Successful Computational Prediction of Novel Imprinted Genes from Epigenomic Features (original) (raw)
Abstract
Approximately 100 mouse genes undergo genomic imprinting, whereby one of the two parental alleles is epigenetically silenced. Imprinted genes influence processes including development, X chromosome inactivation, obesity, schizophrenia, and diabetes, motivating the identification of all imprinted loci. Local sequence features have been used to predict candidate imprinted genes, but rigorous testing using reciprocal crosses validated only three, one of which resided in previously identified imprinting clusters. Here we show that specific epigenetic features in mouse cells correlate with imprinting status in mice, and we identify hundreds of additional genes predicted to be imprinted in the mouse. We used a multitiered approach to validate imprinted expression, including use of a custom single nucleotide polymorphism array and traditional molecular methods. Of 65 candidates subjected to molecular assays for allele-specific expression, we found 10 novel imprinted genes that were maternally expressed in the placenta.
Genomic imprinting refers to genes that are expressed from one of the two parental alleles in a parent-of-origin-specific manner. Thus, far, about 100 mouse imprinted genes have been identified, with many more genes predicted to be imprinted (http://igc.otago.ac.nz/home.html) (25, 32). The identification of novel imprinted genes has become increasingly important with the realization that imprinting defects are associated with a variety of complex disorders, such as obesity, diabetes, and schizophrenia (8, 27, 39, 52).
Given the importance that imprinted genes play in human health, several studies have tackled genome-wide identification of imprinted genes (1, 12, 13, 18, 22, 25, 26, 28, 32-34, 36, 37, 43, 46, 48, 50). These studies have done so mainly by experimental methods, with modest success. Recently, computational prediction of imprinted genes using DNA sequence features alone has been used for both the mouse and the human genomes. Data from these studies resulted in experimental validation of three imprinted genes in the mouse genome, and two candidate genes from analysis of the human genome may also be imprinted (26, 36, 37, 50). However, data from reciprocal crosses are not available for the two genes reported to be imprinted in humans, which is essential to distinguish imprinted genes from expression quantitative trait loci (eQTLs). For example, within the Rasgrf1 imprinted domain, expression of the noncoding AK006067 transcript from the allele found in C57BL/6 mice is more than 100-fold higher than expression from the allele found in PWK mice. Without reciprocal F1 crosses, this bias in expression levels would erroneously be taken as evidence for imprinting (G. Dokshin and P. D. Soloway, unpublished data).
Imprinted gene expression is controlled by allele-specific epigenetic states at imprinting control regions (ICRs), where allele-specific epigenetic modifications are controlled and/or placed and which, in turn, regulate imprinted expression. However, there is little sequence conservation among ICRs, and the DNA sequences that establish epigenetic states have been defined for, at most, three ICRs (3, 19, 40, 53). Given the essential roles for epigenetic mechanisms in imprinting, we reasoned that epigenomic data sets might augment the utility of sequence features to identify novel imprinted genes. In support of this, characteristic epigenetic features have been identified at gene regulatory elements of both nonimprinted and imprinted genes (14, 47). Accordingly, we used a variety of sequence and epigenetic data to train a set of computational prediction models, which we then used to identify a list of candidate imprinted genes. A generalized linear model (GLM), along with a training array of 53 known imprinted genes and 84 nonimprinted genes, was used to select epigenetic and sequence features within each of 11 domains (100, 10, and 1 kb upstream of genes, within genes, 5′ untranslated regions [UTRs], exons, introns, 3′-UTRs, and 1, 10, and 100 kb downstream of genes) that aid in prediction of imprinted status. We next selected 10 genes predicted to be imprinted by 5 or more of the 11 models and determined their imprinting status using molecular methods. This identified one novel imprinted gene. Based on this success, we expanded our candidate list to 1,297 genes that were predicted to be imprinted by 3 or more of the 11 models and then used microarray analysis in a first-tier screen of 563 of them. Based on the microarray results, we tested 32 of the 563 genes for imprinted expression by molecular methods, identifying an additional five novel imprinted genes. After completion of the microarray analysis, we identified an additional 23 genes that were not included on the microarray but could be tested using single nucleotide polymorphisms (SNPs) between available mouse strains. Our multitiered computational and experimental screening directed us to 65 genes that we subjected to stringent molecular testing. Ten of these were novel imprinted loci, indicating that our approach is useful for identifying imprinted loci. Furthermore, the epigenetic signatures that correlate with imprinting may reflect shared epigenetic regulatory mechanisms that control imprinting.
MATERIALS AND METHODS
Sequence data collection.
Genome-wide information regarding the location of CpG islands and microRNA (miRNA) clusters within the Known Genes track was downloaded from the UCSC Genome Browser website (http://genome.ucsc.edu/cgi-bin/hgGateway; February 2006 build), using the table feature. The locations of all nonredundant known gene transcripts, exons, introns, 5′-UTRs, and 3′-UTRs were obtained in the same fashion, and the start and end positions of the domains 1, 10, and 100 kb upstream of each gene, as well as 1, 10, and 100 kb downstream, were calculated from this information. The list of nonredundant transcripts was obtained by filtering the list of 31,752 known gene transcripts so that UCSC Known Gene and RefSeq annotations for a single transcript were not counted twice. This resulted in a list of 29,544 nonredundant gene transcripts. Data regarding the locations of experimentally verified and computationally predicted CTCF binding sites within the mouse genome were obtained from InsulatorDB (http://insulatordb.utmem.edu/).
Genome-wide data reporting the distribution of the histone modifications H3K4me3, H3K9me3, H3K27me3, H3K36me3, and H4K20me3 were downloaded from (ftp://ftp.broad.mit.edu/pub/papers/chipseq/) for two developmental stages: embryonic stem cells and embryonic fibroblasts. The raw data were filtered to include only sites with a read score of 2 or greater. Data describing sites enriched for histone modifications, calculated either by a sliding window method or a hidden Markov model, were also downloaded for embryonic stem cells, embryonic fibroblast cells, and neural progenitor cells (30).
The entire mouse genome was downloaded from the UCSC Genome Browser website (http://genome.ucsc.edu/cgi-bin/hgGateway; February 2006 build), and QUADPARSER (http://www-shankar.ch.cam.ac.uk/quadparser.html) was used to scan the genome for sites with a potential to form G-quartet structures (16). A custom Perl script was designed to calculate the percent GC within each of the domains (gene, exon, intron, 5′-UTR, 3′-UTR, +1 kb, +10 kb, +100 kb, −1 kb, −10 kb, and −100 kb) for each known gene.
For each of the known genes, custom Perl scripts were used to tally the number of times each of the features of interest occurred within each of the 11 domains (gene, exon, intron, 5′-UTR, 3′-UTR, +1 kb, +10 kb, +100 kb, −1 kb, −10 kb, and −100 kb).
Microarray expression data for each of the 155 genes predicted as being imprinted by five or more prediction models were obtained from the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway; February 2006 build).
Calculation of correlation coefficients and P values.
Correlation coefficients were calculated using the cor() function in R (http://www.r-project.org/). P values for the correlation coefficients were calculated using a two-tailed t test and were considered significant if less than 0.000157 [(P value of 0.05)/319 comparisons] after Bonferroni correction for multiple comparisons. A total of 319 comparisons were made when considering 29 sequence and epigenomic features over 11 domains.
Training data set.
A list of 53 genes imprinted in the mouse was compiled from the Imprinted Gene Catalogue (IGC) database (http://igc.otago.ac.nz/home.html) to use in the training set (see Table S1 in the supplemental material). An additional 84 nonimprinted genes were included in the training set (see Table S1). The presumed nonimprinted genes used in our training data set were identified in a systematic search of the Jackson Laboratories MGI database (http://www.informatics.jax.org/) for mice with knockout mutations demonstrating homozygous lethality. We selected those genes that were viable as heterozygotes but that were lethal as homozygotes.
Training procedure.
Using the glm() command in R, we fit a logistic regression model for each of the 11 data sets using the training data. We chose a GLM because our goal was to identify genes that are imprinted based on information already known about these genes. Logistic regression is the standard way to model binary outcomes. In this case the outcome of interest, y_ i, is whether gene i is imprinted or not imprinted. Using stepwise regression to build the model allowed us to identify the best subset of potential predictors with a training data set where the outcome was known. We then applied this model to genes where the imprinting status was still unknown to obtain the predicted probability that the gene is imprinted.
Logistic regression was fit using maximum likelihood estimation (MLE), which avoided assumptions of the ordinary least squares (OLS) method of fitting models used in standard linear regression, such as the outcome following a normal distribution, a linear relationship with the predictors, and a normally distributed error term. Other standard assumptions still apply for our method, such as independent responses, absence of multicolinearity, and sufficient sample sizes. Logistic regression did not require linear relationships between the predictors and the response, as is the case for OLS regression, but it did assume a linear relationship between the response and the log odds (logit) of the predictors. When the assumption of linearity in the logits is violated, then logistic regression underestimates the degree of relationship of the response to the predictors and will lack power, generating type II errors.
The response for our model was whether or not the gene is imprinted, and the potential predictors were the set of 29 sequence and epigenetic features. The predictors to be included in the model were chosen using both forward and backward stepwise selection based on the Akaike information criterion (AIC). The predictors included in each of the 11 resulting logistic regression models varied from domain to domain (see Table 1). The resulting 11 models were then each tested on a corresponding domain data set in which the genes included were known to be imprinted or nonimprinted.
