was corrected for independent cytotoxic effects of olaparib or aphidicolin except in Supplementary Figure S2A where significant toxicity was observed. Linear quadratic (for radiation) or exponential (for MMS) models were fitted to the data sets to generate survival curves. Radiation or MMS doses associated with surviving fractions of 10%, 37% or 50% were calculated from the fitted curves. Sensitizer Enhancement Ratios (SER) (25) for olaparib were calculated as in Equation 1:
| SERx=Dx(no drug)Dx(olaparib) | Equation 1 |
|---|
where D_x_ is the dose of IR or MMS associated with a surviving fraction of x%. Surviving fraction after 2 Gy (SF2) values were obtained from fitted survival data, and SF2 ratios with and without olaparib calculated.
Alkaline single-cell agarose gel electrophoresis (comet) assays
Alkaline comet assays were performed as previously described (26). Cells suspended in medium (c.2.8 × 105 cells/ml) were exposed to 30 Gy γ-irradiation or to 200 μM H2O2 for 20 min on ice and incubated for the indicated repair periods at 37 °C in medium. Where indicated, cells were incubated with 500 nM olaparib for 2 hours prior to DNA damage and during repair incubations. DNA strand breakage was expressed as “comet tail moment” (27). Tail moment was measured for 100 cells per sample using Comet Assay IV software (Perceptive Instruments, UK). Average damage remaining of at least 3 independent experiments was calculated.
γ-H2AX foci assays
γ-H2AX foci assays were performed specifically in G1 cells as previously described (11). Cells were stained with CENP-F (1:100, Santa Cruz CA, USA) for human cells or phospho-histone H3 (1:300, Upstate Biotechnology, Buckingham, UK) for mouse cells to identify G2 phase cells. Mitotic cells were identified by morphology and S-phase cells by diffuse, low-level γ-H2AX staining. Human fibroblasts (Figure 5A) were treated with 3 μM aphidicolin after irradiation to inhibit DNA replication and improve identification of S-phase cells by intensifying γ-H2AX staining (28). Thus DSB repair in G1 cells was monitored by enumerating γ-H2AX foci in cells that were deemed not to be in G2, S or mitosis.
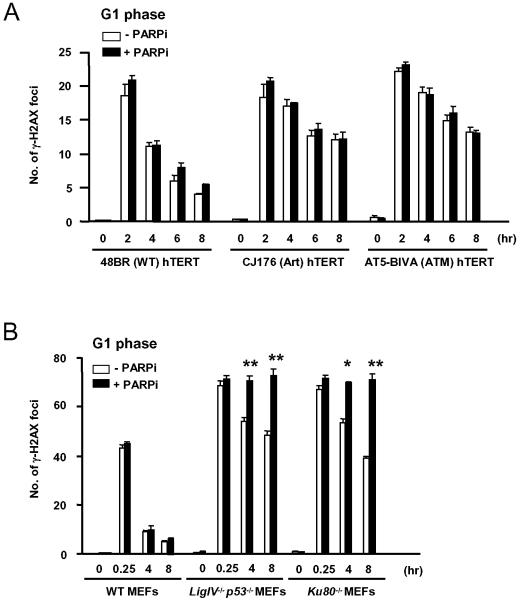
Figure 5.
A. Quantitative analysis of γH2AX foci in G1 phase 48BR hTERT (wild type), CJ176 hTERT (Artemis deficient) and AT5-BIVA hTERT (ATM deficient) fibroblast cell lines exposed to 10 μM PJ-34 (PARP inhibitor) for one hour pre-irradiation (3 Gy, gamma rays), treated with 3 μM aphidicolin then fixed and co-stained for γH2AX and CENP-F. B. Quantitative analysis of γ-H2AX foci in G1 phase MEFs exposed to 10 μM PJ-34 for one hour pre-irradiation (3 Gy, gamma rays), fixed and stained for γ-H2AX and phospho-histone H3. G2 cells distinguished by phospho-histone H3 staining and S phase cells identified by background γ-H2AX staining were excluded. *p<0.05, **p<0.01 for comparison between cells treated or not with PARP inhibitor.
Quantification of γ-H2AX signal and poly(ADP)-ribose signal
Slides stained as above were visualized using a Nikon Eclipse 50i microscope or a Zeiss Axioplan microscope at x40 magnification and image processing performed on Simple PCI software. Signal intensity within selected regions of interest was analyzed using NIH Image-J. MMS-induced γ-H2AX staining intensity was calculated by subtracting mean signal in untreated nuclei from that in MMS treated nuclei. H2O2-induced poly(ADP)-ribose synthesis was measured as mean fluorescence intensity of poly(ADP)-ribose signal per nucleus.
