The Biology and Treatment of EML4-ALK Non-Small Cell Lung Cancer (original) (raw)
. Author manuscript; available in PMC: 2011 Jul 1.
Published in final edited form as: Eur J Cancer. 2010 Apr 24;46(10):1773–1780. doi: 10.1016/j.ejca.2010.04.002
Abstract
The fusion between echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma kinase (ALK) has recently been identified in a subset of non-small cell lung cancers (NSCLCs). EML4-ALK is most often detected in never smokers with lung cancer and has unique pathologic features. EML4-ALK is oncogenic both in vitro and in vivo and ALK kinase inhibitors are quite effective in pre-clinical model systems. More recently ALK inhibitors have entered clinical development and remarkably clinical efficacy has been observed in NSCLC patients harbouring EML4-ALK translocations. This review will focus on the biology, clinical characteristics, diagnosis and treatment of EML4-ALK NSCLC.
Keywords: Non-small cell lung carcinoma, translocation, kinase inhibitor, clinical trial
Introduction
Non-small cell lung cancer (NSCLC) is a major cause of death worldwide, with most of the patients being diagnosed with disease in advanced stage, when treatment is only palliative1. Chemotherapy represents the standard of care for patients with advanced disease but conventional cytotoxic agents has reached a plateau in terms of efficacy in the last few years, encouraging the investigation of new compounds which target proteins that are selectively expressed and/or that undergo genomic alterations in cancer cells2. In the past several years an increase in the molecular understanding of lung cancer has led to a change in the treatment of the disease. This is highlighted by somatic mutations in EGFR where treatment with an EGFR kinase inhibitor (gefitinib) in EGFR mutant NSCLC patients leads to a superior response rate, a prolonged progression free survival and an improved quality of life compared to cytotoxic chemotherapy3.
The fusion of the anaplastic lymphoma kinase (ALK) with the echinoderm microtubule-associated protein-like 4 (EML4) was identified in 2007 in Japanese non-small cell lung cancers (NSCLC) 4. Additional studies, mostly involving East Asian patients, have reported that between 3%–13% of lung tumors harbor EML4-ALK fusions4–11. By extrapolation this would suggest that approximately 5% of all NSCLC cases contain an EML4–ALK translocation, equivalent to over 70,000 patients diagnosed annually worldwide.
Since the ALK tyrosine kinase activity is necessary for its transforming activity and oncogenicity, several ALK kinase inhibitors have been identified and are being evaluated in pre-clinical models in vitro and in vivo as potential clinical therapies7, 12, 13. ALK inhibitors lead to apoptosis in vitro and tumor shrinkage in vivo thus demonstrating the phenomenon of “oncogene addiction” 7. This is further confirmed by the dramatic clinical studies to date. In the phase I trial of PF-02341066, a remarkable 60% radiographic response rate has been observed specifically in EML4-ALK NSCLC patients14. This is a remarkably short period of time from the initial identification of the EML4-ALK translocation as oncogene to validation as a clinical target in NSCLC.
In this reiew, we highlight the clinical, biologic and molecular feature of EML4-ALK NSCLC patients and discuss the use of ALK inhibitors as therapies for this patient population.
Clinical and molecular features of EML4-ALK NSCLC
EML4-ALK NSCLC occurs most commonly in a unique clinical subgroup of NSCLC patients. These patients share many of the clinical features of NSCLC patients likely to harbour EGFR mutations 10, 15. However, for the most part, apart from rare exceptions, EML4-ALK and EGFR mutations are mutually exclusive6, 7, 10, 12. EML4-ALK translocations tend to occur in younger patients and those with more advanced NSCLC while this relationship has not been reported for EGFR mutant NSCLC6, 11.