TABLE 1.
Features included in prediction models, by domaina
| Feature | Significance level for domain | No. of models with indicated P value | No. of positive models | Total no. of models | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100up | 10up | 1up | 5′-UTR | In gene | Exons | Introns | 3′-UTR | 1dn | 10dn | 100dn | <0.001 | <0.005 | <0.05 | |
| %CG | *** | ** | * | ** | * | 1 | 2 | 2 | 0 | 5 | ||||
| G-quartets | + | * | * | 0 | 0 | 2 | 1 | 3 | ||||||
| miRNA | + | + | + | + | + | 0 | 0 | 0 | 5 | 5 | ||||
| CpG island | * | ** | * | + | + | + | 0 | 1 | 2 | 3 | 6 | |||
| Verified CTCF | + | * | 0 | 0 | 1 | 1 | 2 | |||||||
| Predicted CTCF | ** | ** | + | * | 0 | 2 | 1 | 1 | 4 | |||||
| EF H3K4me3 | * | *** | 1 | 0 | 1 | 0 | 2 | |||||||
| EF H3K9me3 | + | * | 0 | 0 | 1 | 1 | 2 | |||||||
| EF H3K27me3 | + | *** | + | * | *** | 2 | 0 | 1 | 2 | 5 | ||||
| EF H3K36me3 | * | ** | *** | ** | *** | *** | * | 3 | 2 | 2 | 0 | 7 | ||
| ES H4K20me3 | ** | * | + | ** | * | * | 0 | 2 | 3 | 1 | 6 | |||
| ES H3K4me3 | ** | ** | + | * | * | 0 | 2 | 2 | 1 | 5 | ||||
| ES H3K9me3 | * | ** | * | ** | * | 0 | 2 | 3 | 0 | 5 | ||||
| ES H3K27me3 | *** | *** | * | ** | ** | *** | *** | 4 | 2 | 1 | 0 | 7 | ||
| ES H3K36me3 | *** | * | + | * | ** | 1 | 1 | 2 | 1 | 5 | ||||
| ES H3K4me3 (HMM) | + | * | *** | + | * | * | * | 1 | 0 | 4 | 2 | 7 | ||
| ES H3K9me3 (HMM) | + | + | ** | + | * | 0 | 1 | 1 | 3 | 5 | ||||
| ES H3K27me3 (HMM) | + | + | ** | + | * | + | + | 0 | 1 | 1 | 5 | 7 | ||
| ES H3K36me3 (HMM) | + | * | + | ** | *** | * | ** | 0 | 3 | 2 | 2 | 7 | ||
| ES H4K20me3 (HMM) | *** | + | + | * | + | ** | + | + | *** | 2 | 1 | 1 | 5 | 9 |
| ES H3K4me3 (WIN) | * | * | * | ** | 0 | 1 | 3 | 0 | 4 | |||||
| ES H3K9me3 (WIN) | + | ** | * | 0 | 1 | 1 | 1 | 3 | ||||||
| ES H3K27me3 (WIN) | * | * | + | * | *** | 1 | 0 | 3 | 1 | 5 | ||||
| EF H3K4me3 (WIN) | * | + | 0 | 0 | 1 | 1 | 2 | |||||||
| EF H3K9me3 (WIN) | * | + | 0 | 0 | 1 | 1 | 2 | |||||||
| EF H3K27me3 (WIN) | * | + | + | 0 | 0 | 1 | 2 | |||||||
| NP H3K4me3 (WIN) | * | + | 0 | 0 | 1 | 1 | 2 | |||||||
| NP H3K9me3 (WIN) | + | 0 | 0 | 0 | 1 | 1 | ||||||||
| NP H3K27me3 (WIN) | ** | * | * | * | 0 | 1 | 3 | 0 | 4 |
Test data set.
A list of nine mouse imprinted genes was compiled from the ICG database (http://igc.otago.ac.nz/home.html) to use in the test set (see Table S4 in the supplemental material). An additional 20 presumed nonimprinted genes were included in the test set (see Table S4). Presumed nonimprinted genes used in our test data set were identified as described above.
Candidate list compilation.
To obtain a list of candidate imprinted genes to subject to the pilot experiment for molecular validation, the predicted imprinted genes identified in each domain were intersected. Only genes predicted to be imprinted by five or more models were considered in the initial candidate list. This list was narrowed further by considering proximity to known imprinted genes (within 1 Mb), number of models predicting each gene to be imprinted, GO classification, and expression levels.
Tissue collection and RNA preparation for confirmation of imprinting status.
AKR/J and PWD/PhJ reciprocal timed matings were performed by placing males and females together overnight and checking for evidence of a copulatory plug the next morning, at day 0.5. For the embryo transfer experiments, FVBn/J females were superovulated and in vitro fertilized with sperm from C3H/HeJ males. The resulting embryos were transferred to a pseudopregnant recipient mother of the strain C57BL/6J. Pregnant females were sacrificed at day 17.5 (E17.5) of pregnancy. Whole brain and placenta (lacking the decidua) were dissected and snap-frozen in liquid nitrogen. Total RNA was extracted from the F1 brain and placenta samples using the guanidium thiocyanate method. For each sample, the RNA concentration and the _A_260/_A_280 ratio were checked using a NanoDrop ND-1000 spectrophotometer.
SNP and restriction site identification.
SNPs were identified in the final candidate imprinted genes using the Jackson Laboratory Mouse Genome Informatics website. In each case, potential SNPs and 10 bp to either side were analyzed using the NEB Cutter tool (New England BioLabs) to select SNPs that overlapped with a restriction site for identification of allele-specific expression.
Confirmation of imprinting by RT-PCR and digestion or sequencing.
For each gene, primers were designed using Primer3 (http://fokker.wi.mit.edu/primer3/input.htm) to overlap an allele-specific restriction site (see Table S6 in the supplemental material). Where possible, primers were placed to span introns. To perform reverse transcription-PCR (RT-PCR), 5 μg of RNA from reciprocal crosses between polymorphic mouse strains (C57BL/6 × Cast [B×C] or PWD × AKR [P×A]) was subjected to random primed reverse transcription to make cDNA, which was PCR amplified using the primers in Table S6 in the supplemental material. Standard PCR was run with GoTaq DNA polymerase (Promega) for 40 cycles (95°C for 30 s, 60°C for 30 s, and 72°C for 50 s) followed by a final extension of 5 min at 72°C. The resulting PCR products (300 to 700 bp) were either digested with an allele-specific restriction enzyme or Sanger sequenced to determine parent-of-origin-specific allelic expression patterns. All PCR products were sequenced to confirm amplification specificity. (see Fig. S2 in the supplemental material.)
Identification of SNPs for array-based allele-specific expression analysis.
Genes predicted to be imprinted by three or more models (1,297 genes) were considered in the initial candidate list. SNPs from mouse strains PWD/PhJ and AKR/J were identified for each of these candidate-imprinted genes in the Jackson Laboratories MGI database (http://www.informatics.jax.org/). Only genes containing SNPs in the 3′-UTR, or within 1 kb of the 3′ end of the gene, were included for experimental validation, narrowing the list to 563 genes. Where possible, multiple SNPs were used for each gene.
Microarray probe design.
A 50-bp sequence to either side of each usable SNP was collected. Using this information, 12 microarray probes were designed for each SNP. Four probes contain the SNP centered on the probe and with base A, T, C, or G at the SNP position. Four probes contain the SNP 1 bp upstream from the center of the probe with base A, T, C, or G at the SNP position. Four probes contain the SNP 1 bp downstream of the center of the probe and with base A, T, C, or G at the SNP position. All 12 probes for each SNP were trimmed to the same length, which was determined by melting temperature. For each SNP, all 12 probes were between 25 and 31 bp, and the length was chosen so that the melting temperature of each of the 12 probes was above 50°C. Probe quality was analyzed using Agilent's e-array website.
Tissue collection and RNA preparation for microarray hybridization.
AKR/J and PWD/PhJ reciprocal timed matings were performed and brain and placentae collected at E17.5 as described above. Total RNA was extracted from two biological replicates of the F1 brain and placenta samples using the Qiagen RNeasy lipid tissue minikit. For each sample, the RNA concentration and the A260/A280 ratio were checked using a NanoDrop ND-1000 spectrophotometer. RNA quality was determined using the Agilent 2100 Bioanalyzer. All of the samples hybridized to the microarray had an RNA integrity number between 9.7 and 10.
Microarray experiment.
RNA was subjected to oligo(dT)-primed cRNA amplification. cRNA was synthesized using cyanine-3-labeled CTP and was hybridized to a custom Agilent 8×15K array by the Cornell Microarray Core facility. Hybridization was carried out at 50°C. Material from reciprocal crosses was included to rule out false positives from expression QTLs. RNA hybridized to the array consisted of two biological replicates of both reciprocal crosses using RNA from E17.5 brain and E17.5 placenta.
Data analysis and candidate imprinted gene identification.
