The Cajal Body and Histone Locus Body (original) (raw)
Abstract
The Cajal body (CB) is a nuclear organelle present in all eukaryotes that have been carefully studied. It is identified by the signature protein coilin and by CB-specific RNAs (scaRNAs). CBs contain high concentrations of splicing small nuclear ribonucleoproteins (snRNPs) and other RNA processing factors, suggesting that they are sites for assembly and/or posttranscriptional modification of the splicing machinery of the nucleus. The histone locus body (HLB) contains factors required for processing histone pre-mRNAs. As its name implies, the HLB is associated with the genes that code for histones, suggesting that it may function to concentrate processing factors at their site of action. CBs and HLBs are present throughout the interphase of the cell cycle, but disappear during mitosis. The biogenesis of CBs shows the features of a self-organizing structure.
Cajal bodies and histone locus bodies act as processing plants for splicing machinery and histone RNAs. Their reassembly after mitosis is a classic example of self-organization.
The CB and the HLB can be considered together for several reasons. First, the canonical marker for the CB, coilin, occurs at high concentration in some HLBs. Second, CBs and HLBs are often physically associated, suggesting some type of interaction between the two. Finally, the very large CBs of the amphibian oocyte, long a favorite for studies of CB function, share properties of both HLBs and CBs.
CAJAL BODIES
As the current name suggests, the CB was first described by Ramon y Cajal, the great Spanish neuroanatomist, who in 1906 shared the Nobel Prize with Camillo Golgi for studies on the cellular architecture of the nervous system. Using a silver impregnation technique, Cajal found a small round body within the nuclei of various nerve cells, which he called the accessory body (cuerpo accessorio) (Cajal 1903; Cajal 1910). His remarkable morphological studies also included observations on splicing speckles, nucleoli, and the nuclear envelope (Lafarga et al. 2009). Bodies that we now know are related to Cajal's cuerpo accessorio were discovered independently in organisms as diverse as mammals, amphibians, insects, and plants. They were given equally diverse names—coiled bodies in mouse, rat, and human cells (Monneron and Bernhard 1969), Binnenkörper or endobodies in insects (Bier et al. 1967), and nucleolus associated bodies in plants (Chamberland and Lafontaine 1993). Order was brought to this somewhat chaotic field with the discovery of a protein named coilin, after its discovery in the coiled bodies of HeLa cells (Andrade et al. 1991; Raska et al. 1991). Antibodies against coilin turned out to be good markers for coiled bodies in vertebrate cells and even cross-reacted with the nucleolus associated bodies of the pea, Pisum sativum (Beven et al. 1995). It thus became clear that homologous nuclear organelles existed in a wide range of eukaryotes (Figs. 1 and 2). To recognize this commonality and to regularize the terminology, the name Cajal body was adopted as a general term for a nuclear body that contains coilin (Gall et al. 1999).
Figure 1.
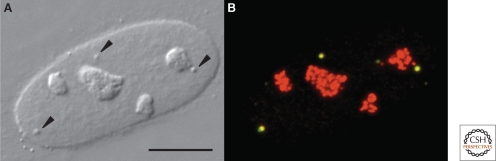
CBs in a HeLa cell nucleus. (A) DIC image showing four prominent nucleoli and three faintly detectable CBs (arrowheads). (B) The same nucleus immunostained with antibodies against coilin (green) and fibrillarin (red). The three CBs in this nucleus appear yellow because they stain with both antibodies. The nucleoli stain only for fibrillarin. Fibrillarin is the methyltransferase for 2′_-O-_methylation of rRNA in the nucleoli and snRNAs in the CBs. Modified from Gall (2003) with permission from Nature Reviews. Bar = 10 µm.
Figure 2.
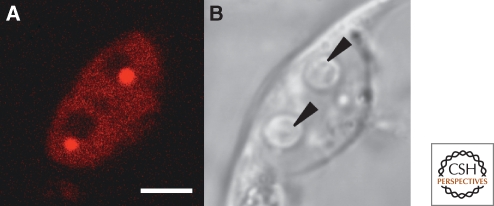
A cell from a protoplast of Arabidopsis after transfection with a construct of U2B″-mRFP, a component of the U2 snRNP that is enriched in the CB. (A) Fluorescence image showing two brightly labeled CBs in the nucleus. Each is associated with a large, unstained nucleolus. (B) DIC image of the same cells. The nucleoli are now visible (arrowheads) but the CBs are not. Bar = 10 µm. Image kindly provided by Zdravko Lorkovic, Medical University of Vienna, Austria.
Coilin
After its discovery in HeLa cells, coilin quickly became the signature marker for CBs in mammalian cells. Human and mouse coilin are similar in size, MW = 62.6 and 62.3 kDa respectively, and their overall sequence identity is high (Andrade et al. 1991; Tucker et al. 2000). Xenopus coilin is slightly smaller, 59.6 kDa, and its sequence diverges considerably from that of the two mammalian proteins (Tuma et al. 1993). Outside the vertebrates it becomes increasingly difficult to recognize coilin by sequence comparison. What are clearly orthologs of coilin have been described from Arabidopsis (Collier et al. 2006) and Drosophila (Liu et al. 2009), but so far coilin has not been identified in Caenorhabditis, Saccharomyces, or other important nonvertebrate model organisms.