Flow cytometric analysis of cell cycle profiles
Cells were harvested by scraping and fixed in ice cold 70% ethanol before staining with propidium iodide (0.45 μg/ml), RNase (0.45 mg/ml) and 0.045% Tween. Resuspended cells were analysed for DNA content on a FACS Canto flow cytometer; data was processed with FACS Diva software (Becton Dickinson).
Statistical analysis
All data were derived from at least three independent experiments. For repair foci, at least 30 nuclei were counted for each experiment except where stated and statistical significance was determined using Student’s two-tailed _t_-test. For clonogenic survival experiments, mean surviving fraction +/− standard error of the mean (SEM) was plotted. SER and SF2 values were derived from individual experiments to enable calculation of mean values and SEM. Mean SER values were assessed for significance by Mann-Whitney U-test.
RESULTS
Sensitization to DNA damaging agents by PARP inhibition is enhanced in cells deficient in Artemis, ATM or Ligase IV
Continuous exposure to PARP inhibition is toxic to HR deficient cells and shorter exposures increase the sensitivity of repair proficient cells to MMS or IR. To investigate the influence of NHEJ on these outcomes we tested the effect of the PARP inhibitor olaparib on survival responses of WT, Artemis, ATM and Ligase IV deficient MEFs. We first demonstrated that all four cell lines were deficient in p53 protein (data not shown) and that asynchronous, undamaged populations showed no significant differences in cell cycle distribution (Figure 1A). Quantitative immunofluorescent detection of pAR synthesis after treatment with hydrogen peroxide was performed (see Supplementary Figure S1), demonstrating no significant difference in PARP activity between cells lines, and that 0.5 μM olaparib inhibited PARP activity to baseline levels (Figure 1B). 24 hour exposure to a range of olaparib doses had minimal effect on clonogenic survival (Figure 1C), but _ATM_−/− cells were significantly more sensitive to prolonged PARP inhibition than the other cell lines (Figure 1D; p<0.01, all doses). This is consistent with previous findings (29, 30) and can be explained by involvement of ATM in the HR pathway (29) and by defects in S-phase checkpoint signalling that have been clearly defined in ATM deficient cells. In contrast, _Artemis_−/− and _Ligase IV_−/− cells were no more sensitive to continuous PARP inhibition than wild type MEFs. Hence, NHEJ does not play an important role in repair of endogenously arising damage in replicating cells, even in the presence of PARP inhibition.
Our previous data supported the hypothesis that PARP inhibition promotes replication-dependent conversion of unrepaired SSB to potentially toxic DSB (11) To investigate whether NHEJ influences the response to these DSB we first measured the effect of PARP inhibition and NHEJ status on sensitivity to the alkylating agent MMS, which induces predominantly SSB (Figure 2A). Wild type MEFs were sensitised by PARP inhibition as expected (SER37 = 2.36), but the magnitude of this effect was markedly increased in _Artemis_−/− (SER37 = 7.90, p<0.05) and _ATM_−/− (SER37 = 4.48, p<0.05) cells (Figure 2A, Table 1). These findings are consistent with a model whereby PARP inhibition promotes the generation of DSB from MMS-induced SSB, and that these lesions are more toxic in the absence of ATM or Artemis. _Ligase IV_−/− cells were markedly sensitized by olaparib (SER37 = 14.29, p<0.05), indicating that core NHEJ is required for repair of lesions arising under these conditions. In the absence of PARP inhibition, Ligase IV deficient MEFs showed mild sensitivity to high doses of MMS, indicating that MMS-induced lesions, when present at high density, can generate DSB that are repaired by core NHEJ. Artemis and ATM deficient cells were no more sensitive to MMS than controls, indicating that MMS-induced lesions do not require processing by these proteins. The mechanisms underlying these findings are explored in more detail later.
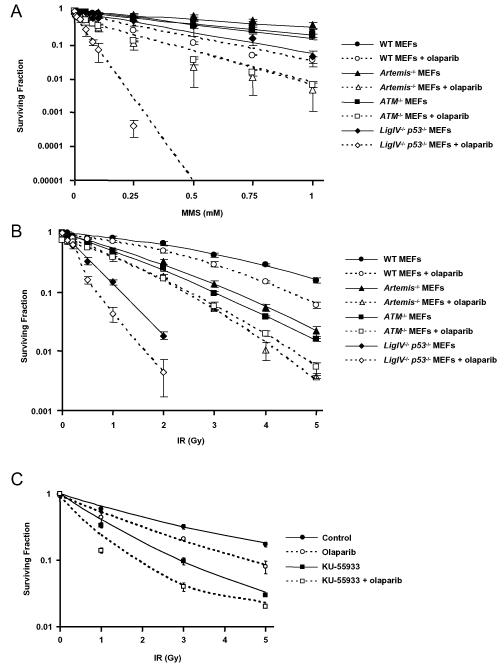
Figure 2.
