Variant of TYR and Autoimmunity Susceptibility Loci in Generalized Vitiligo (original) (raw)
. Author manuscript; available in PMC: 2010 Nov 6.
Published in final edited form as: N Engl J Med. 2010 Apr 21;362(18):1686–1697. doi: 10.1056/NEJMoa0908547
Abstract
BACKGROUND
Generalized vitiligo is an autoimmune disease characterized by melanocyte loss, which results in patchy depigmentation of skin and hair, and is associated with an elevated risk of other autoimmune diseases.
METHODS
To identify generalized vitiligo susceptibility loci, we conducted a genomewide association study. We genotyped 579,146 single-nucleotide polymorphisms (SNPs) in 1514 patients with generalized vitiligo who were of European-derived white (CEU) ancestry and compared the genotypes with publicly available control genotypes from 2813 CEU persons. We then tested 50 SNPs in two replication sets, one comprising 677 independent CEU patients and 1106 CEU controls and the other comprising 183 CEU simplex trios with generalized vitiligo and 332 CEU multiplex families.
RESULTS
We detected significant associations between generalized vitiligo and SNPs at several loci previously associated with other autoimmune diseases. These included genes encoding major-histocompatibility-complex class I molecules (P = 9.05×10−23) and class II molecules (P = 4.50×10−34), PTPN22 (P = 1.31×10−7), LPP (P = 1.01×10−11), IL2RA (P = 2.78×10−9), UBASH3A (P = 1.26×10−9), and C1QTNF6 (P = 2.21×10−16). We also detected associations between generalized vitiligo and SNPs in two additional immune-related loci, RERE (P = 7.07×10−15) and GZMB (P = 3.44×10−8), and in a locus containing TYR (P = 1.60×10−18), encoding tyrosinase.
CONCLUSIONS
We observed associations between generalized vitiligo and markers implicating multiple genes, some associated with other autoimmune diseases and one (TYR) that may mediate target-cell specificity and indicate a mutually exclusive relationship between susceptibility to vitiligo and susceptibility to melanoma.
GENERALIZED VITILIGO IS A DISEASE IN which patchy depigmentation of skin and hair results from autoimmune loss of melanocytes.1,2 It is a complex disorder involving multiple susceptibility genes and unknown environmental triggers. Genetic linkage and candidate-gene association studies have implicated several potentially contributory loci, though few have been consistently supported by the data.3 Patients with generalized vitiligo have elevated frequencies of other autoimmune diseases, including autoimmune thyroid disease, rheumatoid arthritis, psoriasis, adult-onset type 1 diabetes, pernicious anemia, systemic lupus erythematosus, and Addison's disease,4 suggesting that these diseases involve shared genetic components. To identify susceptibility loci for generalized vitiligo, we performed a genome-wide association study of patients with the disease who were of European-derived white (abbreviated as CEU) ancestry.
METHODS
STUDY SUBJECTS
The initial, genomewide association study included 1514 CEU patients from North America and the United Kingdom (see Table 1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). All patients met strict clinical criteria for the diagnosis of generalized vitiligo.5 The single-nucleotide polymorphism (SNP) genotypes of the patients were compared with those of 2813 CEU controls (from data sets obtained from the National Institutes of Health Genotype and Phenotype database [dbGaP]). We also carried out analyses of two independent CEU replication sets: replication set 1 included 677 unrelated patients with generalized vitiligo and 1106 controls, and replication set 2 was a family-based cohort of 183 simplex trios and 332 multiplex families. We obtained written informed consent from all the study participants, and the study was approved by the institutional review board at each participating center. Additional protocol details are provided in the Supplementary Appendix.
GENOTYPING AND QUALITY CONTROL
DNA purification, genotyping, and quality-control procedures are described in the Supplementary Appendix. In the genomewide association study, we determined the genotypes of patients with vitiligo for approximately 610,000 markers by using the Illumina 610-Quad BeadChip. All mitochondrial markers and Y-chromosome markers were excluded, leaving 579,146 SNPs that were present in both patients and controls in the genomewide association data set. Genotype data were then subjected to extensive quality-control filtering. In the two replication studies, genotyping of the SNPs that showed genomewide significance (P<5×10−8) or near significance in the genomewide association study was performed with the use of the Sequenom MassArray iPLEX genotyping system. We also imputed genotypes for rs12206499, which is related to major-histocompatibility-complex (MHC) class I molecules and for technical reasons could not be genotyped, and genotyped several SNPs in the NLRP1 (NLR family, pyrin domain–containing 1 gene) region, which was previously shown to be associated with vitiligo (see the Supplementary Appendix).
STATISTICAL ANALYSIS
After quality-control filtering of the SNPs (see the Supplementary Appendix), exclusion of genetic outliers, and exclusion of data from subjects who could not be matched to an appropriate control or patient,6 a total of 520,460 SNPs in 1392 patients and 2629 controls remained. We compared allele frequencies of these SNPs between the patients and controls by using the unadjusted Cochran–Armitage trend test7 as well as an eigenvector-adjusted Cochran–Armitage trend test.8 Both yielded a genomic inflation factor of 1.048, indicating minimal residual population stratification; we corrected the test statistics accordingly.9 After quality-control filtering of data obtained from the replication sets, for replication set 1 we compared the allele frequencies for the 48 genotyped SNPs, as well as the imputed SNP rs12206499, in 647 patients and 1056 controls by using the unadjusted Cochran–Armitage trend test.7 In replication set 2, we tested for an association with generalized vitiligo by using the family-based association test,10 version 1.5.5 (see the Supplementary Appendix). In the replication and combined analyses, P values of less than 0.05 were considered to indicate statistical significance (as opposed to the P<5×10−8 criterion in the genomewide association analysis).
