NF449 Is a Novel Inhibitor of Fibroblast Growth Factor Receptor 3 (FGFR3) Signaling Active in Chondrocytes and Multiple Myeloma Cells (original) (raw)
Abstract
The FGFR3 receptor tyrosine kinase represents an attractive target for therapy due to its role in several human disorders, including skeletal dysplasias, multiple myeloma, and cervical and bladder carcinomas. By using molecular library screening, we identified a compound named NF449 with inhibitory activity toward FGFR3 signaling. In cultured chondrocytes and murine limb organ culture, NF449 rescued FGFR3-mediated extracellular matrix loss and growth inhibition, which represent two major cellular phenotypes of aberrant FGFR3 signaling in cartilage. Similarly, NF449 antagonized FGFR3 action in the multiple myeloma cell lines OPM2 and KMS11, as evidenced by NF449-mediated reversal of ERK MAPK activation and transcript accumulation of CCL3 and CCL4 chemokines, both of which are induced by FGFR3 activation. In cell-free kinase assays, NF449 inhibited the kinase activity of both wild type and a disease-associated FGFR3 mutant (K650E) in a fashion that appeared non-competitive with ATP. Our data identify NF449 as a novel antagonist of FGFR3 signaling, useful for FGFR3 inhibition alone or in combination with inhibitors that target the ATP binding site.
Keywords: Enzyme Inhibitors, Growth Factors, MAP Kinases (MAPKs), Receptor Tyrosine Kinase, Signal Transduction, FGFR3, NF449, Chondrocyte, Fibroblast Growth Factor Receptor
Introduction
FGFR3 (fibroblast growth factor receptor 3) is a transmembrane tyrosine kinase that serves as a receptor for the members of the fibroblast growth factor (FGF)2 family and functions in many biological processes, including cell proliferation, differentiation, migration, and survival. To date, activating mutations in FGFR3 have been associated with several human disorders, such as skeletal dysplasias, multiple myeloma, and cervical and bladder carcinomas (1–4).
Among the skeletal dysplasias, activating FGFR3 mutations cause achondroplasia, the most common form of human skeletal dysplasia, and thanatophoric dysplasia, the most common form of lethal skeletal dysplasia (1). Long bones of individuals suffering from FGFR3-related skeletal dysplasias show markedly shortened zones of chondrocyte proliferation and differentiation, whereas Fgfr3 knock-out mice and humans without functional FGFR3 demonstrate skeletal overgrowth, together implying the role of FGFR3 as a negative regulator of bone growth (5).
Apart from cartilage, ∼15% of patients suffering from multiple myeloma markedly up-regulate FGFR3 as a consequence of a t(4;14)(p16.3;q32) translocation, with a fraction of patients also harboring activating mutations in FGFR3, identical to those found in the skeletal dysplasias (2, 3). Ectopic expression of FGFR3 enhances multiple myeloma cell proliferation and survival, demonstrating the oncogenic potential of FGFR3 (6).
Given its role in human disease, FGFR3 signaling represents an attractive target for therapy. There is no treatment available for achondroplasia at present, thus inciting the development of novel approaches to target FGFR3. The aim of this study was to identify novel inhibitors of FGFR3 signaling in cellular environments relevant to FGFR3-related disorders. We recently established a chondrocyte cell-based reporter assay suitable for identification of inhibitors of FGFR3 signaling via molecular library screening (7). Here, we used this assay to identify and characterize the compound named NF449 as a novel antagonist of FGFR3 signaling active in both chondrocytes and multiple myeloma cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Growth Assays
Rat chondrosarcoma (RCS) chondrocytes, Chinese hamster ovary (CHO) cells, and multiple myeloma cell lines OPM2 and KMS11 were propagated in Dulbecco's modified Eagle's medium, Opti-MEM, or RPMI medium (Invitrogen), supplemented with 10% fetal bovine serum (Atlanta Biological, Nordcross, GA) and antibiotics. The RCS cell growth arrest experiments utilizing crystal violet staining to quantify cell growth are described in detail elsewhere (7). For the RCS growth arrest experiments here, ∼1 × 104 cells were seeded in 24-well tissue culture plates (Costar, Cambridge, MA), treated as indicated for 72 h, and counted. FGF2 was obtained from R&D Systems (Minneapolis, MN); C-natriuretic peptide, SU5402, suramin, NF023, and NF449 were from Calbiochem; NF007, NF110, NF157, NF279, ATP, MethyleneATP, and BzATP were from Tocris Bioscience (Ellisville, MO).
