The impact of intragenic CpG content on gene expression (original) (raw)
Abstract
The development of vaccine components or recombinant therapeutics critically depends on sustained expression of the corresponding transgene. This study aimed to determine the contribution of intragenic CpG content to expression efficiency in transiently and stably transfected mammalian cells. Based upon a humanized version of green fluorescent protein (GFP) containing 60 CpGs within its coding sequence, a CpG-depleted variant of the GFP reporter was established by carefully modulating the codon usage. Interestingly, GFP reporter activity and detectable protein amounts in stably transfected CHO and 293 cells were significantly decreased upon CpG depletion and independent from promoter usage (CMV, EF1α). The reduction in protein expression associated with CpG depletion was likewise observed for other unrelated reporter genes and was clearly reflected by a decline in mRNA copy numbers rather than translational efficiency. Moreover, decreased mRNA levels were neither due to nuclear export restrictions nor alternative splicing or mRNA instability. Rather, the intragenic CpG content influenced de novo transcriptional activity thus implying a common transcription-based mechanism of gene regulation via CpGs. Increased high CpG transcription correlated with changed nucleosomal positions in vitro albeit histone density at the two genes did not change in vivo as monitored by ChIP.
INTRODUCTION
The increasing demand for pharmaceutical-relevant proteins used in vaccine development, clinical therapy and gene therapy approaches requires improved strategies of production with regard to a sustained and high transgene expression. The recombinant expression of transgenes is often complicated by several factors such as bad solubility and missing posttranslational modifications in a prokaryotic expression background (1) or low-expression yields when using eukaryotic expression systems (2–4). Much effort has been spent on the development of optimized expression platforms, as high-level expression of recombinant proteins strongly depends on the diligent design of the expression cassette (5). The fusion of intronic sequences upstream of the open reading frame and the use of strong hybrid promoters have been demonstrated to be successful in enhancing transgene expression (6,7). The adaptation of the codon usage to avoid rare codon was another progress in yielding higher expression levels (8). With regard to viral sequences it was shown for the human immunodeficiency virus type 1 gag gene that the adaptation to the human codon usage gave a CG-rich sequence and resulted in the constitutive and high-level expression of the otherwise strictly regulated gene in mammalian cell lines (9,10). Optimization of codon usage has therefore substantially encouraged the development of safe lentiviral vectors (11) and highly efficient packaging cell lines for vaccine design.
Yet there is need for further progress considering protein amount and stability of transgene expression to surpass virus-based expression vectors relieving handling and safety issues. The optimization of codon usage has also been applied to plasmid DNA-based gene therapy approaches to take advantage of their safety issues and simultaneously improve protein expression, low expression levels being one of the most prominent disadvantages of plasmid DNA (12). However, plasmid DNA applications using optimized expression cassettes combined with strong viral promoters have been shown to be impaired by downregulation of gene expression via transcriptional silencing in vivo and in vitro (13). The repression of transcription often correlates with methylation of CpG dinucleotides representing an epigenetic control mechanism involved in development, imprinting, X-chromosomal gene inactivation and cancerogenesis (14–16). CpG dinucleotides are statistically underrepresented within the eukaryotic genome as CpG dinucleotides methylated at the carbon 5-position of the cytosine residue are readily deaminated thereby causing a transition mutation from cytosine to thymine (17). However, there are CpG dense genomic regions which are virtually protected from CpG methylation likely due to occupation by nuclear factors or less efficient binding of methyltransferases to CpGs clustering at narrow intervals (18). These CpG islands are almost exclusively either located within or in the proximity of promoter regions and sometimes extend into transcribed areas (19). Whereas unmethylated CpGs are associated with transcriptional activity, their hypermethylated state is often observed in carcinogenesis and mediates gene silencing directly by sterically hindering transcription factor binding or indirectly by interacting with methyl-binding domain repressor proteins which in turn recruit chromatin modifying complexes to achieve a condensed chromatin state (20). The disposition of CpG dinucleotides to cause mutations (21) and gene silencing (22) led to the avoidance of CpGs when designing expression constructs for recombinant protein production (23) and gene therapy (24). So far, it is still unclear whether DNA methylation is a prerequisite or rather a consequence of gene silencing (25). Apart from hypermethylation and silencing of endogenous promoters during cancer progression (26), strong heterogeneous and ubiquitously expressing promoters such as the commonly used immediate-early gene promoter from cytomegalovirus (CMV) have been reported to be susceptible to CpG methylation, in contrast to those derived from housekeeping genes (27). Moreover, methylation of promoter regions is not solely responsible for transcriptional repression. Flanking methylated vector elements including intragenic regions have been shown to contribute to silencing of gene expression, potentially by promoting methylation of adjacent CpGs and establishing transcriptionally inactive chromatin structures (28,29). Especially CpG-rich vector sequences of bacterial origin, which are prone to de novo methylation, have been proven to be significantly involved in silencing of codelivered expression cassettes (30). Accordingly, recent strategies for efficient transgene expression have preferred the usage of CpG depleted vector backbones (24,31). In addition to regulatory vector elements, the transgene encoding region has also been examined for its contribution to transcription in vitro and in vivo. The in vitro methylation of the intragenic region was demonstrated to be sufficient to silence gene expression in the absence of promoter methylation (32,33). In support of these findings, CpG depleted transgenes not susceptible to methylation have been proven to be beneficial for high and sustained levels of protein expression in vivo (22,34). However, gene silencing via de novo methylation has also been described in stably transformed cell lines (35) and the influence of intragenic CpGs on the expression of a transgene might deserve further study in view of efficient production of recombinant proteins. Recent work showed that a CpG-rich reporter gene was silenced in murine cells following _in vitro_-methylation, whereas the unmethylated counterpart was capable of long-term and high-level expression (36). Indeed, other aspects contributing to efficient protein expression in cell culture such as improved mRNA stability or prolonged mRNA half-life as a consequence of increased GC content might put the proposed negative effects of methylation dependent silencing into perspective (37,38).
Nonetheless, the exact contribution of CpG dinucleotides onto transcription is discussed controversially. In this study, the influence of CpG dinucleotides within the open reading frame was addressed in more detail to elucidate their impact on expression and to have the ability to predict essential sequence elements for an appropriate transgene design.
Our results showed a severe reduction in reporter expression following CpG depletion from the reporter encoding region, which clearly correlated with decreased levels of newly synthesized mRNA transcripts thus implying a transcription-based regulation of gene expression via CpG dinucleotides in the absence of methylation.
MATERIALS AND METHODS
Plasmid constructions
A synthetic version of the green fluorescent protein (GFP) encoding gene of Aequorea victoria adapted to human codon-usage (huGFP60) and containing 60 CpG dinuculeotides has been described previously (39). For construction of GFP expression vectors, the huGFP60 gene was amplified by PCR using oligonucleotides 5′-T ACGAAGCTTGCCACCATGGTGAGCAAGGG-3′ and 5′-ACGAGCTGTACAAGTAATAGGATCCTACT-3′. To obtain the pS/huGFP60 construct, the resulting PCR fragment was digested with HindIII and BamHI and inserted into the prokaryotic expression plasmid pPCR-Script (Stratagene) providing expression control by a T7 promoter. Likewise, the fragment was cloned into the eukaryotic expression vector pcDNA5/FRT (Invitrogen) to obtain pc5/huGFP60. A synthetic huGFP gene lacking CpG dinucleotides (huGFP0) was generated by Geneart AG (Regensburg, Germany) via stepwise PCR amplification and inserted into the corresponding expression vectors resulting in plasmids pS/huGFP0 and pc5/huGFP0. Analogously, both GFP genes were cloned into a pcDNA3.1 (Invitrogen)-derived vector devoid of the neomycin resistance gene (pc3.1100) and a derivative thereof exhibiting a 53% reduction in CpG content (pc3.147). Diminishment of CpG numbers was achieved by depleting CpG dinucleotides from the ß-lactamase gene, the multiple cloning site, parts of the pUC origin of replication and other non-coding sequences. To lower CpG content further, the conventional CMV promoter within pc3.147 was replaced by a previously described CpG free promoter version (24) resulting in pc3.132 with an overall CpG content of 32% compared to the original pc3.1100 vector. The gene encoding HIV-1 capsid protein was PCR-amplified from the formerly described syngag gene, which is adapted to the codon usage of highly expressing mammalian genes (9). The cytokine genes murine MIP-1α (GenBank® accession number M23447) and human GMCSF (GenBank® accession number G15899) were RNA- and codon-optimized for expression in mammalian cells with respect to codon adaptation index [CAI,(40)] and synthesized via stepwise PCR from oligonucleotides (Geneart AG, Regensburg, Germany). Based on the resulting codon-optimized genes huCA38, muMIP13 and huGM-CSF12, gene variants completely devoid of CpG dinucleotides were synthesized and designated huCA0, muMIP0 and huGM-CSF0, respectively. All genes were inserted into the eukaryotic expression vector pcDNA3.1 for transient expression experiments.
Calculation of codon frequencies
The ‘Codon Adaptation Index’ CAI represents a mean value that is calculated based on the ‘Relative Adaptiveness’ of each individual codon used for the two CpG high and CpG low alelles encoding GFP, Mip1α, HIV Capsid and GM-CSF, respectively. The Relative Adaptiveness reflects the frequency of individual codons encoding a given amino acid, whereas the most frequently used triplet is put to 1.0 and less frequently used codons are scaled down accordingly: e.g. for His there are two codons (CAC and CAT) used with a frequency of 59 and 41%, respectively, resulting in a Relative Adaptiveness of 1,0 (CAC) and 0,69 (CAT). In the case of the 2 GFP alleles, the Relative Adaptiveness was calculated and plotted on a linear scale for each individual codon position (Figure 1C). To calculate the CAI values, the individual Relative Adaptiveness values determined for each codon within e.g. huGFP0 (modified and non-modified codons) and huGFP60 were added up and divided by the number of amino acid.