Smoking history
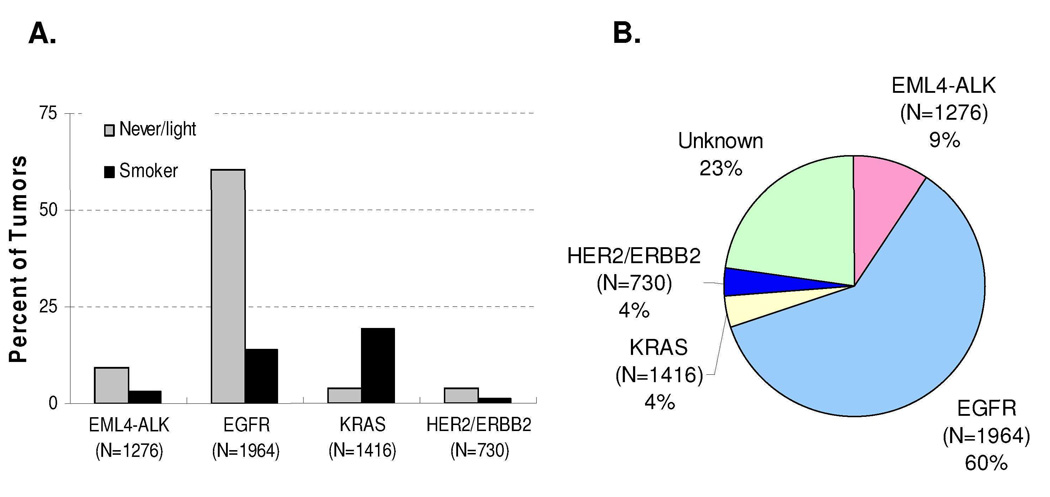
Initially, the EML4-ALK fusion gene was identified in a smoker with lung cancer; however, the accumulating evidence reveals that this genetic alterations is much more common in never/ former light (often defined as ≤ 10 pack years and quite ≥ 1 year ago) smokers with NSCLC 4, 7, 10. As shown in Figure 1A and Table 1, among the NSCLC patients that were never or former light smokers 9.4% of the tumors contained EML4-ALK translocations while the frequency was only 2.9% in current smokers (p<0.0001) 4–11. In this clinical population, never or former light smokers, EGFR mutations still account for the vast majority of patients while a minority contain either KRAS or ERBB2 mutations (Figure 1B)16–28. Of note, genetic alterations have been identified in approximately 25% of never/former light smokers (Figure 1B).
Figure 1. Frequency of somatic genetic changes in NSCLC.
A. EML4-ALK translocations, EGFR, KRAS and ERBB2 mutation frequencies broken down by smoking history. B. Frequency of somatic mutations in never or former light (≤ 10 pack years; quit ≥ 1 year ago) smokers. Data obtained from 16–28. Of note the somewhat higher EGFR mutation frequency is likely a reflection of the predominance of studies from East Asian countries. The EGFR mutation frequency in Caucasian never/former light smokers is ~ 35%21.
Table 1.
Frequency of EML4-ALK translocations broken down based on smoking history.
| No. ofsmokers | No. of neversmokers | EML4-ALK+(smokers) | EML4-ALK+(never smokers) | p | reference | |
|---|---|---|---|---|---|---|
| Soda et al | 24 | 9 | 8.3% | 11.1% | 1.0 | 4 |
| Inamura et al | 84 | 65 | 2.4% | 4.6% | 0.65 | 5 |
| Inamura et al | 147 | 105 | 3.4% | 5.7% | 0.53 | 6 |
| Koivunen et al | 184 | 69 | 1.1% | 8.5% | <.01 | 7 |
| Shinmura et al | 41 | 22 | 4.9% | 0% | 0.54 | 8 |
| Martelli et al | 101 | 16 | 7.9% | 6.3% | 1.0 | 9 |
| Wong | 125 | 141 | 0.8% | 8.5% | < .01 | 11 |
| Shaw et al | 56 | 85 | 0% | 22.4% | <.0001 | 10 |
| TOTAL | 762 | 514 | 2.9% | 9.4% | <.0001 |
Outcomes with current NSCLC therapies
Limited data exists to date on the efficacy of currently available therapies in patients with EML4-ALK NSCLC. In a study by Shaw and colleagues, 12 patients with ALK genomic alterations were treated with platinum based chemotherapy. The response rate, time to progression and overall survivals were similar to NSCLC patients harbouring EGFR mutations or those that were wild for both EML4-ALK and EGFR 10. In contrast, patients with EML4-ALK did not benefit from EGFR tyrosine kinase based therapy; their outcome was similar to patients that lacked EGFR mutations 10. These findings are also mirrored in pre-clinical studies where erlotinib is ineffective in a murine model harbouring of EML4-ALK NSCLC7.