Clonogenic survival of exponential phase populations of WT, _ATM_−/−, _Artemis_−/− and _Ligase IV_−/−_p53_−/− MEFs treated with A. MMS for 1 hour at 37°C +/− 0.5 μM olaparib for 2 hours pre-, during and 21 hours post-treatment B. 250 kV X-rays +/− 0.5 μM olaparib for 2 hours pre- and 22 hours post-irradiation. C. Clonogenic survival curves derived from exponential HeLa cells exposed to 0.5 μM olaparib and/or 10 μM ATM inhibitor, KU55933 for 2 hours pre- and 22 hours post-irradiation.
Table 1.
Mean sensitiser enhancement ratios (SER) for olaparib and mean surviving fraction at 2 Gy (SF2) values (and SEM) calculated from clonogenic survival curves fitted with the linear quadratic model (X-rays) or the exponential model (MMS)
| Treatment | X-ray | X-ray | X-ray | MMS | ||
|---|---|---|---|---|---|---|
| Cell line | SER37 | SER50 | mean SF2 | mean SF2olaparib | ratio | SER37 |
| WT MEFs | 1.30 (0.01) | 1.33 (0.01) | 0.62 (0.04) | 0.49 (0.07) | 1.27 | 2.36 (0.28) |
| _ATM_−/− MEFs | 1.34 (0.11) | 1.56 (0.15) | 0.23 (0.007) | 0.17 (0.02) | 1.35 | 4.48* (0.14) |
| _Artemis_−/−MEFs | 1.50* (0.06) | 1.61* (0.08) | 0.29 (0.02) | 0.16 (0.02) | 1.81 | 7.90* (0.91) |
| _LigIV_−/− _p53_−/−MEFs | 1.66* (0.02) | 1.63* (0.06) | 0.02 (−) | 0.004 (−) | 4.22 | 14.29* (2.99) |
| HeLa control | 1.50 (0.11) | 1.50 (0.11) | 0.45 (0.04) | 0.32 (0.03) | 1.41 | - |
| HeLa +KU-55933 | 1.75 (0.08) | 1.87 (0.07) | 0.19 (0.02) | 0.09 (0.01) | 2.15 | - |
IR induces DNA damage comprising SSB and DSB in a ratio of approximately 25:1, so the cytotoxic effects of DSB arising from SSB are obscured by those of directly induced DSB. Also, radiation induces far fewer SSB than MMS at the doses used in these experiments. Taking this into account, the clonogenic survival data presented in Figure 2B are consistent with the MMS observations. In the absence of olaparib, _Ligase IV_−/− cells were highly sensitive to radiation while _Artemis_−/− and _ATM_−/− cells showed intermediate sensitivity, as expected. Whereas PARP inhibition had a modest radiosensitising effect on wild type MEFs (SER50 = 1.33), consistent with previous reports (31), higher levels of radiosensitization were observed in Artemis (SER50 = 1.61, p<0.05) and Ligase IV deficient cells (SER50 = 1.63, p<0.05) (Figure 2B, Table 1). Sensitization of ATM deficient cells was also greater than in wild type cells, but this did not reach statistical significance (SER50 = 1.56).
To validate the relevance of these findings to the treatment of cancer, the impact of the ATM inhibitor KU-55933 on the radiosensitizing effects of olaparib was measured in HeLa cells. The magnitude of the sensitizing effect of olaparib was enhanced on a background of ATM inhibition (SER50 = 1.87 compared with 1.50, Figure 2C and Table 1). For additional clinical relevance, surviving fractions at 2 Gy (SF2) were calculated for all survival experiments and SF2 ratios in the presence and absence of olaparib derived (Table 1). In all cases, the ratio was markedly increased in DNA repair defective backgrounds, with the greatest effect observed in Ligase IV deficient MEFs.
Radiosensitization associated with PARP inhibition is replication dependent in Artemis and ATM but not Ligase IV deficient cells
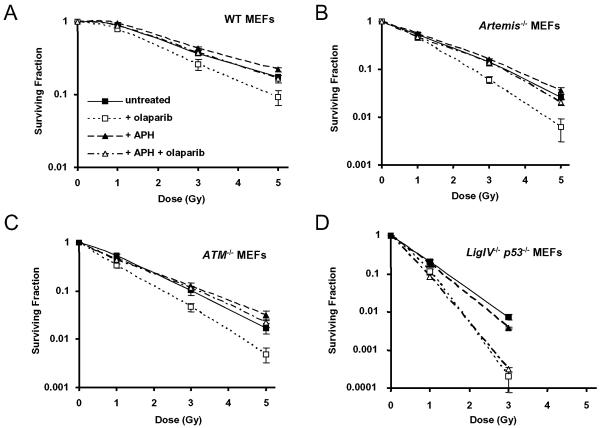
To investigate whether the radiosensitising effects of olaparib in NHEJ deficient cells are mediated by replication-dependent conversion of SSB to DSB, the DNA polymerase inhibitor aphidicolin was used to inhibit DNA replication during the period of PARP inhibition. Radiosensitization by olaparib was completely rescued by aphidicolin in wild type, _Artemis_−/− and _ATM_−/− cells (Figure 3A-C) but was unaffected in Ligase IV deficient cells (Figure 3D). This indicates that DNA replication is not necessary for radiosensitization of Ligase IV defective cells.
