Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model (original) (raw)
Abstract
To investigate the possibility of using commensal bacteria as signal mediators for inhibiting the disease cholera, we stably transformed Escherichia coli Nissle 1917 (Nissle) to express the autoinducer molecule cholera autoinducer 1 (CAI-1) (shown previously to prevent virulence when present with another signaling molecule, autoinducer 2, at high concentrations) and determined the effect on Vibrio cholerae virulence gene expression and colonization in an infant mouse model. We found that pretreatment of mice for 8 h with Nissle engineered to express CAI-1 (Nissle-cqsA) greatly increased the mice’s survival (92%) from ingestion of V. cholerae. Pretreatment with Nissle-cqsA for only 4 h increased survival by 77%, whereas ingesting Nissle-cqsA at the same time as V. cholerae increased survival rates by 27%. Immunostaining revealed an 80% reduction in cholera toxin binding to the intestines of mice pretreated for 8 h with Nissle-cqsA. Further, the numbers of V. cholerae in treated mouse intestines was reduced by 69% after 40 h. This finding points to an easily administered and inexpensive approach where commensal bacteria are engineered to communicate with invasive species and potentially prevent human disease.
Keywords: quorum sensing, probiotic, signal engineering, prophylactic, enteric disease
The human gastrointestinal (GI) tract is home to hundreds of bacterial species required for normal metabolism and comprises the first line of defense against ingested pathogens (1). Commensal bacteria in the GI tract coexist with their human hosts and with each other through a complex system of checks and balances (2). Human defenses against enteric microorganisms include innate and adaptive immunity. Immunoprotection allows commensal populations to colonize both the large and small bowel without causing sepsis. Signaling interactions between bacterial populations within the GI tract are only recently being resolved at the molecular level (3–7). For example, it has been shown that bacterial secretion of formate within the intestinal lumen controls Salmonella typhimurium’s colonization of the GI tract (8). Formate secretion from non-S. typhimurium bacteria allows S. typhimurium to colonize the distal ileum rather than other sections of the bowel that have much lower concentrations of formate.
Some bacterial signaling allows colonies to behave in manners analogous to tissues of multicellular organisms. Termed “quorum sensing” (QS) (9, 10) for its dependence on bacterial population density, this type of signaling relies on extracellular signals (autoinducers) that accumulate as bacteria grow. At high population densities, autoinducers bind to transmembrane-spanning receptors to mediate colony-wide, coordinated gene expression. This phenomenon is seen in both Gram-negative and Gram-positive bacteria (11). One of the most widely studied autoinducers, autoinducer 2 (AI-2), is present in at least 50 different species (11). AI-2 (a furanosyl borate diester; ref. 12) is expressed in Escherichia coli and in that bacterium has been linked to biofilm formation (13) and stress from heterologous protein expression (14, 15). For a review of AI-2 in E. coli see ref. 16.
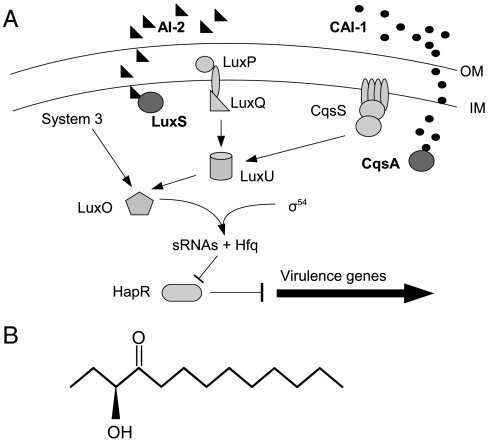
AI-2 is also expressed by the marine bacterium Vibrio cholerae, the causative organism for the enteric disease cholera. In V. cholerae as well as Vibrio harveyi AI-2 is expressed with another externally secreted autoinducer, cholera autoinducer 1[CAI-1, an (S)-3-hydroxytridecan-4-one] (17) (Fig. 1). The final step in CAI-1 synthesis is carried out by the gene product of cqsA (18). For AI-2 synthesis, the final step is mediated by the gene product of luxS (19). AI-2 and CAI-1 work with a third, internal QS circuit to coordinate gene expression when V. cholerae populations reach higher concentrations (20) (Fig. 1). This circuit is almost identical in V. cholerae and V. harveyi. In the latter, the third component is external and involves a _V. harveyi_-specific autoinducer harveyi autoinducer 1 (HAI-1) (21) which activates the lux operon leading to bioluminescence at high cell densities (20). In V. cholerae, high cell densities lead to the cessation of virulence gene expression (22). At low cell density, V. cholerae expresses virulence factors cholera toxin (CT) and the toxin coregulated pilus (TCP) (23). TCP allows for attachment to the intestinal wall (24, 25), whereas CT causes massive diarrhea and dehydration by interrupting cAMP synthesis in intestinal epithelia (26). V. cholerae enters through tainted water into the GI tract where it can grow and continue to express virulence factors. It grows rapidly and can reach very high densities within a few hours (27). Once at high cell density, virulence factor expression is greatly reduced and protease expression allows for degradation of the attachment matrix and movement of V. cholerae out of the host with diarrheal fluids.