Bone Explant Culture
For explant culture, tibiae were harvested from wild type mouse embryos at embryonic day 16.5. Tibiae were dissected out under the microscope, placed in non-treated polystyrene 96-well plates, and cultured in α-minimal essential medium (Invitrogen), supplemented with 0.2% bovine serum albumin, 1 mm β-glycerophosphate, and 50 μg/ml ascorbic acid. Tibiae were further treated with FGF2 in the presence or absence of NF449 for 6 days, fixed in 10% formaldehyde in phosphate-buffered saline, demineralized with 0.5 m EDTA, and embedded in paraffin. Sections were stained with hematoxylin, eosin, and Alcian blue. Images were taken with a Leica microscope and Leica suite software (Leica, Bannockburn, IL). The length of the proximal epiphysis and bone marrow space was measured using PhotoFiltre image editing software.
Alcian Blue Staining and [35S]Sulfate Labeling Assays
For Alcian blue staining, growing RCS cultures were treated with FGF2 (5 ng/ml) alone or in the presence of NF449 (20 μm) for 72 h, fixed with 4% paraformaldehyde, and stained for 30 min with Alcian blue. For quantification of the FGF2-mediated extracellular matrix loss, RCS chondrocytes were treated with FGF2 (5 ng/ml) and NF449 (20 μm) for 72 h in the presence of 10 μCi/ml [35S]sulfate (PerkinElmer Life Sciences). Following the cultivation period, cells were harvested, and the incorporated radioactivity was determined by liquid scintillation.
Preparation of Cell Extracts, Western Blotting (WB), Immunoprecipitation, Signaling, and Kinase Studies
Cells were lysed in ice-cold immunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA, 25 mm NaF, 10 mm Na3VO4) supplemented with proteinase inhibitors. Protein samples were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The following antibodies were used: ERK1/2, phospho-ERK1/2(Thr202/Tyr204), phospholipase Cγ1 (PLCγ1), phospho-PLCγ1(Tyr783), MEK1/2, phospho-MEK1/2(Ser217/221), STAT1, phospho-STAT1(Tyr701), caveolin 1, lamin A/C, ID2, FMS, PDGFRA, AXL, IGF1R, FLT3, and c-Kit (Cell Signaling, Beverly, MA); actin, FGFR1, FGFR2, FGFR3, FGFR4, and TRKA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); and 4G10 (Upstate Biotechnology, Inc., Lake Placid, NY). WB signal was quantified by determining the integrated optical density of a given band using Scion Image software (Scion Corp., Frederick, MA). For cell signaling studies, cells were either treated with NF449 for 30 min to 1 h prior to the FGF2 addition (OPM2 and KMS11) or pretreated with NF449 for 12–24 h before the FGF2 addition (RCS). Heparin (1 μg/ml; Invitrogen) was used together with FGF2 for RCS chondrocyte treatment, and the cell cartilaginous matrix was removed by collagenase (type II; Invitrogen) treatment before the addition of NF449. Immunoprecipitations and FGFR3 kinase assays were performed as described (8). Briefly, the pRK7 vector encoding C-terminally FLAG-tagged K650E-FGFR3 was transfected into CHO cells using FuGENE6 reagent according to the manufacturer's protocol (Roche Applied Science). 48 h after transfection, cells were treated with 20 ng/ml FGF2 for 15 min, and K650E-FGFR3 was immunoprecipitated from ∼800 μg of cell lysate protein using 4 μg of anti-FLAG antibody (Sigma). Cells transfected with a plasmid encoding green fluorescent protein (pCCEY) served as a negative control for the immunoprecipitation. Immunocomplexes were washed two times with kinase buffer (60 mm HEPES-NaOH, pH 7.5, 3 mm MgCl2, 3 mm MnCl2), and the kinase reactions were performed for 30 min at 30 °C in the presence of 2.5 μg of polyethylene glycol, 10 μm ATP, 200–300 ng of recombinant STAT1 (Enzo Life Sciences, Plymouth, PA) or Prolias (Rockville, MD) as a substrate, and inhibitors were added directly to the kinase reaction. Kinase assays utilizing the recombinant tyrosine kinase (TK) domains of FGFR1 to -4, FMS, PDGFRA, TRKA, AXL, IGF1R, FLT3, epidermal growth factor receptor, c-Kit, and DDR2 (SignalChem, Richmond, Canada) were carried out similarly, with 200–300 ng of kinase used in a 50-μl reaction, and inhibitors were added directly to the kinase reaction. Kinase-mediated phosphorylation of STAT1 was detected by WB with phospho-STAT1(Tyr701) antibody.