RESULTS
GENOMEWIDE ASSOCIATION ANALYSIS
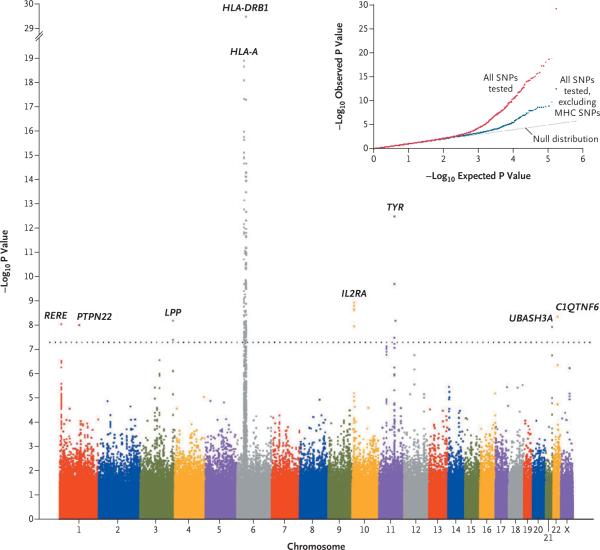
We compared the frequencies of alleles of 520,460 autosomal and X-chromosomal SNPs in 1392 unrelated patients with generalized vitiligo and in 2629 controls. The quantile–quantile plot of the negative logarithms of genomewide P values (excluding 3411 SNPs in the MHC) largely fit the null distribution (predicting no significant difference in P values between the two groups) (Fig. 1), except at the “tail,” where the observed P values were smaller than those expected under the null hypothesis. The most significant results after all corrections, including correction for a genomic inflation factor of 1.048,9 are shown in Table 1. Eight chromosomal regions contained SNPs associated with generalized vitiligo at the genomewide significance level (P<5×10−8).11 Summary statistical results and genotype data can be obtained from dbGaP (accession number, phs000224.v1.p1).
Figure 1. Genomewide Association Results.
The genomewide distribution of –log10 P values from the unadjusted Cochran–Armitage trend test is shown across the chromosomes. Values are shown for 520,460 polymorphic SNPs that were of sufficient quality, after quality-control filtering, in 1392 unrelated CEU patients with generalized vitiligo and 2629 unrelated CEU controls, after genetic matching6 and correction for the inflation factor of 1.048. The dotted line indicates the genomewide significance threshold (P<5×10–8). The inset shows quantile–quantile plots of the observed versus expected –log10 P values from the unadjusted Cochran–Armitage trend test. The plot in red shows P values for all 520,460 SNPs, whereas the plot in blue shows P values for all 520,460 SNPs excluding the 3417 SNPs located across the extended major histocompatibility complex (MHC) (on chromosome 6, spanning 26.0 to 33.5 Mb). C1QTNF6 denotes the C1q and tumor necrosis factor–related protein 6 gene, HLA-A the MHC class I, HLA-A gene, HLA-DRB1 the MHC class II, DR beta 1 gene, IL2RA the interleukin-2–receptor alpha gene, LPP the LIM domain–containing preferred translocation partner in lipoma gene, PTPN22 the lymphoid-specific protein tyrosine phosphatase, nonreceptor type 22 gene, RERE the arginine–glutamic acid dipeptide (RE) repeats gene, TYR the tyrosinase gene, and UBASH3A the ubiquitin-associated and SH3 domain–containing A gene.
Table 1.
Loci Most Strongly Associated with Generalized Vitiligo, According to the Analysis.*
| Locus Region† | SNP | Nucleotide Location | Risk Allele | Genomewide Association Study‡ | Replication Set 1§ | Replication Set 2¶ | Combined Analysis∥ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk-Allele Frequency | EIGENSTRAT P Value | PLINK P Value | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | Pseudo R2 | |||||
| Patients | Controls | ||||||||||||||
| Chromosome 1p36.23 | |||||||||||||||
| RERE | rs301819 | 8424373 | A | 0.485 | 0.417 | 1.65×10−8 | 8.85×10−9 | 1.32 (1.21–1.45) | 5.37×10−7 | 1.42 (1.24–1.63) | 0.06 | 1.17 (0.96–1.43) | 4.88×10−9 | 1.32 (1.20–1.45) | |
| RERE | rs4908760 | 8448729 | G | 0.433 | 0.373 | 6.52×10−7 | 3.48×10−7 | 1.28 (1.17–1.41) | 1.02×10−8 | 1.50 (1.30–1.72) | 0.27 | 1.09 (0.89–1.33) | 7.07×10−15 | 1.36 (1.26–1.48) | 0.008 |
| RERE | rs11121194 | 8492493 | C | 0.435 | 0.375 | 9.35×10−6 | 3.00×10−7 | 1.28 (1.17–1.41) | 4.76×10−8 | 1.47 (1.28–1.69) | 0.26 | 1.09 (0.89–1.33) | 1.33×10−14 | 1.36 (1.26–1.47) | |
| Chromosome 1p13.2 | |||||||||||||||
| PTPN22 | rs2476601 | 114179091 | A | 0.134 | 0.095 | 6.79×10−9 | 9.82×10−9 | 1.54 (1.33–1.78) | 0.59 | 1.07 (0.84–1.69) | 2.18×10−3 | 1.45 (1.04–2.03) | 1.31×10−7 | 1.39 (1.23–1.57) | 0.004 |
| Chromosome 3q28 | |||||||||||||||
| LPP | rs13076312 | 189571948 | T | 0.510 | 0.445 | 3.46×10−8 | 3.96×10−8 | 1.30 (1.19–1.43) | 8.52×10−5 | 1.33 (1.15–1.53) | 2.31×10−3 | 1.23 (1.01–1.50) | 2.70×10−11 | 1.30 (1.20–1.41) | |
| LPP | rs1464510 | 189595248 | T | 0.508 | 0.439 | 5.91×10−9 | 6.45×10−9 | 1.32 (1.21–1.45) | 2.39×10−4 | 1.31 (1.13–1.51) | 1.99×10−3 | 1.26 (1.03–1.54) | 1.01×10−11 | 1.31 (1.21–1.41) | 0.007 |