After hybridization and washing, the microarray slide was scanned with the Axon 4000B scanner, and normalized fluorescence intensities were calculated using GenePix Pro 6.0 software and the background subtraction method. One-way analysis of variance (ANOVA) was performed to compare the averaged normalized fluorescence intensities of the four nucleotides at each SNP position. In total, the ANOVA test was done 12 times: once for each of the three probe sets, using materials from two reciprocal crosses and two tissue types. If the fluorescence intensities for a gene were found to be unequal, the level of the most highly expressed nucleotide was compared to that of the second most highly expressed nucleotide to determine if there was a significant difference. Candidate imprinted genes selected for molecular analyses were those demonstrating reciprocal monoallelic expression of the expected SNP nucleotides. From this list, a subset was selected for further examination based on whether the normalized fluorescence intensity of the most highly expressed nucleotide was significantly higher than that of the second most highly expressed nucleotide and whether results from multiple probe sets were in agreement. After applying these criteria, 32 placenta candidates and 8 brain candidates were subjected to experimental validation by RT-PCR followed by allele-specific restriction digestion or Sanger sequencing.
Quantification of allele-specific expression levels.
The expression levels of the AKR and PWD alleles were determined as previously described using Sanger sequencing and the Peak Picker2 software (10, 35). Briefly, F1 genomic DNA and two to three cDNA biological replicates from both the A×P and the P×A crosses were amplified using the primers shown in Table S6 of the supplemental material. PCR products were Sanger sequenced, and the sequence trace files were analyzed using PeakPicker2 software, which we used to accurately measure peak heights of the two SNP nucleotides in F1 DNA and the cDNA samples. The software normalized the peak height measurements for the cDNA samples in two ways. First, peaks near the SNP positions that carried the same two nucleotides found in the SNP positions were measured for all samples, and these were used to correct for differences in fluorescence intensity between the fluorophores. Second, peak heights from F1 genomic DNA samples, which have equal contributions from the two strains, were used to normalize for any allelic bias introduced by either the amplification or sequencing reactions. From these data, allele-specific expression levels were calculated as a percentage of the total expression.
RESULTS
Selection of features to use for prediction.
To use DNA sequence and epigenomic features for identifying novel imprinted genes, we first selected a set of features we anticipated might correlate with imprinting status and for which genome-wide data sets were available. DNA sequence features we considered included GC content, CpG islands, miRNA clusters, and predicted G-quartet sites (16). The genome-wide epigenetic and chromatin features we considered were predicted and verified CTCF binding sites (http://insulatordb.utmem.edu/help.php) and several histone states, including H3K4me3, H3K9me3, H3K27me3, H3K36me3, and H4K20me3 (30). These histone states were characterized in embryonic stem (ES), mouse embryonic fibroblast (EF), and neuronal progenitor (NP) cells by chromatin immunoprecipitation (ChIP)-chip and ChIP-seq analyses (30). We used three measures of histone modification enrichment; a hidden Markov model (HMM) and the sliding window method (WIN), as originally reported, and the raw data with read scores greater than 2 (30).
These sequence features were selected for specific reasons. GC percentage and CpG islands were examined because the differential methylation that is associated with imprinted genes is usually placed on cytosine residues, specifically, cytosine residues that are followed by guanine residues. In fact, a recent paper compared sequence features within human, mouse, and cattle and correlated these two features with imprinted genes for 20 genes known to be imprinted in all three species (21). miRNA clusters were included because several known imprinted gene regions are associated with miRNA clusters, including the Gtl2/Dlk1 imprinted cluster on mouse chromosome 12 and the well-studied H19/Igf2 imprinted cluster on mouse chromosome 7 (5, 38). Additionally, miRNA clusters have been implicated as having a role in DNA methylation in both plants and mammals (41). CTCF binding sites, both experimentally validated and computationally predicted, were examined because CTCF binds to methylation-sensitive enhancer blocking elements, leading to silencing of the allele to which it is bound. For example, CTCF is associated with allele-specific silencing at imprinted genes such as H19/Igf2 and Rasgrf1 (2, 54). Predicted G-quartet sites were included because the secondary structures that they form can affect the ability of DNA methyltransferases to methylate the underlying DNA sequence, and imprinted expression relies heavily on DNA methylation (44).
Finally, data on the genome-wide localization of five histone modifications in three developmental stages were included in the analysis (30). The specific modification we studied is trimethylation of various lysine residues present in the N-terminal histone tails. The addition of methyl groups to the various lysines can have a dramatic effect on the expression of any genes to which these histones are bound. Of the five modifications examined, H3K4me3 and H3K36me3 are marks of active genes or euchromatin, while H3K9me3, H3K27me3, and H4K20me3 are marks of repressed genes or heterochromatin. Despite the dramatic effect these marks can have on gene expression, little is understood about what controls the placement of these modifications. We focused on these specific histone modifications, because overlapping H3K4me3 and H3K9me3 marks within ICRs, have been correlated with imprinting status in the past and H3K27me3 is known to have an antagonistic relationship with DNA methylation (24, 30). There were a total of 29 sequence and epigenetic features in our analysis.
Histone modifications strongly correlate with known imprinted genes.
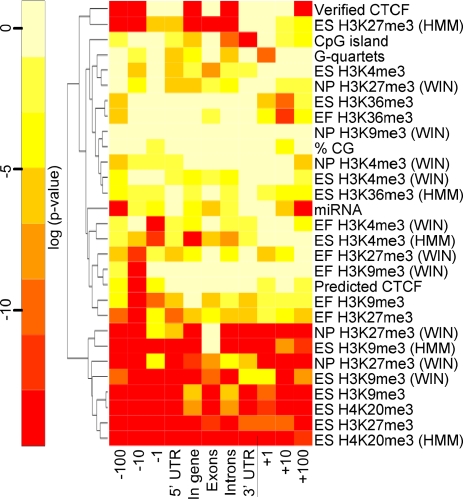
Before any attempts at model training were made, we used R (http://www.r-project.org/) to determine whether any of the epigenomic and sequence features we had collected correlated with the imprinting status of known imprinted genes. For this test, we used a set of 53 known imprinted genes (see Table S1 in the supplemental material), comparing them to 29,544 nonredundant mouse transcripts. The list of nonredundant transcripts was obtained by filtering the list of 31,752 known gene transcripts so that UCSC Known Gene and RefSeq annotations for a single transcript were not counted twice. For each gene, we compared features present in 11 different domains relative to the transcription start site: 100 kb upstream, 10 kb upstream, 1 kb upstream, 5′-UTRs, exons, introns, 3′-UTRs, within genes (including 5′-UTRs, exons, introns, and 3′-UTRs), and 1 kb downstream, 10 kb downstream, and 100 kb downstream. From the correlation analysis, a pattern of histone modifications associated with imprinted genes emerged (Fig. 1). In general, the repressive histone modifications H3K9me3, H3K27me3, and H4K20me3 tended to be associated with imprinted genes in any of the three cell types and over nearly all of the 11 domains. Furthermore, H3K36me3, a histone modification enriched in active chromatin, was associated with nonimprinted genes in the domain 10 kb downstream of genes. The positive association between H3K9me3, H3K27me3, and imprinting is in close agreement with their documented role in mechanisms controlling imprinted DNA methylation at Rasgrf1. At that locus, H3K27me3 excludes DNA methylation from the unmethylated maternal allele and H3K9me3 is needed for optimal placement of DNA methylation on the methylated paternal allele (24). Because the associations we found among H3K9me3, H3K27me3, and imprinting were known to be mechanistically relevant, this provided a measure of confidence in our approach. Additional positive correlates included predicted G-quartet sites, miRNA clusters, and verified CTCF binding sites.
FIG. 1.
Correlation of features in each domain with imprinting. For the 11 domains examined, the correlation coefficients were calculated for the 29 features using the cor() function in R. P values for correlation coefficients were calculated using a two-tailed t test and were considered significant if less than 0.000157, after Bonferroni correction for the 319 comparisons (see Table S2 in the supplemental material for the values). Log-transformed P values were calculated and depicted in a heat map, in which a log-transformed P value less than −8.76 was significant. The significance levels of the log-transformed P values are indicated by a color gradient, with colors at the red end of the gradient representing higher levels of significance. This figure does not indicate the direction of correlation. However, Table S3 in the supplemental material, which lists the correlation coefficients used to derive the P values represented in this figure, provides this information. Features are clustered by similarities in P value distributions according to the dendrogram on the left. The cell sources for each of the histone modifications data sets are indicated (ES, EF, and NP), as is the method used to calculate histone modification enrichment (HMM or WIN) (30).
Histone modifications are important predictors of imprinted status.
Having found significant correlations between imprinting status and several of 29 sequence and epigenetic features across the 11 domains, we sought to use these correlations to identify novel imprinted genes. This required that we develop more refined statistical models. To do this, we trained a set of 11 GLMs, one model for each domain. Model training involved several steps. First we selected a training set of 53 known imprinted genes and 84 likely nonimprinted genes to constrain our modeling. Model development required such a training set. Next, we used R to fit a logistic regression model for each of the 11 domains. For each domain's model, this entailed adding epigenetic and sequence features, one at a time, to identify the features that improved a model's ability to identify the imprinted genes among the training set. In addition, features were considered in combination with one other, meaning that if a given feature by itself was not a good predictor of imprinting status, but in combination with other features improved the predictive power for a given model, then that feature was retained by the model. Finally, we allowed our GLM the choice to discard one feature at the end of each round of analysis, if this improved the predictive power of a model. Because features were eliminated during model development, none of the final models included all sequence or epigenetic features. Also, although the genes included in the training set were the same for each model, the feature density varied for the 11 models because each model represented a different gene domain (e.g., +100 kb, +10 kb, +1 kb, within genes, 5′-UTR, introns, exons, 3′-UTR, −1 kb, −10 kb, or −100 kb). For this reason as well, not all features were equally important in all models.