Morphologic profile of ALK-rearranged NSCLC
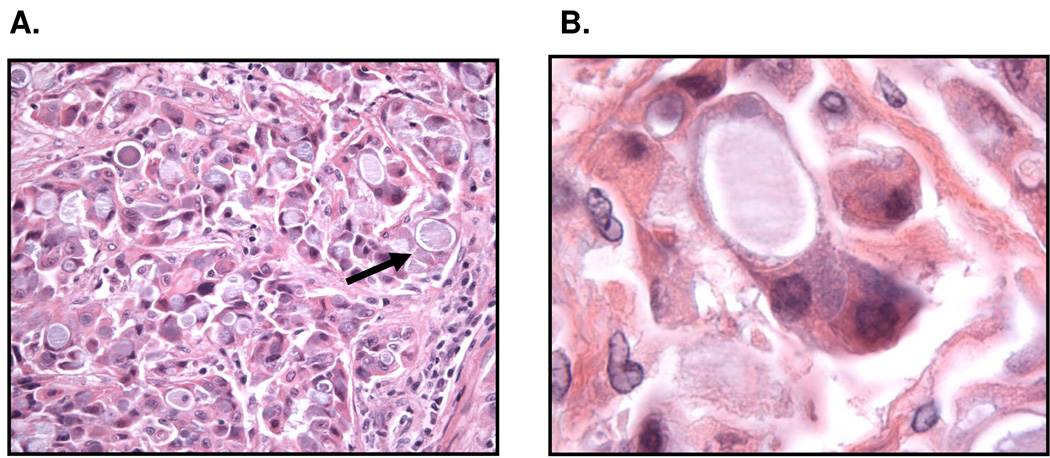
A variety of histologic features are reported to be associated with ALK-rearranged lung adenocarcinomas including acinar (ranging from well-differentiated tubulopapillary and cribriform patterns) to mostly signet-ring cell nests with mucin production 6, 10, 29, 30. Other histologic types such as squamous cell carcinoma and mucoepidermoid carcinoma also rarely contain EML4-ALK translocations4, 10. The acinar pattern is mostly reported to be associated with ALK-rearranged lung adenocarcinomas in Asian populations 6, 30. , whereas the signet-ring cell histology was reported mostly in the Western patients10, 29. The majority of Western patients showed tumor cells with a solid or sheet-like pattern easily distinguishable from the acinar, papillary or bronchioloalveolar patterns. Occasionally, a predominantly acinar pattern and bronchioloalveolar patterns could also be seen29. Even more striking were the cytologic features of tumors harboring ALK rearrangements. _ALK_-rearranged tumors showed at least focally tumor cells with abundant intracellular mucin and small, marginalized nuclei (Figure 2A). In a majority of cases, cells with abundant intracellular mucin comprised >10% of the overall tumor cellularity29. This distinct cytologic characteristic, unusual for lung carcinoma, is reminiscent of the “signet-ring” cells more commonly seen in gastric, colonic, and breast adenocarcinomas (Figure 2B). The majority of ALK-rearranged tumors (> 60%) demonstrate a solid growth pattern with >10% signet-ring cells. In contrast only a small minority of EML4-ALK wild type tumors, including those with EGFR mutations, demonstrate a solid growth pattern with >10% signet ring cells.
Figure 2. Pathologic characteristics of EML4-ALK NSCLC.
EML4-ALK NSCLC demonstrate a signet cell features (arrow). A. 40×X magnification B. 1000 ×X magnification.