| LPP | rs13091753 | 189597283 | T | 0.459 | 0.399 | 7.36×10−7 | 7.83×10−7 | 1.27 (1.16–1.39) | 4.47×10−3 | 1.24 (1.07–1.41) | 0.03 | 1.19 (0.98–1.46) | 1.03×10−8 | 1.26 (1.16–1.36) | |
| LPP | rs1559810 | 189607048 | T | 0.458 | 0.398 | 6.52×10−7 | 7.37×10−7 | 1.27 (1.16–1.39) | 3.87×10−3 | 1.24 (1.07–1.43) | 0.03 | 1.17 (0.96–1.43) | 9.90×10−9 | 1.26 (1.16–1.36) | |
| Chromosome 6p21.3 ** | |||||||||||||||
| HLA-G | rs2975033 | 29930240 | T | 0.382 | 0.289 | 9.12×10−16 | 1.16×10−15 | 1.49 (1.36–1.64) | 7.90×10−4 | 1.29 (1.11–1.49) | 1.20×10−5 | 1.54 (1.23–1.92) | 3.72×10−19 | 1.45 (1.34–1.57) | |
| HLA-A | rs2517715 | 30025418 | G | 0.423 | 0.336 | 1.47×10−12 | 1.66×10−12 | 1.41 (1.29–1.55) | 5.35×10−4 | 1.29 (1.11–1.48) | 6.68×10−4 | 1.36 (1.10–1.68) | 3.64×10−16 | 1.39 (1.28–1.51) | |
| HLA-A–HCG9 | rs3903160 | 30040876 | T | 0.382 | 0.285 | 8.75×10−17 | 1.04×10−16 | 1.52 (1.38–1.67) | 1.43×10−3 | 1.28 (1.10–1.48) | 2.40×10−5 | 1.49 (1.19–1.85) | 3.40×10−19 | 1.45 (1.34–1.57) | |
| HLA-A–HCG9 | rs6457110 | 30041860 | A | 0.487 | 0.397 | 9.51×10−13 | 9.36×10−13 | 1.41 (1.29–1.55) | 5.48×10−5 | 1.34 (1.16–1.54) | 3.27×10−4 | 1.39 (1.11–1.73) | 5.25×10−18 | 1.41 (1.30–1.52) | |
| HLA-A–HCG9 | rs3893464 | 30043229 | C | 0.625 | 0.535 | 1.72×10−12 | 1.44×10−12 | 1.38 (1.20–1.59) | 4.55×10−6 | 1.38 (1.20–1.59) | 8.05×10−4 | 1.44 (1.44–1.81) | 3.54×10−18 | 1.42 (1.31–1.54) | |
| HLA-A–HCG9 | rs12206499 | 30045106 | G | 0.395 | 0.288 | 6.15×10−20 | 1.24×10−19 | 1.58 (1.43–1.74) | 1.47×10−5 | 1.34 (1.17–1.53) | ND | 0.014 | |||
| HLA-A–HCG9 | rs3823355 | 30050062 | T | 0.392 | 0.286 | 1.12×10−19 | 2.13×10−19 | 1.57 (1.43–1.73) | 8.69×10−4 | 1.29 (1.11–1.50) | 2.50×10−6 | 1.52 (1.21–1.91) | 9.05×10−23 | 1.50(1.39–1.63) | |
| HCG9 | rs6904029 | 30051046 | A | 0.391 | 0.287 | 4.06×10−19 | 7.89×10−19 | 1.56 (1.42–1.72) | 1.50×10−3 | 1.27 (1.10–1.47) | 2.40×10−5 | 1.49 (1.18–1.87) | 9.79×10−22 | 1.49 (1.37–1.61) | |
| HCG9 | rs3823375 | 30052137 | C | 0.418 | 0.324 | 1.48×10−15 | 2.15×10−15 | 1.48 (1.35–1.63) | 3.72×10−3 | 1.24 (1.07–1.43) | 1.73×10−4 | 1.39 (1.13–1.72) | 1.26×10−17 | 1.42 (1.31–1.53) | |
| HCG9–HLA-J | rs4947244 | 30062343 | G | 0.367 | 0.273 | 5.34×10−16 | 7.62×10×16 | 1.51 (1.37–1.66) | 6.86×10−3 | 1.23 (1.06–1.43) | 6.45×10−5 | 1.49 (1.19–1.86) | 1.96×10−17 | 1.43 (1.32–1.55) | |
| HLA-J | rs4959039 | 30065048 | G | 0.366 | 0.273 | 8.82×10−16 | 1.28×10−15 | 1.50 (1.36–1.66) | 3.25×10−3 | 1.25 (1.08–1.46) | 1.00×10−5 | 1.58 (1.26–1.97) | 2.23×10−17 | 1.43 (1.32–1.55) | |
| HLA-J | rs9357092 | 30092231 | A | 0.369 | 0.273 | 1.28×10−16 | 1.72×10−16 | 1.52 (1.38–1.68) | 3.92×10−3 | 1.25 (1.07–1.45) | 3.20×10−5 | 1.53 (1.22–1.91) | 3.14×10−18 | 1.44 (1.33–1.57) | |
| ZNRD1 | rs9366752 | 30132656 | T | 0.307 | 0.228 | 6.17×10−13 | 6.59×10−13 | 1.47 (1.33–1.63) | 1.35×10−3 | 1.29 (1.10–1.51) | 2.19×10−4 | 1.45 (1.15–1.83) | 1.56×10−15 | 1.42 (1.30–1.55) | |
| RNF39 | rs4711209 | 30155382 | A | 0.368 | 0.272 | 1.67×10−16 | 2.28×10−16 | 1.52 (1.38–1.67) | 4.53×10−3 | 1.24 (1.07–1.44) | 9.20×10−5 | 1.47 (1.18–1.84) | 5.29×10−18 | 1.44 (1.32–1.56) | |
| RNF39–TRIM31 | rs6909253 | 30163622 | G | 0.508 | 0.414 | 2.51×10−13 | 1.70×10−13 | 1.42 (1.30–1.56) | 5.94×10−4 | 1.28 (1.11–1.50) | 4.13×10−4 | 1.44 (1.17–1.78) | 2.13×10−15 | 1.37 (1.27–1.48) | |
| RNF39–TRIM31 | rs9261394 | 30172541 | A | 0.512 | 0.417 | 1.26×10−13 | 6.99×10−14 | 1.43 (1.31–1.57) | 1.05×10−3 | 1.27 (1.10–1.46) | 8.76×10−4 | 1.41 (1.14–1.75) | 2.20×10−15 | 1.37 (1.27–1.48) | |
| C6orf10–BTNL2 | rs7758128 | 32453261 | A | 0.062 | 0.026 | 3.85×10−15 | 7.21×10−15 | 2.48 (1.97–3.12) | 1.50×10−3 | 1.79 (1.24–2.56) | 5.00×10−7 | 3.66 (2.22–6.04) | 3.29×10−16 | 2.19 (1.80–2.65) | |
| BTNL2–HLA-DRA | rs3806156 | 32481676 | T | 0.474 | 0.370 | 2.31×10−18 | 4.97×10−18 | 1.53 (1.39–1.68) | 0.01 | 1.20 (1.04–1.38) | 5.00×10−7 | 1.72 (1.38–2.13) | 7.22×10−19 | 1.42 (1.32–1.54) | |