Figure 3.
A-D. Clonogenic survival curves derived from MEFs exposed to 0.5 μM olaparib and/or 5 μM aphidicolin (APH) for 1 hour pre- and 2 hours post-irradiation with 250 kV X-rays at the doses shown. A. WT MEFs, B. _Artemis_−/− MEFs, C. _ATM_−/− MEFs, D. _Ligase IV_−/−_p53_−/− MEFs.
To eliminate the possibility that the sensitizing effects of PARP inhibition in ATM and Artemis deficient cells might reflect direct involvement of either protein in SSB repair, alkaline comet assays were performed in _ATM_−/− and _Artemis_−/− MEFs (Supplementary Figure S2). Both cell lines exhibited normal repair kinetics after treatment with IR or the SSB inducing agent hydrogen peroxide. Addition of olaparib delayed repair as expected but the effect was no greater in ATM or Artemis deficient cells than in wild type controls. Together with the replication-dependent effect on survival, these observations implicate increased conversion of SSB to cytotoxic DSB during DNA replication as the primary mechanism underlying the enhanced radiosensitizing effect of PARP inhibition in _ATM_−/− and _Artemis_−/− cells.
Resolution of DNA damage induced by MMS and PARP inhibition during S phase is delayed in Artemis deficient cells
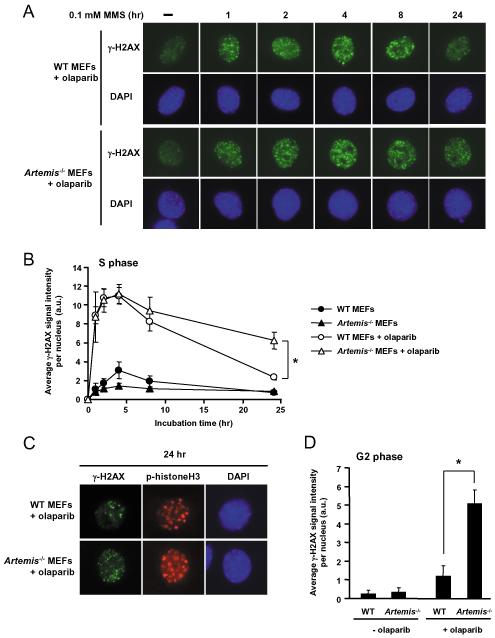
We hypothesized that the increased effects of PARP inhibition in Artemis deficient cells were a consequence of defective repair of replication-dependent DSB. To verify this we exposed cells to MMS and quantified levels of phosphorylated histone H2AX (γ-H2AX) immunofluorescence in S-phase MEFs. In G0 and G1 cells γ-H2AX foci have been shown to correlate well with DSB numbers as measured by neutral comet assay, pulsed field gel electrophoresis and survival after IR (32). However, phosphorylation of H2AX also occurs diffusely in response to damage other than DSB and during apoptosis, and may be stimulated by replication fork collapse. As a result, individual γ-H2AX foci cannot be accurately detected in S-phase cells. To quantify replication-associated DNA damage, therefore, we analysed total γ-H2AX staining intensity in S-phase nuclei. These were identified by negative phospho-histone H3 (pH3) and diffuse γ-H2AX staining (28). In combination with MMS, PARP inhibition increased total γ-H2AX signal in WT and _Artemis_−/− cells, with similar increases occurring up to 4 hours after MMS treatment. At 24 hours, however, S-phase _Artemis_−/− cells exhibited significantly greater γ-H2AX signal than wild type (Figure 4A, quantified in Figure 4B, p<0.05), indicating impaired resolution of damage. By measuring γ-H2AX staining intensity in pH3 positive cells, we showed that the excess damage persisted as cells progressed into G2 (Figure 4C), with _Artemis_−/− cells staining much more intensely than WT (p<0.05 at 24 hours, Figures 4C and D). These findings are consistent with the clonogenic survival data (Figure 2A) and support the hypothesis that PARP inhibition promotes replication-associated conversion of SSB to DSB that require Artemis for efficient repair.
Figure 4.
A. Representative images of γH2AX immunofluorescence in S-phase WT or _Artemis_−/− MEFs. Cells were exposed to 0.5 μM olaparib for one hour prior to treatment with MMS (0.1 mM, 1 hour) and until fixation at the timepoints shown. B. Quantitative analysis of γH2AX signal in S-phase WT or _Artemis_−/− MEFs. Mean fluorescence intensity per nucleus +/− SEM from 3 experiments shown, at least 25 nuclei per timepoint. *p<0.05. C. Representative images of γH2AX and phospho-histone H3 immunofluorescence in G2-phase WT and _Artemis_−/− MEFs 24 hours after MMS +/− olaparib treatment as in A. G2-phase cells were identified by speckled p-histone H3 staining. D. Quantitative analysis of γH2AX signal in G2-phase cells 24 hours after MMS treatment +/− olaparib. *p<0.05.