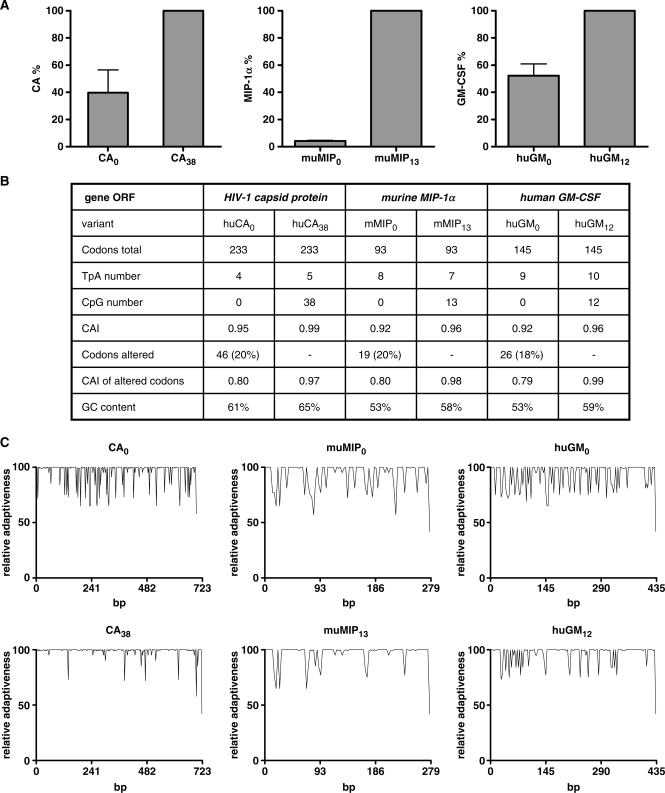
Figure 1.
Comparison of the codon-optimized huGFP reporter genes.(A) Nucleic acid sequence alignment of the unmodified (upper sequence) and CpG-depleted (lower sequence) humanized GFP genes. Sequences were aligned using the DNAman software and exhibit 88% homology. (B) Comparison of sequence-specific parameters. CpG: CpG dinucleotide; CAI: codon adaption index; GC content: percentage of bases C and G within the overall sequence; TpA: TpA dinucleotide. (C) Relative Adaptiveness distribution of the two gfp gene variants. The Relative Adaptiveness reflects the frequency of individual codons, whereas the most frequently used triplet encoding a given amino acid is set to 1.0 and less frequently used codons are scaled down accordingly. (D) In the pc5 vector, huGFP0 and huGFP60 sequences are flanked by the CMV or EF-1α promoter region respectively (CMV/EF-1α) and the BGH poly-adenylation signal (p(A)).
Cell culture, transient transfections and infections
Adherent human lung carcinoma H1299 cells and human embryonic kidney 293 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) heatinactivated fetal calf serum (FCS), 2 mM l-glutamine and 1% penicillin/streptomycin. Flp-In 293 and Flp-In CHO cells (Invitrogen) were cultivated in DMEM or HAM’s F12 medium (Invitrogen) supplemented the same way plus 1% zeocin (Invitrogen). All cell lines were maintained in a 5% CO2 atmosphere at 37°C.
For transient transfections, 2.5 × 105 293 cells or 1.5 × 105 CHO or H1299 cells were plated on six-well culture dishes and transfected after 24 h with 15 µg of plasmid DNA using the calcium phosphate precipitation technique. For infections, 1.5 × 105 293 cells were plated on 12-well dishes. After 24 h, cells were washed with serum-free DMEM and were subsequently infected with a Modified Vaccinia Ankara virus strain expressing the T7-polymerase (MVA-T7) at an MOI of 10. One-hour postinfection, cells were washed and transfected with pS/huGFP60 and pS/huGFP0 constructs, respectively, using FuGENE 6 (Roche). Cells were cultured for another 12 h, and protein expression was analysed by fluorescent-activated cell sorting (FACS) as described.
Generation of stable cell lines
Flp-In CHO and Flp-In 293 cell lines were cotransfected with 1.5 µg of either pc5/huGFP60 or pc5/huGFP0 and 13.5 µg of pOG44 plasmid expressing the Flp recombinase gene, as described in the manufacturer’s instructions (Invitrogen). Cells were incubated for 24 h to allow for expression of the hygromycin resistance gene [hygromycin B phosphotransferase gene (hph)], and cells expressing GFP were selected by increasing concentrations of hygromycin B (PAA) in the medium. Recombinant Flp-In CHO and Flp-In 293 cell lines were maintained under selective pressure in the presence of 1 and 0.2% hygromycin B, respectively. Based upon polyclonal cell cultures expressing either of the huGFP variants, 20 monoclonal cell lines were established by singularizing GFP positive cell clones under the microscope.
Western blot analysis and ELISA
Cells were harvested 48-h posttransfection, washed two times in phosphate buffered saline (PBS) and lysed in RIPA buffer [50 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 1% Nonidet P-40 (w/v), 0.5% sodium desoxycholat (w/v)] supplemented with protease inhibitors (Complete Mini, Roche). Insoluble components were removed by centrifugation (10 000g, 30 min), and the total amount of protein was quantified using the Bio-Rad Protein Assay following the manufacturer’s instructions. For Western blot analysis, 50 μg of total protein were separated by 12.5% SDS polyacrylamid gel electrophoresis (PAGE) and transferred to nitrocellulose. GFP expression was analysed using the GFP-specific polyclonal A. v. peptide antibody sc8334 (Santa Cruz Biotechnology) and a horseradish peroxidase-labelled anti-rabbit antibody (Dako, D0306). Actin expression was detected using a mouse monoclonal antibody (Sigma, A5441) and a horseradish-labelled anti-mouse antibody (Dako, P0260). Cell-associated expression levels of huCA were determined by ELISA using capsid-specific monoclonal antibodies and a capsid protein standard from Polymun (Vienna, Austria). Culture supernatants were collected 48 h posttransfection and cleared from cell debris (300g, 5 min). For quantification of cytokine secretion, 1 μg of total protein diluted in growth medium was used in ELISA development systems from R&D Systems (Minneapolis, USA) for analysis of murine MIP-1α and from BD Biosciences Pharmingen (BD OptEIA Set, San Diego, USA) for analysis of human GM–CSF according to the manufacturer’s protocol. Absorbance of capsid protein and cytokine ELISA was measured at 450 nm on a Micro plate reader 680 from Bio-Rad (Hercules, USA).
FACS analysis
Transiently transfected cells were harvested 48-h posttransfection and resuspended in PBS/1% FCS. Stable cell lines were harvested at different times posttransfection and treated analogously. Fluorescent-activated cell sorting (FACS) was performed with a FACS Calibur from Becton Dickinson or a FACS Canto (BD), respectively. The obtained FACS data were processed using WinMDI 2.8 software (Josef Trotter, La Jolla, USA) or FACSDiva software (BD).
Isolation of genomic DNA and DNA sequencing
Genomic DNA from 5 × 106 recombinant CHO Flp-In cells was isolated with the QIAmp DNA Mini Kit according to the manufacturer’s instructions (Qiagen). 100 ng of genomic DNA were used in a PCR reaction for amplification of the complete expression cassette of the reporter genes stably integrated into the host cells using oligonucleotides 5′-ACGCGTTGACATTGATTATTGAC-3′ and 5′-AATGCGATGCAATTTCCTCA-3′. The resulting products comprised 1632 bp and were sequenced subsequently.
Bisulphite modification and DNA sequencing
Genomic DNA from 5 × 106 recombinant CHO Flp-In cells was isolated with the QIAmp DNA Mini Kit according to the manufacturer’s instructions (Qiagen), and 1 μg of genomic DNA was subsequently subjected to sodium-bisulphite modification using the EpiTect Bisulfite Kit from Qiagen. Ten nanograms of the bisulphite-treated DNA were PCR-amplified in GoTaq® Green Master Mix (Promega) together with the respective oligonucleotides designed to anneal to CpG free regions allowing for equal amplification of methylated and unmethylated DNA. To cover the entire gfp open reading frame, two separate PCR reactions were carried out using oligonucleotides 5′-TTGTTATTATGGTGAGTAAGGG-3′ and 5′-TA ATTATACTCCAACTTATACCCCA-3′ to amplify the 5′ region of huGFP60 and oligonucleotides 5′-AGGAGYGTATTATTTTTTTTAAGGA-3′ and 5′-AATATCTACAAAATTCCACCACACT-3′ to generate the 3′ region. Likewise, the huGFP0 sequence was amplified using oligonucleotides 5′-AGTTTGTTATTATGGTGTTTAAGGG-3′ and 5′-TAAAATCAATACCCTTCAACTCAAT-3′ to generate the 5' moiety and oligonucleotides 5′-A GGAGAGGATTATTTTTTTTAAGGA-3′ and 5′-TAAATATCTACAAAATTCCACCAC A-3′ to obtain the 3′ region. To cover the entire CMV promoter region primers 5′-TTGTATGAAGAATTTGTTTAGGG-3′ and 5′-TAATACCAAAACAAACTCCCAT-3′ were used to amplify the 5′ region and oligonucleotides 5′-GGATTTTTTTATTTGGTAGTATATTTA-3′ and 5′-CTCTAATTAACCAAAAAACTCTACTTATAT-3′ were used to amplify the 3′ region. PCR products were gel-purified (QIAquick Gel Extraction Kit; Qiagen) and sequenced, and sequences were analysed with Sequence Scanner v 1.0 from ABI.