| HLA-DRA–HLA-DRB5 | rs2395185 | 32541145 | T | 0.414 | 0.331 | 6.74×10−13 | 1.04×10−12 | 1.42 (1.29–1.56) | 0.10 | 1.13 (0.97−1.32) | 1.13×10−3 | 1.33 (1.06–1.66) | 2.11×10−13 | 1.35 (1.25–1.47) | |
| HLA-DRB1–HLA-DQA1 | rs2516049 | 32678378 | G | 0.410 | 0.319 | 7.04×10−15 | 1.07×10−14 | 1.47 (1.33–1.61) | 0.02 | 1.19 (1.03–1.39) | 4.71×10−4 | 1.40 (1.12–1.75) | 3.24×10−16 | 1.40 (1.29–1.52) | |
| HLA-DRB1–HLA-DQA1 | rs532098 | 32686030 | T | 0.597 | 0.458 | 2.89×10−30 | 4.83×10−30 | 1.74 (1.59–1.92) | 3.95×10−4 | 1.28 (1.11–1.46) | 6.00×10−9 | 1.87 (1.49–2.36) | 4.50×10−34 | 1.62 (1.50–1.75) | 0.018 |
| HLA-DQA1 | rs34518860 | 32702081 | A | 0.170 | 0.107 | 2.07×10−15 | 2.10×10−15 | 1.72 (1.51–1.96) | ND | ND | |||||
| Chromosome 10p15.1 | |||||||||||||||
| IL2RA | rs706779 | 6138830 | A | 0.607 | 0.535 | 4.61×10−10 | 1.14×10−9 | 1.35 (1.23–1.48) | 0.27 | 1.08 (0.94–1.24) | 8.30×10−5 | 1.32 (1.07–1.62) | 2.78×10−9 | 1.27 (1.17–1.37) | 0.004 |
| IL2RA | rs7090530 | 6150881 | A | 0.669 | 0.599 | 1.07×10−9 | 1.54×10−9 | 1.35 (1.23–1.49) | 0.51 | 1.05 (0.91–1.21) | 4.40×10−5 | 1.29 (1.05–1.58) | 1.21×10−8 | 1.26 (1.17–1.37) | |
| IL2RA–RBM17 | rs12251307 | 6163501 | C | 0.913 | 0.871 | 1.40×10−8 | 1.10×10−8 | 1.59 (1.36–1.86) | 0.22 | 0.87 (0.70–1.09) | 0.06 | 1.19 (0.85–1.66) | 8.10×10−6 | 1.33 (1.18–1.52) | |
| RBM17 | rs4750005 | 6209691 | T | 0.587 | 0.520 | 6.97×10−9 | 1.12×10−8 | 1.32 (1.20–1.45) | 0.38 | 1.09 (0.90–1.30) | 6.97×10−3 | 1.27 (0.97–1.66) | 1.35×10−8 | 1.27 (1.17–1.37) | |
| RBM17–PFKB3 | rs3920615 | 6216786 | A | 0.615 | 0.544 | 1.11×10−9 | 2.11×10−9 | 1.34 (1.22–1.47) | 0.71 | 1.03 (0.89–1.18) | 0.17 | 1.06 (0.84–1.34) | 9.32×10−9 | 1.26 (1.16–1.36) | |
| RBM17–PFKB3 | rs4747887 | 6217688 | T | 0.615 | 0.544 | 1.21×10−9 | 2.34×10−9 | 1.34 (1.22–1.47) | 0.60 | 1.04 (0.90–1.20) | 0.20 | 1.04 (0.83–1.31) | 6.83×10−9 | 1.26 (1.17–1.36) | |
| RBM17–PFKB3 | rs4750012 | 6217800 | C | 0.615 | 0.544 | 1.27×10−9 | 2.42×10−9 | 1.34 (1.22–1.47) | 0.70 | 1.03 (0.89–1.19) | 0.29 | 1.01 (0.80–1.26) | 9.25×10−9 | 1.26 (1.16–1.36) | |
| RBM17–PFKB3 | rs7099083 | 6218242 | G | 0.615 | 0.544 | 1.21×10−9 | 2.27×10−9 | 1.34 (1.22–1.47) | 0.65 | 1.03 (0.90–1.19) | 0.23 | 1.03 (0.82–1.29) | 7.20×10−9 | 1.26 (1.17–1.36) | |
| Chromosome 11q14.3 | |||||||||||||||
| TYR | rs10830236 | 88540464 | C | 0.721 | 0.669 | 1.09×10−6 | 1.15×10−6 | 1.29 (1.17–1.43) | 6.05×10−3 | 1.25 (1.07–1.46) | 0.77 | 1.04 (0.83–1.31) | 1.18×10−8 | 1.28 (1.18–1.40) | |
| TYR | rs11018528 | 88570025 | A | 0.764 | 0.707 | 3.69×10−8 | 3.25×10−8 | 1.36 (1.22–1.51) | 4.00×10−3 | 1.27 (1.08–1.50) | 0.48 | 1.01 (0.81–1.26) | 1.69×10−10 | 1.34 (1.23–1.47) | |
| TYR | rs10765198 | 88609422 | T | 0.771 | 0.715 | 5.36×10−8 | 5.78×10−8 | 1.36 (1.22–1.51) | 1.23×10−3 | 1.31 (1.11–1.55) | 0.36 | 1.04 (0.84–1.29) | 6.37×10−11 | 1.35 (1.23–1.48) | |
| TYR | rs1847134 | 88644901 | A | 0.754 | 0.685 | 2.37×10−10 | 2.01×10−10 | 1.42 (1.28–1.57) | 1.95×10−5 | 1.42 (1.21–1.66) | 0.40 | 1.03 (0.82–1.30) | 3.76×10−15 | 1.42 (1.30–1.56) | |
| TYR | rs1393350 | 88650694 | G | 0.807 | 0.733 | 3.70×10−13 | 3.24×10−13 | 1.54 (1.37–1.72) | 4.37×10−6 | 1.51 (1.26–1.79) | 0.12 | 1.16 (0.91–1.47) | 1.60×10−18 | 1.53 (1.39–1.68) | 0.011 |
| TYR | rs1806319 | 88677584 | A | 0.692 | 0.630 | 7.86×10−8 | 8.52×10−8 | 1.31 (1.19–1.45) | 1.47×10−4 | 1.34 (1.15–1.56) | 0.83 | 1.04 (0.83–1.30) | 2.12×10−11 | 1.33 (1.22–1.44) | |
| Chromosome 14q12 | |||||||||||||||
| GZMB | rs8192917 | 24172000 | G | 0.282 | 0.236 | 3.46×10−6 | 5.54×10−6 | 1.28 (1.15–1.42) | 4.87×10−4 | 1.33 (1.13–1.55) | 0.03 | 1.35 (1.09–1.67) | 3.44×10−8 | 1.28 (1.17–1.39) | 0.005 |
| GZMB | rs2273844 | 24173254 | A | 0.280 | 0.236 | 5.74×10−6 | 9.24×10−6 | 1.28 (1.15–1.42) | 4.04×10−4 | 1.33 (1.14–1.56) | 0.02 | 1.40 (1.13–1.75) | 6.78×10−8 | 1.27 (1.17–1.39) | |
| Chromosome 21q22.3 | |||||||||||||||
| UBASH3A | rs11203203 | 42709255 | A | 0.432 | 0.373 | 1.56×10−7 | 1.72×10−7 | 1.30 (1.18–1.42) | 2.58×10−3 | 1.24 (1.08–1.43) | 0.04 | 1.15 (0.93–1.43) | 1.26×10−9 | 1.27 (1.18–1.38) | 0.005 |