Despite its usefulness as a marker for the CB, relatively little is known about coilin as a protein, particularly what biochemical function(s) it might play in the CB. Coilin binds to the survival motor neuron (SMN) protein and to various Sm and Lsm proteins, suggesting that it may function in assembly or modification of snRNPs (Hebert and Matera 2000; Hebert et al. 2001; Xu et al. 2005). There is strong genetic evidence from the mouse, Arabidopsis, and Drosophila that coilin is required for formation of CBs. Knockout of the coilin gene in the mouse leads to a semi-lethal phenotype. Some homozygous individuals die as embryos, and those adults that do survive have significant fertility and fecundity defects (Walker et al. 2009). Cultured cells derived from the knockouts do not have typical CBs. Instead, they show three types of “residual” bodies, each of which contains a subset of CB components (Tucker et al. 2001; Jády et al. 2003). In Arabidopsis the no cajal body 1 (ncb-1) mutant involves a single base substitution in the coilin gene, although it is not certain that the mutant is completely null for coilin (Collier et al. 2006). Homozygous ncb-1 plants are fully viable, but CBs are not detectable with antibodies against other CB components (U2B″ and fibrillarin) or by electron microscopy. In Drosophila two independent null mutants for coilin are fully viable as homozygotes (Beumer et al. 2008; Liu et al. 2009). Cells from coilin-null flies lack CBs detectable by immunostaining or in situ hybridization for several typical CB components. Thus, in the three organisms in which coilin mutations have been studied, coilin is required for normal CB formation, but neither coilin nor a typical CB is essential for viability.
Despite the long-standing use of coilin as a unique marker for CBs, and the critical role that coilin plays in maintaining structural integrity of CBs, we now know that coilin can also occur in HLBs, as discussed later in this article.
Small Nuclear Ribonucleoproteins (SnRNPs)
As soon as CBs were identified by immunostaining with antibodies against coilin, it became easy to use other antibodies and in situ hybridization probes to make a catalog of common CB components. It was quickly realized that CBs contain a variety of proteins and RNAs involved in RNA processing, particularly the splicing snRNAs (U1, U2, U4, U5, and U6) and their associated proteins (Lamond and Carmo-Fonseca 1993; Matera and Ward 1993; Spector 1993). Because splicing itself does not occur in CBs, it was suggested that CBs play some role in the assembly or modification of the splicing snRNPs before their recruitment to the chromosomes. The biogenesis of splicing snRNPs is a complex process involving both nuclear and cytoplasmic steps (Will and Lührmann 2001; Patel and Bellini 2008). In brief, transcription of the snRNAs occurs in the nucleus followed by their export to the cytoplasm. In the cytoplasm the monomethyl guanosine cap at the 5′ end becomes trimethylated, and each snRNA is assembled into a complex with seven conserved Sm proteins. Finally, the assembled snRNPs are imported back into the nucleus. Because snRNAs in the CB are associated with snRNP proteins and have a trimethylguanosine cap, it is assumed that they have already returned from the cytoplasm. This conclusion is reinforced by kinetic studies showing that newly imported snRNPs travel first to the CB (Sleeman and Lamond 1999) and only later appear in the speckles (interchromatin granule clusters) and finally on the chromosomes, where splicing itself takes place. Modification of specific nucleotides on the snRNAs probably occurs in the CB. It is less clear how much assembly of the splicing machinery takes place in the CB. A role for CBs has been suggested in the final steps of U2 snRNP biogenesis (Nesic et al. 2004) and a particularly compelling case has been made that assembly of the U4/U6-U5 tri-snRNP occurs in the CB (Stanek et al. 2003; Schaffert et al. 2004). Evidence has also been presented that snRNPs recycle through CBs (Stanek et al. 2008). The splicing snRNPs very probably pass from CBs to the nuclear speckles (interchromatin granule clusters) on their way to the sites of RNA synthesis and splicing on the chromosomes. However, the extent to which individual snRNPs are organized into higher complexes in the speckles is not known (Lamond and Spector 2003). A recent study on the amphibian oocyte suggests that snRNPs may be recruited to lampbrush chromosomes independently of their assembly into mature spliceosomes (Patel et al. 2007). If this is generally true for other cells, then CBs may have only a limited role in assembly of snRNPs into higher order complexes.