RNA isolation, cDNA synthesis and real-time PCR
Nuclear and cytoplasmic RNA species were prepared using the RNeasy-Kit following the manufacturer’s instructions (Qiagen). RNA samples were then treated with RNase-free DNase (Roche) and subsequently used in a PCR with transgene-specific primers to exclude DNA contamination. One microgram of DNA-free RNA was reverse transcribed using the first strand cDNA synthesis kit for real-time PCR (RT-PCR) (AMV) from Roche together with 3.2 µg p(dN)6 random primer in the presence of RNase inhibitor. To detect alternative splice products, 1 µl of resulting cDNA samples was used in a PCR reaction together with primers 5′-AAGCTTGCCACCATGGTG-3′ and 5′-GAGCTGTACAAGTGATAGGATCC-3′ amplifying the complete open reading frame of huGFP60 and huGFP0. RT-PCR was carried out in a LightCycler (Roche) with the LC Fast Start DNA Master SYBR Green kit according to the manufacturer’s instructions (Roche) using 1 µl of cDNA corresponding to 50 ng of total RNA. gfp-specific cDNA was amplified with oligonucleotides 5′-CCCTGAAGTTCATCTGCACC-3′ and 5′-GATCTTGAAGTTCACCTTGATG-3', whereas oligonucleotides 5′-GAGCGGGTTCGGCCCATTC-3′ and 5′-GTATTGGGAATCCCCGAACATCG-3′ served for amplification of the hph cDNA. Beta-actin cDNA was quantified with oligonucleotides 5′-GGTGGGCATGGGCCAGAAGG-3′ and 5′-CGCGTAGCCCTCGTAGATGG-3′. Product specificity was assessed based on melting curves following the manufacturer’s protocol. SYBR Green fluorescence was analysed and expressed as number of crossing points (cp) by LightCycler Software 3.5 (Roche).
In relative quantification analyses, the amount of gfp-specific transcripts was related to hph transcripts, which served as internal control in the same run. PCR efficiencies (E) of the respective templates were determined according to the manufacturer’s protocol (Roche) with E (huGFP60) = 1.94, E (huGFP0) = 1.94, E (hygr) = 1.89, and resulting error values ranged between 0.01 and 0.03. The relative transcription level of the gfp transgenes was calculated as described previously (41):
For absolute quantification, mRNA amounts were extrapolated from an external standard curve generated with 10-fold serial dilutions (102–1010 copies/µl) of the respective plasmid DNA template. Transcript copy numbers were then calculated from the external standards via an internal calibrator by LightCycler Software 3.5.
Analysis of mRNA half-life
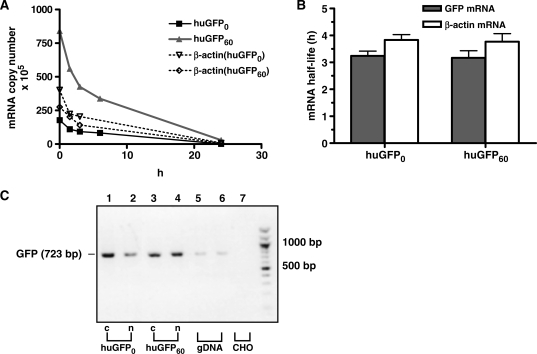
Decay of respective mRNA transcripts was measured as described previously (42). Briefly, Actinomycin D (BioCat GmbH) was added to cell culture supernatants of stable GFP expressing CHO cells at different time points (0, 1.5, 3, 6, 12, 24 h) prior to cell harvest. Total RNA was isolated, reverse transcribed and quantified for each given time point via RT-PCR using an external standard as described above. After determination of decay constant k, the respective mRNA half-life was calculated.
Nuclear run-on
Nuclear run-on was performed according to a previously described protocol (43). Briefly, nuclei of 3 × 107 stably transfected cells were prepared on ice, supplemented with biotin-16-UTP for 30 min at 29°C, and labelled transcripts were bound to streptavidin-coated magnetic beads. Total cDNA was then synthesized by random hexamer reverse transcription of captured molecules. gfp- and hph-specific transcripts were quantified via RT–PCR using external standards as described above.
Nucleosome reconstitution by salt gradient dialysis
Nucleosome reconstitution by salt gradient dialysis was performed as described by Längst et al. (44). Shortly, salt dialysis was performed in Sartorius collodion bags, which were rinsed with water and blocked with HI salt buffer (10 mM Tris/HCl [pH 7.6], 2 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.05% Nonidet P40), containing 200 ng/ml BSA. A typical assembly reaction (500 μl) contained 60 μg DNA, 40 μg BSA and 60 μg histones in HI salt buffer, adjusted to a final concentration of 2.5 M NaCl. The salt was continuously reduced to 50 mM NaCl during 16–24 h. Chromatin assembly extracts were derived from 3- to 6-h-old Drosophila embryos (45) and core histones were purified by affinity chromatography on hydroxylapatite columns (46). Primer used for the generation of according PCR fragments were: anterior: huGFP60_fwd 5′-CGACTCACTATAGGGAGACCCA-3′; huGFP60_rev 5′-TGCTGCTTCATGTGGTCGGG-3′, huGFP0_fwd 5′-CGACTCACTATAGGGAGACCCA-3′; huGFP0_rev 5′-TGCTGCTTCATGTGGTCTGG-3′; central: huGFP60_fwd 5′-GTGCAGTGCTTCAGCCGC-3′; huGFP60_rev 5′-TGCCGTTCTTCTGCTTGTCG-3′; huGFP0_fwd 5′-GTGCAGTGCTTCAGCAGATACC-3′; huGFP0_rev 5′-TGCCATTCTTCTGCTTGTCTG-3′; posterior: huGFP60_fwd 5′-ACGTCTATATCATGGCCGACA-3′; huGFP60_rev 5′-TCCACCACACTGGACTAGTGG-3′; huGFP0_fwd 5′-ATGTGTACATCATGGCAGACAAG-3′; huGFP0_rev 5′-TCCACCACACTGGACTAGTGG-3′. After completion of the salt dialysis, DNA–protein complexes were resolved by native polyacrylamid gel electrophoresis using a polyacrylamid content of 5%.
Chromatin immunoprecipitation
Exponentially growing CHO cells were cross-linked using 1% formaldehyde for 5 min.Cross-linking reactions were quenched with 0.125 M glycine. Cells were washed three times in 1× PBS. Hypotonic lysis of cells was performed by 15 min incubation on ice in 1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1, EDTA-free complete protease inhibitor cocktail (Roche) containing buffer. Samples were sonicated using a Bioruptor sonicator (Diagenode) to approximately 500 bp DNA fragments. Cell debris was spun at 16100g for 5 min at room temperature and the supernatant fraction was transferred into a new tube. This chromatin fraction was stored at 4°C and used as input for immunoprecipitations. Chromatin samples were diluted 1:10 with 150 mM NaCl, 20 mM Tris–HCl pH 8.1, 2 mM EDTA, 1% Triton X-100, EDTA-free complete protease inhibitor cocktail (Roche) containing buffer. 5 µg of a polyclonal histone H3 specific antibody (Abcam, ab1791) was added to 500 µg chromatin and incubated with gentle rotation overnight at 4°C. Protein-A-sepharose beads were preblocked in the above buffer supplemented with 500 µg/ml sonicated salmon sperm DNA and 100 µg/ml BSA. About 10 µl blocked beads were added to antibody-chromatin complexes and incubated for a further 90 min at 4°C with gentle rotation. Beads were washed subsequently with 1 ml each of the immunoprecipitation buffer (5x), in 0.25 M LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1 containing buffer (1×), and in 1×TE (1×). Elution was performed using 1% SDS, 0.1 M NaHCO3 at 37°C for 2 × 15 min in a shaker. Eluted chromatin fractions were subjected to RNaseA treatment for 1 h at 37°C followed by proteinase K digestion at 37°C, 8 h and reversal of cross-linking at 65°C for a further 8 h. DNA was precipitated and dissolved in 50 µl of H2O. DNA samples were then diluted 1:10 with water and used as a template in quantitative PCR reactions. Chromatin immunoprecipitations (ChIPs) were performed from different chromatin preparations and chromatin samples incubated with normal rabbit IgG (SantaCurz, sc-2027) served as negative control.
RESULTS
The GFP reporter genes
Although CpG dinucleotides have been shown to affect efficiency of transgene expression in vivo, the consequences of CpG-mediated regulation on high-level expression of foreign genes in established cell lines still remain unknown. In order to analyse the influence of intragenic CpG content on transgene expression, a humanized version of the GFP (huGFP) was used as reporter gene (39). Apart from quantitative determination of reporter expression, GFP was selected to allow evaluation of transfection efficiency (47). Based on the huGFP sequence comprising 60 CpG dinucleotides (huGFP60), a novel synthetic huGFP0 construct lacking CpG dinucleotides was generated (Figure 1A). Both reporter genes do not contain any introns and are referred to as huGFP60 or huGFP0 respectively hereafter. By using alternative codons to modify the open reading frame, the GFP amino acid sequence was maintained. Despite the fact that 75 codons within huGFP0 (31%) were substituted mostly by the second frequently used codon, the overall frequency of all codons in the two GFP alleles as expressed by the Codon Adaptation Index CAI (40) was not significantly impacted compared to the codon optimized huGFP60 sequence. The Relative Adaptiveness (codon frequency) was calculated for each individual and consecutive codon and plotted on a linear scale (Figure 1C; see also Supplementary Data, Figure 1). Even though huGFP60 and huGFP0 sequences strongly differ in their intragenic CpG amounts, they exhibit similar overall GC contents and comprise equal TpA/UpA dinucleotide numbers (Figure 1B), the latter representing putative targets for endonucleases mediating RNA cleavage (38). The creation of additional negative _cis_-acting elements like cryptic splice sites, internal polyadenylation sites or TATA-boxes was avoided if possible. Additionally, the gfp sequences were analysed for Sp1-binding sites and neither generation nor destruction of Sp1-binding sites due to sequence modifications could be detected.