Variants of EML4-ALK and non-EML4 translocation partners
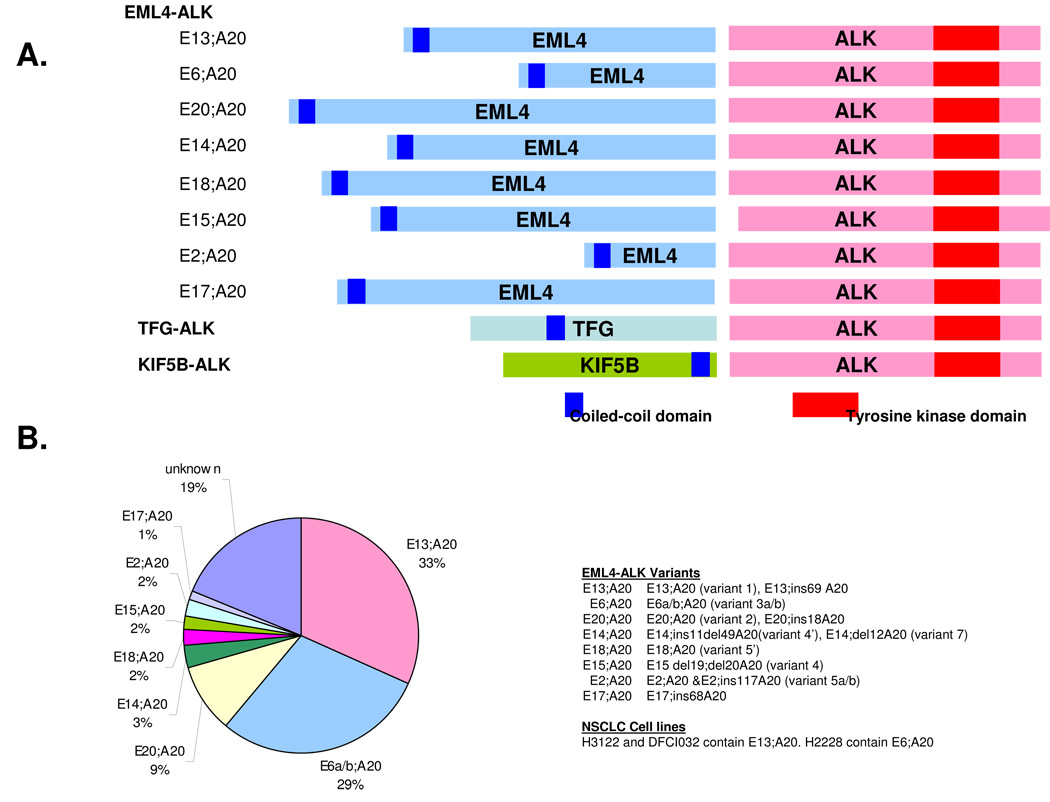
The inversion on chromosome 2 p, leading to the formation of the EML4-ALK fusion oncogene, is most commonly found in lung cancer patients and lung cancer cell lines. A few reports have also identified EML4-ALK in other cancers including breast and colorectal cancers31. The chromosomal inversion does not always occur in the same location and multiple EML4-ALK variants have been identified (Figure 3A). All involve the intracellular tyrosine kinase domain of ALK beginning at the portion encoded by exon 20. EML4, however, is variably truncated and gives rise to various variants of EML4-ALK. The amino-terminal coiled-coil domain within EML4 is necessary and sufficient for the transforming activity of EML4-ALK, probably through dimerization of the fusion proteins and hence is contained in all of the variants (Figure 3A) 4. At least 11 variants have been reported to date and most of them are oncogenic as assayed in NIH-3T3 cells or in Ba/F3 cells 4–11, 30, 32–36. The most common variants were E13;A20 (the nomenclature refers to the exons in EML4 (E) that are fused to ALK (A)) and E6a/b;A20, which are also referred to as variants 1 and 3a/b, respectively. These two are the most common variants of EML4-ALK and have been detected in 33% and 29% of NSCLC patients, respectively (Figure 3B). The NSCLC cell lines, H3122 and DFCI032 contain the E13;A20 variant while H2228 contains the E6a/b;A20 variant7. The clinical significance, if any, of the different variants is currently not defined.
Figure 3. Different variants of EML4-ALK and non-EML4 fusion partners.
A. Different variants of EML4-ALK are depicted. The nomenclature refers to the exon in EML4 translocated to the exon in ALK. B. Frequency of different EML4-ALK variants. The most common variants are E13;A20 (variant 1) and E6a/b; A20 (variant 3). Data obtained from 4–11, 30, 32–36. Of note not all studies list the specific EML4-ALK variant.