Fig. 1.
Quorum sensing in V. cholerae. V. cholerae QS is comprised of three systems (A). These converge at the response regulator LuxO which serves to downregulate virulence expression at high cell densities. Two external autoinducers, AI-2 (black triangles, synthesized by LuxS) and CAI-1 (black circles, synthesized by CqsA) bind to their respective sensor proteins (LuxP/LuxQ for AI-2 and CqsS for CAI-1) at high concentrations outside the cell. Autoinducer binding converts internal kinases to phosphatases and prevents the transcription of sRNAs responsible for blocking virulence inhibition. The result is the cessation of virulence gene expression at high cell densities. Arrows indicate regulation direction at low cell densities (i.e., when autoinducer is unbound). V. cholerae CAI-1 is an (S)-3-hydroxytridecan-4-one (B). OM, outer membrane; IM, inner membrane.
As temperatures around the world increase, it is expected that threats from water borne illnesses will increase as well (28). Cholera poses a particularly strong threat to developing nations (29). South America, Africa, and the Indian subcontinent are hardest hit with an estimated annual death toll between 120,000 and 200,000 (30). To investigate the possibility of using commensal bacteria as a prophylactic against V. cholerae, we stably transformed the commercially available probiotic E. coli Nissle 1917 (Nissle) (31) to express CAI-1 (Nissle already expresses AI-2) and determined the effect on V. cholerae virulence gene expression and colonization. Previously we reported reduced CT and TCP expression in cocultures of V. cholerae and Caco-2 human epithelia (32). In those experiments, Caco-2 cells were first incubated with cell-free media from Nissle expressing CAI-1 before being challenged with V. cholerae. The result was a reduction in V. cholerae virulence gene expression. The objective for the work described here was to determine if simple feeding of engineered Nissle to infant mice could reduce CT levels and V. cholerae colonization in mouse intestines.
Results
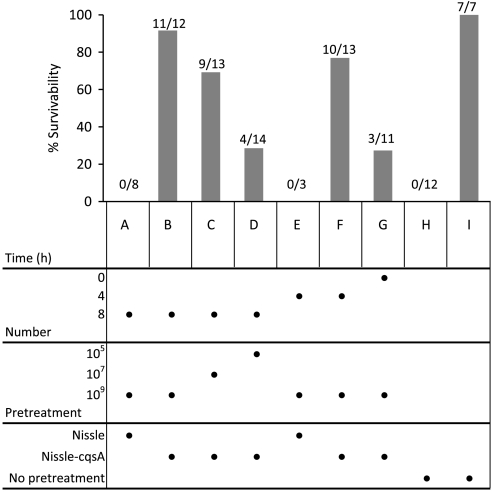
Nissle, a model commensal bacterium which has been proven safe and effective as a delivery vehicle for therapeutics (31), was stably transformed to express the gene cqsA (the gene encoding the final CAI-1 synthesis enzyme; Fig. 1) under control of the native constitutive promoter fliC to make the strain Nissle-cqsA. AI-2 and CAI-1expression comparable to V. cholerae was confirmed for this strain via bioassay (Figs. S1 and S2, respectively). As a pretreatment against cholera, 2–3-day-old mice were fed at three different times preinfection (0, 4, and 8 h) with Nissle, Nissle-cqsA, or no pretreatment and subsequently challenged with V. cholerae. To determine the limits of protection in this model, various numbers of Nissle and Nissle-cqsA were fed to the mice as a pretreatment. Survivability of mice was assayed 48 h after V. cholerae challenge (Fig. 2). Mice fed the highest number of Nissle-cqsA (109 cells) 8 h before V. cholerae ingestion exhibited a 92% survival rate (Fig. 2B). When less Nissle-cqsA was fed in the pretreatment at 8 h, lower survivability was seen: 69% for 107 cells (Fig. 2C) and 29% for 105 cells (Fig. 2D). Similarly, when mice were fed Nissle-cqsA at 109 cells for shorter times, protection was lessened to 77% for 4 h and 27% for mice fed Nissle-cqsA at the same time as V. cholerae (Fig. 2 F and G, respectively). The unmodified Nissle strains gave no protection from V. cholerae at the concentrations tested (Fig. 2 A and E). No mice fed V. cholerae survived without pretreatment (Fig. 2H) and all mice not fed V. cholerae survived without pretreatment (Fig. 2I).