Both reporter variants were inserted into various eukaryotic expression vectors for transient or stable expression under transcriptional control of the CMV major immediate early promoter or the human elongation factor 1 α promoter, respectively, and the bovine growth hormone polyadenylation signal (BGH) (Figure 1D).
The CpG quantity in the open reading frame influences the reporter activity in transiently transfected mammalian cells
To investigate the influence of intragenic CpG content on protein expression in cell lines, H1299 cells were transiently transfected with the respective huGFP60 and huGFP0 constructs, and GFP expression was determined by FACS analysis. Interestingly, CpG depletion in huGFP0 consistently resulted in a 1.6 or 2-fold decrease in reporter activity as compared to huGFP60, likewise in the context of the commercial plasmids pc3.1100 or pc5, both of which support transgene expression via the CMV immediate early promoter (Figure 2A). Since transfection efficiencies were comparable for both GFP variants irrespective of the vector backbone used (Figure 2B), the huGFP0-derived reduction of auto-fluorescence presumably results directly from the specific depletion of intragenic CpGs or the general alterations in overall plasmid CpG content. To address this question, we analysed expression of the huGFP gene variants within vectors pc3.1100 and pc3.147 comprising CpG contents in the backbone of 100 and 47%, respectively. As displayed in Figure 2C, reporter activities of both huGFP variants were not altered significantly when expressed from different vector backbones. Beyond the CpG reduction within the vector backbone, the conventional CMV promoter within pc3.147 was replaced by a previously described CpG free CMV promoter version (24) resulting in an overall CpG content within the complete plasmid of 32%. This promoter-specific CpG reduction slightly decreased the absolute expression levels of both reporter genes (data not shown) while the relative difference in expression was maintained. Thus, the decrease in huGFP0-derived fluorescence apparently is directly associated with intragenic CpG depletion. To examine whether the observed effect was universally valid rather than restricted to the gfp gene, three more RNA- and codon-optimized reporter genes and their CpG-depleted counterparts were tested. The viral gene encoding the capsid protein of human immunodeficiency virus type-1 (huCA; 699 bp) and two endogenous cytokine genes encoding murine macrophage inflammatory protein 1α (muMIP; 279 bp) and human granulocyte–macrophage colony-stimulating factor (huGM-CSF; 435 bp) were analysed. Reinforcing the results with GFP and irrespective of their origin, ELISA analyses revealed that all reporter genes lacking intragenic CpG dinucleotides showed a significant decrease of expression ranging between 95 (muMIP0) and 52% (huGM-CSF0) as compared to the CpG containing equivalents (Figure 3A). Interestingly, the number of CpGs deleted from the open reading frame did not directly correlate with the loss in expression, since huCA0 lacking 38 CpG dinucleotides still reached 40% of huCA38 expression level, whereas synthesis of muMIP0 lacking only 13 CpGs was heavily reduced to 5%. However, neither the overall GC content nor the codon quality as indicated by CAI (Figure 3B and C; Supplementary Figure S2) were significantly altered in these CpG-free gene variants suggesting that apart from codon-optimization the amount of intragenic CpG dinucleotides might be of special relevance for efficient protein synthesis in mammalian cells.
Figure 2.
Influence of intragenic and plasmid backbone CpG content on transient GFP expression in eukaryotic cells. H1299 cells were transiently transfected (A) with 15 μg of the eukaryotic expression vectors pc5 or pc3.1100 carrying the huGFP0 or huGFP60 reporter genes or (C) with pc3.1100 and pc3.147-based reporter constructs. Cells were harvested 48-h posttransfection and subjected to FACS analysis counting 10 000 events per gate. An exemplary FACS analysis of cells transfected with pcDNA3.1 (mock) or huGFP0 and huGFP60 in pc3.1100 is shown for evaluation of transfection efficiencies (B). The average mfi is indicated as percentage of huGFP60 derived fluorescence (A) or as percentage of pc3.1100-mediated GFP expression (C), respectively. The mean of three independent transfection experiments is given, and standard deviations are indicated.
Figure 3.
Influence of intragenic CpG depletion on transient expression of viral and cytokine reporter genes. (A) H1299 cells were transiently transfected with 15 μg of the indicated constructs in pcDNA3.1. Forty-eight-hours posttransfection, normalized cell lysates (CA) and supernatants (muMIP and huGM) were screened for protein content by ELISA as described in materials and methods. Protein amounts obtained from transfections with the CpG-rich genes were set to 100%, and values for the CpG lacking variants were related accordingly. The mean of four independent transfection experiments is shown, and standard deviations are indicated. (B) Comparison of sequence-specific parameters. CpG: CpG dinucleotide; CAI: codon adaption index; GC content: percentage of bases C and G within the overall sequence; TpA: TpA dinucleotide. (C) Relative Adaptiveness distribution of the CA, muMIP and huGM gene variants. The Relative Adaptiveness reflects the frequency of individual codons, whereas the most frequently used triplet encoding a given amino acid is set to 1.0 and less frequently used codons are scaled down accordingly.
Intragenic CpG depletion reduces expression in stably transfected mammalian cells
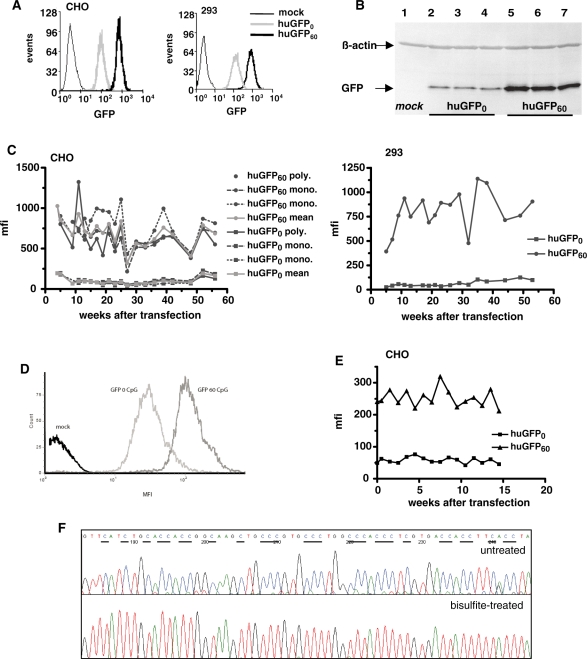
Due to the apparent advantages concerning information content of GFP read out, the gfp reporter genes were selected to further evaluate potential effects of CpG depletion on reporter expression. For this purpose we used CHO and 293 cell lines, both being widely employed for recombinant protein production (48). Corresponding cell lines stably expressing the reporter gene variants within otherwise identical genetic backgrounds were established using the Flp-In recombination system. The integrity of the reporter gene expression cassettes was verified by PCR of isolated genomic DNA and sequencing. Flp-In CHO and 293 cell clones expressing single copies of either huGFP60 or huGFP0 genes were selected in the presence of hygromycin and analysed by fluorescence microscopy (data not shown) and FACS analysis (Figure 4A). In accordance with data obtained from transient expression, cells stably expressing huGFP0 showed a significant loss in GFP-mediated fluorescence with mean fluorescence intensity (mfi) values determined to be 7-fold (CHO) and 10-fold (293) lower than measured for the huGFP60 expressing counterparts. This effect was observed in all established monoclonal cell lines (15 monoclonal cell lines for each gfp gene variant) irrespective of the cell type analysed (data not shown), indicating that cell-specific effects did not play a role within this context. To further exclude that compromised expression of huGFP0 was due to incomplete production of the full-length gene product or rapid protein degradation, various cell samples derived from stably transfected polyclonal and monoclonal CHO cell lines were subjected to western blot analysis with a GFP-directed antibody. As expected, detectable GFP amounts differed significantly in cells expressing huGFP60 or huGFP0 while truncated proteins could not be detected in corresponding cell lysates (Figure 4B), suggesting correct protein synthesis of the CpG-depleted GFP variant.
Figure 4.
Influence of intragenic CpG content on long-term stable GFP expression in mammalian cells. (A) CHO (left) and 293 cells (right) stably expressing huGFP0 or huGFP60 were subjected to FACS analysis counting 10 000 events per gate. The number of scored events (_y_-axis) versus the respective fluorescence intensity of GFP (_x_-axis) is shown for huGFP0 and huGFP60 as well as for untransfected cells (mock) to monitor background fluorescence. (B) Stably transfected CHO cells were harvested and 50 µg of total protein were subjected to western blot analysis using a GFP-specific antibody. Two monoclonal and one polyclonal CHO cell line expressing either huGFP0 (lanes 2–4) or huGFP60 (lanes 5–7) were analysed. The monoclonal cell lines for huGFP0 are shown in lanes 2 and 3, the monoclonal cell lines for huGFP60 are depicted in lane 5 and 6. The positions of the 27-kDa GFP protein and the 42-kDa ß-Actin protein are indicated by arrows. Untransfected cells (mock, lane 1) served as negative control. (C) Polyclonal (poly) and monoclonal (mono) CHO cell lines (left) and polyclonal 293 cells (right) stably expressing either huGFP0 or huGFP60 were quantified for expression of the GFP reporter genes by FACS analysis over a period of 56 weeks (_x_-axis) indicated as mfi values (_y_-axis). Mocktransfected cells were used for background subtraction. (D) CHO cells stably expressing huGFP0 or huGFP60 under control of the EF-1α promoter were subjected to FACS analysis counting 10 000 events per gate. The number of scored events (_y_-axis) versus the respective fluorescence intensity of GFP (_x_-axis) is shown for huGFP0 and huGFP60 as well as for untransfected cells (mock) to monitor background fluorescence. (E) Polyclonal CHO cells stably expressing either huGFP0 or huGFP60 under control of the EF-1α promoter were quantified for expression of the GFP reporter genes by FACS analysis over a period of 15 weeks (_x_-axis) indicated as mfi values (_y_-axis). (F) Genomic DNA from stably transfected CHO cells was isolated, bisulphite-treated and amplified via PCR with huGFP specific oligonucleotides. PCR products were sequenced, and chromatograms were analysed as described in materials and methods. A section of huGFP60 sequence prior to (upper panel) and post bisulphite treatment (lower panel) is depicted, and cytosine residues potentially available for methylation are underlined. Unmethylated cytosines are represented by thymines the chromatogram, whereas methylated cytosines are not converted. The first nucleotide (G) of the extracted sequence corresponds to position 138 of the gfp open reading frame.