The model significance level assigned by R for each of the features of interest in each domain examined is shown in Table 1. From this modeling, several features of interest stand out as effective predictors of imprinting. Six histone features were predictive of imprinted status in at least 7 of the 11 GLM: ES H4K20me3 (HMM) in 9 of 11 models; EF H3K36me3, ES H3K27me3, ES H3K4me3 (HMM), ES H3K27me3 (HMM), and ES H3K36me3 (HMM) in 7 of 11 models. The most included nonhistone features were CpG islands clusters, used in 6 of the 11 prediction models, and percent CG and miRNA clusters, used in 5 of the 11 prediction models. Likewise, the features that were identified as being highly significant in the greatest number of prediction models were the histone modifications ES H3K27me3 and EF H3K36me3.
Model sensitivity and specificity.
To assess the effectiveness of our prediction method, we analyzed a separate set of 29 genes using our 11 logistic regression models. This set included 9 known imprinted and 20 likely nonimprinted genes, none of which was included in our training data set (see Table S4 in the supplemental material). From our analysis of the test set, we determined the sensitivity of our models, calculated as the number of known imprinted genes in the test data set that were correctly identified as imprinted, and their specificity, calculated as the number of nonimprinted genes in the test data set that were correctly identified as nonimprinted. Within each model, we identified genes as predicted to be imprinted if P, the probability the gene is imprinted, was greater than or equal to 0.8.
When we used stringent prediction criteria, requiring that five or more models predict a gene to be imprinted, sensitivity was 66.7% (six of nine known imprinted genes were called imprinted), while specificity was 100% (none of 20 nonimprinted genes was called imprinted). The six genes correctly identified in the test data set were Air, Ddc, Inpp5f_ v2, Peg10, Sfmbt2, and Th. The sensitivity did not increase when we used less stringent criteria, requiring that only three or more models predicted imprinting, although the specificity dropped to 95%. In this case, 1 of 20 nonimprinted genes, Myh6, was incorrectly predicted to be imprinted, assuming our criteria for identifying nonimprinted genes were valid. If the stringency was increased, requiring that six or more models predict a gene to be imprinted, the sensitivity fell to 33.3% (three of nine known imprinted genes were called imprinted). Since no sensitivity was gained with a stringency lower than prediction based on five or more models, we used this stringency level for our first-tier analysis.
Genome-wide prediction of imprinted status reveals models are not biased.
For our first tier of the genome-wide analysis, we used our 11 models to query 29,544 mouse transcripts for their predicted imprinting status. When genes predicted as imprinted by five or more models were considered, we identified a candidate list of 155 genes (Table 2). We were concerned that this list of genes was biased by the presence of an adjacent known imprinted gene, whose ICR may act over a large genomic domain. This did not appear to be the case, as there was at least one gene that was predicted as nonimprinted between each of our 155 candidates and the nearest known imprinted gene. Reassuringly, four of the five known mammalian microimprinted genes were correctly predicted as imprinted: Nap1l5, Nnat1, Inpp5f_ v2, and U2af1rs1, and the imprinting status of three of the host genes in which the microimprinted genes are located was correctly classified as well (see Table S5 in the supplemental material). In the two instances where the host gene was incorrectly classified, they were classified as nonimprinted.
TABLE 2.
Genes predicted as being imprinted by five or more models
| Genea | No. of models |
|---|---|
| Gpa33 | 10 |
| 6430706D22Rik | 10 |
| Dusp27 | 9 |
| Mid1 | 9 |
| BC096660 | 8 |
| Rpo1-4 | 8 |
| Thb | 8 |
| 9530015I07Rik | 7 |
| A530088H08Rik | 7 |
| AK016821 | 7 |
| AK046026 | 7 |
| BC005471 | 7 |
| BC099500 | 7 |
| C030011J08Rik | 7 |
| Hoxc13 | 7 |
| Slc38a1 | 7 |
| Sytl3 | 7 |
| Ugt1a1 | 7 |
| Zfp629 | 7 |
| 1700029J07Rik | 6 |
| 4930478A21Rik | 6 |
| 9130017C17Rik | 6 |
| A830031A19Rik | 6 |
| AK143924 | 6 |
| Ank1 | 6 |
| BC046305 | 6 |
| BC048950 | 6 |
| BC065085 | 6 |
| BC089472 | 6 |
| Bcmo1 | 6 |
| Cdh15 | 6 |
| Cmah | 6 |
| Cyp2j13 | 6 |
| Dynlt1 | 6 |
| Edar | 6 |
| Gm944 | 6 |
| Hmox2 | 6 |
| II3ra | 6 |
| Kcnq2 | 6 |
| LOC629678 | 6 |
| LOC633640 | 6 |
| Nef3 | 6 |
| Neurod2 | 6 |
| Odz4 | 6 |
| Ugt1a10 | 6 |
| Ugt1a2 | 6 |
| Ugt1a5 | 6 |
| Ugt1a6a | 6 |
| Ugt1a6b | 6 |
| Ugt1a7c | 6 |
| Ugt1a9 | 6 |
| Umodl1 | 6 |
| Vil2 | 6 |
| Vnn3 | 6 |
| Wdr27 | 6 |
| Wdr27 | 6 |
| Zmat4 | 6 |
| 3110070M22Rik | 5 |
| 4921537P18Rik | 5 |
| 4930417M19Rik | 5 |
| 4930539E08Rik | 5 |
| 4933403G14Rik | 5 |
| 5330420D20Rik | 5 |
| 6430573F11Rik | 5 |
| A030007L22 | 5 |
| A630008I04 | 5 |
| AB125595 | 5 |
| Adam18 | 5 |
| Agrn | 5 |
| AK004563 | 5 |
| AK016553 | 5 |
| AK029828 | 5 |
| AK052253 | 5 |
| AK131836 | 5 |
| AK133237 | 5 |
| AK138193 | 5 |
| AK140614 | 5 |
| AK146072 | 5 |
| AK147510 | 5 |
| AK154031 | 5 |
| AK158329 | 5 |
| Amph | 5 |
| B230363K08Rik | 5 |
| BC006684 | 5 |
| BC007165 | 5 |
| BC054080 | 5 |
| Bcl11a | 5 |
| Bsnd | 5 |
| Cd244 | 5 |
| Cd59b | 5 |
| Cdh13 | 5 |
| Cflar | 5 |
| Chrnb3 | 5 |
| Cntn3 | 5 |
| Cryba2 | 5 |
| Cyp4f15 | 5 |
| Dennd1a | 5 |
| Dspp | 5 |
| Enpp3 | 5 |
| Fbxo40 | 5 |
| Fyco1 | 5 |
| Gm889 | 5 |
| Gnao1 | 5 |
| Gng2 | 5 |
| Grhl2 | 5 |
| Hlcs | 5 |
| Hoxa10 | 5 |
| Hoxc4 | 5 |
| Hps3 | 5 |
| Hunk | 5 |
| Ifitm7 | 5 |
| Irf8 | 5 |
| Kcnmb4 | 5 |
| Kit | 5 |
| Lair1 | 5 |
| Lgi1 | 5 |
| LOC432436 | 5 |
| LOC432823 | 5 |
| Mam13 | 5 |
| Metapl1 | 5 |
| Mst1r | 5 |
| Neu4 | 5 |
| Nfasc | 5 |
| Nhlrc1 | 5 |
| Oasl1 | 5 |
| Otx2 | 5 |
| Oxsm | 5 |
| Pag1 | 5 |
| Parva | 5 |
| Pcgf4 | 5 |
| Pigl | 5 |
| Pitx2 | 5 |
| Piwil1 | 5 |
| Pkp1 | 5 |
| Pnpt1 | 5 |
| Prss35 | 5 |
| Ptf1a | 5 |
| Ptprn2 | 5 |
| Rgs8 | 5 |
| Rnf36 | 5 |
| Scin | 5 |
| Shcbp1 | 5 |
| Slc6a17 | 5 |
| Sspn | 5 |
| Syt9 | 5 |
| Tcam1 | 5 |
| Tmprss2 | 5 |
| Trpc2 | 5 |
| Ube2l6 | 5 |
| Xkr5 | 5 |
| Zfp160 | 5 |
| Zfp180 | 5 |
| Zfp445 | 5 |
| Zfp706 | 5 |
Experimental testing revealed one novel maternally expressed gene.