Fig. 2.
Survival of infant mice fed V. cholerae with different pretreatments. Infant mice were fed with E. coli Nissle 1917 (Nissle), E. coli Nissle 1917 expressing the V. cholerae autoinducer gene cqsA chromosomally (Nissle-cqsA), or no bacteria (no pretreatment) before being challenged with V. cholerae (A–H). “No pretreatment” mice were either challenged with V. cholerae (H) or not (I). Pretreatment times, numbers of bacteria fed, and bacterial strains used are shown in the figure under the bar graph. Mice were fed with bacterial treatments either 0, 4, or 8 h before V. cholerae challenge. Survival of mice after 48 h is shown. The total amount of bacteria fed each mouse (number) is also given. Directly below each bar on the chart is the set of conditions for that experiment. For example, in B, 109 cells of Nissle-cqsA were fed to mice 8 h before V. cholerae challenge and 11 out of 12 mice survived for a survivability of 92%. In D, 105 cells of Nissle-cqsA were fed to mice 8 h before V. cholerae challenge and 4 out of 14 mice survived for a survivability of 29%. The numbers of mice surviving in each group is shown over the total number of mice in that group.
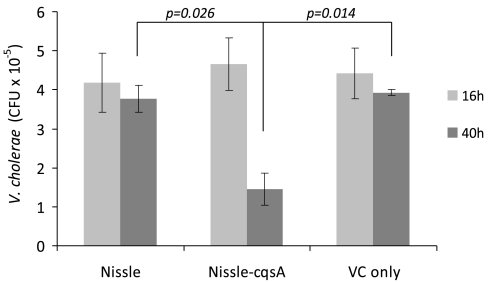
At 16 and 40 h after infection with V. cholerae, some of the mice were killed and their intestinal tracts were homogenized. After 16 h, plate counts indicated that the numbers of V. cholerae bacteria surviving in the GI tract were the same for mice fed Nissle (109 cells 8 h prior to challenge), Nissle-cqsA (109 cells 8 h prior to challenge) and a control that had no pretreatment (Fig. 3). After 40 h, the amounts of V. cholerae in plate counts from homogenized intestinal tissue was relatively unchanged for mice fed Nissle or only V. cholerae. However, mice fed Nissle-cqsA had significantly fewer V. cholerae colonies (69% on average) than they had at 16 h (Fig. 3). For mice fed 109 Nissle-cqsA 8 h prior to V. cholerae challenge, plate counts indicated that Nissle-cqsA survivability was on average 6.5 × 106 cfu (per mouse after 16 h and 7.2 × 106 CFU per mouse after 40 h) (Fig. S3).
Fig. 3.
Survival of V. cholerae in mouse intestinal tracts. Infant mice were fed with E. coli Nissle 1917 (Nissle), E. coli Nissle 1917 expressing the V. cholerae autoinducer gene cqsA (Nissle-cqsA), or no bacteria (VC only) before being challenged with V. cholerae. Plate counts determined the cfu of V. cholerae present in the intestines after 16 h and again after 40 h. Values are averages of duplicate mice. Error bars represent 1 SD. The p values are for a Student t test with n = 2.