In ALK translocated NSCLC, EML4 does not appear to be the exclusive fusion partner with ALK. Two other fusions have been described, TFG and KIF5B, and both were identified as an ALK-fusion partner from NSCLC tumor samples30, 37 these 2 proteins also fuse with intracellular domain of ALK. Intriguingly TFG-ALK has also been described in anaplastic large cell lymphoma38. The presence of these non-EML4 fusion partners for ALK has implications for the method used for clinical detection of ALK translocated NSCLC.
Diagnosis of EML4-ALK NSCLC
_ALK_-rearrangements in a subset of anaplastic large cell lymphomas (ALCL) have been recognized for over 15 years and a variety of diagnostic techniques, currently employed in clinical practice, have been validated as sensitive and specific for detecting the genetic lesions characteristic of this tumor type39. However, there is currently no standard method for detecting EML4-ALK NSCLC. Several methods including polymerase chain reaction (PCR), immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) are currently being evaluated.
PCR based identification of EML4-ALK
Reverse transcriptase (RT)-PCR is a potentially rapid diagnostic method for identifying ALK translocated NSCLCs. A theoretical advantage of this technique is its extreme sensitivity for detecting mutant transcript and the presence of any amplification product implies an ALK rearrangement. However in practice, the technique faces several challenges. First, the RT-PCR analysis must be multiplexed. As mentioned above there are at least 11 variant EML4-ALK fusions, and non-_EML_4 translocation partners, therefore any PCR based strategy must incorporate validated primer pairs for all known ALK fusions. Second, the vast majority of patient biopsy specimens from lung cancer patients are stored as formalin fixed paraffin embedded (FFPE) tissues. RNA extracted from FFPE is highly degraded and, in general, more difficult to PCR relative to non-fixed, fresh-frozen tissue. Third, there is published evidence indicating that RT-PCR based detection of EML4-ALK can yield positive results in the absence of detectable _ALK-_rearrangements in both tumor, and non-tumor tissues9. Although the interpretation of these findings is still open to debate, it suggests a propensity for false positive results. Despite these disadvantages, there are advocates for using RT-PCR based screening methods32. However, this method may be difficult to implement in a routine clinical diagnostic laboratory.
FISH based methods for identification of EML4-ALK
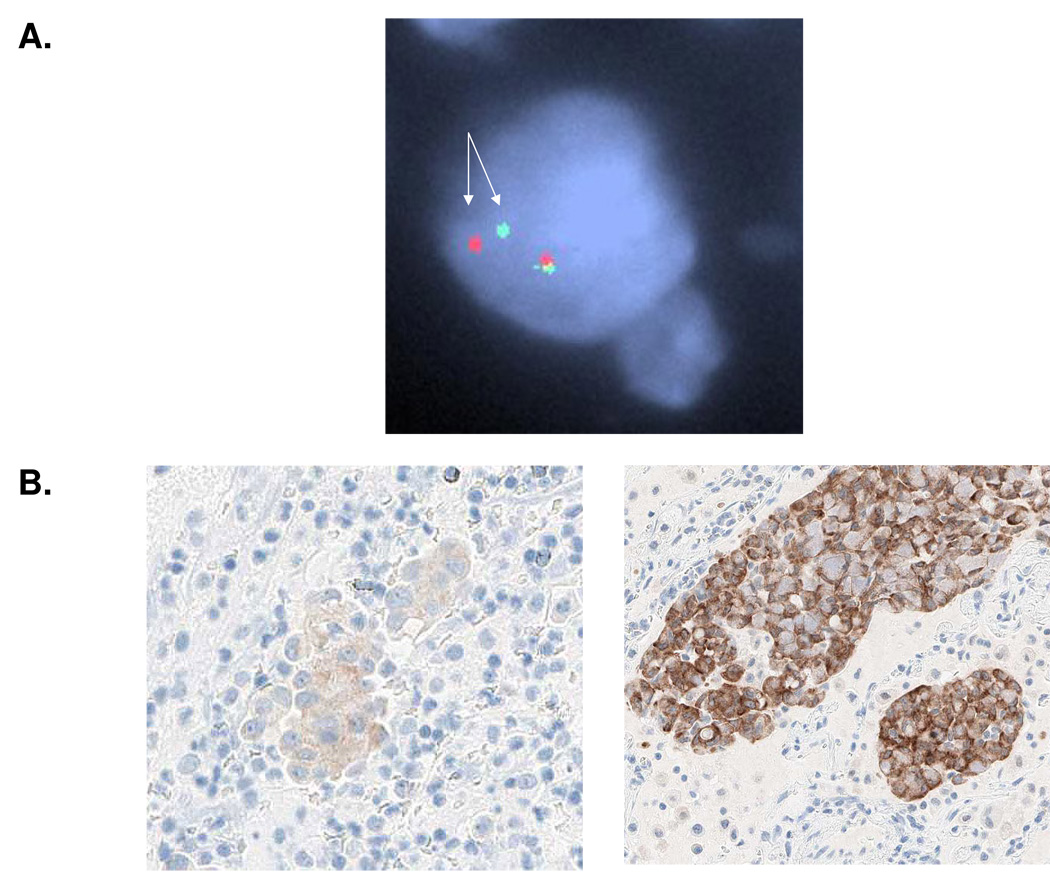
More specific detection of _ALK_-rearrangements can be achieved by the fluorescence in-situ hybridization (FISH) of probes flanking the ALK breakpoint with tumor cell nuclei10. A big advantage of FISH is that a commercially available probe set, developed for the diagnosis of _ALK_-rearranged ALCLs, is applicable for the diagnosis of _ALK_-rearranged lung adenocarcinomas. The test employs one probe 5' of the ALK locus and one probe within the ALK gene, which when hybridized against normal nuclei, yield a merged (green-orange fluorescent) signal that is easily visualized microscopically. However when the probe set is hybridized against nuclei with a rearrangement involving the 5' portion of the ALK locus the result is a "split" (green and orange fluorescent) signal (Figure 4). In theory, any inter-chromosomal or intra-chromosomal lesion involving ALK (including cancers harboring non-EML4 fusion partners) will be detected by this test. However a cautionary finding is that the "split" signal characteristic of an EML4-ALK fusion can be subtle, due to the loss and inversion of only a small amount of genetic material on chromosome 2. Also the 5' probe occasionally fails to hybridize, presumably due a loss of the target locus in the tumor. These patterns contrast the widely split, dual hybridization pattern found in tumors with an inter-chromosomal rearrangement of the ALK locus- such as _ALK_-rearranged ALCLs. An additional complicating factor with FISH is the destruction of tissue morphology when formalin-fixed, paraffin embedded (FFPE) biopsy specimens are analyzed in this manner. Thus, although FISH is a sensitive and specific means to detect _ALK_-rearrangements in lung adenocarcinoma, it is not infallible. In fact, in a recent study, an _ALK_-rearranged lung adenocarcinoma by immunohistochemical testing for ALK protein expression that was, initially, mistakenly diagnosed as ALK wild type by prior FISH analysis29. Furthermore, unlike PCR, FISH cannot distinguish between the different EML4-ALK fusion variants. It is currently not clear whether there are any functional or therapeutic differences amongst the different variants to warrant more specific knowledge. FISH is the diagnostic method used as an eligibility criterion in the current clinical trials of PF-02341066. Current studies use ≥ 15% split nuclei as indicative of an ALK rearrangement40. However, the therapeutic implications, if any, of the frequency of split signals and the number of nuclei evaluated remains to be determined.
Figure 4. Diagnostic methods for EML4-ALK NSCLC.
A. FISH analysis using the ALK split apart probe. The arrows depict the split signals indicative of a chromosome 2 inversion. The tumor is heterozygous – the signals remain together on the other allele. B. IHC analysis for ALK using the D5F3 (Cell Signaling Technology 41) antibody. Shown are examples of low (left) and high (right) ALK expressing tumors.