To prove the promoter independency of this CpG-mediated expression effect, we also tested EF-1α driven GFP expression. The choice for the human elongation factor 1α promoter was based on its mammalian origin and its strong and at the same time gene silencing evading nature. Expression analysis confirmed the CMV derived results and showed a clear correlation between intragenic CpG content and protein expression levels (Figure 4D).
As silencing of gene expression is rather a gradual process manifested in the course of time (49), we monitored whether depletion of CpGs from the GFP coding region is beneficial for long-term protein expression in mammalian cells. Therefore, CHO cells stably expressing the huGFP variants under control of the CMV and the EF-1α promoter, respectively and under constant selective pressure were weekly subjected to FACS analysis. During this observation period, all cell lines showed relatively constant huGFP60 expression levels with average mfi values of 750 for the CMV-driven GFP expression in CHO and 293 cells and 250 for the EF-1α-driven protein expression in CHO cells, respectively (Figure 4C and E). Likewise, expression of CpG-depleted huGFP0 reporter genes remained constant, however, at lower levels which were reduced by 6- to 9-fold when compared with huGFP60. Although a steady expression under selective pressure is not surprising, these data assured unchanging experimental conditions for further studies, as neither huGFP60 nor huGFP0 reporter expression was silenced over the observed period.
Expression of the CpG-rich gfp gene is not impaired by intragenic CpG methylation
It is now firmly established, that deficient gene expression often correlates with CpG hypermethylation within and around promoter regions. However, long-term effects of intragenic CpG methylation have been explored less extensively. Since the established cell lines were under constant selective pressure and huGFP60 showed a constant expression profile over >56 weeks, silencing via CpG methylation is unlikely. Nonetheless, we found it intriguing to what extent CpG dinucleotides within huGFP60 might trigger cytosine hypermethylation in the genomic background. Thus, huGFP60 DNA from stably transfected CHO cells cultivated for >50 weeks was subjected to bisulphite genomic sequencing. Sodium bisulphite selectively deaminates unmethylated cytosines to uraciles, whereas methylated cytosines are not modified (50). In the subsequent PCR reaction, uraciles are replaced by thymines resulting in a final C to T conversion. To validate the method, the pc3.1100/huGFP60 plasmid was subjected to quantitative in vitro methylation prior to bisulphite sequencing and resulting chromatograms indicated that _in vitro_-methylated cytosines were not affected by bisulphite treatment (data not shown). In contrast, analysis of the genome-derived huGFP60 sequence revealed a quantitative conversion of all cytosines to thymines following the bisulphite reaction. This clearly shows that none of the CpG dinucleotides within the huGFP60 open reading frame was originally methylated (Figure 4F). As methylation of CpG dinucleotides within promoter regions is also known to diminished transgene expression we analysed the methylation status of the integrated CMV promoter of the stable cell lines, promoting transcription of either huGFP0 or huGFP60. The results clearly showed that under the tested assay conditions neither the promoter of huGFP0 nor of huGFP60 exhibited CpG methylation, thus excluding a promoter-methylation specific influence on the transcription of huGFP0 and huGFP60 (Supplementary Data; Figure 3).
As expected, these results imply that under selective conditions the genome-integrated CpG-rich reporter gene was not susceptible for intragenic hypermethylation within the observed period.
Intragenic CpG depletion has no effect on translational efficiency
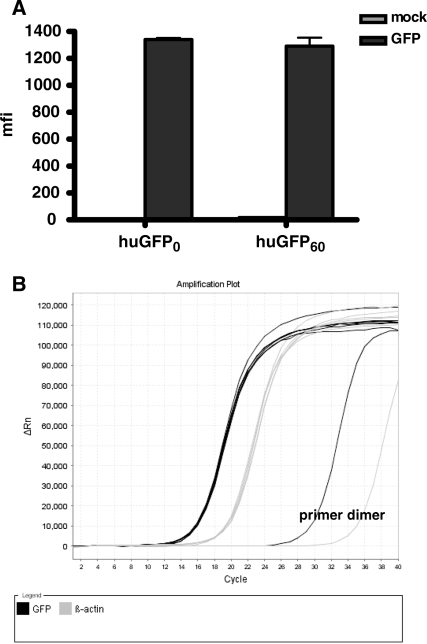
Since CpG depletion from the huGFP reading frame was associated with a clear loss of reporter expression, we next examined, to what extent this phenomenon might result from decreased efficiency of protein translation due to the changes in codon usage. In order to exclude potential effects of the modified reporter gene sequence on posttranscriptional events like RNA export, we infected 293 T cells with a modified vaccinia ankara strain known to replicate exclusively in the cytoplasm and expressing the T7 polymerase (MVA-T7). Upon infection this system allows the cytoplasmic transcription of a transfected reporter gene controlled by the T7 promoter. The translation efficiency of the pS/huGFP60 and pS/huGFP0 constructs following transfection of the MVA-T7-infected cells was quantified by FACS analysis (Figure 5A) while the amount of _gfp_-specific transcripts was assessed by quantitative PCR (Figure 5B). Both reporter genes produced almost equal amounts of gfp mRNA copy numbers and also comparable amounts of GFP in this artificial system, widely excluding a major impact of CpG content on translation although possible saturated translation conditions in the artificial T7/vaccinia system cannot be completely ruled out.
Figure 5.
Influence of intragenic CpG depletion on translational efficiency. 293 cells were infected with MVA-T7 at an MOI of 10 and transfected with pS/huGFP0 and pS/huGFP60 1-h postinfection. After 48 h, GFP-specific transcripts were accurately measured via qPCR (B) and GFP expression in transfected cells was quantified by FACS analysis (A). The mfi of three independent transfection experiments is shown.
Intragenic CpG content correlates with levels of steady-state RNA
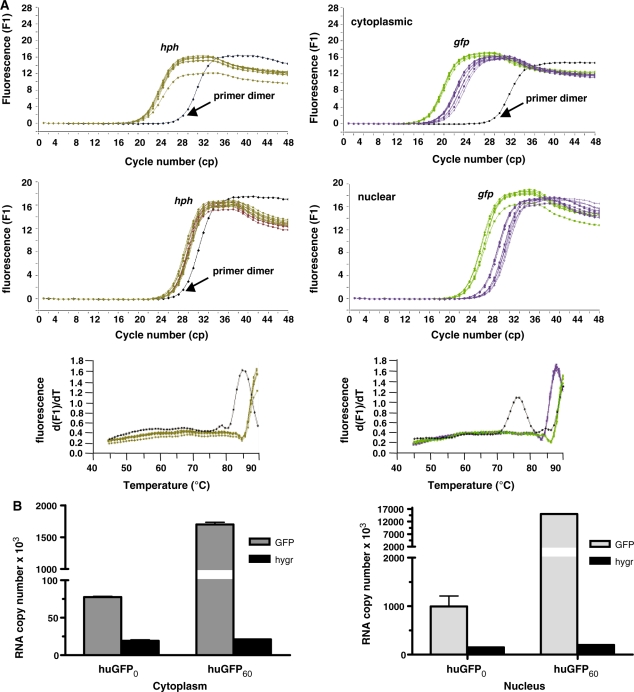
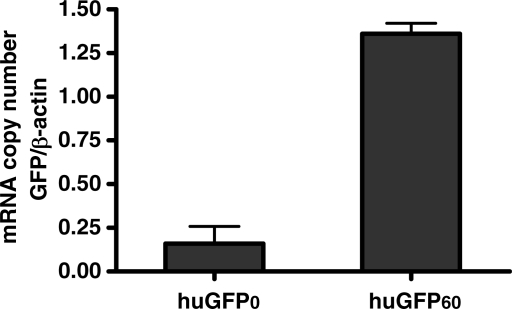
To further elucidate the mechanism underlying the significant decrease in huGFP0 expression, we asked, whether CpG depletion had a major influence on related mRNA amounts. To quantify the steady-state level of huGFP transcripts, cytoplasmic and nuclear RNA fractions from stably transfected CHO and 293 cells were subjected to reverse transcription and quantified via RT-PCR. Whereas the overall number of hph transcripts within cytoplasmic and nuclear fractions of CHO cells remained constant (Figure 6A), relative quantification analysis, the most accurate method to quantify cDNA (41), yielded a 7-fold cytoplasmic and 13-fold nuclear reduction of huGFP0 transcripts compared to huGFP60 mRNA levels. Even more striking effects were obtained from stably transfected 293 cells, where the total amount of huGFP60 transcripts exceeded those of huGFP0 by 29-fold (data not shown). The specificity of obtained PCR products was evaluated by sequence verification and melting curve analysis. To confirm the data gained by relative quantification, absolute amounts of gfp and hph transcripts in corresponding fractions of CHO cells were determined using external standard curves (Figure 6B). According to this method, the CpG depleted mRNAs were decreased by 5-fold in the nucleus and 8-fold in the cytoplasm compared to the CpG containing counterparts. In sum, these data provide evidence that the observed reduction of huGFP0 reporter expression strictly correlates with decreased steady-state mRNA levels. Since ratios of cytoplasmic versus nuclear transcript numbers calculated from absolute quantification data were similar for both reporter species (12- and 9-fold for huGFP60 and huGFP0, respectively), we can exclude that effects associated with mRNA nuclear export contribute to differential GFP expression.