PARP inhibition increases radiation sensitivity in NHEJ deficient cells by obstructing an alternative end-joining pathway
Survival data indicated that DNA replication was not required for radiosensitization of Ligase IV deficient cells by olaparib (Figure 3D). To explore replication-independent mechanisms we measured the effect of PARP inhibition on induction and resolution of γ-H2AX foci after IR specifically in G1 phase nuclei. Cells were co-stained for centromere protein F (CENP-F, human fibroblasts) or pH3 (MEFs) to identify G2 (CENP-F or pH3 positive) and G1 (CENP-F or pH3 negative) nuclei (28). S-phase nuclei were identified by intermediate CENP-F staining (human cells) and diffuse background γH2AX staining. Mitotic or apoptotic cells were identified by their distinctive DAPI staining pattern. By excluding these nuclei the specificity of the γH2AX foci assay for true DSB was increased. To further substantiate DSB specificity we demonstrated that induction of γ-H2AX foci by IR was abolished by simultaneous inhibition of the DSB dependent kinases ATM and DNA-PK, both in human fibroblasts and MEFs (Supplementary Figure S3A and B). Hence, although IR induces high levels of both SSB and DSB, H2AX phosphorylation occurs only in response to signalling events that are specifically activated by DSB.
As reported previously (33), G1 phase Artemis and ATM deficient cells exhibited a partial DSB repair defect (Figure 5A). PARP inhibition did not affect induction or resolution of γ-H2AX foci, consistent with the model that DNA replication is required to generate excess double stranded lesions in these cell lines.
G1 phase Ligase IV and Ku80 deficient cells exhibited a more marked but not complete DSB repair defect (Figure 5B). In these cells, PARP inhibition significantly increased the number of persistent γ-H2AX foci (Figure 5B, p<0.01 at 4 and 8 hours for _Ligase IV_−/− cells; p<0.05 and p<0.01 at 4 and 8 hours respectively for _Ku80_−/− cells). Indeed residual DSB repair function appeared to be abolished by PARP inhibition in these NHEJ deficient cells. This supports the hypothesis that activity of an alternative end-joining pathway is promoted in the absence of either Ku80 or Ligase IV and eliminated by inhibition of PARP (20, 34).
To exclude the possibility that these observations reflected effects on H2AX phosphorylation or focus dynamics rather than DSB repair, we demonstrated that treatment with the PARP inhibitor did not affect the number of foci induced by irradiation (Supplementary Figure S4A), the rate at which they resolved (Figure 5A and Supplementary Figure S4A) or the signal intensity per focus at 30 minutes or 8 hours after irradiation (Supplementary Figures S4B and C).
PARP inhibition promotes replication independent accumulation of DSB in MMS treated Ligase IV deficient cells
Our observation that Ligase IV −/− cells were sensitive to high doses of MMS in the absence of PARP inhibition (Figure 2A), and were more profoundly sensitized to MMS by PARP inhibition than other repair defective cells raised the possibility that additional mechanisms might be operating. We hypothesized that PARP inhibition would delay repair of SSB induced by MMS and increase the probability of unrepaired lesions giving rise to DSB by interacting either with adjacent SSB or with transcription machinery (35). Either mechanism might be exacerbated by persistent binding of inhibited PARP at the damaged sites. To investigate this hypothesis we monitored the appearance of γ-H2AX foci in wild type and repair deficient cells. Effects of DNA replication were eliminated by restricting the analysis to cells in G1 phase and the specificity of γ-H2AX foci for DSB was validated as described above and illustrated in Supplementary Figure 3.