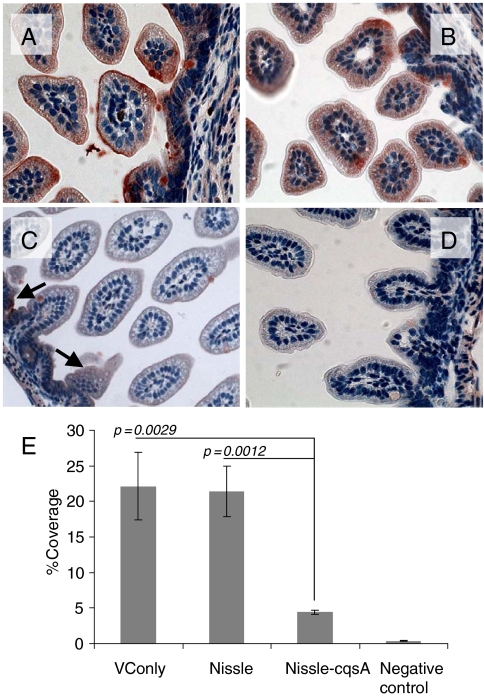
After 48 h of V. cholerae challenge dissected mouse GI tracts were subject to immunohistochemical staining for CT (Fig. 4). Red staining indicated the presence of CT which mostly accumulated on the epithelial surface of infected cells as expected (Fig. 4 A_–_D). To quantify the amount of CT bound, images of intestinal slices from three mice per treatment were analyzed for percent coverage by CT using ImageJ software from the National Institutes of Health (NIH) (Fig. 4E). For mice fed only V. cholerae (without Nissle) the amount of CT coverage was approximately 22% (Fig. 4A). Nissle-fed (109 cells, 8 h prior to V. cholerae challenge) mice also had close to 22% coverage with CT following V. cholerae challenge (Fig. 4B). Mice fed Nissle-cqsA (109 cells, 8 h prior to V. cholerae challenge) had significantly lower CT (4% coverage with CT on average) after V. cholerae challenge (Fig. 4 C and E) than all other mice except those not challenged with V. cholerae which exhibited less than 1% of red staining (Fig. 4D). Although there was no significant difference in coverage between nonpretreated mice and mice fed Nissle, there was a significant reduction (80%) in CT coverage for mice pretreated with Nissle-cqsA.
Fig. 4.
CT binding to mouse intestines. Infant mice were given the following pretreatments: no treatment (A), E. coli Nissle 1917 (Nissle) (B), E. coli Nissle 1917 engineered to express V. cholerae cqsA (Nissle-cqsA) (C). Mice in A_–_C were then challenged with V. cholerae. Mice in D were not pretreated and were not challenged with V. cholerae. Intestines were stained for the presence of CT. Red staining indicates the presence of CT. Arrows in C indicate red staining. Slides from triplicate mice for each treatment were analyzed using ImageJ software (NIH) to quantify the amount of red staining (CT) present. The percentage of red coverage for each slide was determined and the values averaged (D). Error bars represent 1 SD. The p values are for a Student t test with n = 3. Images in A_–_D were taken at 40× magnification.
Discussion
The data in Fig. 2 demonstrate a dose-dependent benefit to feeding with Nissle-cqsA prior to challenge with V. cholerae. Of interest as well is the time dependence of the protection. For mice whose pretreatment was only 4 h later (from 8 to 4 h and from 4 to 0 h) there was a drop in protection. In time-dependent experiments, it became clear that the duration and quantity of Nissle-cqsA fed will be crucial to its efficacy as a prophylactic. Further, it could be surmised from the data in Figs. 2 and 4 that less CT binding to the intestinal epithelia was correlated with increased survivability. The red staining (less than 1%) for noninfected cells was attributed to nonspecific staining of mucin-containing goblet cells (Fig. 3D). In comparing these results with those of our previous work (32), we found that the reduced amount of CT detected in the infant mouse to be similar to what was seen in human epithelial cells (an 80% reduction in mice and an almost 70% reduction in human cells). This protective effect, although not 100%, appears to be sufficient to allow for a high level of survival (92%).