| UBASH3A | rs2839511 | 42721590 | A | 0.287 | 0.227 | 9.71×10−9 | 1.17×10−8 | 1.36 (1.23–1.51) | 0.13 | 1.13 (0.96–1.32) | 0.02 | 1.21 (0.95–1.53) | 5.14×10−9 | 1.30 (1.19–1.42) | |
| Chromosome 22q13.1 | |||||||||||||||
| C1QTNF6 | rs229527 | 35911431 | T | 0.490 | 0.419 | 4.06×10−9 | 4.36×10−9 | 1.33 (1.21–1.46) | 6.26×10−9 | 1.54 (1.33–1.78) | 2.88×10−3 | 1.15 (0.94–1.40) | 2.21×10−16 | 1.38 (1.28–1.50) | 0.009 |
| C1QTNF6 | rs5756546 | 35919751 | T | 0.271 | 0.219 | 6.35×10−7 | 4.34×10−7 | 1.33 (1.19–1.48) | 1.37×10−5 | 1.44 (1.22–1.70) | 0.49 | 1.12 (0.91–1.40) | 1.93×10−11 | 1.35 (1.24–1.48) |
MHC Loci
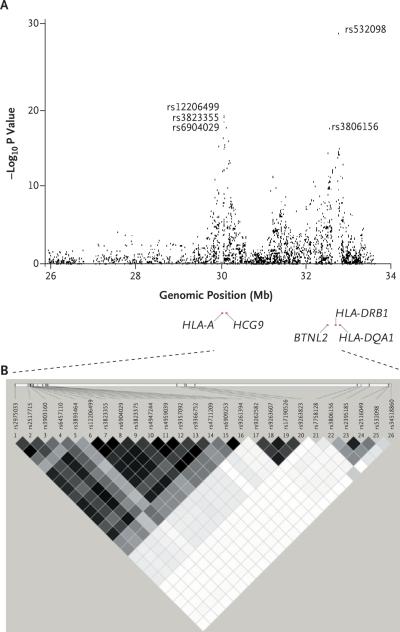
The most strongly associated SNPs were distributed across the MHC on chromosome 6p21.3 (Fig. 1), with 124 MHC SNPs exceeding the threshold for genomewide significance (P<5×10−8) and 689 MHC SNPs ranked among the 2000 SNPs with the highest genomewide significance. We found two major MHC association peaks (Fig. 2). One peak was in the class I gene region, between HLA-A (encoding the MHC class I,HLA-A molecule) and HCG9 (encoding the HLA complex group 9), characterized by three SNPs: rs12206499 (P = 1.24×10−19 for the association with generalized vitiligo; odds ratio for the presence of the risk allele in patients vs. controls, 1.58), rs3823355 (P = 2.13×10−19; odds ratio, 1.57), and rs6904029 (P = 7.89×10−19; odds ratio, 1.56). The second peak was in the class II gene region, between HLA-DRB1 (the MHC class II, DR beta 1 gene) and HLA-DQA1 (the MHC class II, DQ alpha 1 gene), characterized by rs532098 (P = 4.83×10−30; odds ratio, 1.74) and rs3806156 (P = 4.97×10−18; odds ratio, 1.53). The highest odds ratio in the genomewide association study was 2.48, for rs7758128 (P = 7.21×10−15). A third, lower peak was located at the proximal end of the MHC class I gene region (Fig. 2A).
Figure 2. Detailed Results from the Genomewide Association Study and the Linkage-Disequilibrium Plot across the Major Histocompatibility Complex (MHC) on Chromosome 6p21.3.
Panel A shows the distribution of –log10 P values from the unadjusted Cochran–Armitage trend test in the genomewide association analysis, corrected by the genomic inflation factor of 1.048, for the 3417 SNPs located across the extended MHC region (on chromosome 6, spanning 26.0 to 33.5 Mb). Panel B shows the plot of linkage disequilibrium (r2) for the 22 NPs in the MHC region that were highly associated with generalized vitiligo and that were included in the logistic-regression analysis. The plot gives the physical positions of the markers (according to build 36.1 from the National Center for Biotechnology Information). Darker boxes indicate stronger linkage disequilibrium (range of possible r2 values, 0.0 to 1.0). BTNL2 denotes the butyrophilin-like protein 2 gene, HCG9 the HLA complex group 9 gene, HLA-A the MHC class I, HLA-A gene, HLA-DQA1 the MHC class II, DQ alpha 1 gene, and HLA-DRB1 the MHC class II, DR beta 1 gene.
SNPs in the two major association peaks, one in the MHC class I region and one in the MHC class II region, corresponded with different linkage-disequilibrium blocks (Fig. 2, and Table 2 in the Supplementary Appendix). To determine whether these two peaks represent independent associations with generalized vitiligo, rather than secondary associations due to long-range linkage disequilibrium across the MHC, we applied logistic-regression analysis12 to the 22 MHC-region SNPs that showed the strongest association with generalized vitiligo in the genomewide association study (Table 3 in the Supplementary Appendix). We observed that the major peak in the MHC class I region, represented by SNP rs12206499, and the major peak in the MHC class II region, represented by rs532098, constitute independent association signals, with no apparent epistasis between them (P = 0.38), indicating that susceptibility to vitiligo that is attributable to the MHC involves more than one locus.