Small CB-associated RNAs (scaRNAs)
An important advance in the understanding of CB function came with the discovery of CB-specific RNAs (scaRNAs). Sca-RNAs are closely related to small nucleolar RNAs (snoRNAs) in both structure and function. Both share the so-called box C/D and box H/ACA motifs and both are involved in the posttranscriptional modification of other RNAs. Box C/D snoRNAs act as guides for the insertion of 2′_-O-_methyl groups on specific ribose moieties of rRNA, whereas the box H/ACA class mediates the conversion of specific uridines to pseudouridine (Bachellerie and Cavaille 1997; Tollervey and Kiss 1997). Fibrillarin is the methyl transferase and dyskerin/NAP57/CBF5 is the pseudouridine synthase; each is complexed with three additional proteins to form the active enzyme. ScaRNAs carry out similar reactions on snRNAs. The first discovered and the best-studied scaRNA is U85 (Darzacq et al. 2002; Kiss et al. 2002). It is an unusual guide RNA that mediates two modifications: 2′_-O-_methylation of C45 and pseudouridylation of U46 in human U5 snRNA (C46 and U47 in Drosophila). Cell fractionation and in situ hybridization experiments demonstrate that U85 scaRNA is strongly localized in CBs in both HeLa cells and Drosophila (Figs. 3A and 4B). This localization contrasts with that of its substrate, U5 snRNA, which is concentrated in CBs, but like other snRNAs is widely distributed throughout the nucleus. The localization of U85 and other scaRNAs in the CB differs from the localization of most box C/D and box H/ACA guide RNAs, which concentrate in the nucleolus. It has been shown that CB localization in vertebrate cells is dependent on a relatively short consensus sequence termed the CAB box (Richard et al. 2003). A related but somewhat different motif has been described for Drosophila scaRNAs. The CAB box of both human and Drosophila scaRNAs binds a conserved WD40-repeat protein, which is required for their CB localization (Tycowski et al. 2009).
Figure 3.
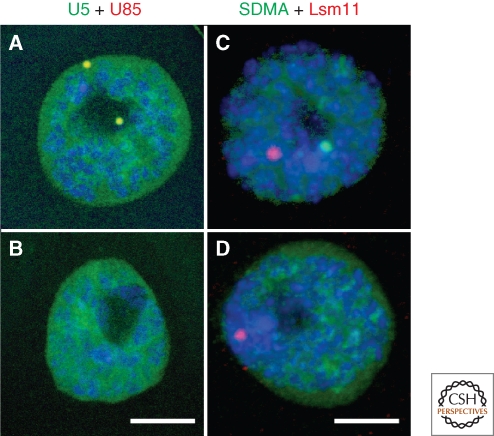
Malpighian tubule nuclei of Drosophila after fluorescent in situ hybridization (A and B) or immunostaining (C and D) for CB components. (A) A nucleus from a wild type fly showing two CBs. The CBs appear yellow because they contain both U85 scaRNA (red) and U5 snRNA (green). There is a high level of U5 throughout the nucleoplasm. (B) A nucleus from a coilin-null fly hybridized as in A. A CB is no longer detectable, although U5 label in the nucleoplasm is unaffected. (C) A nucleus from a wild type fly showing a CB immunostained for symmetric dimethylarginine, a marker for snRNP proteins (green), and an HLB labeled for Lsm11, a component of the U7 snRNP (red). (D) A nucleus from a coilin-null fly labeled as in C. A CB is no longer detectable, but the HLB is apparently unaffected. Modified from Liu et al. (2009) with permission from Molecular Biology of the Cell. Bar = 10 µm in A and B, 5 µm in C and D.
Figure 4.
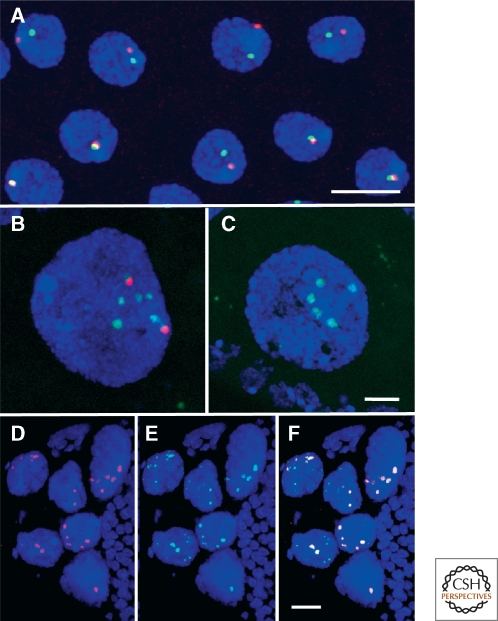
CBs and HLBs in Drosophila. (A) Each nucleus of the ejaculatory duct of the male fly usually has a single CB, immunostained green with an antibody against coilin, and a single HLB, immunostained red with an antibody against Lsm11. Often the two bodies are close to one another or touching. Bar = 5 µm. (B and C) In Drosophila and many other insects, highly polyploid nurse cells supply proteins and RNAs to the transcriptionally silent oocyte. Their nuclei display prominent CBs and HLBs, here shown after double in situ hybridization for U85 scaRNA (red) and U7 snRNA (green). (B) shows a stage 8 egg chamber of a wild type fly. C shows the same stage from a coilin-null fly. CBs are not detectable in the mutant, whereas HLBs appear unaffected. Bar = 5 µm. (D_–_F) Nurse cell nuclei from a stage 11 egg chamber showing prominent HLBs that contain coilin. Immunostained with antibodies against Lsm11 (D, red) and coilin (E, green). The overlay in F shows colocalization of Lsm11 and coilin. Modified from Liu et al. (2009) with permission from Molecular Biology of the Cell. Bar = 20 µm.