Real-time Reverse Transcription-PCR
Total RNA was isolated using the RNeasy minikit (Qiagen, Valencia, CA), and poly(dT)-primed cDNA was synthesized from 3 μg of RNA using the Omniscript RT kit (Qiagene). Real-time reverse transcription-PCRs were carried out as described in detail elsewhere (9), using Dynamo SYBR Green qPCR chemistry (Finnzymes, Espoo, Finland). The PCR primers were the following (5′ to 3′, followed by product size): CCL3, CACATTCCGTCACCTGCTCAG and TGGCTGCTCGTCTCAAAGTAGTC, 192 bp; CCL4, TCTCAGCACCAATGGGCTCAG and CAGTTCAGTTCCAGGTCATACACG, 214 bp; actin, GATTCCTATGTGGGCGACGAG and ATGGCTGGGGTGTTGAAGGTCTC, 245 bp. The results are expressed as -fold difference relative to control (2_ddCt_).
RESULTS
NF449 Inhibits FGFR3 Signaling in Chondrocytes
The RCS is an FGFR3-expressing chondrocytic cell line that represents a well characterized in vitro cellular model for FGFR3-related skeletal dysplasias (10). RCS chondrocytes respond to FGFR3 activation (via exogenous FGF2 addition) with potent growth arrest, loss of the cartilage-like extracellular matrix, and marked alteration of their cellular shape (9, 11, 12). We have taken advantage of this response and adapted the FGF2/FGFR3-mediated growth arrest for rapid screening of chemical compounds for their activity against FGFR3 signaling (7).
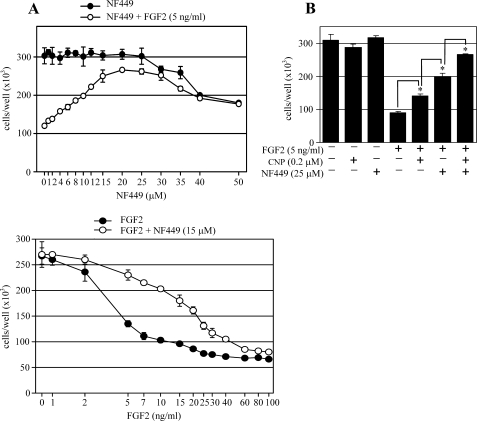
We used the RCS growth arrest assay to screen a molecular library consisting of 1120 molecules (Tocris Bioscience). Initial screenings performed at 5, 10, and 20 μm scales identified a compound named NF449 as a potent antagonist of the FGF2/FGFR3-inhibitory effect on RCS proliferation.3 This effect was confirmed in detailed cell growth experiments, where 20 μm NF449 caused significant rescue of the growth arrest phenotype without apparent cellular toxicity throughout its active concentration range (∼6–30 μm; Fig. 1A). This activity of NF449 is remarkable, considering the robust nature of the RCS growth arrest phenotype (7).
FIGURE 1.