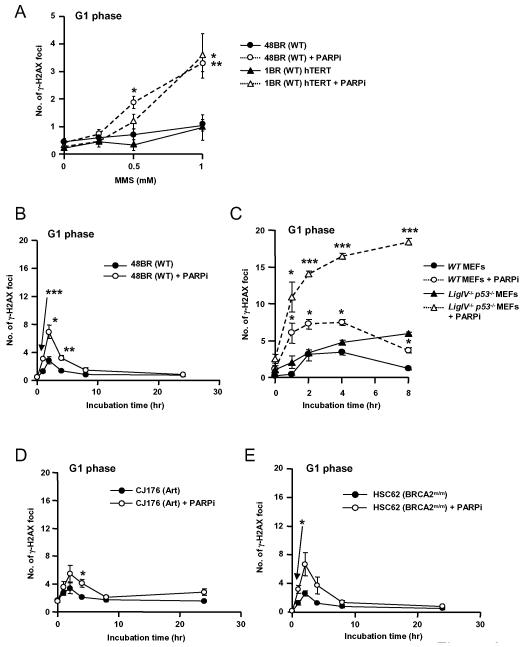
PARP inhibition was required for generation of γ-H2AX foci in G1 phase WT fibroblasts exposed to MMS for 1 hour (Figure 6A, Supplementary Figure S5). PARP inhibition significantly increased DSB formation between 1 and 4 hours after MMS treatment (p<0.001, p<0.05, p<0.01 at 1, 2 and 4 hours respectively, Figure 6B). In WT cells, γ-H2AX foci were few (6 – 8 per cell) and were almost completely repaired within 8 hours (Figure 6B) but in Ligase IV deficient MEFs foci continued to accumulate over 8 hours (Figure 6C). This accumulation was observed both in the presence and absence of the PARP inhibitor, but the increase in foci associated with PARP inhibition was much greater than in wild type cells, and was highly significant (p<0.05 at 1 hour, p<0.001 at 2, 4 and 8 hours). At 24 hours, cells exhibited intense γ-H2AX staining indicative of apoptosis and foci could not be counted. In G1 phase Artemis deficient human fibroblasts (CJ176), the kinetics of induction and repair of γ-H2AX foci were essentially normal, and the effect of PARP inhibition was no greater than in WT fibroblasts (Figure 6D), indicating that DNA replication is required to generate DSB that are substrates for Artemis. Similarly, BRCA2 mutant HR defective cells (HSC62) showed normal DSB repair kinetics and the same response to PARP inhibition as WT (Figure 6E). Hence DSB arising from MMS-induced damage in the absence of replication are repaired by Ligase IV dependent NHEJ.
Figure 6.
A. Quantitative analysis of γH2AX foci in G1 phase 48BR primary and 1BR hTERT fibroblasts exposed to 10 μM PJ-34 for one hour prior to treatment with MMS at the indicated doses. B-E. Quantitative analysis of γH2AX foci in 48BR (B), _Ligase IV_−/−_p53_−/− MEFs (C), CJ176 (Artemis deficient) (D) and HSC62 (BRCA2 deficient) (E) primary fibroblast cell lines and exposed to 10 μM PJ-34 for one hour prior to and during damage induction with 1 mM MMS and for the indicated repair periods. Cells were fixed and co-stained for γH2AX and CENP-F or phospho-histone H3. *p<0.05, **p<0.01, ***p<0.001 for comparison between cells treated or not with PARP inhibitor. Representative images are shown in Supplementary Figure S5.
These data show that treatment with MMS yields low levels of DSB, most of which are not normally detected because they are rapidly repaired by core NHEJ. In the presence of a PARP inhibitor, DSB induction is greatly increased and these lesions are also repaired by Ligase IV dependent NHEJ. These novel observations provide a possible explanation for the relative sensitivity of Ligase IV −/− MEFs to high doses of MMS and the gross sensitizing effect of PARP inhibition (Figure 2A).
DISCUSSION
PARP inhibitors have clinical potential as sensitizers to be used in combination with IR or with alkylating agents. It is important to establish whether these effects will be tumour specific, and whether therapeutic benefit can be predicted by the integrity of DNA repair pathways in the target tumour. Our previous study showed that radiosensitization of human tumour cells is replication dependent and mediated primarily via conversion of unrepaired SSB to DSB during DNA replication (11). Here we show that sensitization to IR and the alkylating agent MMS is enhanced in DSB repair defective cells. Both findings predict that the sensitizing effects of PARP inhibitors will be more pronounced in tumours than in normal tissues. Furthermore we demonstrate that the mechanisms responsible for sensitization differ between _Ligase IV_−/− cells and _Artemis_−/− or _ATM_−/− cells and provide evidence for distinct mechanisms by which PARP inhibition increases unrepaired DSB after induction of single stranded DNA damage.
1) Replication dependent effects of PARP inhibition: IR and MMS
In this study we examined the effect of PARP inhibition on MMS sensitivity and observed a markedly greater effect in _Artemis_−/− or _ATM_−/− cells than in DSB repair proficient cells. The radiosensitizing effects of PARP inhibition were also enhanced in Artemis and ATM deficient cells than wild type, but the differential effect was less marked, probably because ionizing radiation induces a spectrum of damage that includes both SSB and DSB. Radiosensitization was replication dependent in wild type, _Artemis_−/− or _ATM_−/− cells. These observations are consistent with a model in which the primary effect of PARP inhibition is to abrogate SSB repair leading to the replication dependent generation of cytotoxic DSB, repair of which is inhibited in the absence of Artemis or ATM.