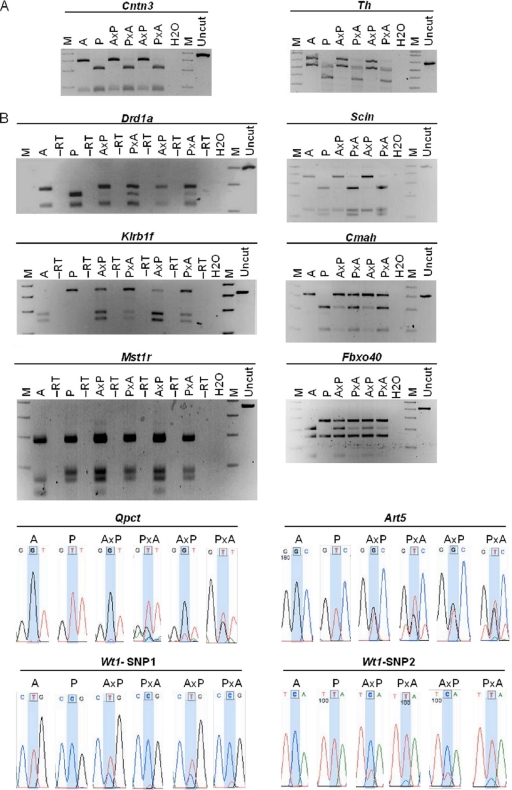
We then set up a pilot experiment to test a subset of the 155 candidate genes predicted as imprinted by five or more models for experimental evidence of imprinting. Genes were selected for experimental validation using a variety of criteria. First, we selected genes that were categorized as imprinted by 8 or more of the 11 prediction models. Second, since many imprinted genes occur in clusters, we selected the genes predicted as imprinted by five or more models that also fell within 1 Mb of known imprinted genes. Third, we selected genes predicted as imprinted by five or more models that also fell into GO categories that are significantly overrepresented in imprinted genes (see Fig. S1 in the supplemental material). Finally, genes that were not only expressed but also were expressed at high levels in either brain or placenta were selected for further testing. From the selected genes, we identified those containing SNPs between the AKR/J (AKR) and PWD/PhJ (PWD) mouse strains. A total of 11 out of the 155 candidate genes were ultimately selected for experimental validation (Table 2, genes shown in boldface). One of these was Th, which was not listed as imprinted in the Otago database (http://igc.otago.ac.nz/home.html) at the time we assembled our sequence data for the 29,544 mouse transcripts but was subsequently reported as imprinted (37). The remaining 10 genes selected for experimental validation were tested for evidence of imprinting in E17.5 mouse placenta. For each test, we used biological replicates of F1 crosses between the polymorphic mouse strains AKR and PWD. Importantly, we analyzed materials from reciprocal crosses to control for strain-specific expression QTL. This differs from previous attempts to validate predicted imprinted genes in humans, which could not use reciprocal F1 crosses to assess imprinting status and hence could not distinguish imprinting from expression QTL (26). We dissected the embryonic component of the placenta for this analysis and also did controls to verify that our dissections effectively excluded maternal tissue contamination (see below). RT-PCR primers were designed to span SNPs that allowed us to determine which allele(s) was expressed (see Table S6 in the supplemental material). We used two different approaches to identify expressed alleles. One entailed sequencing the RT-PCR product; the other relied on amplifying SNPs within restriction sites and digesting the PCR products. In cases where imprinting is absolute, we expect to see no expression whatsoever from the silenced allele. However, many imprinted loci exhibit a bias in allelic expression, for which both alleles are expressed but one parental allele is preferentially expressed (46). In addition, for some genes where imprinting is absolute in some tissues, there is a bias in allelic expression in other tissues (33). Furthermore, according to the widely accepted conflict theory describing the emergence of imprinting, biased expression is expected as an intermediate phenotype during selection for imprinted status (31). It is easy to discern genes with absolute imprinted expression. We identified genes with imprinting biases on the basis of consistently more intense bands or higher sequencing peaks from one of the parental alleles in our biological replicates and our reciprocal crosses. One of the 10 genes tested in placenta, Cntn3, showed clear and exclusive maternal allele-specific expression by restriction digestion of PCR products from AKR and PWD reciprocal crosses, consistent with the imprinted status predicted by our models (Fig. 2A). PCR amplification specificity was confirmed by sequencing the PCR product (see Fig. S2 in the supplemental material). This gene is not likely to be regulated by ICRs in known imprinting domains; the imprinted gene closest to Cntn3 on chromosome 6 is 44 Mb away (Nap1l5) (http://igc.otago.ac.nz/home.html). We also tested each of these 10 genes for imprinted expression in E17.5 brain, but we saw no evidence of imprinted expression in brain at this developmental time point.
FIG. 2.
Maternal allele-specific expression of 10 novel imprinted genes. We amplified placental cDNA from E17.5 embryos prepared using reciprocal crosses between AKR/J (A) and PWD/PhJ (P) mice, as well as from the parental strains. For the labels shown in the figure, the maternal strain is written first. PCR primers were specific to Cntn3 and Th, 2 of the 10 candidate genes from our pilot study (A), and the nine candidate genes from the expanded analysis, Drd1a, Scin, Klrb1f, Cmah, Mst1r, Fbxo40, Qpct, Art5, and Wt1 (B). PCR products were digested overnight with restriction enzymes specific for one parental allele and run on 3% agarose gels. All 10 genes, and Th, showed expression patterns consistent with maternal allele expression. For those genes lacking restriction enzymes at SNP positions, we sequenced the PCR products. Traces overlapping the SNP positions (blue shading) are shown. M, 1-kb DNA marker.
Microarray validation of 563 candidate imprinted genes yielded 9 additional novel imprinted genes.
Having demonstrated that the computational prediction method greatly increased our ability to identify imprinted genes (1 of 10 tested in our pilot experiment), we designed a custom microarray to experimentally test a larger number of candidate genes for evidence of imprinting. The 1,297 genes considered for testing by this approach were predicted as imprinted by three or more of the prediction models. For each gene, 3′-biased SNPs were identified between the AKR and PWD mouse strains. Appropriate SNPs were available for 563 of the 1,297 candidate genes. We designed 12 probes for each SNP. Each probe carried one of the four possible nucleotides in the SNP location, which was at three positions (−1, 0, and +1) relative to the center of the probe. When possible, we used multiple SNPs per gene.
Each slide contained eight identical arrays, to which we hybridized two biological replicates each of E17.5 brain and placenta RNA from AKR/PWD reciprocal crosses, for a total of one sample per array. After hybridization, washing, and scanning the array, we performed a one-way ANOVA to compare the normalized fluorescent intensities of the four nucleotides at each SNP position. This first-tier analysis provided suggestive evidence for imprinting of 40 genes (32 in placenta, 8 in brain). For each of these genes, the microarray expression results agreed for more than one of the probe sets described above. We examined all 40 genes for definitive second-tier testing using RT-PCR followed by allele-specific restriction digestion or Sanger sequencing. In addition to these 40 genes, we also performed direct molecular analysis on an additional 23. Those 23 genes were predicted as imprinted by five or more of our models; however, there were no 3′ biased SNPs, which were needed for optimal analysis on the microarray. Of the 63 genes we tested by these definitive molecular methods, we discovered 9 more novel genes imprinted in placenta (Art5, Cmah, Drd1a, Fbxo40, Klrb1f, Mst1r, Qpct, Scin, and Wt1), for a total of 10 new imprinted loci (Fig. 2B). Expression patterns of each of these nine genes were also examined in E17.5 brain from reciprocal crosses, but none showed evidence of imprinting in this tissue at this developmental time point.
Scin showed strong imprinting with exclusive expression from the maternal allele in placenta, as we found for Cntn3 and Th. The remaining genes were partially imprinted, with biased and not exclusive expression from one parental allele. The bias toward maternal allele expression was apparent for Klrb1f, for which the slowly migrating maternal PWD band was the major species in P×A crosses, whereas the faster migrating maternal AKR bands were the major species in the reciprocal A×P crosses. A maternal expression bias was also seen for Qpct and Wt1, for which the sequence traces showed large peaks that corresponded to maternal expression, but small peaks indicating paternal expression were not completely silenced. This was also true for Art5; however, the bias toward maternal expression was slight.
Maternal expression bias is due to imprinting and not amplification bias or eQTL.
Our analyses of Drd1a, Cmah, Mstr1, and Fbxo40 also showed evidence of maternally biased expression, but the evidence was more apparent for one direction of the cross than the other. This result is expected for partially imprinted quantitative trait loci for which allelic expression level varies with the strain of origin (eQTL), in addition to varying with parent of origin. It is also expected for partially imprinted genes when there are strain-specific differences in the efficiency of RT-PCR amplification, which can arise because of particular SNPs or mRNA secondary structures. Both of these cases represent strain biases in amplification that should not be confused with a parent-of-origin bias, which defines imprinting. For genes that are both partially imprinted and influenced by strain biases, the evidence for imprinting will be strongest in the reciprocal cross for which the partially silenced allele is from the strain with the lower level of expression or amplification. When the partially silenced allele is from the strain with the higher level of expression or amplification, there will be clear evidence for expression of both alleles. It is important to note that for genes with strain biases and no imprinting, both directions of the reciprocal cross should produce equal intensities of the bands. Drd1a and Cmah showed predominantly maternal allele expression in A×P crosses. The apparent expression from the paternal AKR allele in the reciprocal P×A cross is consistent with both biased imprinted expression from the maternal allele and biased amplification of AKR alleles for both genes. The weakly amplified PWD allele showed more prominent expression when maternally inherited, consistent with maternally biased imprinted expression for these genes. Mstr1r showed nearly exclusive expression from the maternal PWD allele in P×A crosses, as indicated by the lack of the fastest migrating band, diagnostic of expression from the paternal AKR allele. The AKR-specific band was apparent only in the reciprocal A×P crosses. In A×P, the expression from the paternal PWD allele is consistent with partial imprinted expression from the maternal allele and a strain bias that generates quantitatively higher PWD amplification. Fbxo40 also shows partial imprinting with maternally biased expression in the P×A cross. This, too, is consistent with preferential expression of the maternal allele and higher amplification of the PWD allele relative to AKR.