Figure 6.
Influence of CpG content on steady-state RNA levels. Cytoplasmic and nuclear RNA fractions prepared from 3 × 107 stably transfected CHO cells were subjected to reverse transcription and quantified via LightCycler analyses as described in materials and methods. (A): Relative quantification. The amount of _gfp_-specific transcripts (right) was related to hph (hygromycin resistance gene) transcripts (left), which served as internal controls in the same run. The _x_-axis denotes the cycle number (cp) necessary to significantly detect the SYBR Green fluorescence signal (_y_-axis) of the respective cDNA sample referring to cytoplasmic (upper panel) or nuclear RNA (lower panel). Melting point analysis for hph- (left) and _gfp_-specific RT-PCR products (right) compared to primer dimer formation is indicated below. Colour code: brown, hph; purple, huGFP0; green, huGFP60; black, primer dimer. (B): Absolute quantification. The amount of RNA transcripts was extrapolated from an external standard curve as described in ‘Materials and Methods’ section, and the number of cytoplasmic (left) and nuclear (right) RNA transcripts derived from three independent experiments is given.
CpG variations do not affect stability or splicing of reporter transcripts
Next we examined, whether CpG reduction in the huGFP0 reporter gene influenced the stability of corresponding RNA messages. Thus, we determined the half-lives of huGFP60 and huGFP0 transcripts in stably transfected CHO cells. For this purpose, de novo RNA synthesis was blocked by Actinomycin D for different time periods, and total RNA was subjected to reverse transcription and quantified (Figure 7A). As further depicted in Figure 7B, β-actin mRNA isolated from the huGFP expressing cells exhibited consistent half-life values of 3.5 h, whereas half-lives of ∼3 h were determined for both gfp reporter transcript species, indicating that codon substitutions within huGFP0 did not impact transcript stability. Despite diligent design of the huGFP0 variant, CpG depletions by means of introducing silent mutations generated four additional cryptic splice sites [huGFP60 (8) versus huGFP0 (12)], which might in theory cause covert alternative splicing and thereby reduce the amount of detectable full-length transcripts. To deal with this issue, respective cytoplasmic and nuclear mRNA fractions were subjected to reverse transcription, and resulting cDNA samples were used in qualitative PCR with oligonucleotides amplifying the whole GFP coding region. Resulting PCR products were analysed and sequence-verified. As indicated in Figure 7C, only full-length messenger transcripts were detected for either GFP variant, which could be confirmed in northern blot analyses using a BGH-specific probe (data not shown). The corresponding results strongly argue against alternative splicing events as a cause for decreased huGFP0 transcript levels. In sum, these data imply that reduced protein and mRNA levels observed for a CpG depleted GFP version in vitro cannot be ascribed to reduced mRNA stability or erroneous splicing.
Figure 7.
Influence of intragenic CpG content on stability and alternative splicing of the GFP transcripts. (A) Stably GFP expressing CHO cells were treated with 2.4 μM Actinomycin D at different time points prior to cell harvest. Total RNA was isolated, reverse transcribed, and the resulting cDNA samples were quantified via LightCycler using SYBR Green technology. (B) RNA half-lives (_y_-axis) of the respective transcripts (_x_-axis) were determined as described in ‘Material and Methods’ section. Shown data represent the mean of two independent experiments performed in triplicates using ß-actin RNA as a control. (C) To detect alternative splice products, RNA samples from cytoplasmic and nuclear fractions of stably transfected CHO cells were subjected to reverse transcription and qualitative PCR analysis. Obtained PCR products referring to cytoplasmic (c) or nuclear (n) huGFP0 (lanes 1 and 2) or huGFP60 (lanes 3 and 4) transcripts were analysed by 1% agarose gel electrophoresis. Genomic DNA (gDNA) and RNA from untransfected CHO cells (CHO) (lane 7) were used as positive and negative PCR controls, respectively. Nucleotide positions are indicated on the right.
A loss of de novo transcriptional activity is responsible for moderate expression of the CpG depleted GFP variant
Apart from posttranscriptional processes modulating transgene expression, transcriptional dynamics crucially contribute to transgene-specific RNA synthesis. To investigate, whether differential transcriptional activity may account for decreased huGFP0 mRNA amounts, a nuclear run-on assay was performed using nuclei from stably transfected CHO cells. Captured de novo synthesized RNA molecules were subjected to reverse transcription and resulting cDNA samples were quantified by qPCR analysis and referred to de novo synthesized ß-actin transcripts (Figure 8). Interestingly, the CpG depleted huGFP0 construct yielded a 7-fold reduced amount of de novo synthesized GFP mRNA as compared to huGFP60. This thoroughly reflects the observed discrepancies in RNA and protein expression levels. Taken together, our data propose a clear correlation of intragenic CpG content and de novo transcriptional activity, implying a transcription-based regulation of transgene expression via CpG dinucleotides.
Figure 8.
Influence of CpG depletion on de novo synthesis of gfp-specific transcripts. The nuclear run-on assay was performed with stably transfected CHO cells by supplying nuclei with biotin-16-UTP. Labelled transcripts were bound to streptavidin-coated magnetic beads, and total cDNA was synthesized by means of random hexamer-primed reverse transcription of captured molecules. Absolute cDNA copy numbers obtained from newly synthesized mRNA transcripts were quantified via LightCycler and normalized to ß-actin transcripts. The mean of two independent experiments performed in duplicates is shown.
Intragenic CpG dinucleotides affect nucleosome positioning
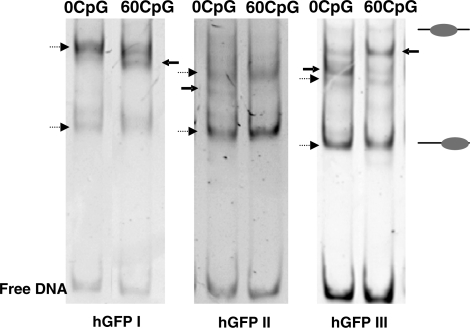
Having experienced the relation between CpG dinucleotides and RNA de novo synthesis, we assumed a CpG mediated effect on chromatin structure, which affects the activity of gene expression. To initially test the impact of CpG dinucleotides and sequence variation on nucleosome positioning, we performed in vitro nucleosome reconstitution assays with the variant huGFP DNA sequences. As the open reading frame of the huGFP gene variants comprises 720 bp and hence is too large to reveal the positions of individual nucleosomes by native electrophretic mobility shift assays (EMSAs), the huGFP coding sequence was partitioned into three overlapping DNA fragments of 280–300 bp and prepared by PCR amplification. Mononucleosomes were reconstituted by the salt dialysis method and comparative analysis of the nucleoprotein complexes revealed distinct band patterns for each of the analysed DNA fragments of the two gene variants. This suggests that changing the level of intragenic CpG content directly affects sequence dependent nucleosome positioning characteristics within the huGFP constructs in vitro (Figure 9). Whereas the proximal DNA element of the huGFP0 construct shows a decreased number of different nucleosome positions around the centre of the DNA fragment as compared to the huGFP60 construct, the central and the terminal DNA elements exhibit a more distributed pattern with additional nucleosome positioning sites. Changes in the DNA sequences do directly affect the histone-binding properties of the DNA which were shown to contribute to nucleosome positioning in vivo (51). The observed changes in in vitro nucleosome positioning could affect chromatin structure and gene expression in vivo.
Figure 9.
Influence of intragenic CpG content on nucleosome positioning in vitro. Three fragments of similar length, huGFPI (300 bp), huGFPII (280 bp) and huGFPIII (299 bp), were amplified from pcDNA5/FRT, containing the respective huGFP variants. The huGFP fragments were reconstituted with defined histone concentrations in the presence of competitor DNA (pUC 19), followed by salt dialysis, PAGE and detection by UV. The pattern of nucleoprotein bands fractionated by PAGE is characteristic for each of the fragments. Bands which represent nucleosome positions that are not specified for a single gene variant are indicated as dashed arrows. Bands characteristic for only one specific gene variant are highlighted by arrows.
CpG dinucleotides do not alter nucleosome density
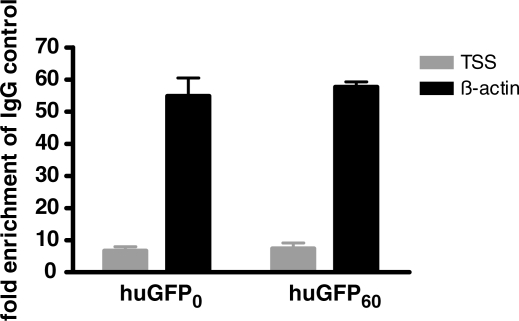
Considering that nucleosome positions in vitro often correlate with the chromatin structure in vivo (52), we examined a possible influence of intragenic CpG dinucleotides on the chromatin structure in vivo. Chromatin structure plays a major role in the regulation of gene expression. Transcriptionally active genes are generally located in an open, euchromatic chromatin configuration and vice versa (53). We monitored nucleosome density within the coding region of the stable cell lines expressing huGFP0 and huGFP60 by ChIP of the histone H3. Nucleosome occupancy of the GFP constructs integrated at the same genomic locus was quantified and compared to the the ß-actin gene_._ The global histone H3 density at the transcription start site (TSS) and at the control region within the ß-actin gene was assessed via quantitative PCR of precipitated genomic DNA, and expressed as enrichment of precipitated DNA molecules compared to the negative control. The evaluation of the results showed that taking the internal ß-actin control into account, the overall histone H3 occupancy among the two gene variants huGFP0 and huGFP60 is similar (Figure 10). For the control gene ß-actin a 55-fold enrichment of precipitated DNA was achieved, contrasting the 5-fold enrichment of DNA molecules covering the TSS. Consequently reduced nucleosome levels (95%) were detected for the TSS of the reportergenes as compared to ß-actin. Both reporter gene variants show a low histone H3 density as compared to the ß-actin gene but their divergent content of intragenic CpG dinucleotides has no influence on the global histone H3 occupancy. In addition it is surprising to see that a 5-fold activity in gene transcription can be achieved without altering the nucleosome density on the active Pol II gene. It suggests that active chromatin assembly mechanisms exist that allow efficient nucleosome deposition upon the passage of the RNA Polymerase.