NF449 inhibits FGF2-mediated growth arrest in chondrocytes. A, RCS chondrocytes were treated with 5 ng/ml FGF2 and various concentrations of NF449 (upper graph) or with 15 μm NF449 and various FGF2 concentrations (lower graph; samples containing no FGF2 were given an artificial value of 0. 8 on the logarithmic x axis) for 72 h and counted. Data represent an average from four wells with the indicated S.D. (error bars). Note the potent growth arrest induced by FGF2 as well as the NF449-mediated reversal of this phenotype. Also note the lack of significant NF449 toxicity throughout its active range (8–30 μm; upper graph). B, RCS chondrocytes were treated as indicated, grown for 72 h, and counted. Note the cumulative effect of both C-natriuretic peptide (CNP) and NF449 on the FGF2-mediated growth arrest. Data represent an average from four wells with the indicated S.D. Statistical differences are indicated (Student's t test; *, p < 0.01).
We next compared the NF449 effect with that of C-natriuretic peptide, which is a recently discovered inhibitor of FGFR3 signaling (12, 13). Fig. 1B shows that C-natriuretic peptide caused a ∼20% rescue of the FGF2-mediated RCS growth arrest, consistent with our previous data (12). This effect was significantly exceeded by NF449, which caused a ∼50% rescue of the growth arrest phenotype. When used together, C-natriuretic peptide and NF449 acted synergistically, rescuing nearly 80% of the growth arrest phenotype.
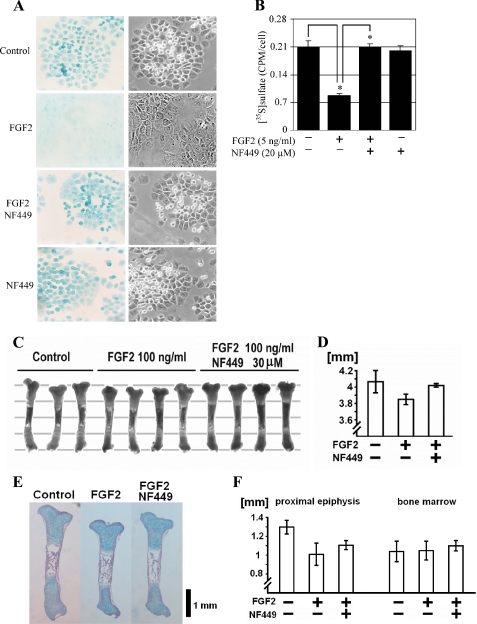
Apart from the growth inhibition, aberrant FGFR3 signaling in cartilage results in the loss of the extracellular matrix, rich in sulfated proteoglycans (12). Growing RCS cultures produce abundant amounts of cartilage-like sulfated proteoglycan matrix, as demonstrated by Alcian blue staining (Fig. 2A). This staining is lost after the 72 h of FGF2 treatment as a result of both inhibition of matrix production and induction of matrix metalloproteinase-mediated degradation (12). In cells treated with FGF2 together with NF449, the intensity of Alcian blue staining equaled that of the control cells, suggesting that NF449 rescued the FGF2-mediated loss of the extracellular matrix (Fig. 2A). To further confirm this observation, we incubated RCS cells with [35S]sulfate for 72 h to label the extracellular matrix and determined the effect of FGF2 and NF449 on the cell-bound radioactivity. [35S]sulfate incorporation was significantly inhibited by FGF2, and this effect was completely rescued by NF449 (Fig. 2B). Altogether, these data demonstrate that NF449 rescues the FGFR3-mediated effects on both chondrocyte proliferation and extracellular matrix loss in vitro.
FIGURE 2.