To assess directly if there were any strain biases in amplification, we included two controls. In the first, we performed quantitative PCR analysis to determine if each of the four genes had expression differences in the two strain backgrounds we used. Our results showed that for three of the genes, Cmah, Fbxo40, and Mst1r, expression levels were equivalent in the two strain backgrounds (see Fig. S3A in the supplemental material), indicating strain amplification biases were probably not due to eQTL. In the second control, we mixed cDNA prepared from the two pure inbred strains at 3:1 (AKR:PWD), 1:1 (AKR:PWD), and 1:3 (AKR:PWD) ratios, PCR amplified each of these mixtures using the same primer pairs used for the imprinting analysis, and digested the resulting PCR products with the restriction enzymes used for the imprinting analysis. For Cmah and Drd1a, an AKR strain amplification bias was seen, which explains why evidence for imprinting was most apparent in the A×P cross (see Fig. S3B). It is worth noting that expression of Drd1a in the PWD pure inbred strain was approximately 1.5 times higher than in the AKR pure inbred strain based on quantitative real-time PCR analysis, indicating that there is a very strong amplification bias for the AKR allele. Fbxo40 and Mst1r did not show any amplification or expression bias, so we added a third biological replicate for these genes, as well as for Cmah and Drd1a, and the results of the third biological replicate confirmed the results seen with the other two biological replicates (see Fig. S4 in the supplemental material). This provides additional support for our conclusions that Drd1a, Cmah, Mstr1, and Fbxo40 are in fact imprinted, with maternally biased expression.
Quantification of allele-specific expression confirmed imprinting.
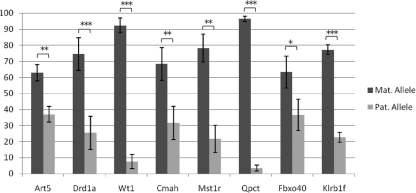
Because a subset of our novel imprinted genes demonstrated evidence of amplification bias or eQTL, based on allele-specific restriction digestion, quantitative PCR, or mixing experiments, we quantified the expression levels from both parental alleles for each of the genes that did not show rigid imprinting. We did this by PCR amplifying two cDNA biological replicates from both the A×P and the P×A crosses, gel purifying the PCR products, and performing Sanger sequencing, and then the sequence trace files were analyzed using the PeakPicker2 software. This allowed us to quantify the percent expression from the two parental alleles (10, 35). As a control, we also did this analysis with DNA from F1 mice, which have precisely equal allelic contributions from the AKR and PWD backgrounds. Each of the novel imprinted genes analyzed by this method showed a clear and significant imprinting bias, with expression predominantly from the maternally inherited allele (Fig. 3; see also Fig. S5 in the supplemental material).
FIG. 3.
Quantitation of maternal and paternal allele expression levels. The expression levels of the maternal and paternal alleles were determined as previously described using Sanger sequencing and the Peak Picker2 software (10, 35). For each gene, the expression level for both parental alleles is reported as a percentage. Error bars represent standard errors. Each gene was queried by at least one SNP and, whenever possible, data from multiple SNPs were used. For Wt, Cmah, and Fbxo40, two SNPs were available. Three and six SNPs were available for Qpct and Klrb1f, respectively. The data reported represent combined results from all available SNPs. Differences in expression levels of the maternal and paternal allele were significant in every case, as determined by a one-tailed t test. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Novel imprinted genes are not likely to be regulated by known ICRs.
These novel imprinted genes are not likely to be regulated by known ICRs, as they are all located greater than 3 Mb from known imprinted clusters. Furthermore, Cmah, Drd1a, and Fbxo40 are located on chromosomes not previously known to contain imprinted genes (Table 3). This gave us further assurance that our model was not biased by nearby known imprinted genes and was capable of identifying novel imprinted genes outside of known imprinted clusters.
TABLE 3.
Summary of 10 novel imprinted genes
| Novel imprinted gene | Expressed allele | Chromosomal locationa | Distance (Mb) to nearest known imprinted gene (gene name) |
|---|---|---|---|
| Art5 | Maternal (bias) | Chr 7, 101970700-101974050 | 26 (Inpp5f) |
| Cmah | Maternal (bias) | Chr 13, 24334885-24484750 | —b |
| Cntn3 | Maternal | Chr 6, 102129404-102430765 | 43 (Nap1l5) |
| Drd1a | Maternal | Chr 13, 54060805-54065279 | — |
| Fbxo40 | Maternal (bias) | Chr 16, 36885312-36898375 | — |
| Klrb1f | Maternal | Chr 6, 128672127-128688665 | 70 (Nap1l5) |
| Mst1r | Maternal | Chr 9, 107764990-107778482 | 18 (Rasgrf1) |
| Qpct | Maternal | Chr 17, 78956964-78995301 | 66 (Air) |
| Scin | Maternal | Chr 12, 40570196-40644651 | 27 (Mirn337) |
| Wt1 | Maternal | Chr 2, 104927368-104974453 | 17 (Gatm) |
Maternal tissue contamination was negligible.
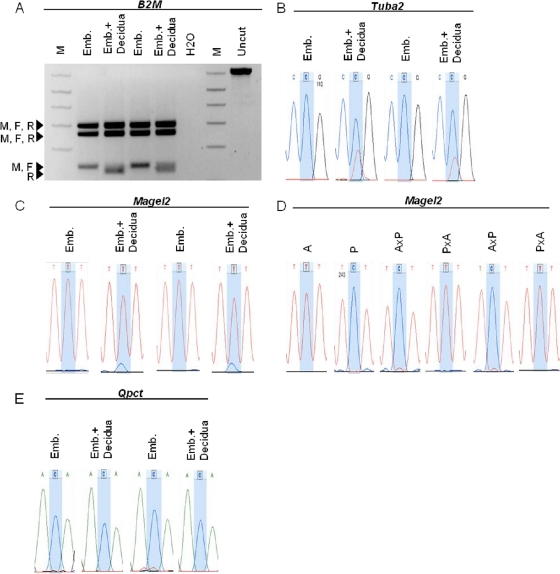
Because all the novel imprinted genes we discovered were maternally expressed in placenta, we were concerned that our dissections may not have rigorously excluded maternal material and that the genes were in fact expressed from contaminating tissue from the inbred mother and not from the F1 embryonic placenta. To rule out such false positives, we included several controls. In the first, we assayed the placental samples used in our imprinting assays for allele-specific expression of Magel2, which is reported to be expressed exclusively from the paternal allele in placenta and is also expressed in the maternal decidua (4). Our analysis showed Magel2 expression was exclusively from the paternal allele of the embryo (Fig. 4C). There was no expression from maternal sources, indicating that maternal tissue contamination was negligible (Fig. 4D). We also performed a second more rigorous control. We transferred F1 embryos from an FVBn/J (biological mother) × C3H/HeJ (father) cross to a pseudopregnant recipient mother of the strain C57BL/6J, and at E17.5 we dissected part of the embryonic portion of the placenta from the remaining tissue that included maternal contributions to the placenta. This was precisely how we prepared the embryonic placental samples used in our earlier assays. We then performed allele-specific expression analysis of two housekeeping genes (B2M and Tuba2) that carry polymorphisms that distinguish expression from the C57BL/6J (recipient mother), FVBn/J (biological mother), and C3H/HeJ (father) alleles, using two biological replicates. If our dissections of embryonic F1 placental tissue effectively excluded the maternal deciduae, then we would expect to detect expression of only FVBn/J and C3H/HeJ alleles and not from C57BL/6J alleles in the RNA we isolated. Placental portions that included the maternal deciduae should have expression from C57BL/6J alleles. This is what we observed in each test, indicating there was no evidence of maternal tissue contamination and that our dissections effectively excluded maternal tissue from the fetal portions of the placentae (Fig. 4A and B). Using the same cDNAs from the embryo transfer experiment above, we were able to directly test for maternal contamination of one of our novel maternally expressed imprinted genes, Qpct. In the Qpct assay, the FVBn/J biological mother and the C3H/HeJ father share one SNP (a C allele in sequence traces), while the C57BL/6J recipient mother contains a different SNP (T allele in sequence traces). Therefore, if this gene is truly imprinted, we would expect to see expression from the C allele shared by the FVBn/J and C3H/HeJ mouse strains and no expression from the C57BL/6J T allele. The results from this analysis, shown in Fig. 4E, confirmed that our claim that Qpct is imprinted was not confounded by maternal tissue contamination. There was no detectable expression of maternal Qpct in material rich in decidual RNA, again providing confidence that our observation of Qpct imprinting was unaffected by decidual contamination (Fig. 4E) and that our dissections effectively eliminated maternal tissue contamination.
FIG. 4.