Although longer experiments would likely have been informative, one of the limitations of the infant mouse model is the 6–9 day duration for V. cholerae susceptibility in infant mice (33). Adult mice are resistant to long-term V. cholerae colonization. Hence, the duration of protection that could be expected from feeding Nissle-cqsA to infant mice could not be determined. However, these data suggest that in the event of a cholera outbreak feeding of Nissle-cqsA would have to already be undertaken at least 8 h prior to exposure to provide a high level of protection. This implies a preventative that would become part of the diet in impoverished areas or something that would be distributed as part of aid to areas where a recent natural disaster would make the likelihood of cholera outbreak high.
That V. cholerae were significantly cleared from the mice (Fig. 3) indicates feeding Nissle-cqsA was effective at both reducing CT accumulation and reducing V. cholerae binding to the GI tract. Although less CT secretion from V. cholerae was demonstrated previously to be a result of Nissle-cqsA activity (32), it could not be determined from the results presented here if the reduction in CT was caused by lower CT secretion from V. cholerae or from there being fewer V. cholerae adhered to the mouse small intestines. The amount of time that was required to see a difference in V. cholerae colonization was expected as the mechanism for removal of nonadherent bacteria from the GI tract would be normal bowel movements in a mouse not experiencing diarrhea. V. cholerae can appear in human stools for weeks and even years after ingestion (27). Here again, a longer experiment would have been useful but not possible with the infant mouse model. Future work may make use of a recently developed adult mouse model that requires the use of several exogenous factors to facilitate colonization (34) or a Drosophila melanogaster model (35). Drosophila reportedly exhibits a much more human-like colonization pattern than do nonhuman mammals.
Engineering probiotic bacteria to prevent or treat cholera has been described previously. Focareta and coworkers engineered commensal bacteria to express the GM1 ganglioside (the target of CT) as a method for effectively sorbing CT and reducing V. cholerae virulence. However, there is significant potential to illicit immune responses to GM1 (a ubiquitous ganglioside in the GI tract) using this approach (36). By using a bacterial autoinducer as a preventative, immune responses to the host’s own biochemistry are avoided. Still, there is potential for stimulating immune responses (specifically, IgA) against CAI-1 in an intestinal tract containing bacteria secreting CAI-1. IgA are secreted into the mucosa and lumen of the small intestine in response to ligands from intestinal bacteria, food, and other intestinal contents. Although the pathways leading to IgA secretion against specific ligands are not fully understood (37), the result is that secreted IgA can bind to ligands within the lumen and render them ineffective. This allows for noninflammatory immunoprotection of the host (38). If CAI-1 were targeted by IgA it could lead to heightened pathogenicity in the presence of V. cholerae as CAI-1 is bound by IgA and thereby potentially prevented from reentering V. cholerae. Without effective CAI-1 accumulation, V. cholerae virulence would not abate at high cell densities. The result to the host would be sustained CT expression from V. cholerae still expressing binding protein (TCP) and not expressing the proteases needed to release it from the intestinal wall. Clearly, further study is required to determine if CAI-1 elicits immunoresponses in the host. In order to best answer this question, human subjects fed bacteria secreting CAI-1 for extended periods of time would have to be tested for expression of IgA-decorated CAI-1.
Reported here is an important step toward determining if commensal bacteria could be engineered to serve as a prophylactic against cholera. Although further study is needed to elucidate the long-term effects of feeding engineered bacteria to human hosts, our findings are encouraging. They highlight the potential of engineering commensal bacteria to serve as in vivo signal mediators. In this case, pathogen-specific communication is used to lower pathogenic bacterial virulence. In other work, we have shown how signaling between commensal bacteria and the host’s epithelial cells can potentially treat autoimmune disease (39). These examples point to an exciting and versatile approach to preventing and treating disease where the body’s own bacteria are engineered to communicate on the body’s behalf.
Materials and Methods
Unless otherwise indicated all chemicals and reagents were purchased from VWR International. All cloning was carried out using standard techniques (40). A construct was prepared for chromosomal insertion of the cqsA gene under control of the native fliC promoter. The construct was used to chromosomally insert the cqsA gene into Nissle using established methods (41). Briefly, one-step inactivation was used to insert the cqsA gene in place of fliC downstream of the fliC promoter region in the Nissle chromosome. This technique uses two plasmids, pKD3 (conferring chloramphenicol resistance) and pKD46 (conferring ampicillin resistance) (41).