The specific localization of scaRNAs in the CB suggests that methylation and pseudouridylation of snRNAs occur in the CB, after import of the assembled snRNPs into the nucleus. This hypothesis is strongly supported by experiments in cultured cells showing that artificial substrates of scaRNAs became modified when targeted to the CB but not when targeted to the nucleolus (Jády et al. 2003). This hypothesis is also consistent with the well-known concentration of fibrillarin in the CB (Fig. 1). At the same time, it is unlikely that modification of snRNAs is limited to CBs, because coilin-null flies, which lack CBs (Fig. 3 B,D; Fig. 4C), have normal levels of scaRNAs, and all of their snRNAs are correctly modified (Deryusheva and Gall 2009). It seems probable that scaRNAs and other CB components normally exist in the nucleoplasm as macromolecular complexes that are too small to be resolved individually by conventional light microscopy. Coilin is essential for these complexes to assemble into detectable CBs, but assembly of such bodies is not required for function, at least not for the scaRNA-dependent modification of splicing snRNAs.
A scaRNA of unusual interest is the RNA component of telomerase, the enzyme responsible for maintaining the ends or telomeres of eukaryotic chromosomes. Telomerase RNA has been shown by in situ hybridization in CBs in human cancer cell lines (Zhu et al. 2003; Jády et al. 2004), but it is at low or undetectable levels in noncancer cells. Telomerase RNA has a box H/ACA motif as well as a CAB box. As with other scaRNAs, the CAB box is required for CB localization (Theimer et al. 2007). The same WD40-repeat protein that binds other scaRNPs is part of the human telomerase holoenzyme and is required for telomere synthesis in HeLa cells (Venteicher et al. 2009).
CBs, Gems, and the SMN Protein
An unusually interesting component of CBs is the survival motor neuron protein (SMN). When the intracellular localization of SMN was first studied by immunofluorescence, the protein was seen throughout the cytoplasm and in a nuclear body similar in size to the CB and often close to it. For this reason the body was called the “Gemini of the CB” or simply gem (Liu and Dreyfuss 1996). By chance the HeLa line in which the discovery was made is unusual: In other human cell lines, including different HeLa strains, in primary neurons, and in Drosophila, SMN is colocalized with coilin in the CB (Matera and Frey 1998; Carvalho et al. 1999; Liu et al. 2006). Hence SMN can generally be treated as an important component of the CB, not as a marker for a separate nuclear body.
As the name implies, SMN is required in mammals for proper functioning of motor neurons, especially those of the spinal cord. In the mouse and Drosophila, null mutations in the single-copy smn gene are lethal (Miguel-Aliaga et al. 2000; Chan et al. 2003; Rajendra et al. 2007). The situation in humans is somewhat different, because there are two copies of the gene, one of which has an altered splice site that leads to inefficient processing of its transcript. Without going into the somewhat complex genetics of the human smn gene, suffice it to say that mutations often lead to the condition known as spinal muscular atrophy (SMA). SMA occurs in approximately 1 in 6000 newborns and results in early death.
Biochemical studies show that SMN in vertebrate cells occurs in a macromolecular complex called the assemblysome, consisting of SMN itself, seven gemins, and several other factors. This complex functions in the cytoplasm as the chaperone for assembly of the splicing snRNAs with the seven-member Sm ring (Meister et al. 2002; Pellizzoni et al. 2002; Shpargel and Matera 2005; Battle et al. 2006). SMN accompanies the assembled snRNPs on their return to the nucleus (Narayanan et al. 2004), but whether SMN has a specific nuclear function in the CB is not known.
Expression of GFP-tagged human SMN in budding yeast led to specific localization of the probe in a small structure inside the nucleolus, which the authors called the nucleolar body (Verheggen et al. 2001; Verheggen et al. 2002). Some steps in the maturation of U3 snoRNA also occur in this body. The association with the nucleolus, the targeting of SMN, and the maturation of U3 all suggest that the yeast nucleolar body is equivalent to the CB of more complex eukaryotes.
Association of CBs with Specific Gene Loci
Because nucleoli are associated with a specific locus or loci on chromosomes, it is natural to ask whether similar associations exist for CBs and other nuclear organelles. In the case of the CB, there is no evidence that transcription occurs in the body itself and so there is no reason to suppose that a CB corresponds to a highly active gene locus like the nucleolus. Nevertheless, CBs might form at specific loci or travel there by some unknown mechanism as a means of bringing needed factors to those loci. Such a relationship is suggested by the fact that CBs in cultured vertebrate cells show preferential association with snRNA gene loci. CBs in these cells associate not only with U1, U2, and U4 gene clusters but also with the loci of the minor U11 and U12 snRNAs (Smith et al. 1995; Frey et al. 1999; Jacobs et al. 1999; Frey and Matera 2001). Matera suggested that snRNAs in the CB somehow exert a feedback regulation on snRNA transcription at these gene loci. Whatever the reason for the association, the relationship between CBs and snRNA loci is both dynamic and dependent on transcription, as shown in a recent experimental analysis (Dundr et al. 2007). An array of inducible U2 snRNA genes was cotransfected into cultured cells along with fluorescently labeled coilin. So long as the U2 array was transcriptionally silent, the array showed no specific relationship to CBs in the same nucleus. However, upon induction of transcription, the U2 array moved toward the closest CB and eventually came in contact with it. This remarkable movement was disrupted in the presence of a dominant negative mutant of β-actin, suggesting a role for nuclear actin in repositioning chromosomal loci in response to transcriptional activation.
Another special relationship exists between CBs and the telomeres. During most of the cell cycle telomerase RNA is demonstrable only in CBs. However, during S phase, when telomeres are elongated, telomerase RNA can also be detected in a subset of telomeres (Jády et al. 2006; Tomlinson et al. 2006; Tomlinson et al. 2008). These telomeres contain other components of the telomerase holoenzyme, which do not normally accumulate there. In addition, CBs were seen to make transient associations with telomeres during the S phase. These results suggest that specific interactions occur between CBs and telomeres during telomere elongation. The functional relevance remains to be determined.
HISTONE LOCUS BODIES
The HLB was described several times in Drosophila and human cells before a clear distinction was made between it and the CB (Frey and Matera 1995; Calvi et al. 1998; Liu et al. 2000; Ma et al. 2000). The existence of two separate bodies within the same nucleus was shown in experiments designed to identify the Drosophila CB in tissues of the fly (Liu et al. 2006). Because Drosophila coilin was not available at that time, our laboratory decided to use two other probes that we thought would serve equally well: the U7 snRNP and U85 scaRNA. Based on studies of the Xenopus oocyte, the U7 snRNP seemed to be a good marker for the CB (Wu and Gall 1993), and U85 scaRNA had been shown to label the CB in Drosophila S2 cells (Darzacq et al. 2002). To our surprise we found that the U7 snRNP and U85 scaRNA detected two independent nuclear organelles in various tissues of the fly (Fig. 3C,D; Fig. 4A,B). The organelle that contained U85 scaRNA was designated the CB, based on the presence of other common CB components. Because the organelle that contained the U7 snRNP was invariably associated with the histone genes on chromosome 2, we called it the HLB. The CB and HLB lie close together or actually touch one another in many Drosophila nuclei, suggesting some still unknown functional relationship between the two. Studies from other laboratories soon showed that HLBs could be identified as organelles distinct from CBs in the nuclei of cultured mammalian cells (Bongiorno-Borbone et al. 2008; Ghule et al. 2008).
Most of the factors that have been shown in HLBs are known to be involved in processing of histone pre-mRNAs. The genes coding for the major histones do not contain introns and hence their pre-mRNAs are not spliced. However, the pre-mRNAs carry a 3′ extension that is cut off before export to the cytoplasm (reviewed in Marzluff 2005). The site of cleavage is determined by the U7 snRNP acting in concert with a variety of other factors (Dominski et al. 2005; Kolev and Steitz 2005; Sullivan et al. 2009). In addition to U7 snRNA and the two U7 specific snRNP proteins Lsm10 and Lsm11, a number of other proteins have recently been detected in HLBs, including stem-loop binding protein (SLBP), FLASH, p220NPAT, negative elongation factor (NELF), symplekin, and the unknown protein recognized by mAb MPM-2 (Narita et al. 2007; White et al. 2007; Bongiorno-Borbone et al. 2008; Ghule et al. 2008; Ghule et al. 2009; Sullivan et al. 2009; Yang et al. 2009).
The behavior of CBs and HLBs during oogenesis in Drosophila turned out to be particularly interesting. The polyploid nurse cell nuclei in the youngest egg chambers usually display a single CB and a single HLB. As oogenesis proceeds, the number of HLBs increases until about stage 8, when there are usually 8–16 HLBs. As the nurse cells continue to grow, their CBs break up into tiny fragments that eventually disappear. In contrast, the HLBs retain their size and number and begin to accumulate coilin. After the nurse cells have dumped their cytoplasm into the oocyte and their nuclei begin to degenerate, the HLBs are especially prominent. They display the usual HLB markers like the U7 snRNP, but now stain for coilin as brightly as typical CBs in other cells (Fig. 4D–F). The net result is that at the end of oogenesis nurse cell nuclei lack typical CBs but have prominent coilin-containing HLBs.
AMPHIBIAN OOCYTE NUCLEI: CBs OR HLBs?
The giant oocyte nucleus or germinal vesicle (GV) of amphibians such as Xenopus houses several thousand extrachromosomal bodies. Among these are 50–100 bodies originally called spheres on the basis of their shape, but now referred to as CBs, because they contain a high concentration of coilin (Fig. 5). Although most of these bodies “float” freely in the nucleoplasm, a few are attached to the lampbrush chromosomes at the histone gene loci (Gall et al. 1981; Callan et al. 1991). There is no question that the free and attached bodies are equivalent structures, because they are identical in all features of morphology and molecular composition, including a high concentration of the U7 snRNP and other factors involved in histone pre-mRNA processing (Wu and Gall 1993; Abbott et al. 1999). Thus, by composition and association with the histone genes, these amphibian oocyte bodies are HLBs. It is tempting to equate them with the late-stage HLBs of the Drosophila ovary, which also have a high concentration of coilin. If this analogy is carried further, one might expect to find both CBs and HLBs in early amphibian oocytes, with loss of the CBs during oogenesis. The distribution of scaRNAs should shed light on this issue, because they are not in the HLBs of Drosophila, and hence may provide a more definitive marker to distinguish the two types of bodies. We know that the Xenopus early oocyte contains several types of nuclear bodies (besides the amplified nucleoli), but it is not yet clear what happens to them during the midstages of oogenesis.
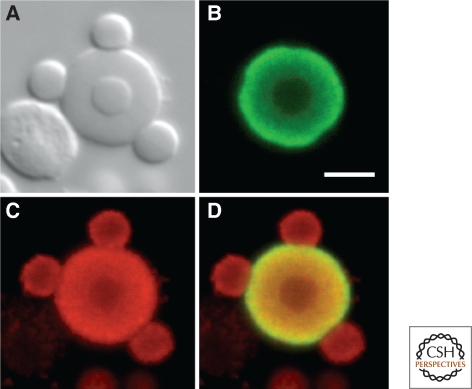
Figure 5.
A single large nuclear body from a Xenopus germinal vesicle after immunostaining for coilin (green) and trimethylguanosine (red). Because of their high concentration of coilin, these bodies have been described as CBs. However, they contain the U7 snRNP and some are attached to the histone gene loci, making them HLBs by definition. (A) DIC image showing one speckle inside the CB/HLB and three speckles attached to its periphery. The irregular object to the lower left is an extrachromosomal nucleolus. (B) Green channel, showing that coilin is limited to the CB/HLB, being absent from the speckles. (C) Red channel showing trimethylguanosine at high concentration in the CB/HLB and lower concentration in the speckles. (D) Overlay of B and C. Although trimethylguanosine occurs in the nucleolus (caps on U3 and U8 snoRNAs), its concentration is too low to detect in this image. Modified from Bellini and Gall (1998) with permission from Molecular Biology of the Cell. Bar = 5 µm.
DYNAMICS AND ASSEMBLY OF CBs AND HLBs
The rate at which components of a cellular structure turn over can be a useful indicator of metabolic activity and ultimately of function. Broadly speaking, one can distinguish storage particles, whose components are expected to be relatively immobile, and metabolically active bodies that exchange molecules with their environment on a short time scale. Various studies, especially those involving fluorescence recovery after photobleaching (FRAP), suggest that CBs and HLBs fall into the latter category. FRAP studies performed on CBs in cultured cells show rapid exchange of coilin, fibrillarin, and other CB components between the body and the nucleoplasm, with half-times for recovery of the order of a few minutes (Snaar et al. 2000; Sleeman et al. 2003; Dundr et al. 2004). Somewhat slower kinetics were found for coilin, U7 snRNA, and the TATA-binding protein in the large CBs/HLBs of Xenopus oocytes, of the order of 30 min (Handwerger et al. 2003; Deryusheva and Gall 2004). These studies suggest that macromolecules have relatively short residence times inside CBs and HLBs, consistent with these bodies having metabolic functions independent of simple storage.
All subcellular organelles show a certain degree of Brownian movement, depending primarily on their size and the viscosity of the surrounding medium. CBs show Brownian motion, but careful time-lapse measurements made on CBs in both mammalian and plant cells show more complex behavior as well, including anomalous diffusion and associations with chromatin (Boudonck et al. 1999; Platani et al. 2000; Carmo-Fonseca et al. 2002; Platani et al. 2002). Occasionally CBs have been seen to move in a directed manner, even traversing a nucleolus (Boudonck et al. 1999).
Despite considerable knowledge about the composition of CBs and HLBs, it remains unclear what function is served by the concentration of their components in structures large enough to be seen by conventional microscopy. Because coilin null mutants either lack CBs entirely (Drosophila and Arabidopsis) or show unusual distribution of CB components, one cannot argue that CBs are absolutely essential for nuclear physiology. A possible scenario is that CBs bring macromolecular components together at higher concentrations than ordinarily found in the nucleus, thus favoring reactions that otherwise might proceed at unacceptably slow rates. Alternatively, one could argue that CBs might serve to sequester splicing components and hence act to regulate the rate of RNA processing in the cell.
In a sense, it is easier to speculate about the function of HLBs. In this case, factors that process histone pre-mRNAs are concentrated at the sites where the pre-mRNAs are transcribed. Whether the bodies are the actual sites of processing is not known. HLBs are found throughout the cell cycle in Drosophila (Liu et al. 2006; White et al. 2007), even though histone pre-mRNA transcription and processing is limited to the S phase.