NF449 inhibits FGF2-mediated extracellular matrix loss in chondrocytes and inhibition of growth in murine bone explant cultures. A, RCS chondrocytes were treated with FGF2 (5 ng/ml) alone or in the presence of NF449 (20 μm) for 72 h, and the extracellular matrix was visualized by Alcian blue staining (left panel; magnification, ×200). The right panel shows the corresponding photograph taken at bright field with contrast. Note the FGF2-mediated loss of Alcian blue staining that was prevented by NF449. B, RCS chondrocytes were treated as indicated in the presence of [35S]sulfate for 72 h, and the amount of incorporated radioactivity was determined as described under “Experimental Procedures.” Note the FGF2-mediated loss of sulfated proteoglycans, which was rescued by NF449. Data are an average from four wells with the indicated S.D. (error bars). Statistically significant differences are indicated (Student's t test; *, p < 0.01). Results are representative of three experiments. C–F, NF449 counteracts the growth-inhibitory effects of FGF2 in cultured tibiae. Tibiae were harvested from embryonic day 16.5 murine embryos, treated with 100 ng/ml FGF2 in the presence or absence of 30 μm NF449, cultured for 6 days, and measured (C). Parallel lines in the background are at 1-mm increments. D, length of the tibiae after 6 days in culture. The results are representative of three experiments (supplemental Fig. S1). E, tibia sections stained with hematoxylin, eosin, and Alcian blue. Note the inhibitory FGF2 effect on tibia growth that is rescued by NF449 via increasing the length of both growth plate cartilage and bone elements (F).
We next determined the effect of NF449 on FGFR3-mediated inhibition of cartilage development in bone organ culture system employing limb rudiments isolated from developing murine embryos. Such rudiments maintain their growth in culture for several days, which can be attenuated by continuous stimulation of endogenous FGFR3 via the addition of the FGF ligand, thus exploiting the role of FGFR3 as a negative regulator of cartilage growth (5, 14). Here, the addition of exogenous FGF2 inhibited growth of tibiae isolated from embryonic day 16.5 wild type embryos. When used together with FGF2, NF449 rescued the FGF2-mediated growth inhibition in all three experiments carried out (Figs. 2, C and D, and supplemental Fig. S1). Tibia histology showed that NF449 rescued the FGF2 effect by increasing the length of both growth plate cartilage and bone elements (Fig. 2, E and F).
NF449 Inhibits FGFR3 Signaling in Multiple Myeloma Cell Lines OPM2 and KMS11
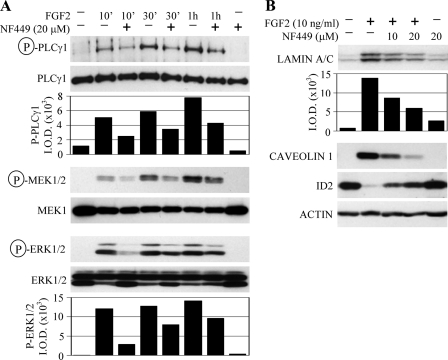
Because multiple myeloma represents another important area of pathological FGFR3 signaling, we asked whether NF449 inhibits FGFR3 signaling in a multiple myeloma cellular environment. OPM2 and KMS11 represent multiple myeloma-derived cells that up-regulate FGFR3 carrying the strongly activating mutations K650E or Y373C, respectively (15). As a surrogate for FGFR3 activation, we used the activatory, Thr202/Tyr204 phosphorylation of ERK MAPK, which is induced by the addition of exogenous FGF2 to the OPM2 or KMS11 cells (16). Fig. 3A demonstrates that FGF2 treatment led to ERK phosphorylation, which was almost completely blocked by NF449. Next, we analyzed the effect of NF449 on transcription of the genes induced by FGFR3 signaling in multiple myeloma cells. Because the CCL3 (MIP-1α) and CCL4 (MIP-1β) chemokines were previously identified as transcriptional targets of FGFR3 signaling in multiple myeloma (17),3 we determined the effect of NF449 on the FGF2-mediated accumulation of the CCL3 and CCL4 transcripts. Real-time reverse transcription-PCR analysis showed the partial NF449-mediated rescue of the CCL3 and CCL4 accumulation induced by FGF2 treatment in the OPM2 cells (Fig. 3B).
FIGURE 3.