Nissle 1917 and all other E. coli strains were maintained in LB at 37 °C, with shaking at 225 rpm. For infection experiments, a streptomycin-resistant strain of V. cholerae El Tor C6706_str_ (kind gift from Ronald Taylor, Dartmouth Medical School, Hanover, NH) was used. V. cholerae were maintained at 30 °C without shaking in either LB or AKFD (15 g/L peptone, 4 g/L yeast extract, 10 g/L sodium chloride, pH 7.4) media. V. harveyi strains BB120 (wild type) and BB170 (Δ_luxS_) were used as positive control and reporter strain for AI-2 assays, respectively. Both strains were maintained in AB medium (0.3 M NaCl, 0.05M MgSO4, 0.2% vitamin-free casamino acids (Difco), adjusted to pH 7.5 with KOH. The medium was sterilized and then 10 mL 1 M potassium phosphate (pH 7.0), 10 mL of 0.1 M L-arginine, 20 mL of Glycerol, 1 mL of 10 μg mL-1 riboflavin, and 1 mL of 1 mg mL-1 thiamine was added per liter) (42) at 30 °C with shaking at 225 rpm. V. cholerae MM920 [Δ_cqsA_ Δ_luxQ_ pBB1 (luxCDABE from V. harveyi)] was used as the reporter strain for CAI-1 and maintained in LB medium at 30 °C with shaking at 225 rpm. BB120, BB170, and MM920 were kind gifts from Bonnie Bassler (Princeton University, Princeton, NJ).
E. coli DH5α (DH5α), Nissle, and Nissle with a cqsA chromosomal insertion (Nissle-cqsA) were grown in AKFD at 37 °C shaking at 225 rpm for 8 h. V. cholerae was grown in AKFD with 10 μg/mL streptomycin at 30 °C at 225 rpm and V. harveyi BB120 was grown in AB medium, at 225 rpm and 30 °C, both for 8 h. After 8 h, all bacteria were spun down and washed three times with the corresponding culture medium. All cultures were adjusted to the same OD600 and inoculated into the same amount of culture medium. After inoculation, DH5α, Nissle, and Nissle-cqsA were grown overnight in AKFD at 37 °C shaking at 200 rpm. V. cholerae was grown overnight at 30 °C shaking at 200 rpm in AKFD and V. harveyi BB120 was grown overnight at 30 °C shaking at 200 rpm in AB medium. After growing 14–16 h, overnights were centrifuged at 4,000 × g for 30 min at 4 °C. The supernatant was filtered (0.2 μm, PALL Life Sciences). The cell-free culture medium was diluted to OD600 = 1 with AKFD, and 10 ng/mL leupeptin was added to inhibit proteases before storage at 4 °C.
To test for AI-2 activity V. harveyi BB170 was grown overnight in AB medium and diluted 1∶3,000 in AB medium. Overnights of strains to be tested for AI-2 activity were centrifuged (4,000 × g) and 10 μL of their cell-free supernatant was added to 90 μL of diluted V. harveyi BB170 in a sterile 96-well plate and incubated at 30 °C with shaking at 225 rpm. Luminescence from the reporter strain was measured in a microtiter plate reader (FLX800, BIO-TEK Instruments, Inc.) every 0.5 h until the luminescence of the control increased. As controls, we tested the strains DH5α [an AI-2 mutant strain (19) that has no CAI-1 activity] and V. harveyi BB120 (which has both CAI-1 and AI-2 activity).
To test for CAI-1 activity, V. cholera MM920 was grown to a high density overnight and diluted 1∶10 in LB with 5 μg mL-1 tetracycline. Overnights of strains to be tested for CAI-1 activity were centrifuged (4,000 × g) and 30 μL of cell-free supernatant was added to 70 μL of diluted V. cholera reporter MM920 (diluted in LB) in a sterile 96-well plate and incubated at 30 °C with shaking at 225 rpm. Luminescence was measured by microtiter plate reader (FLX800, BIO-TEK Instruments, Inc.) every 0.5 h until the luminescence decreased. As controls we tested the strains DH5α (an AI-2 mutant strain that has no CAI-1 activity) and V. harveyi BB120 (which has both CAI-1 and AI-2 activity).