Without knowing the biological role of nuclear bodies in detail, one can still ask how such bodies assemble and disassemble during the cell cycle. Two broad categories of explanation can be distinguished, one being that a body must be built step by step, using cellular machinery in an energy-dependent fashion, the other being that a body will nucleate spontaneously when and if its macromolecular components occur at high enough concentration (Misteli 2001). A recent study provides strong support for this second self-assembly mechanism. In an experiment designed to ask whether coilin can act as a nucleating site for CB formation, Dundr and his colleagues made a construct in which GFP-coilin was linked to the Lac repressor and then transfected into cells that carried an array of 250 repeats of the Lac operator at a specific chromosome site (Kaiser et al. 2008). As expected, an accumulation of GFP-coilin appeared at the site of the Lac operator array. Remarkably, however, this accumulation not only had the size and shape of a typical CB but it contained a variety of other CB components, including SMN, snRNP proteins, and U85 scaRNA. Thus a genuine CB had formed at the chromosomal locus, morphologically and compositionally identical to other CBs found elsewhere in the same nucleus. To test whether coilin was required for the nucleation process, 17 other CB components were targeted one by one to the Lac operator. In each case a CB was formed essentially identical to the one formed by coilin. In fact, several other components were more efficient than coilin in terms of the fraction of transfected cells that displayed a CB. These experiments demonstrate clearly that formation of a CB is not a step-wise process involving the hierarchical addition of components starting, for instance, with coilin. Instead assembly of the CB has the hallmarks of a self-organizing system, taking place whenever the concentration of one of the major macromolecules, or macromolecular complexes, is high enough. The process continues, sweeping up other components until some still undefined end point is reached.
CONCLUDING REMARKS
In the 18 years since the identification of coilin as a marker for the CB, this nuclear organelle has gone from an almost completely neglected curiosity to a well-studied nuclear body whose involvement in snRNP biogenesis has been clearly shown. Because CBs are found in all eukaryotes that have been carefully examined, it is probable that new and unexpected roles for them will be uncovered in the near future, when additional genetic and molecular studies are undertaken. The relationship of the newly recognized HLB to the CB remains to be worked out. As additional organisms are examined, it seems likely that the HLB, like the CB, will prove to have a wide phylogenetic distribution. Because of its intimate relationship with the histone genes, the HLB promises to give new insight into the control of histone pre-mRNA processing during the cell cycle.
ACKNOWLEDGMENTS
We thank former members of our laboratory for their contributions to the understanding of CB and HLB structure and function. Original research cited here was supported by National Institutes of Health grant GM 33397 from the National Institute for General Medical Studies. JGG is American Cancer Society Professor of Developmental Genetics.
Footnotes
Editors: David Spector and Tom Misteli
REFERENCES
- Abbott J, Marzluff WF, Gall JG 1999. The stem loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell 10:487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM 1991. Human autoantibody to a novel protein of the nuclear coiled body: Immunological characterization and cDNA cloning of p80-coilin. J Exp Med 173:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J-P, Cavaille J 1997. Guiding ribose methylation of rRNA. Trends Biochem Sci 22:257–261 [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G 2006. The SMN complex: An assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71:313–320 [DOI] [PubMed] [Google Scholar]
- Bellini M, Gall JG 1998. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell 9:2987–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu J-L, Rutter J, Gall JG, Carroll D 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA 105:19821–19826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven AF, Simpson GG, Brown JWS, Shaw PJ 1995. The organization of spliceosomal components in the nuclei of higher plants. J Cell Sci 108:509–518 [DOI] [PubMed] [Google Scholar]
- Bier K, Kunz W, Ribbert D 1967. Struktur und Funktion der Oocytenchromosomen und Nukleolen sowie der Extra-DNS während der Oogenese panoistischer und meroistischer Insekten. Chromosoma 23:214–254 [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V 2008. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 7:2357–2367 [DOI] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ 1999. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell 10:2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SRy 1903. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol Univ Madrid 2:129–221 [Google Scholar]
- Cajal SRy 1910. El núcleo de las células piramidales del cerebro humano y de algunos mamiferos. Trab Lab Invest Biol Univ Madrid 8:27–62 [Google Scholar]
- Callan HG, Gall JG, Murphy C 1991. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma 101:245–251 [DOI] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC 1998. Cell cycle control of chorion gene amplification. Genes Dev 12:734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Platani M, Swedlow JR 2002. Macromolecular mobility inside the cell nucleus. Trends Cell Biol 12:491–495 [DOI] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M 1999. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 147:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland H, Lafontaine JG 1993. Localization of snRNP antigens in nucleolus-associated bodies: Study of plant interphase nuclei by confocal and electron microscopy. Chromosoma 102:220–226 [DOI] [PubMed] [Google Scholar]
- Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trulzsch B, Sattelle DB, Davies KE, van den Heuvel M 2003. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet 12:1367–1376 [DOI] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P 2006. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell 17:2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T 2002. Cajal body-specific small nuclear RNAs: A novel class of 2′-_O_-methylation and pseudouridylation guide RNAs. EMBO J 21:2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG 2004. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci USA 101:4810–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG 2009. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 20:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF 2005. The polyadenylation factor CPSF-73 is Involved in histone-pre-mRNA processing. Cell 123:37–48 [DOI] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T 2004. In vivo kinetics of Cajal body components. J Cell Biol 164:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179:1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA 92:5915–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG 2001. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 154:499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG 1999. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Current Biol 9:126–135 [DOI] [PubMed] [Google Scholar]