NF449 inhibits FGFR3 signaling in the multiple myeloma cell lines KMS11 and OPM2. A, KMS11 and OPM2 cells were serum-starved for 12 h and treated with NF449 for 1 h before treatment with FGF2, and the levels of ERK MAPK phosphorylation were determined by WB with the phospho-ERK1/2(Thr202/Tyr204) antibody. As a loading control, the membranes were reprobed with antibody recognizing ERK regardless of its phosphorylation. Note the FGF2-mediated ERK phosphorylation, which is inhibited by NF449 in both KMS11 and OPM2 cells. B, OPM2 cells were serum-starved for 12 h, pretreated with NF449 for 1 h before FGF2 treatment, and analyzed for CCL3 and CCL4 expression by real-time RT-PCR, as described under “Experimental Procedures.” The results are expressed as the -fold difference relative to untreated cells (2_ddCt_) (9). Note the FGF2-mediated induction of CCL3 and CCL4 that is inhibited by NF449. The data represent two independent samples, each run as a technical duplicate, with the indicated range (error bars). The results are representative of three experiments.
NF449 Inhibits Tyrosine Kinase Activity of FGFR3
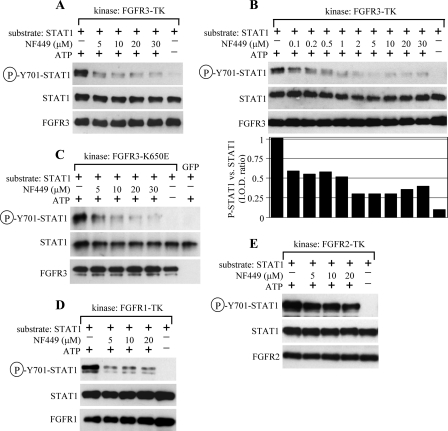
To gain better insight into the mechanism of NF449-mediated inhibition of FGFR3 signaling, we focused on its effect on the mediators of downstream FGFR3 signaling as well as other molecular markers characteristic of FGFR3-mediated RCS chondrocyte growth arrest. Fig. 4A shows that FGF2-mediated activation of PLCγ as well as two kinases belonging to the Ras-ERK MAPK pathway (i.e. MEK and ERK) was sensitive to NF449. Furthermore, we analyzed the effect of NF449 on FGF2-mediated modulation of the senescence markers in RCS chondrocytes. The induction of caveolin 1 and lamin A/C and suppression of ID2 expression, which is triggered by 24 h of FGF2 treatment, was rescued by NF449 (Fig. 4B).
FIGURE 4.
NF449 inhibits FGF2-mediated activation of intracellular mediators of FGFR3 signaling. A, RCS chondrocytes were pretreated with NF449 for 24 h before treatment with FGF2 (20 ng/ml for PLCγ and MEK experiment; 2 ng/ml for ERK experiment) and heparin (1 μg/ml), and the levels of activatory PLCγ, MEK, and ERK phosphorylation were determined by WB. As a loading control, the membranes were reprobed with antibodies recognizing PLCγ, MEK, or ERK regardless of their phosphorylation. Phospho-PLCγ and phospho-ERK signals were quantified by densitometry and graphed. Note the FGF2-mediated PLCγ, MEK, and ERK phosphorylation, which is partially rescued by NF449. B, RCS chondrocytes were treated as indicated for 24 h, and amounts of lamin A/C, caveolin 1, and ID2 were determined by WB. Lamin A/C signal was quantified by densitometry and graphed. Actin served as a loading control. Note the FGF2-mediated modulation of quantities of all three proteins that was rescued by NF449.
Because NF449 was originally designed as an inhibitor of P2X adenine receptors (29), we next asked whether P2X receptors are involved in growth-inhibitory FGFR3 signaling. Treatment of RCS chondrocytes with three different P2X agonists (i.e. ATP, MethyleneATP, and BzATP (1, 10, and 50 μm scale)) failed to inhibit RCS proliferation or sensitize cells to FGF2 in a growth arrest assay (data not shown), suggesting that NF449 action might be independent of P2X receptors. To test whether NF449 inhibits the kinase activity of FGFR3, we used a cell-free FGFR3 kinase assay that employs a recombinant TK domain of FGFR3 as a kinase and STAT1 as a substrate (18). In the initial experiment, we used a 5–30 μm NF449 concentration range and found that FGFR3-mediated tyrosine phosphorylation of STAT1 was inhibited at all concentrations. In a broader 0.1–30 μm range, NF449 caused some inhibition of FGFR3 kinase activity at all concentrations, although ≥2 μm NF449 was necessary to achieve more than 50% inhibition (Fig. 5, A and B).