Maternal tissue contamination was negligible. Placentae from E17.5 embryos were dissected to either retain or eliminate the maternal decidua. RNA was extracted from tissues, cDNA was synthesized, and RT-PCR was performed using primers specific to four genes: B2M, Tuba2, Magel2, and Qpct. Whenever possible, primers were designed to span an intron and, in each case, amplified an SNP between the strains AKR/J and PWD/PhJ. The available SNPs for Magel2 and Tuba2 did not overlap with allele-specific restriction enzyme sites, so PCR products were gel purified (Qiaex Quick Spin; Qiagen) and sequenced. (A, B, and E) The placentae were from embryo transfer experiments where the biological mother was FVBn/J (M), the father was C3H/HeJ (F), the recipient mother was C57/BL6J (R), and both biological parents shared an SNP, which differed from the recipient mother. For both Tuba2 and Qpct, the biological mother and the father share the C allele while the recipient mother contains the T allele. In each case, there was no evidence of expression from the recipient mother in the embryo-derived portion of the placenta, indicating that our dissection method eliminated maternal tissue contamination. (C) We tested for imprinting of a known paternally expressed gene, Magel2. The placentae used for this analysis were from natural matings between PWD/PhJ mothers (C allele) and AKR/J fathers (T allele). In the samples containing both the embryo-derived portion of the placenta as well as the maternal decidua, expression from the maternal PWD/PhJ allele was evident, indicating that Magel2 is expressed in the maternal decidua. However, the samples containing only embryo-derived portions of the placenta showed no evidence of expression from the maternal PWD/PhJ allele, as expected for a paternally expressed imprinted gene, and indicated that maternal tissue did not contaminate our dissections of the embryo-derived portion of the placenta. (D) Placentae from the same reciprocal F1 samples used in Fig. 2. Magel2 expression was exclusively from the paternal allele, indicating that maternal tissue contamination in these samples was negligible. Lane M, 1-kb DNA marker.
DISCUSSION
Computational prediction using sequence and epigenomic data identified 10 novel imprinted genes.
Here we have described a computational approach to identify novel imprinted genes in mice. This differs from other sequence-based prediction algorithms in that it includes epigenetic features as well as sequence features. Using a series of 11 prediction models that correspond to 11 domains surrounding 29,544 transcripts, we identified a list of 155 candidate imprinted genes and, in a pilot experiment, we experimentally tested 10 candidates for evidence of genomic imprinting in placenta. Of the 10 genes tested, one gene showed maternal allele-specific expression in placenta. Based on this initial success, which demonstrated the utility of our algorithm, we expanded our analysis, using a microarray-based approach to screen a larger list of 563 candidate genes. From the first-tier microarray approach, we identified 32 genes warranting molecular verification of their imprinting status and identified an additional five imprinted loci. Twenty-three of the remaining genes that were predicted as imprinted by five or more models, but that were not included on the microarray, were tested as well. This final level of analysis yielded another 4 novel imprinted genes, for a total of 10 novel imprinted loci revealed by this approach. In each case, we performed molecular verification using RNA from reciprocally crossed strains in biological replicates, which is the most rigorous test for imprinting.
The success rate of this method is 40-fold greater than expected by chance.
We expect that there are more than 10 genes that are imprinted among the 1,297 genes on the candidate list and the 563 genes tested by microarray. There were at least three reasons to expect this. First, our microarray and molecular analyses used a single strain combination, AKR and PWD. This combination provided an abundance of SNPs and genes we could test, but we were unable to test several of our strongest candidates predicted by the greatest number of models. Second, as our molecular validation was limited by both tissue type and experimental time point, it is possible that genes we tested and classified as nonimprinted may be imprinted in other tissues or at other developmental times. Third, the microarray signals for genes with low levels of expression may have been below the threshold needed to detect significant first-tier evidence for allele-specific expression, which excluded those genes from the more sensitive and definitive second-tier molecular testing. Nonetheless, because past molecular analyses revealed 100 of ∼30,000 genes are imprinted in mouse, we would have expected fewer than 1 of the 65 genes we subjected to molecular analysis would be imprinted if our prediction algorithms were no better than a random guess. We found 10 new imprinted genes among these 65, which is significantly more than expected from a random guess (Fisher's exact test, P = 7.892 × 10−16), indicating our approach is a valid method for the identification of novel imprinted genes.
Histone modifications important for imprinted gene predictions have biological relevance.
Our observations are consistent with two recent papers reporting associations between imprinted genes and histone modifications. The first report found that regions containing the overlapping marks of DNA methylation and H3K4me2 showed an approximately 5-fold enrichment for imprinted genes (47). The second study looked at enrichment of H3K4me3 and H3K9me3 in imprinted gene regions and found that of the top 20 regions enriched for both H3K4me3 and H3K9me3, 13 of these mapped to ICRs or imprinted gene promoters (30).
Our analysis included a variety of sequence features and histone modifications as potential predictors of imprinting. Because histone modification status not only correlated with imprinting but also proved to be a valid predictor of imprinting, our results provide insights into regulatory mechanisms controlling imprinting. H3K9me3, H3K27me3, and H4K20me3 were the best positive correlates with, and predictors of, imprinting. Notably, this is consistent with our experimental work demonstrating that H3K9me3 and H3K27me3, respectively, enforce the methylated and unmethylated states on the parental alleles that are essential to Rasgrf1 imprinting, and it is also consistent with other studies demonstrating that histone-modifying factors, including PRC2 components and KDM1B, are important for imprinting (7, 24, 29). Furthermore, others have demonstrated the presence of H4K20me3 on the maternal, but not on the paternal, allele at Rasgrf1 (9). These results are also consistent with work from others showing that H3K9me3 and H4K20me3 are needed for proper DNA methylation of the maternal allele of Snrpn (15, 51). H3K36me3 was the best negative correlate with imprinting. This mark is associated with repression of intergenic transcripts in yeast and is largely absent from the region surrounding the imprinted Igf2r promoter in mice (6, 17, 20, 30). This is significant, as imprinting of Igf2r is dependent on transcription of the 103-kbp noncoding antisense Air transcript across the Igf2r promoter (42). It is likely that the imprinting mechanisms operating at these loci are widely followed at many other imprinted loci that share these four commonly held epigenetic marks.
Histone modification data sets may bias prediction toward placenta-specific imprinted genes.
It is important to note that these histone modification data are not allele specific. Allele-specific epigenomic maps will reveal if the histone modifications important for imprinted prediction occur in combination, either on both alleles or on a single allele, or whether one modification marks the active allele and the other marks the repressed allele. However, our data strongly indicate that the marks we found to be correlated with imprinting could predict the imprinting status of novel imprinted genes and are probably involved in the imprinting mechanism, at least for a subset of imprinted genes. It is also worth mentioning that our analysis using histone marks may be biased toward the identification of genes imprinted in placenta. At least one gene cluster imprinted in placenta (Kcnq1/Kcnq1ot1) depends heavily on the presence of histone modifications, which make up the majority of features tested in these models, for imprinted expression (23). Likewise, G9a mutants deficient in both H3K9me2 and H3K9me3 demonstrate placenta-specific imprinting defects, while the embryo proper maintains proper imprinting (45).
It is not clear why each of the novel imprinted genes that we identified is maternally expressed in placenta and none showed imprinted expression in the brain. The epigenomic features we queried may be the best predictors of placental imprinting. It is worth noting that of 22 genes known to be imprinted in placenta, 19 are maternally expressed (http://igc.otago.ac.nz/home.html).
Although epigenomic data have been used to predict particular classes of transcripts and genomic regulatory elements (11, 14, 49), this is the first study to link histone modification data with the successful prediction of novel imprinted genes. As data become available that describe placement of additional epigenetic marks, these methods can complement transcriptome sequencing to identify imprinted genes and provide insights into mechanisms controlling imprinting (1, 46).
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Andy Clark and Xu Wang for helpful discussions and sharing reagents. We thank Robert Bukowski, Lalit Ponnala, and the Cornell Theory Center for automating scripts and help with statistical analyses. We also thank the Cornell Microarray Core and Sequencing Core Facilities for microarray support.
This work was supported by NIH funding to P.D.S. (CA120870, CA98596, and DA025722). C.M.B. was supported in part by NIH training grant T32GM007617.
We declare no competing interests.
Footnotes
▿
Published ahead of print on 26 April 2010.