We analyzed data for 180 participants from 27 CEU Centre d'Etude du Polymorphisme Humain (CEPH) reference families for whom class I and class II HLA genotypes, as well as SNP genotypes,13,14 were available and for 203 participants from 78 families with generalized vitiligo for whom class II HLA and SNP genotypes were available.15 We found strong linkage disequilibrium between the risk allele (G) at rs12206499 and HLA-A*02 (r2 = 0.964 and linkage-disequilibrium coefficient [D′] = 1.0 in the CEPH reference families) and moderate linkage disequilibrium between the risk allele (T) at rs532098 and HLADRB1*04 (r2 = 0.43 and D′ = 1.0 in the CEPH reference families; r2 = 0.19 and D′ = 0.83 in the families with generalized vitiligo). These findings were consistent with previous reports of an association of generalized vitiligo with both the HLA-A*02 allele16 and the HLA-DRB1*04 allele.15
Non-MHC Loci
Outside the MHC region, SNPs in seven different chromosomal regions showed a significant association with generalized vitiligo (Table 1 and Fig. 1 in the Supplementary Appendix). Five of these regions contain genes previously shown to be associated with other autoimmune diseases; for each region, the same high-risk SNP alleles associated with vitiligo were also associated with other autoimmune diseases.
We found an association between generalized vitiligo and SNP rs2476601 (P = 9.82×10−9; odds ratio for the presence of the risk allele in patients vs. controls, 1.54), on chromosome 1p13.2. This SNP is located in PTPN22, which encodes the lymphoid-specific protein tyrosine phosphatase, nonreceptor type 22, involved in T-cell–receptor signaling. The implicated variant is associated with many autoimmune diseases, including generalized vitiligo, and specifies an amino acid substitution, R620W, thought to have a direct effect on autoimmune susceptibility (rather than a benign effect through linkage disequilibrium with the “causal” SNP).17
We also found an association between generalized vitiligo and 11 SNPs on chromosome 3q28. These SNPs span the LPP gene region (including the LPP gene, encoding the LIM domain–containing preferred translocation partner in lipoma). Of the 11 SNPs, rs1464510 (P = 6.45×10−9; odds ratio, 1.32) and rs13076312 (P = 3.96×10−8; odds ratio, 1.30) showed genomewide significance. We observed tight linkage disequilibrium among most of the 11 SNPs; no haplotype was more strongly associated with disease than any component SNP. SNPs in the LPP gene region have also been associated with celiac disease and rheumatoid arthritis18 the function of the LPP protein is unknown.
Generalized vitiligo was also associated with 25 SNPs in the region of IL2RA (encoding the interleukin-2–receptor alpha chain) on chromosome 10p15.1, 8 of which showed genomewide significance. Of these, rs706779 (P = 1.14×10−9; odds ratio, 1.35) and rs7090530 (P = 1.54×10−9; odds ratio, 1.35) had the strongest association. Variants of IL2RA have been shown to be associated with type 1 diabetes,19 Graves' disease,20 multiple sclerosis,21 rheumatoid arthritis,22 and systemic lupus erythematosus.23 Elevated serum interleukin-2-receptor levels, which indicate T-cell activation,24 occur in many autoimmune diseases, including generalized vitiligo.25
There was also an association between generalized vitiligo and nine SNPs in the region spanning UBASH3A (the ubiquitin-associated and SH3 domain–containing A gene) on chromosome 21q22.3; of these, rs2839511 showed genomewide significance (P = 1.17×10−8; odds ratio, 1.36). UBASH3A encodes a regulator of T-cell–receptor signaling and is associated with type 1 diabetes.26 Also associated with type 1 diabetes27.28 (as well as rheumatoid arthritis29) are variants of C1QTNF6 (the C1q and tumor necrosis factor–related protein 6 gene), located on chromosome 22q13.1. The SNP rs229527 at this locus showed an association with generalized vitiligo (P = 4.36×10−9; odds ratio, 1.33).
In addition, we observed an association between generalized vitiligo and two non-MHC regions that have not previously been reported to be associated with autoimmune disease. The first was the RERE gene region (encoding the arginine–glutamic acid dipeptide [RE] repeats protein, also known as atrophin-like protein 1) on chromosome 1p36.23, in which 40 SNPs were identified as having an association, 1 of which showed genome-wide significance (rs301819; P = 8.85×10−9; odds ratio, 1.32). The most strongly associated SNPs in the RERE region were in tight linkage disequilibrium; no haplotype showed a significantly stronger association than any component SNP. The RERE protein is a transcriptional corepressor30 that is highly expressed in lymphoid cells and is thought to regulate apoptosis.31
Perhaps the most interesting association that we found was between generalized vitiligo and a locus on chromosome 11q14.3, consisting of 37 SNPs spanning the TYR gene region. The major allele of each SNP was associated with vitiligo. Three of these SNPs showed genomewide significance; the strongest associations were with rs1847134 (P = 2.01×10−10; odds ratio, 1.42) and rs1393350 (P=3.24×10−13; odds ratio, 1.54). Haplotype analysis revealed a strong association with a block of six SNPs in tight linkage disequilibrium — rs1018528, rs10765198, rs1847134, rs1393350, rs1126809, and rs1806319 — although no haplotype had a significantly stronger association with generalized vitiligo than any component SNP. TYR encodes tyrosinase, a melanosomal enzyme that catalyzes the rate-limiting steps of melanin biosynthesis32 and constitutes a major autoantigen in generalized vitiligo.1,2
Finally, we detected a suggestive association signal at chromosome 14q12, involving 11 SNPs spanning the GZMB (the granzyme B gene) region, particularly intragenic SNPs rs8192917 (P=5.54× 10−6; odds ratio, 1.28) and rs2273844 (P=9.24× 10−6; odds ratio, 1.28). Granzyme B is a caspaselike serine protease that mediates two processes: immune-induced target-cell apoptosis mediated by cytotoxic T cells and natural killer cells33 and activation-induced cell death of effector type 2 helper T cells, which terminates the immune response.34
Independent Replication and Combined Analyses
We aimed to replicate the results of the genomewide association analysis in two replication sets of independently collected samples from CEU patients and controls (replication set 1) and CEU simplex trios and multiplex families (replication set 2) (Table 1 in the Supplementary Appendix). Specifically, we tested 50 SNPs in nine chromosomal regions that had the strongest associations with generalized vitiligo, as detected in the genomewide association analysis (Table 1). In one or both replication sets, the results for most of these SNPs, and all the corresponding genomic regions tested, replicated the associations observed in the genomewide association analysis. Furthermore, a combined meta-analysis of all three study sets (by means of a Cochran–Mantel–Haenszel test) provided strong evidence of an association of all of these loci with generalized vitiligo (Table 1).
In the MHC region, both replication analyses and the combined analyses supported strong association in both the distal class I region (rs3823355; in the combined analysis, P=9.05× 10−23; odds ratio, 1.50) and the HLA-DRB1_–_DQA1 class II region (rs532098; in the combined analysis, P=4.50×10−34; odds ratio, 1.62). SNPs in the proximal class I region were not tested in the replication studies.