FIGURE 5.
NF449 inhibits FGFR3 kinase activity in a cell-free kinase assay. A and B, cell-free kinase assays were carried out as described under “Experimental Procedures,” with the recombinant TK domain of FGFR3 as a kinase, recombinant STAT1 as a substrate, and NF449 added directly to the kinase reaction. FGFR3-mediated phosphorylation of STAT1 was detected by WB with phospho-STAT1(Tyr701) antibody. The membrane was reprobed with FGFR3 and STAT1 antibodies to control for kinase and substrate quantity. The sample with ATP omitted serves as a negative control for the kinase reaction. Note the FGFR3-mediated phosphorylation of STAT1 that is inhibited by NF449. The phospho-STAT1 and STAT1 signal was quantified by densitometry and expressed as a ratio between P-STAT1 and STAT1 signal for each given sample (B, lower graph). C, the cell-free FGFR3 kinase assay was carried out using a full-length FLAG-tagged K650E-FGFR3 mutant that was expressed in CHO cells, purified by immunoprecipitation with FLAG antibody, and subjected to a kinase reaction with recombinant STAT1 as a substrate and NF449 added to the kinase reaction. Cells transfected with green fluorescent protein-encoding plasmid serve as a negative control for the immunoprecipitation. D and E, cell-free kinase assays were carried out as described in A, except that the recombinant TK domains of FGFR1 (D) or FGFR2 (E) were used as a kinase. Note the poor NF449 inhibition of FGFR2 kinase activity when compared with both FGFR1 and FGFR3.
The K650E substitution is one of the most strongly activating FGFR3 mutations known to date, present in both multiple myeloma and thanatophoric dysplasia (1–3). Because the OPM2 cells express K650E-FGFR3 and their FGFR3 signaling is inhibited by NF449 (Fig. 3), we used the cell-free kinase assay to determine whether NF449 inhibits the kinase activity of K650E-FGFR3. As described in detail elsewhere (8), FLAG-tagged full-length K650E-FGFR3 was expressed in CHO cells for 48 h, activated by brief FGF2 treatment, and purified by immunoprecipitation. Immunocomplexes were subjected to a kinase assay with STAT1 as a substrate and NF449 added directly to the kinase reaction. Fig. 5C shows that NF449 inhibits K650E-FGFR3 kinase activity with an efficiency similar to TK-FGFR3 (Fig. 5A).
We next determined the activity of NF449 against other receptor tyrosine kinases. NF449 inhibited FGFR1 kinase activity but showed very weak activity against FGFR2, demonstrating some selectivity of NF449 action within the FGFR family of kinases (Fig. 5, D and E). We were unable to evaluate NF449 activity against FGFR4 because the recombinant FGFR4 used here showed no activity in a kinase assay (data not shown). We next probed a panel of nine other receptor tyrosine kinases for their phosphorylation of STAT1 in a kinase assay and measured sensitivity of this phosphorylation to NF449. Recombinant FMS, PDGFRA, TRKA, AXL, IGF1R, FLT3, epidermal growth factor receptor, and c-Kit induced strong STAT1(Tyr701) phosphorylation in a kinase assay, which was very weakly inhibited by NF449 in the case of TRKA, AXL, and FLT3. In contrast, IGF1R-mediated phosphorylation of STAT1 was strongly inhibited by NF449, comparable with FGFR3 (supplemental Fig. S2_A_). As recombinant DDR2 used here did not phosphorylate STAT1 in a kinase assay, we employed DDR2 autophosphorylation as a reporter of its kinase activity. Supplemental Fig. S2_B_ shows that NF449 did not inhibit DDR2 autophosphorylation in contrast to FGFR3 autophosphorylation, which was inhibited by NF449 similarly to FGFR3-mediated phosphorylation of STAT1 (Fig. 5, A and B, and supplemental Fig. S2_A_).