IHC detection methods for EML4-ALK
Immunohistochemical (IHC) analysis of FFPE tissue specimens remains a mainstay of routine surgical pathology practice. The major advantage of this approach is an ability to assay for tumor-specific antigen expression without loss of the cytologic and architectural features that distinguish normal from pathologic tissue. Several antibodies specific for the human ALK protein have been developed and a few validated in IHC tests that are widely used to diagnose _ALK_-rearranged ALCLs today39. The sensitivity and specificity of the IHC test is such that genetic testing is considered unnecessary and redundant when it is employed during the routine evaluation of an ALCL of unknown ALK status. However the IHC tests used to diagnose _ALK_-rearranged ALCLs in clinical laboratories worldwide is inadequate for the detection of the majority of _ALK_-rearranged lung adenocarcinomas. This is due to the lower level of ALK expression in _ALK_-rearranged NSCLCs compared with _ALK_-rearranged ALCLs. In an initial survey of 10 FISH-confirmed, _ALK_-rearranged lung adenocarcinomas, Rodig and colleagues found that only 4 stained positively for ALK protein expression by the standard clinical test29. They and others have been able to increase the number of positive staining cases using an additional, non-traditional amplification step in the immunostaining protocol29, 30. However, not all cases stained positive by even this method. These findings initially raised the question of whether a subset of _ALK_-rearranged lung adenocarcinomas fail to express the ALK protein, or whether the levels of ALK protein expression in these cases was simply too low to be detected using standard reagents.
More recently Mino-Kenudson and colleagues reported on an IHC test based on novel antibodies with increased sensitivity and specificity for detecting ALK protein expression in FFPE (Figure 4B)41. Using this test, they detected ALK protein expression in all 22 _ALK_-rearranged lung adenocarcinomas tested, and found no expression in 131 ALK wild type lung adenocarcinomas. ALK protein expression was detected in all _ALK_-rearranged lung adenocarcinomas to be substantially lower than the expression in _ALK_-rearranged ALCLs, and in 13 of 22 cases (59%), ALK protein expression could only be detected using our novel, highly sensitive IHC test (Figure 4B)41. These findings support that ALK expression is restricted to lung cancers that harbor _ALK_-rearrangements. Furthermore they open up the possibility of being able to diagnose such cancers using routine IHC based methods which, unlike FISH, is available in every pathology laboratory. This may allow pathologist a means to rapidly screen for patients harboring an ALK translocation who may be candidates for ALK targeted therapies. A caveat however is that tissue staining for ALK, even with the most sensitive of IHC tests, may be weak and focal in the biopsy sections (Figure 4B), in which case confirmatory FISH studies should be considered.
ALK-targeted therapy in NSCLC
Preclinical studies
The initial studies reporting on the discovery of EML4-ALK raised the possibility that inhibiting the kinase activity of ALK may be an effective clinical therapy 4. Furthermore, transgenic mice expressing EML4-ALK in the lung epithelium develop numerous lung adenocarcinomas demonstrating the oncogenic nature of this fusion gene12. Preclinical studies demonstrate that EML4-ALK NSCLC cell lines undergo downregulation of critical survival signalling pathways and apoptosis when treated with an ALK kinase inhibitor 7, 13. This is analogous to what has been observed with EGFR inhibitors in EGFR mutant NSCLC 42. Similarly, ALK inhibitors have been evaluated in vivo, both in xenograft models generated from EML4-ALK NSCLC cell lines, and lead to effective tumor regressions of established tumors7, 12. Currently, only one agent targeting ALK, PF-02341066 initially designed as an inhibitor of MET, is in clinical use although others have been examined in pre-clinical model systems7, 12, 43. This orally bioavailable small molecule inhibitor has been shown to inhibit the growth of ALK translocated cancer cell lines including EML4-ALK NSCLC13, 43.
Clinical studies
PF-02341066 (crizotinib) is an orally bioavailable ALK inhibitor currently under clinical development. The phase I study of this agent started in May 2006. After the discovery of EML4-ALK, and that PF-02341066 also inhibits ALK, the dose expansion cohorts in this study included patients with either genomic alterations in MET (amplifications and/or mutations) or ALK14. The initial findings were presented at ASCO 2009 and demonstrated a remarkable 53 % response rate (10/19 patients) and a disease control rate (partial response & stable disease) of 79% (15/19)14. Additional responses continue to be reported. These dramatic findings have led to two subsequent clinical trials of PF-02341066. The first is a randomized phase III trial of PF-02341066 compared with standard second line chemotherapy (pemetrexed or docetaxel) in second line EML4-ALK NSCLC. The primary endpoint of this trial is progression free survival. The second is a phase II clinical trial of single agent PF-02341066 in EML4-ALK NSCLC designed for patients not eligible for the phase III trial or patients randomized to chemotherapy who subsequently developed progressive disease. In a remarkably short period of time – from initial discovery to clinical validation – ALK targeted therapies are in advanced clinical development for EML4-ALK NSCLC. It is anticipated, that if EML4-ALK NSCLC behaves in an analogous manner to EGFR mutant NSCLC, that ALK targeted therapies will quickly emerge as the standard systemic therapy for this subset of NSCLC patients.