REFERENCES
- 1.Babak, T., B. Deveale, C. Armour, C. Raymond, M. A. Cleary, D. van der Kooy, J. M. Johnson, and L. P. Lim. 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 18**:**1735-1741. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405**:**482-485. [DOI] [PubMed] [Google Scholar]
- 3.Birger, Y., R. Shemer, J. Perk, and A. Razin. 1999. The imprinting box of the mouse Igf2r gene. Nature 397**:**84-88. [DOI] [PubMed] [Google Scholar]
- 4.Boccaccio, I., H. Glatt-Deeley, F. Watrin, N. Roëckel, M. Lalande, and F. Muscatelli. 1999. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum. Mol. Genet. 8**:**2497-2505. [DOI] [PubMed] [Google Scholar]
- 5.Cai, X., and B. R. Cullen. 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13**:**313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123**:**581-592. [DOI] [PubMed] [Google Scholar]
- 7.Ciccone, D. N., H. Su, S. Hevi, F. Gay, H. Lei, J. Bajko, G. Xu, E. Li, and T. Chen. 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461**:**415-418. [DOI] [PubMed] [Google Scholar]
- 8.Crespi, B. 2008. Genomic imprinting in the development and evolution of psychotic spectrum conditions. Biol. Rev. Camb. Philos. Soc. 83**:**441-493. [DOI] [PubMed] [Google Scholar]
- 9.Delaval, K., J. Govin, F. Cerqueira, S. Rousseaux, S. Khochbin, and R. Feil. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 26**:**720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge, B., S. Gurd, T. Gaudin, C. Dore, P. Lepage, E. Harmsen, T. J. Hudson, and T. Pastinen. 2005. Survey of allelic expression using EST mining. Genome Res. 15**:**1584-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman, M., I. Amit, M. Garber, C. French, M. F. Lin, D. Feldser, M. Huarte, O. Zuk, B. W. Carey, J. P. Cassady, M. N. Cabili, R. Jaenisch, T. S. Mikkelsen, T. Jacks, N. Hacohen, B. E. Bernstein, M. Kellis, A. Regev, J. L. Rinn, and E. S. Lander. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458**:**223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatada, I., T. Sugama, and T. Mukai. 1993. A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res. 21**:**5577-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashizaki, Y., H. Shibata, S. Hirotsune, H. Sugino, Y. Okazaki, N. Sasaki, K. Hirose, H. Imoto, H. Okuizumi, M. Muramatsu, H. Komatsubara, T. Shiroishi, K. Moriwaki, M. Katsuki, N. Hatano, H. Sasaki, T. Ueda, N. Mise, N. Takagi, C. Plass, and V. M. Chapman. 1994. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 6**:**33-40. [DOI] [PubMed] [Google Scholar]
- 14.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39**:**311-318. [DOI] [PubMed] [Google Scholar]
- 15.Henckel, A., K. Nakabayashi, L. A. Sanz, R. Feil, K. Hata, and P. Arnaud. 2009. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum. Mol. Genet. 18**:**3375-3383. [DOI] [PubMed] [Google Scholar]
- 16.Huppert, J. L., and S. Balasubramanian. 2007. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35**:**406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20**:**971-978. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko-Ishino, T., Y. Kuroiwa, N. Miyoshi, T. Kohda, R. Suzuki, M. Yokoyama, S. Viville, S. C. Barton, F. Ishino, and M. A. Surani. 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11**:**52-59. [DOI] [PubMed] [Google Scholar]
- 19.Kantor, B., K. Makedonski, Y. Green-Finberg, R. Shemer, and A. Razin. 2004. Control elements within the PWS/AS imprinting box and their function in the imprinting process. Hum. Mol. Genet. 13**:**751-762. [DOI] [PubMed] [Google Scholar]
- 20.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123**:**593-605. [DOI] [PubMed] [Google Scholar]
- 21.Khatib, H., I. Zaitoun, and E. S. Kim. 2007. Comparative analysis of sequence characteristics of imprinted genes in human, mouse, and cattle. Mamm. Genome 18**:**538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroiwa, Y., T. Kaneko-Ishino, F. Kagitani, T. Kohda, L. L. Li, M. Tada, R. Suzuki, M. Yokoyama, T. Shiroishi, S. Wakana, S. C. Barton, F. Ishino, and M. A. Surani. 1996. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat. Genet. 12**:**186-190. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36**:**1291-1295. [DOI] [PubMed] [Google Scholar]
- 24.Lindroth, A. M., Y. J. Park, C. M. McLean, G. A. Dokshin, J. M. Persson, H. Herman, D. Pasini, X. Miró, M. E. Donohoe, J. T. Lee, K. Helin, and P. D. Soloway. 2008. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 4**:**e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luedi, P. P., A. J. Hartemink, and R. L. Jirtle. 2005. Genome-wide prediction of imprinted murine genes. Genome Res. 15**:**875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luedi, P. P., F. S. Dietrich, J. R. Weidman, J. M. Bosko, R. L. Jirtle, and A. J. Hartemink. 2007. Computational and experimental identification of novel human imprinted genes. Genome Res. 17**:**1723-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackay, D. J., J. L. Callaway, S. M. Marks, H. E. White, C. L. Acerini, S. E. Boonen, P. Dayanikli, H. V. Firth, J. A. Goodship, A. P. Haemers, J. M. Hahnemann, O. Kordonouri, A. F. Masoud, E. Oestergaard, J. Storr, S. Ellard, A. T. Hattersley, D. O. Robinson, and I. K. Temple. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 40**:**949-951. [DOI] [PubMed] [Google Scholar]
- 28.Maeda, N., and Y. Hayashizaki. 2006. Genome-wide survey of imprinted genes. Cytogenet. Genome Res. 113**:**144-152. [DOI] [PubMed] [Google Scholar]
- 29.Mager, J., N. D. Montgomery, F. P. de Villena, and T. Magnuson. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33**:**502-507. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448**:**553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, T., and D. Haig. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7**:**45-49. [DOI] [PubMed] [Google Scholar]
- 32.Nikaido, I., C. Saito, Y. Mizuno, M. Meguro, H. Bono, M. Kadomura, T. Kono, G. A. Morris, P. A. Lyons, M. Oshimura, Y. Hayashizaki, Y. Okazaki, et al. 2003. Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res. 13**:**1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plass, C., H. Shibata, I. Kalcheva, L. Mullins, N. Kotelevtseva, J. Mullins, R. Kato, H. Sasaki, S. Hirotsune, Y. Okazaki, W. A. Held, Y. Hayashizaki, and V. M. Chapman. 1996. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 14**:**106-109. [DOI] [PubMed] [Google Scholar]
- 34.Pollard, K. S., D. Serre, X. Wang, H. Tao, E. Grundberg, T. J. Hudson, A. G. Clark, and K. Frazer. 2008. A genome-wide approach to identifying novel-imprinted genes. Hum. Genet. 122**:**625-634. [DOI] [PubMed] [Google Scholar]
- 35.Qiu, P., G. J. Soder, V. J. Sanfiorenzo, L. Wang, J. R. Greene, M. A. Fritz, and X. Y. Cai. 2003. Quantification of single nucleotide polymorphisms by automated DNA sequencing. Biochem. Biophys. Res. Commun. 309**:**331-338. [DOI] [PubMed] [Google Scholar]
- 36.Ruf, N., S. Bähring, D. Galetzka, G. Pliushch, F. C. Luft, P. Nürnberg, T. Haaf, G. Kelsey, and U. Zechner. 2007. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum. Mol. Genet. 16**:**2591-2599. [DOI] [PubMed] [Google Scholar]
- 37.Schulz, R., T. R. Menheniott, K. Woodfine, A. J. Wood, J. D. Choi, and R. J. Oakey. 2006. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 34**:**e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitz, H., H. Royo, M. L. Bortolin, S. P. Lin, A. C. Ferguson-Smith, and J. Cavaillé. 2004. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14**:**1741-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao, W. J., L. Y. Tao, C. Gao, J. Y. Xie, and R. Q. Zhao. 2008. Alterations in methylation and expression levels of imprinted genes H19 and Igf2 in the fetuses of diabetic mice. Comp. Med. 58**:**341-346. [PMC free article] [PubMed] [Google Scholar]
- 40.Shemer, R., A. Y. Hershko, J. Perk, R. Mostoslavsky, B. Tsuberi, H. Cedar, K. Buiting, and A. Razin. 2000. The imprinting box of the Prader-Willi/Angelman syndrome domain. Nat. Genet. 26**:**440-443. [DOI] [PubMed] [Google Scholar]
- 41.Sinkkonen, L., T. Hugenschmidt, P. Berninger, D. Gaidatzis, F. Mohn, C. G. Artus-Revel, M. Zavolan, P. Svoboda, and W. Filipowicz. 2008. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 15**:**259-267. [DOI] [PubMed] [Google Scholar]
- 42.Sleutels, F., R. Zwart, and D. P. Barlow. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415**:**810-813. [DOI] [PubMed] [Google Scholar]
- 43.Smith, R. J., W. Dean, G. Konfortova, and G. Kelsey. 2003. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 13**:**558-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, S. S., A. Laayoun, R. G. Lingeman, D. J. Baker, and J. Riley. 1994. Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J. Mol. Biol. 243**:**143-151. [DOI] [PubMed] [Google Scholar]
- 45.Wagschal, A., H. G. Sutherland, K. Woodfine, A. Henckel, K. Chebli, R. Schulz, R. J. Oakey, W. A. Bickmore, and R. Feil. 2008. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 28**:**1104-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X., Q. Sun, S. D. McGrath, E. R. Mardis, P. D. Soloway, and A. G. Clark. 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One 3**:**e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen, B., H. Wu, H. Bjornsson, R. D. Green, R. Irizarry, and A. P. Feinberg. 2008. Overlapping euchromatin/heterochromatin-associated marks are enriched in imprinted gene regions and predict allele-specific modification. Genome Res. 18**:**1806-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf, J. B., J. M. Cheverud, C. Roseman, and R. Hager. 2008. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 4**:**e1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Won, K. J., I. Chepelev, B. Ren, and W. Wang. 2008. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics 9**:**547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood, A. J., R. G. Roberts, D. Monk, G. E. Moore, R. Schulz, and R. J. Oakey. 2007. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 3**:**e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, M. Y., T. F. Tsai, and A. L. Beaudet. 2006. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 20**:**2859-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie, T., M. Chen, O. Gavrilova, E. W. Lai, J. Liu, and L. S. Weinstein. 2008. Severe obesity and insulin resistance due to deletion of the maternal Gsα allele is reversed by paternal deletion of the Gsα imprint control region. Endocrinology 149**:**2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon, B. J., H. Herman, A. Sikora, L. T. Smith, C. Plass, and P. D. Soloway. 2002. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30**:**92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon, B., H. Herman, B. Hu, Y. J. Park, A. Lindroth, A. Bell, A.G West, Y. Chang, A. Stablewski, J. C. Piel, D. I. Loukinov, V. V. Lobanenkov, and P. D. Soloway. 2005. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell. Biol. 25**:**11184-11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]