Outside the MHC, associations in the LPP region of chromosome 3q28, the GZMB region of chromosome 14q12, the UBASH3A region of chromosome 21q22.3, and the C1QTNF6 region of chromosome 22q13.1 were observed in both replication sets; the combined analysis likewise provided strong support for the associations (for LPP rs1464510, P=1.01×10−11; odds ratio, 1.31; for GZMB rs8192917, P=3.44×10−8; odds ratio, 1.28; for UBASH3A rs11203203, P=1.26×10−9; odds ratio 1.27; and for C1QTNF6 rs229527, P=2.21×10−16; odds ratio, 1.38). For PTPN22 on chromosome 1p13.2 and the IL2RA region of chromosome 10p15.1, associations were replicated in the family-based set (replication set 2) but not in the case–control set (replication set 1). Conversely, in the RERE region of chromosome 1p36.23 and the TYR region of chromosome 11q14.3, associations were replicated in the case–control set but not in the family-based set (Table 1). Nevertheless, for all these loci, the combined analysis provided strong support for an overall association (RERE rs4908760, P=7.07×10−15; odds ratio, 1.36; PTPN22 rs2476601, P=1.31×10−7; odds ratio, 1.39; IL2RA rs706779, P=2.78×10−9; odds ratio, 1.27; and TYR rs1393350, P=1.60×10−18; odds ratio, 1.53).
Thus, the replication studies showed consistent support for an association of generalized vitiligo with SNPs within or near LPP, the HLA class I and HLA class II genes, GZMB, UBASH3A, and C1QTNF6 and less consistent, although significant, support for an association with SNPs within or near RERE, PTPN22, IL2RA, and TYR.
Tyrosinase is naturally presented by HLAA*0201 and is recognized by autologous cytotoxic T lymphocytes on HLA-A2–positive melanomas, which tend to show good response to immunotherapy. Presentation of TYR peptide by HLA-A*0201 may also mediate the loss of partial self-tolerance to tyrosinase.35 We therefore tested epistasis between HLA-A SNP rs12206499 and TYR SNP rs1393350 with logistic regression analysis, using SNP genotypes from the genomewide association study and replication set 1, thereby detecting evidence of significant epistasis between HLA-A and TYR (P=0.03).
DISCUSSION
We identified 10 independent susceptibility loci for generalized vitiligo. In general, these associations were observed in both the subgroup of patients with vitiligo only and the subgroup with vitiligo and concomitant autoimmune diseases (Table 4 in the Supplementary Appendix). PTPN22 (rs2476601) was the exception; the risk allele was significantly more prevalent among patients with generalized vitiligo and other autoimmune diseases. In the MHC region, we detected two major association signals, one in the class I gene region, between HLA-A and HCG9, and the other in the class II gene region, between HLA-DRB1 and HLA-DQA1 (Fig. 2). This finding is consistent with previous reports of an association of vitiligo with the HLA-A*02 allele16 and the HLA-DRB1*04 allele.15 Indeed, the near-perfect linkage disequilibrium between HLA-A*02 and the risk variant at rs12206499 suggests that HLA-A*02 may be the etiologic variant at this locus that confers susceptibility to vitiligo.
Outside the MHC, we identified signals of an association with generalized vitiligo within or near the RERE, PTPN22, LPP, IL2RA, TYR, GZMB, UBASH3A, and C1QTNF6 genes. PTPN22, LPP, IL2RA, UBASH3A, and C1QTNF6 are also associated with genetic susceptibility to other autoimmune diseases, several of which are epidemiologically associated with generalized vitiligo. These findings support the long-standing hypothesis that generalized vitiligo involves genetic susceptibility loci shared with other autoimmune diseases.3 Most of these genes encode immune-system proteins involved in biologic pathways that probably influence the development of autoimmunity.
In contrast, TYR encodes tyrosinase, which is not a component of the immune system. Rather, it is an enzyme of the melanocyte that catalyzes the rate-limiting steps of melanin biosynthesis.32 It is also a major autoantigen in generalized vitiligo.1,2 Generalized vitiligo is thus genetically analogous to type 1 diabetes, in which variation in the INS gene, encoding insulin (an important autoantigen in diabetes), contributes to disease susceptibility. Vitiligo is associated with the major alleles of SNPs in the TYR region, particularly rs1393350 (in the combined analysis, P=1.60×10−18; odds ratio, 1.53) and the nearby nonsynonymous (R402Q) SNP rs1126809 (Table 5 in the Supplementary Appendix), with which rs1393350 is in tight linkage disequilibrium (Table 6 in the Supplementary Appendix). The minor alleles of both these SNPs are associated with susceptibility to malignant melanoma,36,37 suggesting that susceptibility to _TYR_-related generalized vitiligo and susceptibility to _TYR_-related malignant melanoma are mediated by different or perhaps even inverse biologic mechanisms. The 402Q variant of the TYR protein (the variant associated with melanoma) is thermosensitive38 and at 37°C tends to be misfolded, aberrantly glycosylated, retained in the endoplasmic reticulum, and subject to proteosomal degradation,39 resulting in steady-state 402Q tyrosinase activity that is only 25% of that of the major 402R polypeptide (which is associated with susceptibility to generalized vitiligo).38 Correct glycosylation of tyrosinase augments the generation of an MHC class I–restricted tyrosinase epitope that is presented by HLA-A*0201,40 the predominant HLA-A*02 allele, and indeed, we detected significant epistasis between HLA-A SNP rs12206499 and TYR SNP rs1393350 (P=0.03).
Thus, 402R tyrosinase not only is expressed at higher levels than the 402Q variant but also may be more efficiently presented to the immune system by HLA-A*02. It is therefore plausible that the 402R variant makes a greater contribution than the 402Q variant to immune surveillance against neoplastic melanocytes and susceptibility to vitiligo. On the other hand, in persons carrying one or two copies of the 402Q variant, immune surveillance may fail to detect neoplastic melanocytes, owing to the reduced expression, and thus the reduced immune presentation, of tyrosinase.
Generally, there is limited correspondence between the associations shown in the present study and genetic linkage and candidate-gene association signals reported previously for generalized vitiligo,3 though a detailed analysis supported an association with NLRP1 in multiplex families and specific subgroups of patients (see the Supplementary Appendix). Moreover, none of the significant associations reported in this study correspond to the principal up-regulated or down-regulated transcripts identified with the use of gene-expression profiling of melanocytes from patients with generalized vitiligo.41 Although the implicated loci that we have described here together account for only approximately 7.4% of the total genetic risk for generalized vitiligo, they provide insights into disease pathogenesis and implicate potential targets for therapeutic intervention.