All mice used in these experiments were treated in accordance with protocols approved by the Cornell University Institutional Animal Care and Use Committee. All mice were housed at the Transgenic Mouse Core Facility at the Cornell University Veterinary School. Pregnant mice (CD-1) used in this study were purchased from Charles River Laboratories. Briefly, 2–3-day-old suckling CD-1 mice were separated from their mothers for 1 h. Each mouse was fed with cultures of Nissle (Nissle or Nissle-cqsA) that had been grown 10 h in LB and then washed with 1 × PBS and concentrated to either 105, 107, or 109 cells in 20 μL 20% sucrose and blue food-coloring dye (Evans Blue, VWR; the dye provided visual confirmation of bacteria through the mouse skin). Nissle-fed mice were put back with their mothers.
After 0, 4, or 8 h, each mouse was fed with 2 × 107 V. cholerae except for the blank control group. Some of the mice were euthanized after 16 or 40 h of V. cholerae challenge and their guts were removed and mechanically homogenized in 2 mL of LB. For colony counting at 16 and 40 h, serial dilutions of extracts from homogenized guts of those mice receiving either 109 cells of Nissle, Nissle-cqsA, or no treatment were plated onto thiosulfate citrate bile salts sucrose agar supplemented with 100 μg/mL streptomycin to enumerate V. cholerae cfu. To count Nissle-cqsA survival at 16 and 40 h, homogenized guts were plated onto MacConkey agar supplemented with 10 μg/mL chloramphenicol.
The numbers of mice still living after 48 h was recorded before all of the remaining mice were euthanized. The gut tissues of mice sacrificed after 48 h were fixed in 4% paraformaldehyde (Mallinckrodt) overnight and washed three times with 1 × PBS and soaked in 70% ethanol. Fixed gut tissues were then dissected. After deparaffinization, fixed tissue slides were steamed in 0.01 M citrate buffer pH 6.0 and immersed in 0.5% hydrogen peroxide (Fisher) in methanol for 10 min to block endogenous peroxidase. After washing in 0.01 M PBS (pH 7.2), 10% normal blocking goat serum (Invitrogen) was applied for 20 min at room temperature in a humid chamber. Anticholera toxin subunit B (US Biological) diluted in PBS plus 1× casein (Vector) was applied to blocked samples that were then incubated in a humid chamber for 1.5 h at 37 °C. After washing four times in PBS, a biotinylated secondary antibody (Vector) diluted 1∶200 in PBS was applied to samples for 20 min at room temperature in a humid chamber. Samples were incubated with streptavidin peroxidase (Invitrogen) for 20 min at room temperature in a humid chamber and washed three times with PBS. Samples were incubated with 3-amino-9-ethylcarbazole chromogen/substrate solution (Invitrogen) at room temperature. Color development was monitored under ordinary light microscopy for approximately 5–15 min. A distilled H2O rinse was used to stop the reaction. Brightfield slides were counterstained with hematoxylin (Fisher) for 30 s before rinsing in tap H2O for 5 min. Samples were mounted using an aqueous mounting medium Fluoromount (Fisher). Pictures were taken with a color camera under an ordinary light microscope (Leica). The stained gut tissue pictures were analyzed by Image J software (NIH-National Center for Biotechnology Information) for the percent coverage by CT as estimated from red coloring.
Supplementary Material
Supporting Information
Acknowledgments.