ATM might promote repair of replication dependent DSB by a variety of mechanisms. These include initiation of cell cycle checkpoints, activation of HR repair of replication coupled DSB (29) and promotion by phosphorylation of the end-processing activity of Artemis (33). Likewise, Artemis has established roles in end-processing of complex DSB and resolving hairpin structures during V(D)J recombination (36). Since PARP inhibition causes persistent binding of PARP to damaged DNA (37), it is also possible that the nuclease activity of Artemis is required to remove the PARP-DNA complex and allow repair. Our data indicate that such a requirement may be particularly marked in the context of DNA replication.
2) Replication-independent effects of PARP inhibition: ionizing radiation
It has been proposed that an alternative end-joining pathway functions in the absence of classical NHEJ (20). To consolidate the concept that this pathway is compromised by PARP inhibition, we analysed induction and repair of γ-H2AX foci after low doses of radiation. To eliminate the effects of HR in S and G2 phase, foci were enumerated in G1 cells only. Consistent with previous findings, PARP inhibition blocked DSB repair in _Ku80_−/− and _Ligase IV_−/− cells. These data support the previously proposed model whereby Ku and PARP compete for binding at DSB ends (20) and provide an explanation for our observation that radiosensitization of Ligase IV deficient cells by PARP inhibition is more pronounced than in WT cells and is independent of DNA replication.
In this study, we also show for the first time that PARP inhibition has no direct impact on DSB repair in Artemis and ATM defective G1 cells following exposure to IR. This is consistent with our observation that sensitization to DNA damaging agents by PARP inhibition is replication dependent in _Artemis_−/− and _ATM_−/− cells and demonstrates that activity of the alternative end-joining pathway is not promoted by the absence of Artemis or ATM. It also supports the idea that Ku binding activity at DSB is unaffected by the absence of these repair factors. Of note, these results do not necessitate that PARP plays a functional role in alternative end-joining. The data show only that inhibition of PARP can block the activity of this process.
3) Replication-independent effects of PARP inhibition: MMS
PARP inhibition caused gross sensitization of _Ligase IV_−/− cells to MMS. This was unexpected because MMS is not thought to induce significant numbers of DSB directly. To investigate whether MMS treatment generates DSB in the absence of DNA replication, and the role of PARP inhibition in this process, γ-H2AX foci were enumerated in G1 cells. Ligase IV deficient cells exhibited significant accumulation of DSB (c.18 per G1 nucleus) following exposure to 1 mM MMS in the presence of a PARP inhibitor. This indicates that PARP inhibition promotes replication independent generation of DSB from MMS-induced lesions, and that these DSB require core NHEJ for repair. Such DSB might arise from overlapping SSB or from interactions between transcription and obstructed SSB. Low numbers of γ-H2AX foci (c.7 per G1 nucleus) were detected in wild type cells exposed to MMS in combination with the PARP inhibitor, again indicating that MMS is capable of inducing replication-independent DSB when BER is impaired. In NHEJ proficient cells, however, these DSB were rapidly repaired.
A previous study failed to detect induction of DSB following exposure to MMS (38). This discrepancy may be explained by the increased sensitivity of the γ-H2AX foci assay compared with pulsed-field gel electrophoresis, which has a detection limit of approximately 300 DSB (equivalent to at least 10 Gy in G1 cells). The γ-H2AX foci assay is highly sensitive but must be used with caution because of the ability of DNA lesions other than DSB to stimulate phosphorylation of H2AX. Its validity in these experiments is supported by the fact that induction of foci by MMS was entirely abolished by inhibition of ATM and DNA-PK, since phosphorylation of H2AX by these two proteins occurs only when they are activated by the presence of DNA DSB (39). Also, resolution of the γ-H2AX foci induced by MMS was Ligase IV dependent. In support of our observations, Woodhouse and colleagues have shown that hydrogen peroxide treatment generates DSB in PARP-1 deficient but not control cells (40). Our study extends this finding by demonstrating that SSB are converted into DSB in the absence of DNA replication, and that the resulting DSB require NHEJ for repair. Artemis and BRCA2 deficient cells exhibited normal repair of DSB under these conditions. Hence in Ligase IV deficient cells, inhibition of PARP acts via two distinct, replication independent mechanisms to increase the cytotoxic effects of SSB inducing agents such as MMS and IR.
4) Relevance to cancer therapy
The capacity of PARP inhibitors to increase tumour radiosensitivity has been demonstrated in a number of in vitro and in vivo models (41, 42), and the low toxicity of these compounds predicts a beneficial clinical response. However, the major impediment to clinical use of radiosensitising agents is parallel sensitisation of normal tissues (43). In this study we have demonstrated that the sensitising effects of PARP inhibitors are manifest only in replicating cells or in non-replicating cells that are deficient in core NHEJ proteins. Sensitization to IR and particularly to the alkylating agent MMS was enhanced in cells deficient in the ‘non-core’ NHEJ repair proteins ATM and Artemis. Tumours exhibiting defective NHEJ repair may therefore be particularly sensitive to PARP inhibitors in combination with radiation or alkylating chemotherapy agents. In general, tumour cells are characterized by higher rates of replication than normal tissues and are much more likely to exhibit defective DNA repair (5, 44). Furthermore, specific defects in NHEJ have been documented in high-grade malignancies such as carcinoma of the bladder (45) and glioblastoma multiforme (46), and ATM deficiency is a feature of mantle cell lymphoma (47). Thus it is likely that the radiosensitising effects of PARP inhibitors such as olaparib will be more marked in tumour cells than in adjacent normal tissues. The resulting therapeutic benefit may be particularly apparent in the case of brain tumours, where the critical normal tissue – the brain – is composed predominantly of non-replicating cells with intact DNA repair pathways (48).