Figure 10.
Influence of intragenic CpG dinucleotides on histone density in vivo. The ChIP experiment was performed by cross-linking DNA of stably transfected CHO cells, incubating the DNA with a pan H3 specific antibody and collecting DNA with protein A sepharose. Precipitated DNA was quantified via qPCR. Data obtained for DNA amounts at the TSS of the two gfp gene variants were normalized to the corresponding ß-actin (actin) amounts. The mean of two independent experiments is shown.
DISCUSSION
Various strategies have been exploited to improve and optimize efficacy of plasmid DNA vectors for recombinant protein production in vitro. Amongst others, fusion of intronic sequences 5′ to the coding region, usage of strong hybrid or cell-specific promoters or insertion of elements promoting the formation of transcriptionally active chromatin-like structures may serve as potent tools to increase protein synthesis in established cell lines (6,23,54).
Beyond, adaptation of the codon usage to highly expressing genes of the respective organism has been shown to significantly augment transgene expression (8). However, conflicting results have been reported for the endogenous content of CpG dinucleotides potentially modulating transgene expression in vivo as well as in cell culture. Whereas usage of CpG depleted expression cassettes has clearly shown to boost expression levels in transgenic mice and somatic cells, arguing that corresponding sequences are not prone to methylation-dependent gene silencing (24,28,31), the presence of CpG-rich regions might otherwise be beneficial for posttranscriptional processes based on the significant increase in RNA stability (9,38,39). Interestingly, expression of transgenes in stable cell lines has been shown to be critically influenced by differentiation status of the used cell type, integration locus and orientation, the usage of weak or strong promoter/enhancer units or simply maintenance of selective pressure (34,35,55,56) which altogether might result in controversial interpretation of CpG-mediated effects.
To explore whether the intragenic CpG content directly influences transgene expression in established mammalian cells, we synthesized a codon-optimized GFP reporter gene completely devoid of CpG dinucleotides and compared expression efficiency to its CpG-rich equivalent. Our data demonstrated a clear and direct correlation of intragenic CpG content with reporter activity in vitro, resulting in significantly reduced expression and fluorescence of the CpG lacking GFP variant. The associated phenotype was observed irrespective of the used cell type and promoter, and could not be ascribed to aberrant protein production. Furthermore, this CpG-specific effect was successfully transferred to unrelated genes of viral or mammalian origin which points to a general mechanism driven by CpG dinucleotides.
Transcriptional silencing via extensive CpG hypermethylation associated with chromatin condensation is a well-documented phenomenon in therapeutic DNA applications but has also been reported to interfere with cell culture-based production of recombinant proteins over time (35). Different strategies such as inserting CpG island fragments into expression vectors (57) or supplementing culture medium with demethylating agents (58) have been applied to avoid methylation-dependent silencing thereby improving expression efficiencies of foreign transgenes in CHO cells. To further reveal any chromatin structure-dependent consequences of DNA methylation, we compared transient GFP production with expression in stably transfected cell lines, where transgenes are integrated at defined sites and may be potentially influenced by adjacent genomic CpG hypermethylation patterns. As compared to transient experiments, differential expression of the gfp genes was even more pronounced upon stable transfection, where reporter activity of the CpG-rich GFP variant exceeded the CpG lacking counterpart by 6–9-fold and 10–20-fold in the analysed cell lines, respectively.
Interestingly, decreased reporter activity of similar CpG free gfp genes expressed in cultured cells or in transduced mice has also been observed by others suggesting that deficiencies in reporter activity might originate from either posttranscriptional events or less efficient translation of the CpG-depleted transcript due to modified codon usage (34,59). In order to avoid effects of codon quality on protein production, we have used gfp genes comprising similar CAI values as indicators of codon quality (40) and comparable codon distributions. Indeed, equal _gfp_-specific transcripts and GFP amounts were obtained for both reporter genes in a MVAT7-based cytoplasmic transcription/translation system, widely excluding a contribution of differential translational activities to the loss of reporter expression seen in huGFP0 stable cell lines.
In contrast we could provide clear evidence that diminished levels of the CpG-depleted GFP strictly correlated with a significant decrease in steady-state RNA copy numbers. Since downmodulation of reporter gene expression as reflected by decreased RNA amounts might yet become manifest at the posttranscriptional level, we analysed potential effects of CpG depletion on splicing, translocation or stability of the respective RNA messages. Our results, however, clearly demonstrated that reduced RNA levels could not be ascribed to erroneous splicing events, nuclear export restrictions or diminished RNA half-life, indicating that corresponding mechanisms were not significantly influenced by alterations of the intragenic CpG content. In contrast to this finding, compromised RNA stability was formerly shown to be responsible for the low levels of steady-state RNA detected upon CpG depletion (38). Noteworthy the study at hand was performed with GFP variants comprising equal TpA numbers, while the changed stability was detected upon maximization of TpA numbers in the CpG-free gene variant. Since UpA sites emerging in the corresponding transcripts serve as preferential targets for cellular endonucleases (37,60), an excess of these motifs is likely associated with rapid RNA degradation, thus explaining the reported effects.
Since we could not provide any evidence for altered posttranscriptional regulation of CpG-depleted genes, decreased RNA levels rather pointed to altered transcriptional events. This hypothesis was confirmed by nuclear run-on analysis revealing a clear positive correlation of intragenic CpG content and the amount of de novo synthesized mRNA transcripts. The in vitro analysis of nucleosome positioning gave first hints, that the variations of CpG dinucleotide content has an influence on the chromatin structure along the open reading frames, since nucleosome reconstitution exhibited distinct nucleosome positioning patterns on the related DNA elements. Taken this observation together with the finding that DNA sequence is sufficient to direct nucleosome positioning in vivo (51) it may well be that sequence dependent nucleosome positioning and stability may contribute to the observed transcriptional disparity. Yet, ChIP experiments could not reveal differences in the overall histone H3 occupancy of the reporter gene variants. However, ChIP experiments only resolved histone H3 density, but did so far not address the presence of histone modifications, histone variants or even the presence of transcription factors. Hence, future experiments have to deal with the detection of histone modifications as well as the analysis of transcription factors present at the transcription start sites of the gfp gene variants and RNA Polymerase II at the coding region.
In light of these findings it is worth mentioning that the ubiquitously expressed and CpG binding protein CFP1 (CXXC finger protein 1) has previously been shown to transactivate exclusively CpG containing promoter regions (61). This nuclear transcriptional activator specifically binds to unmethylated CpG dinucleotides via a CXXC zinc finger motif, the affinity for target sequences increasing with accumulative numbers of CpGs (62). Hence, CFP1 might play a role as an epigenetic regulator in modulating gene expression via CpG dinucleotide methylation and histone modifications (63–65). Furthermore, consensus CFP1 binding motifs specified as (A/C)CpG(A/C) (62) could be mapped within the CpG rich gfp reporter gene, promoting a possible interaction with CFP1. Electrophoretic mobility supershift assays of a recombinant CFP1 protein suggest binding to the CpG-containing gfp gene but not to the CpG-free gfp allele (data not shown). However, upon the availability of appropriate antibodies ChIP experiments may be well suited to elaborate different gene occupancies of CFP1 between CpG high and CpG low reporter gene containing cell lines. Besides, it remains to be examined whether high-level transcription of a CpG-rich gene as compared to its CpG depleted counterpart is directly associated with efficient intragenic binding of another corresponding trans-activator or a related cellular protein.
Herein we have shown that depletion of CpGs from the coding region of a reporter gene correlated with a clear loss of expression originating from attenuated transcriptional activity. In further support of these findings, reduced de novo transcription could also be observed for other CpG-free gene variants (unpublished data) indicating that this effect was not gene specific. Concerning the interaction of the huGFP gene variants with chromatin related factors, it could be shown that site-specific preferences for nucleosome formation were determined among the huGFP variants. It is suggested that the distribution of CpG dinucleotides, which differs in the huGFP fragments is responsible for variable binding efficiencies and positions. The presence of a certain sequence pattern, predicted to enable efficient nucleosome binding, however have yet to be identified. A 3 bp periodicity of CG and GC dinucleotides has been found to be a highly nucleosome favoured sequence (66). It is suggested that different nucleosome preferences displayed by nucleosome reconstitutions are at least partly due to altered bending flexibility.
It remains to be seen whether transgene expression can be further increased by introduction of additional CpGs into otherwise moderately expressing genes. In sum, these findings provide a new perspective for the application of transgenes comprising a high intragenic CpG density for the rational design of recombinant expression cassettes.
FUNDING
BMBF grant 01KI0211 (to R.W.). Funding for open access charge: BMBF grant 01KI0211 (to R.W.).
Conflict of interest statement. None declared.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
The authors thank Gerd Sutter (Paul Ehrlich Institute, Langen, Germany) for kindly providing the MVA-T7 virus strain and the infection protocol. They also thank Marcus Graf (GENEART AG) for calculation of codon frequencies.