Supplementary Material
Supp
Acknowledgments
Supported by grants (AR45584 and AR056292) from the National Institutes of Health and a grant from the Anna and John Sie Foundation.
We thank Deborah Herbstman, Amanda Tran, the University of Colorado Microarray Core, the Genotyping Core in the Division of Human Genetics at Washington University School of Medicine in St. Louis, and the membership of Vitiligo Support International, the Vitiligo Society, the National Vitiligo Foundation, the American Vitiligo Research Foundation, and Associazione Ricerca Informazione per la Vitiligine for their participation.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Hann S-K, Nordlund JJ. Vitiligo: a monograph on the basic and clinical science. Blackwell Science; Oxford, England: 2000. [Google Scholar]
- 2.Rezaei N, Gavalas NG, Weetman AP, Kemp EH. Autoimmunity as an aetiological factor in vitiligo. J Eur Acad Dermatol Venereol. 2007;21:865–76. doi: 10.1111/j.1468-3083.2007.02228.x. [DOI] [PubMed] [Google Scholar]
- 3.Spritz RA. The genetics of generalized vitiligo. Curr Dir Autoimmun. 2008;10:244–57. doi: 10.1159/000131501. [DOI] [PubMed] [Google Scholar]
- 4.Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–14. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 5.Taïeb A, Picardo M. The definition and assessment of vitiligo: a consensus report of the Vitiligo European Task Force. Pigment Cell Res. 2007;20:27–35. doi: 10.1111/j.1600-0749.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 6.Luca D, Ringquist S, Klei L, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82:453–63. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 9.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–66. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 10.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 11.Ioannidis JPA, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–29. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70:124–41. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bakker PIW, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map or disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fain PR, Babu SR, Bennett DC, Spritz RA. HLA class II haplotype DRB1*04-DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset. Pigment Cell Res. 2006;19:51–7. doi: 10.1111/j.1600-0749.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu J-B, Li M, Chen H, et al. Association of vitiligo with HLA-A2: a meta-analysis. J Eur Acad Dermatol Venereol. 2007;21:205–13. doi: 10.1111/j.1468-3083.2006.01899.x. [DOI] [PubMed] [Google Scholar]
- 17.Gregersen PK, Lee H-S, Batliwalla F, Begovich AB. PTPN22: setting thresholds for autoimmunity. Semin Immunol. 2006;18:214–23. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Coenen MJ, Trynka G, Heskamp S, et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum Mol Genet. 2009;18:4195–203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 19.Vella A, Cooper JD, Lowe CE, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–9. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand OJ, Lowe CE, Heward JM, et al. Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007;66:508–12. doi: 10.1111/j.1365-2265.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 21.The International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 22.Hinks A, Ke X, Barton A, et al. Association of the IL2RA/CD25 gene with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:251–7. doi: 10.1002/art.24187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carr EJ, Clatworthy MR, Lowe CE, et al. Contrasting genetic association of IL2RA with SLE and ANCA-associated vasculitis. BMC Med Genet. 2009;10:22. doi: 10.1186/1471-2350-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Wicker LS, Santamaria P. IL-2 and its high-affinity receptor: genetic control of immunoregulation and autoimmunity. Semin Immunol. 2009;21:363–71. doi: 10.1016/j.smim.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Honda Y, Okubo Y, Koga M. Relationship between levels of soluble interleukin-2 receptors and the types and activity of vitiligo. J Dermatol. 1997;24:561–3. doi: 10.1111/j.1346-8138.1997.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 26.Concannon P, Onengut-Gumuscu S, Todd JA, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes. 2008;57:2858–61. doi: 10.2337/db08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallis RH, Wang K, Marandi L, et al. Type 1 diabetes in the BB rat: a polygenic disease. Diabetes. 2009;58:1007–17. doi: 10.2337/db08-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julià A, Ballina J, Cañete JD, et al. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58:2275–86. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Tsai C-C. Atrophin proteins: an overview of a new class of nuclear receptor corepressors. Nucl Recept Signal. 2008;6:e009. doi: 10.1621/nrs.06009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waerner T, Gardellin P, Pfizenmaier K, Weith A, Kraut N. Human RERE is localized to nuclear promyelocytic leukemia oncogenic domains and enhances apoptosis. Cell Growth Differ. 2001;12:201–10. [PubMed] [Google Scholar]
- 32.Spritz RA, Chiang PW, Oiso N, Alkhateeb A. Human and mouse disorders of pigmentation. Curr Opin Genet Dev. 2003;13:284–9. doi: 10.1016/s0959-437x(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 33.Trapani JA, Sutton VR. Granzyme B: pro-apoptotic, antiviral and antitumor functions. Curr Opin Immunol. 2003;15:533–43. doi: 10.1016/s0952-7915(03)00107-9. [DOI] [PubMed] [Google Scholar]
- 34.Devadas S, Das J, Liu C, et al. Granzyme B is critical for T cell receptor-induced cell death of type 2 helper T cells. Immunity. 2006;25:237–47. doi: 10.1016/j.immuni.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Lotz C, Ferreira EA, Drexler I, et al. Partial tyrosinase-specific self tolerance by HLA-A*0201-restricted cytotoxic T lymphocytes in mice and man. Int J Cancer. 2004;108:571–9. doi: 10.1002/ijc.11602. [DOI] [PubMed] [Google Scholar]
- 36.Bishop TD, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–5. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 38.Tripathi RK, Giebel LB, Strunk KM, Spritz RA. A polymorphism of the human tyrosinase gene is associated with temperature-sensitive enzymatic activity. Gene Expr. 1991;1:103–10. [PMC free article] [PubMed] [Google Scholar]
- 39.Toyofuku K, Wada I, Spritz RA, Hearing VJ. The molecular basis of oculocutaneous albinism type 1 (OCA1): sorting failure and degradation of mutant tyrosinases results in a lack of pigmentation. Biochem J. 2001;355:259–69. doi: 10.1042/0264-6021:3550259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostankovitch M, Altrich-Vanlith M, Robila V, Engelhard VH. N-glycosylation enhances presentation of a MHC class I-restricted epitope from tyrosinase. J Immunol. 2009;182:4830–5. doi: 10.4049/jimmunol.0802902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strömberg S, Björklund MG, Asplund A, et al. Transcriptional profiling of melanocytes from patients with vitiligo vulgaris. Pigment Cell Melanoma Res. 2008;21:162–71. doi: 10.1111/j.1755-148X.2007.00429.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp