The Integrator Complex Recognizes a New Double Mark on the RNA Polymerase II Carboxyl-terminal Domain (original) (raw)
Abstract
The carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (pol II) comprises multiple tandem repeats of the heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. This unusual structure serves as a platform for the binding of factors required for expression of pol II-transcribed genes, including the small nuclear RNA (snRNA) gene-specific Integrator complex. The pol II CTD specifically mediates recruitment of Integrator to the promoter of snRNA genes to activate transcription and direct 3′ end processing of the transcripts. Phosphorylation of the CTD and a serine in position 7 are necessary for Integrator recruitment. Here, we have further investigated the requirement of the serines in the CTD heptapeptide and their phosphorylation for Integrator binding. We show that both Ser2 and Ser7 of the CTD are required and that phosphorylation of these residues is necessary and sufficient for efficient binding. Using synthetic phosphopeptides, we have determined the pattern of the minimal Ser2/Ser7 double phosphorylation mark required for Integrator to interact with the CTD. This novel double phosphorylation mark is a new addition to the functional repertoire of the CTD code and may be a specific signal for snRNA gene expression.
Keywords: Gene Regulation, Gene Transcription, Protein Kinases, RNA Polymerase II, RNA Processing, CTD, Integrator
Introduction
The CTD5 of the largest subunit of pol II is an evolutionary conserved structure, specific to eukaryotic organisms, which plays a fundamental role in transcription and RNA processing (1). It comprises multiple tandemly repeated heptapeptides with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, with 52 repeats in the mammalian protein (2). The CTD serves as a scaffold for the interaction of a wide range of nuclear factors involved in various processes linked to transcription. The ability of the CTD to interact specifically with appropriate proteins during the transcription cycle in vivo is determined largely by the phosphorylation status of the three serine residues within the consensus heptapeptide (Ser2/Ser5/Ser7) (3). Different patterns of CTD serine phosphorylation correlate with the position of pol II along transcribed protein-coding genes and allow the recruitment of the appropriate factors at different stages of the transcription cycle. Differential modification thus enables a wide range of signaling combinations to be read as a code (3–5). Phosphorylation of Ser2 and phosphorylation of Ser5 are the best studied CTD modifications. Ser5 phosphorylation is directed by the CDK7 kinase as part of the TFIIH complex. Ser5 phosphorylation occurs early in the transcription cycle, is highest near the promoter, and activates capping of mRNAs. Ser2 phosphorylation, directed by the CDK9 kinase subunit of the positive transcription elongation factor b (P-TEFb) complex, occurs later in the transcription cycle and is generally highest toward the 3′ end of the genes. This modification plays roles in splicing and 3′ end processing of transcripts. There is some evidence that CTD heptapeptides can bear both Ser2 and Ser5 marks as factors implicated in transcription such as Set2 preferentially recognize a combination of both in vitro (3, 6). Some Ser2 phosphorylation must therefore occur before the removal of the phosphate on Ser5. Recently, it has been shown that Ser7 of the heptapeptide repeat is also phosphorylated during transcription (7–10), further expanding the number of possible phosphorylation combinations on the CTD. The CDK7 component of the general transcription factor TFIIH has been implicated in Ser7 phosphorylation both in vivo and in vitro (7, 10, 11). The CDK9 component of P-TEFb has also been shown to have Ser7 kinase activity (10). Ser7 phosphorylation has been found on both protein-coding genes and the pol II-transcribed snRNA genes (7–11). Human snRNA genes transcribed by pol II are structurally different from protein-coding genes (12, 13). snRNAs are neither spliced nor polyadenylated, and instead of a polyadenylation signal, the genes contain a conserved 3′ box RNA-processing element downstream of the snRNA-encoding region (14). However, these two different gene types share a requirement for the pol II CTD for efficient transcription and RNA processing (15–19). The demonstration that the snRNA gene-specific Integrator complex, which is required for 3′ box recognition, binds to the pol II CTD (20) provided a molecular link between transcription and 3′ processing of snRNA gene transcripts. More recently, we have shown that the serine in position 7 of the CTD heptapeptide plays a pivotal role in expression of snRNA genes (9). Mutation of Ser7 to alanine abolishes the association of the snRNA gene-specific Integrator complex to snRNA promoters and drastically affects transcription of snRNA genes and 3′ end processing of transcripts. In addition, we have shown that CTD phosphorylation is critical for efficient binding of the Integrator complex to the CTD (9). However, the precise mark on the CTD required for binding of the Integrator complex was not fully defined. Here, we precisely identify the CTD mark required for Integrator binding. We show that phosphorylation of both Ser2 and Ser7 is necessary for efficient binding of Integrator to the CTD. Interestingly, efficient binding requires two heptapeptide repeats and occurs when Ser7 in the first repeat and Ser2 in the second repeat are phosphorylated, providing a clear demonstration that recognition by some CTD-binding proteins spans more than a single heptapeptide repeat. This doubly phosphorylated mark may represent a novel gene type-specific CTD mark as Integrator complex recruitment is restricted to snRNA genes (20). In addition, we demonstrate that Ser7 kinase activity in HeLa cell nuclear extracts is largely attributable to DNA-PK and that recombinant DNA-PK specifically phosphorylates Ser7 of GST-CTD fusion proteins with 25 heptapeptide repeats in vitro.
EXPERIMENTAL PROCEDURES
Superose 6 Fractionation
200 μl of HeLa nuclear extract in 20 mm Hepes, pH 7.9, 100 mm KCl, 20% glycerol, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, 0.2 mm EDTA (Buffer D) (21) was loaded onto a Superose 6 column, and 0.5-ml fractions were collected. Molecular weight protein standards were run through the same column under the same conditions.
GST-CTD Phosphorylation
0.1 μg of GST-CTD (25 repeats) was incubated with 20 μl of fraction or 0.2–1 μl of HeLa nuclear extract in a 40-μl final volume of 20 mm Hepes-NaOH, pH 7.9, 8 mm MgCl2, 20 mm creatine phosphate, 5 mm β-glycerol phosphate, 50 mm KCl, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 1 mm ATP, 0.4 μl of Sigma phosphatase inhibitor mixture 1 (kinase buffer) for 15 h at 30 °C. For recombinant DNA-PK (Promega) studies, 50 units were incubated with 0.1 mg of GST-CTD (25 repeats) according to the manufacturer's instructions. For inhibitor studies, wortmannin (Invitrogen) was preincubated with the reaction mix for 10 min at 30 °C prior to the addition of ATP and GST-CTD substrate. Activated calf thymus DNA (Sigma) was added to the reaction mix as indicated. Reactions were stopped by boiling in SDS buffer, and phosphorylation was analyzed by Western blot with phospho-CTD-specific antibodies.
GST-CTD Pulldown
GST-CTD pulldowns were performed as described previously (9).
CTD Peptide Binding Assay
CTD peptides were synthesized (Cambridge Peptides) with one, two, or four repeats of YSPTSPS or YAPTSPA and amino-terminal biotinylation. Ser2/Ser7 phosphorylated peptides were also made. For the Integrator binding assay, 20 μl of streptavidin-coated Sepharose beads was resuspended in 0.1 m KCl HEGN, mixed with 10 μg of biotinylated peptides, and incubated at 4 °C for 1 h. After washing four times with 0.1 m KCl HEGN, 1 ml of the P11 0.5 m KCl fraction (precleared with streptavidin beads for 1 h) was added, and the mixture was incubated at 4 °C for 3 h. After washing the beads five times with 1 ml of 0.3 m KCl HEGN, protein-bound beads were boiled and loaded on SDS-PAGE gels directly.
Western Blot Analysis
Western blotting was carried out as described (22) using antibodies against Int11 (23), CTD-Ser(P)2, Ser(P)5, and Ser(P)7 (8), CDK9 (Santa Cruz Biotechnology, SC484), cyclin T1 (Santa Cruz Biotechnology, SC8128), Int7 (Bethyl Laboratories, DKFZP434B168), CDK7 (Santa Cruz Biotechnology, SC529), and DNA-PKcs (Santa Cruz Biotechnology, SC1552).
RESULTS
Both Ser2 and Ser7 of the CTD Are Required for Binding of the Integrator Complex
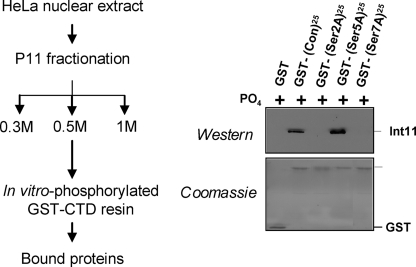
The Integrator complex comprises 12 subunits totaling ∼2 MDa and has been shown to interact with the CTD of pol II in vitro (9, 20). It is associated with U1 and U2 snRNA genes, as shown by chromatin immunoprecipitation, and is required for recognition of the snRNA gene-specific 3′ box (9, 20), which directs 3′ end formation of pre-snRNA (13, 14). The Integrator complex may also play a role in activating transcription of snRNA genes, at a step downstream of pol II recruitment (9). The role of most of the 12 Integrator subunits is unclear, but two subunits, Int9 and Int11, display sequence homology to the CPSF-100 and CPSF-73 subunits of the cleavage and polyadenylation specificity factor, respectively (20, 23). Int11 is therefore thought to carry the endonuclease activity responsible for the first cleavage during the formation of the 3′ end of pre-snRNAs (20). We have previously shown that in vitro binding of Int11 to the CTD requires phosphorylation of the CTD (9). Mutation of Ser7 in the heptapeptide consensus sequence affects the interaction between the CTD and Integrator in vitro and prevents its recruitment on snRNA genes in vivo, suggesting that Ser7 phosphorylation participates in Integrator recruitment to snRNA genes (9). However, the CTD is the site of multiple phosphorylations, with all three serines phosphorylated in vitro (9). To investigate the role of Ser2, Ser5, and Ser7 phosphorylation, we performed glutathione _S_-transferase (GST) pulldown analysis with GST-CTDs carrying 25 consensus (wild type), S2A, S5A, or S7A heptapeptide repeats (Fig. 1). For detection of the catalytic subunit, Int11 was used as a diagnostic for Integrator binding to the CTD. As expected, Integrator interacts strongly with the consensus repeats after phosphorylation of the CTD. Mutation of Ser2 and Ser7 to alanine has a drastic effect on Integrator binding, demonstrating that these residues are critical for this interaction. In contrast, mutation of Ser5 does not reduce the binding of Integrator but rather enhances it, excluding any positive role of Ser5 and Ser5 phosphorylation in this process. Together, these results demonstrate that Integrator binding to the CTD requires a serine in positions 2 and 7 of the CTD repeat and that phosphorylation of Ser2 and/or Ser7, but not Ser5, participates in this interaction.
FIGURE 1.
Both Ser2 and Ser7 of the CTD are required for binding of the Integrator complex. The 0.5 m fraction of P11 fractionation containing the Integrator complex was incubated with in vitro phosphorylated GST-CTD as indicated by the scheme on the left. The panel on the right shows the results of Integrator binding to the wild type CTD (consensus (Con)) and mutant (S2A/S5A/S7A (indicated by Ser2A, Ser5A, and Ser7A)) CTDs assayed by Western blot analysis with antibodies to Int11 (top) and Coomassie Blue staining of the input GST-CTD (bottom).
Ser2 and Ser7 Kinase Activities Can Be Separated in Vitro
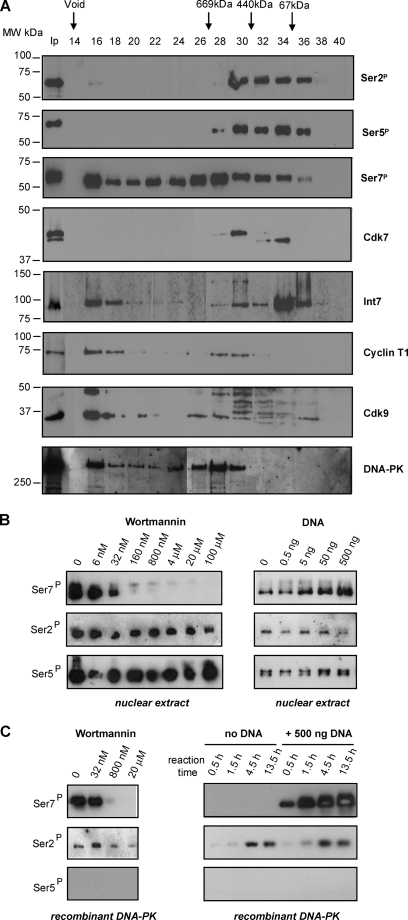
The requirement for CTD phosphorylation and serines at positions 2 and 7 can be explained by three distinct scenarios. Binding of the Integrator complex to the CTD requires: 1) a phospho-serine in position 2 and a serine in position 7; 2) a serine in position 2 and a phospho-serine in position 7; or 3) phospho-serines in positions 2 and 7. To discriminate between these possibilities, we separated Ser2 and Ser7 kinase activities in vitro. HeLa nuclear extract was fractionated on a Superose 6 size exclusion column, and the ability of each fraction to phosphorylate GST-CTD containing 25 consensus repeats in vitro was tested by Western blot with antibodies specific to phospho-Ser2, phospho-Ser5, and phospho-Ser7 (8) (Fig. 2A). The fractions were also tested for the presence of the CTD kinases CDK7 and CDK9, the cyclin T1 partner of CDK9, and the Int7 subunit of the Integrator complex. Interestingly, Ser7 kinase activity, which is present in fractions 16–36 and peaks in fractions 16 and 26–28, can be separated from Ser2 and Ser5 kinase activities, which are present in fractions 28–36, with only a small amount of Ser2 kinase activity in fraction 16. Thus, a high level of Ser7 kinase activity fractionates together with Cdk9, cyclin T1, and the large Integrator complex with an apparent molecular mass of >2 MDa (20). A second peak of Ser7 kinase activity fractionates with an apparent molecular mass of ∼500–700 kDa, whereas Ser2 and Ser5 kinases fractionate between 50 and 500 kDa. As expected, the pattern of Ser5 phosphorylation closely follows the two isoforms of Cdk7. In contrast, the two major isoforms of the Ser2 kinase Cdk9 do not closely correlate with the Ser2 phosphorylation pattern, although the antibody detects some bands between the two isoforms in the fractions active for Ser2 phosphorylation. Similarly, Ser7 phosphorylation does not correlate closely with the presence of either CDK7 or CDK9, as might be expected (7, 10, 11), although some Ser7 kinase activity is present in the fractions where CDK7 and CDK9 are detected. However, Ser7 kinase activity in fractions 16–30 correlates closely with the presence of the 460-kDa catalytic subunit of the DNA-activated kinase DNA-PK, which has CTD kinase activity (24) and has been shown to phosphorylate CTD peptides on Ser7 (25). In addition, the DNA-PK inhibitor, wortmannin (26), effectively and selectively inhibits Ser7 phosphorylation by nuclear extracts (Fig. 2B) and fractions 16, 26, 28, and 30 (data not shown). Furthermore, Ser7 phosphorylation by nuclear extracts (but not Ser2 or Ser5 phosphorylation) is enhanced by ∼4-fold by adding double-stranded DNA, as expected for DNA-PK (27) (Fig. 2B). In addition, recombinant DNA-PK phosphorylates the GST-CTD fusion protein on Ser7, and this activity is inhibited by wortmannin and activated by double-stranded DNA (Fig. 2C). DNA-PK was also previously found to phosphorylate CTD peptides on Ser2 (25). Recombinant DNA-PK also has some Ser2 kinase activity toward the GST-CTD substrate. However, this is neither inhibited by wortmannin nor activated by double-stranded DNA and is significantly lower than Ser7 kinase activity (Fig. 2C). Taken together, these results indicate that under the conditions used, DNA-PK is the major Ser7 kinase in HeLa nuclear extract.
FIGURE 2.
Ser2 and Ser7 kinase activities can be separated in vitro. A, fractions from HeLa nuclear extract fractionation on a Superose 6 column were tested for the ability to phosphorylate the CTD. The results of Western blot analysis using the phospho-specific antibodies (8) to the modifications noted at the right are shown in the top three panels. The results of Western blot analysis of the fractions using antibodies to the proteins noted at the right are shown in the bottom five panels. The fraction numbers and size of molecular weight (MW) standards are shown at the top. Ip indicates input. Ser2P, Ser(P)2; Ser5P, Ser(P)5; Ser7P, Ser(P)7. B, Western blot analysis of GST-CTD phosphorylation by nuclear extract supplemented with wortmannin (left panel) or DNA (right panel) using the phospho-specific antibodies noted at the left. C, Western blot analysis of GST-CTD phosphorylation by recombinant DNA-PK supplemented with wortmannin (left panel) or DNA (as a time course) (right panel) using the phospho-specific antibodies noted at the left.
Phosphorylation of Both Ser2 and Ser7 Is Required for Integrator Binding
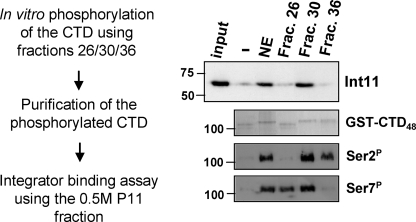
Separation of Ser2 and Ser7 kinase activities by Superose 6 fractionation facilitated the study of Integrator binding to the CTD using fractions containing the Ser7 kinase but not the Ser2 kinase (fraction 26), the Ser2 kinase but not the Ser7 kinase (fraction 36), or both kinase activities (fraction 30). Fractions 26, 30, and 36 were independently used to phosphorylate a GST-CTD containing 48 consensus repeats. As expected, the GST-CTD is differentially modified depending on the fraction used for in vitro phosphorylation (Fig. 3). CTDs were then tested for their ability to interact with Integrator using the binding assay described in Fig. 1. Interestingly, Int11 does not interact with the CTD phosphorylated with fractions 26 and 36, showing that Ser2 and Ser7 phosphorylation cannot independently support efficient Integrator binding (Fig. 3). In contrast, simultaneous phosphorylation of Ser2 and Ser7 results in a strong interaction with Integrator. Taken together, these results demonstrate that Integrator binding to the CTD requires phosphorylation of both Ser2 and Ser7, raising the possibility that together, these modifications create a new CTD mark specifically recognized by the Integrator complex.
FIGURE 3.
Phosphorylation of both Ser2 and Ser7 is required for Integrator binding. Fractions (Frac.) containing Ser2 kinase activity, Ser7 kinase activity, or both were used to phosphorylate GST-CTD and tested for their ability to bind the Integrator complex, as indicated in the scheme on the left. The results of Western blot analysis using anti-Int11 antibodies (23) (top), Coomassie Blue staining of the input GST-CTD (middle), and Western blot analysis using antibodies to phospho-Ser2 (Ser2P) and phospho-Ser7 (Ser7P) (bottom) are shown on the right. NE = nuclear extract.
A Specific Mark on the CTD Directs the Binding of Integrator
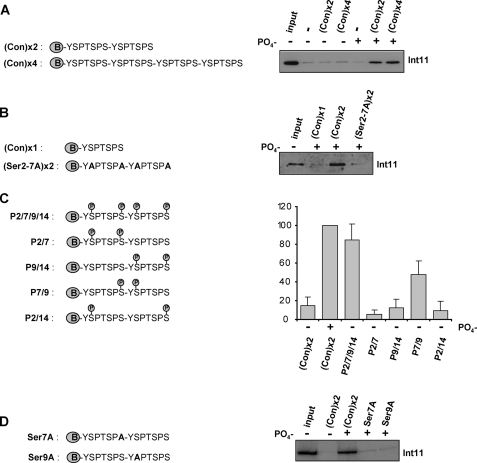
As a final step, we designed a set of N-terminally biotinylated peptides to further characterize the Integrator binding motif on the CTD (Fig. 4). First, we determined the minimal number of CTD repeats required for efficient Int11 binding. CTD peptides consisting of 2–4 heptapeptide repeats were tested for their ability to interact with Integrator before or after in vitro phosphorylation (Fig. 4A). As expected, non-phosphorylated peptides were unable to bind Integrator. In contrast, Int11 interacts with in vitro phosphorylated CTD peptides comprising two and four repeats. This result demonstrates that two repeats are sufficient for Integrator binding and is in accordance with the previous demonstration in yeast that the basic unit of the CTD comprises two repeats (28). Importantly, an in vitro phosphorylated CTD peptide containing only one repeat failed to interact with Int11, as did an in vitro phosphorylated peptide carrying two repeats with alanine instead of serine at positions 2 and 7 (Fig. 4B). These controls demonstrate the specificity of the binding assay and confirm the requirement for Ser2 and Ser7 for binding. To more precisely characterize the motif required for Integrator binding, five CTD peptides consisting of two repeats with different combinations of phospho-Ser2/phospho-Ser7 were tested for their ability to bind Integrator (Fig. 4C). The peptide phosphorylated on both Ser2 and Ser7 residues (phosphates in positions 2, 7, 9, and 14) binds Integrator almost as well as the in vitro phosphorylated CTD peptide, suggesting that this peptide contains all the modifications required for Integrator binding. Although the peptides with phosphates in positions 2/7, 9/14, or 2/14 do not appreciably interact with Integrator, the peptide with phosphates in positions 7/9 interacts with Integrator, although less well than the double repeat with both Ser2 and Ser7 residues phosphorylated. These results demonstrate that the minimal mark required for Integrator binding is a phospho-Ser7 on the first CTD repeat followed by a phospho-Ser2 on the second CTD repeat. None of the other combinations of two phosphates are sufficient for Integrator binding. In addition, Integrator binding to peptides phosphorylated by incubation with nuclear extract is lost when either Ser7 in the first repeat or Ser2 in the second repeat is replaced by alanine, S7A and S9A, respectively (Fig. 4D), demonstrating the specificity of this new CTD motif. However, phosphorylation of an additional Ser2, Ser7, or both enhances binding.
FIGURE 4.
A specific double mark on the CTD directs the binding of Integrator. A–D, the structures of the biotinylated peptides used are shown on the left. The results of Western blot analysis using anti-Int11 (23) of proteins pulled down by the peptides noted above the lanes are shown on the right. In panel C, quantitation is shown underneath with error bars indicating the S.D. from four independent experiments. Con, consensus.
DISCUSSION
The RNA pol II CTD plays the role of scaffold for the recruitment of numerous factors involved in regulation of transcription and co-transcriptional RNA processing (3). The recent discovery of a new phosphorylation mark on serine 7 of the YSPTSPS CTD heptapeptide (3, 8) extends the repertoire of CTD modifications and raises the possibility of the existence of new regulatory phosphorylation combinations. The requirement for a double mark has only previously been demonstrated for factors that require phosphorylation of both Ser2 and Ser5, for example, Set2 (3, 6). The double mark comprising Ser7 phosphorylation on the first repeat of a CTD heptapeptide pair, followed by Ser2 phosphorylation on the second, is therefore the first example involving Ser7 phosphorylation and the first where different marks on two repeats are required for protein binding. Thus, this discovery emphasizes the increasing complexity of CTD modifications during RNA pol II transcription. Phosphorylation of both Ser2 and Ser7 occurs on snRNA genes (9, 22, 29). Although the currently available antibodies specific for phosphorylation on each of the three serines are very useful in studying transcription, they are unable to give precise information about the juxtaposition of modifications on individual heptads (or pairs). It would therefore be of great interest to raise an antibody against the Ser7/Ser2-specific mark and test its presence on snRNA genes by chromatin immunoprecipitation. Interestingly, phosphorylation of Ser2 and Ser7 is also detected at the 3′ end of human protein-coding genes (8, 29), but the Integrator complex is not. It is therefore possible that within the CTD, the location of phosphorylation of Ser2 and Ser7 relative to each other differs from snRNA to protein-coding genes or that this mark works in synergy with promoter-specific factors/elements to specifically recruit Integrator to snRNA genes but not protein-coding genes. In addition, Ser5 and its phosphorylation appear to have a negative impact on the recognition of the double mark by Integrator, indicating that some phosphorylation marks interfere with CTD-protein interaction, adding a further layer of complexity to the CTD code.
Surprisingly, our study also indicates that the majority of the in vitro Ser7 kinase activity in HeLa nuclear extract is attributable to the large kinase, DNA-PK. So far, no role for DNA-PK as a CTD Ser7 kinase in vivo has been demonstrated, and further investigation of this possibility is warranted. Nevertheless, the Ser7-specific activity obtained by fractionation of nuclear extract and recombinant DNA-PK will provide useful tools to selectively phosphorylate CTD heptapeptides in GST fusion proteins on Ser7, which would, for example, facilitate further studies of the role of this modification in recruitment of proteins to the CTD.
Acknowledgments
We thank Zbig Dominski for the Int11 antibody and Dirk Eick for the phospho-Ser2, phospho-Ser5, and phospho-Ser7 antibodies.
*
This work was supported by a grant from the Medical Research Council (UK) and Wellcome Trust grants (to S. M.).
5
The abbreviations used are:
CTD
carboxyl-terminal domain
pol II
RNA polymerase II
snRNA
small nuclear RNA
DNA-PK
DNA-dependent protein kinase
GST
glutathione _S_-transferase.
REFERENCES
- 1.Hirose Y., Ohkuma Y. (2007) J. Biochem. 141, 601–608 [DOI] [PubMed] [Google Scholar]
- 2.Chapman R. D., Heidemann M., Hintermair C., Eick D. (2008) Trends Genet. 24, 289–296 [DOI] [PubMed] [Google Scholar]
- 3.Egloff S., Murphy S. (2008) Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S. (2003) Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 5.Corden J. L. (2007) Science 318, 1735–1736 [DOI] [PubMed] [Google Scholar]
- 6.Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005) Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 9.Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007) Science 318, 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R. P., Bentley D. L. (2009) Mol. Cell. Biol. 29, 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M., Suh H., Cho E. J., Buratowski S. (2009) J. Biol. Chem. 284, 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez N. (2001) J. Biol. Chem. 276, 26733–26736 [DOI] [PubMed] [Google Scholar]
- 13.Egloff S., O'Reilly D., Murphy S. (2008) Biochem. Soc. Trans. 36, 590–594 [DOI] [PubMed] [Google Scholar]
- 14.Hernandez N. (1985) EMBO J. 4, 1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egloff S., Murphy S. (2008) Biochem. Soc. Trans. 36, 537–539 [DOI] [PubMed] [Google Scholar]
- 16.Jacobs E. Y., Ogiwara I., Weiner A. M. (2004) Mol. Cell. Biol. 24, 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S. D., Wickens M., Bentley D. L. (1997) Nature 385, 357–361 [DOI] [PubMed] [Google Scholar]
- 18.Medlin J. E., Uguen P., Taylor A., Bentley D. L., Murphy S. (2003) EMBO J. 22, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 20.Baillat D., Hakimi M. A., Näär A. M., Shilatifard A., Cooch N., Shiekhattar R. (2005) Cell 123, 265–276 [DOI] [PubMed] [Google Scholar]
- 21.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medlin J., Scurry A., Taylor A., Zhang F., Peterlin B. M., Murphy S. (2005) EMBO J. 24, 4154–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominski Z., Yang X. C., Purdy M., Wagner E. J., Marzluff W. F. (2005) Mol. Cell. Biol. 25, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvir A., Peterson S. R., Knuth M. W., Lu H., Dynan W. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11920–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trigon S., Serizawa H., Conaway J. W., Conaway R. C., Jackson S. P., Morange M. (1998) J. Biol. Chem. 273, 6769–6775 [DOI] [PubMed] [Google Scholar]
- 26.Izzard R. A., Jackson S. P., Smith G. C. (1999) Cancer Res. 59, 2581–2586 [PubMed] [Google Scholar]
- 27.Lees-Miller S. P., Chen Y. R., Anderson C. W. (1990) Mol. Cell. Biol. 10, 6472–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiller J. W., Cook M. S. (2004) Eukaryot Cell 3, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egloff S., Al-Rawaf H., O'Reilly D., Murphy S. (2009) Mol. Cell. Biol. 29, 4002–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]