REFERENCES
- 1.Valente CA, Monteiro GA, Cabral JM, Fevereiro M, Prazeres DM. Optimization of the primary recovery of human interferon Alpha2b from Escherichia Coli inclusion bodies. Protein Expr. Purif. 2006;45:226–234. doi: 10.1016/j.pep.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Barrett JW, Sun Y, Nazarian SH, Belsito TA, Brunetti CR, Mcfadden G. Optimization of codon usage of poxvirus genes allows for improved transient expression in mammalian cells. Virus Genes. 2006;33:15–26. doi: 10.1007/s11262-005-0035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes LM, Dickson AJ. Mammalian cell factories for efficient and stable protein expression. Curr. Opin. Biotechnol. 2006;17:381–386. doi: 10.1016/j.copbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins N, Murphy L, Tyther R. Post-Translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol. Biotechnol. 2008;39:113–118. doi: 10.1007/s12033-008-9049-4. [DOI] [PubMed] [Google Scholar]
- 5.Ill CR, Chiou HC. Gene therapy progress and prospects: recent progress in transgene and Rnai expression cassettes. Gene Ther. 2005;12:795–802. doi: 10.1038/sj.gt.3302524. [DOI] [PubMed] [Google Scholar]
- 6.Chapman BS, Thayer RM, Vincent KA, Haigwood NL. Effect Of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeshita F, Takase K, Tozuka M, Saha S, Okuda K, Ishii N, Sasaki S. Muscle creatine kinase/SV40 hybrid promoter for muscle-targeted long-term transgene expression. Int. J. Mol. Med. 2007;19:309–315. [PubMed] [Google Scholar]
- 8.Graf M, Deml L, Wagner R. Codon-optimized genes that enable increased heterologous expression in mammalian cells and elicit efficient immune responses in mice after vaccination of naked DNA. Methods Mol. Med. 2004;94:197–210. doi: 10.1385/1-59259-679-7:197. [DOI] [PubMed] [Google Scholar]
- 9.Graf M, Bojak A, Deml L, Bieler K, Wolf H, Wagner R. Concerted Action of multiple cis-acting sequences is required for rev dependence of late human immunodeficiency virus type 1 gene expression. J. Virol. 2000;74:10822–10826. doi: 10.1128/jvi.74.22.10822-10826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, et al. Codon usage optimization of HIV Type 1 subtype C Gag, Pol, Env, And Nef Genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses. 2003;19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- 11.Wagner R, Graf M, Bieler K, Wolf H, Grunwald T, Foley P, Uberla K. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus Type 1 and Simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum. Gene Ther. 2000;11:2403–2413. doi: 10.1089/104303400750038507. [DOI] [PubMed] [Google Scholar]
- 12.Tejeda-Mansir A, Montesinos RM. Upstream Processing of plasmid DNA for vaccine and gene therapy applications. Recent Pat. Biotechnol. 2008;2:156–172. doi: 10.2174/187220808786241015. [DOI] [PubMed] [Google Scholar]
- 13.Bestor TH. Gene silencing as a threat to the success of gene therapy. J. Clin. Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Sidhu SK, Minks J, Chang SC, Cotton AM, Brown CJ. X chromosome inactivation: heterogeneity of heterochromatin. Biochem. Cell Biol. 2008;86:370–379. doi: 10.1139/o08-100. [DOI] [PubMed] [Google Scholar]
- 16.Lopez J, Percharde M, Coley HM, Webb A, Crook T. The context and potential of epigenetics in oncology. Br. J. Cancer. 2009;100:571–577. doi: 10.1038/sj.bjc.6604930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 18.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling Cpg Islands. J. Cell Biochem. 2005;94:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 19.Larsen F, Gundersen G, Lopez R, Prydz H. Cpg Islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 20.Davey C, Pennings S, Allan J. Cpg methylation remodels chromatin structure in vitro. J. Mol. Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer GP. Mutagenesis At methylated Cpg sequences. Curr. Top. Microbiol. Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 22.Hisano M, Ohta H, Nishimune Y, Nozaki M. methylation of Cpg dinucleotides in the open reading frame of a testicular germ cell-specific intronless gene, Tact1/Actl7b, Represses its expression in somatic cells. Nucleic Acids Res. 2003;31:4797–4804. doi: 10.1093/nar/gkg670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yew NS, Wysokenski DM, Wang KX, Ziegler RJ, Marshall J, Mcneilly D, Cherry M, Osburn W, Cheng SH. Optimization Of plasmid vectors for high-level expression in lung epithelial cells. Hum. Gene Ther. 1997;8:575–584. doi: 10.1089/hum.1997.8.5-575. [DOI] [PubMed] [Google Scholar]
- 24.Yew NS, Zhao H, Przybylska M, Wu IH, Tousignant JD, Scheule RK, Cheng SH. Cpg-Depleted plasmid dna vectors with enhanced safety and long-term gene expression in vivo. Mol. Ther. 2002;5:731–738. doi: 10.1006/mthe.2002.0598. [DOI] [PubMed] [Google Scholar]
- 25.Richards EJ. Chromatin methylation: Who's on first? Curr. Biol. 2002;12:R694–R695. doi: 10.1016/s0960-9822(02)01208-3. [DOI] [PubMed] [Google Scholar]
- 26.Deng G, Chen A, Pong E, Kim YS. Methylation in Hmlh1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncogene. 2001;20:7120–7127. doi: 10.1038/sj.onc.1204891. [DOI] [PubMed] [Google Scholar]
- 27.Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J. Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- 28.Chevalier-Mariette C, Henry I, Montfort L, Capgras S, Forlani S, Muschler J, Nicolas JF. Cpg Content affects gene silencing in mice: evidence from novel transgenes. Genome Biol. 2003;4:R53. doi: 10.1186/gb-2003-4-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 30.Chen ZY, He CY, Meuse L, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- 31.Hodges BL, Taylor KM, Joseph MF, Bourgeois SA, Scheule RK. Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by Cpg dinucleotides. Mol. Ther. 2004;10:269–278. doi: 10.1016/j.ymthe.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Graessmann A, Sandberg G, Guhl E, Graessmann M. Methylation of single sites within the Herpes Simplex virus tk coding region and the Simian virus 40 T-antigen intron causes gene inactivation. Mol. Cell Biol. 1994;14:2004–2010. doi: 10.1128/mcb.14.3.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keshet I, Yisraeli J, Cedar H. Effect Of regional DNA methylation on gene expression. Proc. Natl Acad. Sci. USA. 1985;82:2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalle B, Rubin JE, Alkan O, Sukonnik T, Pasceri P, Yao S, Pawliuk R, Leboulch P, Ellis J. Egfp reporter genes silence Lcrbeta-Globin Transgene expression via Cpg dinucleotides. Mol. Ther. 2005;11:591–599. doi: 10.1016/j.ymthe.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Doerfler W, Schubbert R, Heller H, Kammer C, Hilger-Eversheim K, Knoblauch M, Remus R. Integration of foreign DNA and its consequences in mammalian systems. Trends Biotechnol. 1997;15:297–301. doi: 10.1016/S0167-7799(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 36.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA Methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 37.Beutler E, Gelbart T, Han JH, Koziol JA, Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc. Natl Acad. Sci. USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan J, Antezana MA. Mammalian mutation pressure, synonymous codon choice, and Mrna degradation. J. Mol. Evol. 2003;57:694–701. doi: 10.1007/s00239-003-2519-1. [DOI] [PubMed] [Google Scholar]
- 39.Graf M, Ludwig C, Kehlenbeck S, Jungert K, Wagner R. A quasi-lentiviral green fluorescent protein reporter exhibits nuclear export features of late human immunodeficiency virus type 1 transcripts. Virology. 2006;352:295–305. doi: 10.1016/j.virol.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2:1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Ravazzolo R. Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques. 2000;29:1012–1017. doi: 10.2144/00295st02. [DOI] [PubMed] [Google Scholar]
- 44.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 45.Becker PB, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon RH, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander L, Lee H, Rosenzweig M, Jung JU, Desrosiers RC. EGFP-containing vector system that facilitates stable and transient expression assays. Biotechniques. 1997;23:64–66. doi: 10.2144/97231bm12. [DOI] [PubMed] [Google Scholar]
- 48.Baldi L, Hacker DL, Adam M, Wurm FM. Recombinant protein production by large-scale transient gene expression in mammalian cells: state of the art and future perspectives. Biotechnol. Lett. 2007;29:677–684. doi: 10.1007/s10529-006-9297-y. [DOI] [PubMed] [Google Scholar]
- 49.Loser P, Jennings GS, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J. Virol. 1998;72:180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson RT. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 53.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 54.van Gaal EV, Hennink WE, Crommelin DJ, Mastrobattista E. Plasmid engineering for controlled and sustained gene expression for nonviral gene therapy. Pharm. Res. 2006;23:1053–1074. doi: 10.1007/s11095-006-0164-2. [DOI] [PubMed] [Google Scholar]
- 55.Clark SJ, Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21:5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 56.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 57.Antoniou M, Harland L, Mustoe T, Williams S, Holdstock J, Yague E, Mulcahy T, Griffiths M, Edwards S, Ioannou PA, et al. Transgenes encompassing dual-promoter Cpg islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics. 2003;82:269–279. doi: 10.1016/s0888-7543(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 58.Cihak A, Weiss JW, Pitot HC. Effects of 5-azacytidine on hepatic polyribosomes and maturation of ribosomal RNA. Acta Biol. Med. Ger. 1974;33:859–865. [PubMed] [Google Scholar]
- 59.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 60.Newbury SF. Control Of mRNA stability in eukaryotes. Biochem. Soc. Trans. 2006;34:30–34. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 61.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 63.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 64.Carlone DL, Skalnik DG. CpG binding protein is crucial for early embryonic development. Mol. Cell Biol. 2001;21:7601–7606. doi: 10.1128/MCB.21.22.7601-7606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carlone DL, Lee JH, Young SR, Dobrota E, Butler JS, Ruiz J, Skalnik DG. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol. Cell Biol. 2005;25:4881–4891. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta S, Dennis J, Thurman RE, Kingston R, Stamatoyannopoulos JA, Noble WS. Predicting human nucleosome occupancy from primary sequence. PLoS Comput. Biol. 2008;4:e1000134. doi: 10.1371/journal.pcbi.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]