Conclusions
EML4-ALK NSCLC represents a unique subset of NSCLC patients for whom ALK inhibitors may represent a very effective therapeutic strategy. The challenge remains to incorporate and disseminate widespread use of diagnostic testing for EML4-ALK to identify this patient subset. As we learn more about EML4-ALK NSCLC we will continue to uncover unique biological and molecular features of this patient subset also undoubtedly encounter drug resistance to ALK targeted therapies.
Acknowledgments
This study was supported by the National Institutes of Health R01CA136851 (P.A.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflicts of interest
References
- 1.Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31(1):100–110. doi: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007 doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3(1):13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 6.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22(4):508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 7.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14(13):4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinmura K, Kageyama S, Tao H, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61(2):163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Martelli MP, Sozzi G, Hernandez L, et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol. 2009;174(2):661–670. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115(8):1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 12.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105(50):19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68(9):3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 14.Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. Journal of Clinical Oncology. 2009;27 15s:abstract 3509. [Google Scholar]
- 15.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99(2):245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 17.Tomizawa Y, Iijima H, Sunaga N, et al. Clinicopathologic significance of the mutations of the epidermal growth factor receptor gene in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11(19 Pt 1):6816–6822. doi: 10.1158/1078-0432.CCR-05-0441. [DOI] [PubMed] [Google Scholar]
- 18.Shih JY, Gow CH, Yu CJ, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer. 2006;118(4):963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh RK, Lim KH, Kuo HT, Tzen CY, Huang MJ. Female sex and bronchioloalveolar pathologic subtype predict EGFR mutations in non-small cell lung cancer. Chest. 2005;128(1):317–321. doi: 10.1378/chest.128.1.317. [DOI] [PubMed] [Google Scholar]
- 20.Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11(3):1167–1173. [PubMed] [Google Scholar]
- 21.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 22.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 23.Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65(12):5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 24.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26(6):983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 25.Blons H, Cote JF, Le Corre D, et al. Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol. 2006;30(10):1309–1315. doi: 10.1097/01.pas.0000213285.65907.31. [DOI] [PubMed] [Google Scholar]
- 26.Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92(6):1525–1530. doi: 10.1002/1097-0142(20010915)92:6<1525::aid-cncr1478>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 27.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65(5):1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 28.Mounawar M, Mukeria A, Le Calvez F, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67(12):5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- 29.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15(16):5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15(9):3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 31.Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7(9):1466–1476. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14(20):6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 33.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68(13):4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 34.Fukuyoshi Y, Inoue H, Kita Y, Utsunomiya T, Ishida T, Mori M. EML4-ALK fusion transcript is not found in gastrointestinal and breast cancers. Br J Cancer. 2008;98(9):1536–1539. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10(3):298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17(3):889–897. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 37.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez L, Pinyol M, Hernandez S, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94(9):3265–3268. [PubMed] [Google Scholar]
- 39.Cataldo KA, Jalal SM, Law ME, et al. Detection of t(2;5) in anaplastic large cell lymphoma: comparison of immunohistochemical studies, FISH, and RT-PCR in paraffin-embedded tissue. Am J Surg Pathol. 1999;23(11):1386–1392. doi: 10.1097/00000478-199911000-00009. [DOI] [PubMed] [Google Scholar]
- 40.Shaw RJ, Paez JG, Curto M, et al. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell. 2001;1(1):63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 41.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16(5):1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Janne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64(20):7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 43.Christensen JG, Zou HY, Arango ME, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6(12):3314–3322. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]