Yung-Fu Chang at Cornell University advised mouse experiments. This work was funded by the Gates Foundation and the Life Sciences Initiative at Cornell University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 3.Bourlioux P, Koletzko B, Guarner F, Braesco V. The intestine and its microflora are partners for the protection of the host: Report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am J Clin Nutr. 2003;78:675–683. doi: 10.1093/ajcn/78.4.675. [DOI] [PubMed] [Google Scholar]
- 4.Antunes LC, Ferreira RB. Intercellular communication in bacteria. Crit Rev Microbiol. 2009;35:69–80. doi: 10.1080/10408410902733946. [DOI] [PubMed] [Google Scholar]
- 5.Canny G, Swidsinski A, McCormick BA. Interactions of intestinal epithelial cells with bacteria and immune cells: Methods to characterize microflora and functional consequences. Methods Mol Biol. 2006;341:17–35. doi: 10.1385/1-59745-113-4:17. [DOI] [PubMed] [Google Scholar]
- 6.Boontham P, et al. Significant immunomodulatory effects of Pseudomonas aeruginosa quorum-sensing signal molecules: Possible link in human sepsis. Clin Sci. 2008;115(11–12):343–351. doi: 10.1042/CS20080018. [DOI] [PubMed] [Google Scholar]
- 7.Corthesy B, Gaskins HR, Mercenier A. Cross-talk between probiotic host immune system. J Nutr. 2007;137:781S–790S. doi: 10.1093/jn/137.3.781S. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol. 2008;190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nealson KH, Markovitz A. Mutant analysis and enzyme subunit complementation in bacterial bioluminescence in Photobacterium fischeri. J Bacteriol. 1970;104:300–312. doi: 10.1128/jb.104.1.300-312.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun. 2005;73:3197–3209. doi: 10.1128/IAI.73.6.3197-3209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez Barrios AF, et al. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLisa MP, Valdes JJ, Bentley WE. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J Bacteriol. 2001;183:2918–2928. doi: 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLisa MP, Valdes JJ, Bentley WE. Quorum signaling via AI-2 communicates the “metabolic burden” associated with heterologous protein production in Escherichia coli. Biotechnol Bioeng. 2001;75:439–450. doi: 10.1002/bit.10034. [DOI] [PubMed] [Google Scholar]
- 16.Ahmer BM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 17.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 18.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 19.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrid cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 25.Herrington DA, et al. Toxin, toxin-coregulated pili, and the toxr regulon are essential for Vibrio-cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominguez P, Barros F, Lazo PS. The activation of adenylate cyclase from small intestinal epithelium by cholera toxin. Eur J Biochem. 1985;146:533–538. doi: 10.1111/j.1432-1033.1985.tb08684.x. [DOI] [PubMed] [Google Scholar]
- 27.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelle K, Pascual M, Yunus M. Pathogen adaptation to seasonal forcing and climate change. Proc Roy Soc B-Biol Sci. 2005;272:971–977. doi: 10.1098/rspb.2004.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerrant RL, Carneiro-Filho BA, Dillingham RA. Cholera, diarrhea, and oral rehydration therapy:triumph and indictment. Clin Infect Dis. 2003;37:398–405. doi: 10.1086/376619. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez J, Holmgren J. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr Opin Immunol. 2005;17:388–398. doi: 10.1016/j.coi.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Westendorf AM, et al. Intestinal immunity of Escherichia coli NISSLE 1917: A safe carrier for therapeutic molecules. FEMS Immunol Med Mic. 2005;43:373–384. doi: 10.1016/j.femsim.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Duan F, March JC. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol Bioeng. 2008;101:128–134. doi: 10.1002/bit.21897. [DOI] [PubMed] [Google Scholar]
- 33.Pitkin D, Actor P. Immunity to Vibrio cholerae in the mouse. I. Passive protection of newborn mice. Infect Immun. 1972;5(4):428–432. doi: 10.1128/iai.5.4.428-432.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivier V, Queen J, Satchell KJ. Successful small intestine colonization of adult mice by Vibrio cholerae requires ketamine anesthesia and accessory toxins. PLoS One. 2009;4:e7352. doi: 10.1371/journal.pone.0007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blow NS, et al. Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 2005;1:e8. doi: 10.1371/journal.ppat.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Focareta A, Paton JC, Morona R, Cook J, Paton AW. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology. 2006;130:1688–1695. doi: 10.1053/j.gastro.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Fagarasan S, Honjo T. Intestinal IgA synthesis: Regulation of front-line body defences. Nat Rev Immunol. 2003;3:63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 39.Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: Rewiring the gut to treat diabetes. Appl Environ Microbiol. 2008;74:7437–7438. doi: 10.1128/AEM.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 41.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenberg EP, Hastings JW, Ulitzur S. Induction of luciferase synthesis in Beneckea-Harveyi by other marine-bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information