ACKNOWLEDGEMENTS
Thanks to Graeme Smith and Niall Martin at KuDOS Pharmaceuticals (AstraZeneca) for helpful discussion and for providing olaparib and KU-55933.
This work was supported by a Medical Research Council Clinician Scientist Fellowship (G108/589 to AC, DL), a Medical Research Council Programme Grant (G0500897 to PJ, AS) and a JSPS Research Fellowship for Young Scientists (AS).
REFERENCES
- 1.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 2.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch CA, Agyei R, Galicia S, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–85. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radford IR. The level of induced DNA double-strand breakage correlates with cell killing after X-irradiation. Int J Radiat Biol Relat Stud Phys Chem Med. 1985;48:45–54. doi: 10.1080/09553008514551051. [DOI] [PubMed] [Google Scholar]
- 5.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–6. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 6.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- 7.Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–69. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 8.Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–9. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dungey FA, Loser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys. 2008;72:1188–97. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Shi Y, Guan R, et al. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol Cancer Res. 2008;6:1621–9. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 13.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 14.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 15.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–15. [PubMed] [Google Scholar]
- 16.Tong WM, Cortes U, Hande MP, et al. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 2002;62:6990–6. [PubMed] [Google Scholar]
- 17.Noel G, Giocanti N, Fernet M, Megnin-Chanet F, Favaudon V. Poly(ADP-ribose) polymerase (PARP-1) is not involved in DNA double-strand break recovery. BMC Cell Biol. 2003;4:7. doi: 10.1186/1471-2121-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner syndrome protein complex with Ku70/80 and PARP-1. J Biol Chem. 2004 doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 19.Corneo B, Wendland RL, Deriano L, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–6. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–33. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 22.Moshous D, Callebaut I, de Chasseval R, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–86. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 23.Rooney S, Alt FW, Lombard D, et al. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J Exp Med. 2003;197:553–65. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 25.McCulloch EA, Till JE. The sensitivity of cells from normal mouse bone marrow to gamma radiation in vitro and in vivo. Radiat Res. 1962;16:822–32. [PubMed] [Google Scholar]
- 26.Breslin C, Clements PM, El-Khamisy SF, Petermann E, Iles N, Caldecott KW. Measurement of chromosomal DNA single-strand breaks and replication fork progression rates. Methods Enzymol. 2006;409:410–25. doi: 10.1016/S0076-6879(05)09024-5. [DOI] [PubMed] [Google Scholar]
- 27.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 28.Deckbar D, Birraux J, Krempler A, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–55. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–91. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutation Research. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 32.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–62. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riballo E, Kuhne M, Rief N, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Hochegger H, Dejsuphong D, Fukushima T, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. Embo J. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–6. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 37.Godon C, Cordelieres FP, Biard D, et al. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–64. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundin C, North M, Erixon K, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 40.Woodhouse BC, Dianova II, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair (Amst) 2008;7:932–40. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Albert JM, Cao C, Kim KW, et al. Inhibition of poly(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin Cancer Res. 2007;13:3033–42. doi: 10.1158/1078-0432.CCR-06-2872. [DOI] [PubMed] [Google Scholar]
- 42.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–68. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Groves MD, Maor MH, Meyers C, et al. A phase II trial of high-dose bromodeoxyuridine with accelerated fractionation radiotherapy followed by procarbazine, lomustine, and vincristine for glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 1999;45:127–35. doi: 10.1016/s0360-3016(99)00122-4. [DOI] [PubMed] [Google Scholar]
- 44.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 45.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–59. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Zhou K, Zhang H, et al. Polymorphisms of LIG4 and XRCC4 involved in the NHEJ pathway interact to modify risk of glioma. Hum Mutat. 2008;29:381–9. doi: 10.1002/humu.20645. [DOI] [PubMed] [Google Scholar]
- 47.Greiner TC, Dasgupta C, Ho VV, et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2006;103:2352–7. doi: 10.1073/pnas.0510441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Kogel AJ. Radiation-induced damage in the central nervous system: an interpretation of target cell responses. Br J Cancer Suppl. 1986;7:207–17. [PMC free article] [PubMed] [Google Scholar]