- Gall JG 2003. The centennial of the Cajal body. Nature Rev Mol Cell Biol 4:975–980 [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C 1999. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell 10:4385–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G 1981. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma 84:159–171 [DOI] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS 2009. The subnuclear organization of histone gene regulatory proteins and 3' end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol 220:129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS 2008. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci USA 105:16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Murphy C, Gall JG 2003. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol 160:495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG 2000. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 11:4159–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG 2001. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 15:2720–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG 1999. Coiled bodies preferentially associate with U4, U11 and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell 10:1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164:647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22:1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17:944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M 2008. De novo formation of a subnuclear body. Science 322:1713–1717 [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Darzacq X, Verheggen C, Bertrand E, Kiss T 2002. A Cajal body-specific pseudouridylation guide RNA is composed on two box H/ACA sno-RNA-like domains. Nucleic Acids Res 30:4643–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA 2005. Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs. Genes Dev 19:2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT 2009. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma 118:437–443 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nature Rev Mol Cell Biol 4:604–612 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M 1993. Localisation of splicing snRNPs in mammalian cells. Mol Biol Reports 18:127–133 [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J 15:3555–3565 [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Hebert MD, Ye Y, Templeton DJ, Kung H-J, Matera AG 2000. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci 113:1543–1552 [DOI] [PubMed] [Google Scholar]
- Liu J-L, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG 2006. The Drosophila melanogaster Cajal body. J Cell Biol 172:875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-L, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Gao H, Beumer KJ, Carroll D, Matera AG, Gall JG 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell 20:1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW 2000. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev 14:2298–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF 2005. Metazoan replication-dependent histone mRNAs: A distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol 17:274–280 [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR 1998. Coiled bodies and gems: Janus or Gemini? Amer J Hum Gen 63:317–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Ward DC 1993. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol 121:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U 2002. SMN-mediated assembly of RNPs: A complex story. Trends Cell Biol 12:472–478 [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Chan YB, Davies KE, van den Heuvel M 2000. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett 486:99–102 [DOI] [PubMed] [Google Scholar]
- Misteli T 2001. The concept of self-organization in cellular architecture. J Cell Biol 155:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneron A, Bernhard W 1969. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res 27:266–288 [DOI] [PubMed] [Google Scholar]
- Narayanan U, Achsel T, Lührmann R, Matera AG 2004. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell 16:223–234 [DOI] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26:349–365 [DOI] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Kramer A 2004. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 117:4423–4433 [DOI] [PubMed] [Google Scholar]
- Patel SB, Bellini M 2008. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 36:6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SB, Novikova N, Bellini M 2007. Splicing-independent recruitment of spliceosomal small nuclear RNPs to nascent RNA polymerase II transcripts. J Cell Biol 178:937–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L 2007. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep 8:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298:1775–1779 [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Lamond AI, Swedlow JR 2002. Cajal body dynamics and association with chromatin are ATP-dependent. Nature Cell Biol 4:502–508 [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond A 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 151:1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra TK, Gonsalvez GB, Walker MP, Shpargel KB, Salz HK, Matera AG 2007. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol 176:831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM 1991. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 195:27–37 [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T 2003. A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J 22:4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Lührmann R 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J 23:3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Matera AG 2005. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci USA 102:17372–17377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI 1999. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Current Biol 9:1065–1074 [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI 2003. Cajal body proteins SMN and coilin show differential dynamic behaviour in vivo. J Cell Sci 116:2039–2050 [DOI] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB 1995. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 59:473–485 [DOI] [PubMed] [Google Scholar]
- Snaar S, Wiesmeijer K, Jochemsen A, Tanke H, Dirks R 2000. Mutational analysis of fibrillarin and its mobility in living human cells. J Cell Biol 151:653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL 1993. Nuclear organization of pre-mRNA processing. Curr Opin Cell Biol 5:442–447 [DOI] [PubMed] [Google Scholar]
- Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM 2008. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 19:2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Rader SD, Klingauf M, Neugebauer KM 2003. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 160:505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34:322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Jády BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J 2007. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol Cell 27:869–881 [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T 1997. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol 9:337–342 [DOI] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP 2008. Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19:3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17:955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EKL, Lafarga M, Conlon RA, Matera AG 2001. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 154:293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Massello LK, Gao L, Barber TJ, Hebert MD, Chan EKL, Matera AG 2000. Structure and characterization of the murine p80 coilin gene, Coil. J Struct Biol 129:269–277 [DOI] [PubMed] [Google Scholar]
- Tuma RS, Stolk JA, Roth MB 1993. Identification and characterization of a sphere organelle protein. J Cell Biol 122:767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DLJ, Samarsky D, Mouaikel J, Blanchard J-M, Bordonné R, Bertrand E 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 21:2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Mouaikel J, Thiry M, Blanchard J-M, Tollervey D, Bordonné R, Lafontaine DLJ, Bertrand E 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J 20:5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG 2009. Reduced viability, fertility and fecundity in mice lacking the Cajal body marker protein, coilin. PLoS One 4:e6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ 2007. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell 18:2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13:290–301 [DOI] [PubMed] [Google Scholar]
- Wu C-HH, Gall JG 1993. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA 90:6257–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schümperli D, Matera AG 2005. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 114:155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36:267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP 2003. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 15:81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]