NF449 Cooperates with SU5402 in FGFR3 Inhibition
To test whether NF449 targets the ATP binding site on FGFR3, we performed cell-free FGFR3 kinase assays with a range of ATP concentrations (1–50 μm) in the presence of 20 μm NF449. The amounts of STAT1(Tyr701) phosphorylation were detected by WB, quantified by densitometry, and graphed. In the presence of NF449, increasing amounts of ATP did not elevate the rate of the kinase reaction to the control levels (no NF449 in the reaction) (Fig. 6A), suggesting that NF449 inhibits FGFR3 non-competitively with ATP. This opened the possibility that NF449 could act synergistically with the well established FGFR inhibitors that target the ATP binding site, such as SU5402 (19). Indeed, NF449 and SU5402 had additive effects in the FGFR3 kinase assay (Fig. 6B). Similar data were achieved in the RCS growth arrest assay, where NF449 and SU5402 together caused a significantly greater rescue of the FGF2-mediated growth arrest compared with each inhibitor alone (Fig. 6C).
FIGURE 6.
NF449 and SU5402 act synergistically to inhibit FGFR3. A, cell-free kinase assays were carried out as described under “Experimental Procedures,” with recombinant TK domain of FGFR3 as a kinase, recombinant STAT1 as a substrate, and NF449 added directly to the kinase reaction. FGFR3-mediated phosphorylation of STAT1 was detected by WB with phospho-STAT1(Tyr701) antibody (not shown), quantified by densitometry and graphed. The lower rate of the kinase reaction in the presence of NF449, apparent at all used ATP concentrations, suggests that NF449 does not compete with ATP for FGFR3 binding. The results are representative of five experiments. B, the cell-free FGFR3 kinase assay was carried out as described in A with NF449 and/or SU5402 (19) added directly to the kinase reaction. FGFR3-mediated phosphorylation of STAT1 was detected by WB with phospho-STAT1(Tyr701) antibody, quantified by densitometry, and graphed. The background of the P-STAT1 blot was uniformly subtracted before quantification using Scion Image densitometric software. C, RCS chondrocytes were treated as indicated, grown for 72 h, and counted. The data represent an average from two wells with the indicated range (error bars). Results are representative of three experiments. Note the additive effect of NF449 and SU5402 on FGFR3 inhibition evident in both the kinase assay (B) and growth arrest assay (C).
Suramin Analogs Inhibit FGFR3 Kinase Activity
Finally, we evaluated other compounds that share structural similarity to NF449 (i.e. suramin and its analogs NF007, NF023, NF110, NF157, and NF279) (Fig. 7) for their activity against FGFR3. In the RCS growth arrest assay, suramin, NF157, and NF279 rescued the FGF2-mediated inhibition of cell proliferation in a manner similar to NF449, whereas NF007, NF023, and NF110 had no effect (Fig. 8A). Similar data were obtained in a kinase assay, where suramin, NF157, NF279, and NF449 but not NF007, NF023, and NF110 inhibited FGFR3 kinase activity (Fig. 8B).
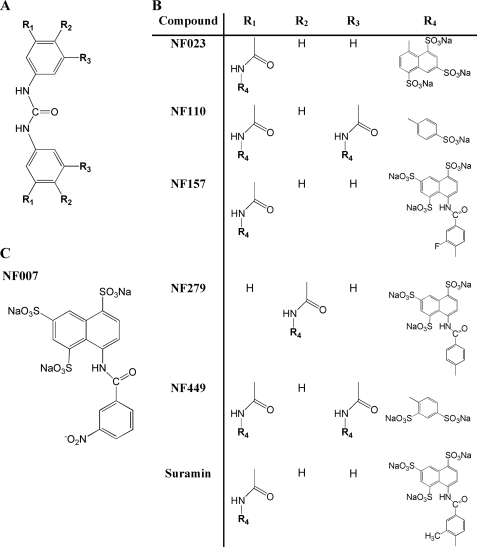
FIGURE 7.
Chemical structures of suramin and its analogs NF007, NF023, NF110, NF157, NF279, and NF449. A, symmetrically phenyl-substituted urea moiety, which represent the backbone of all assayed compounds (B) except for NF007 (C). B, suramin (8,8′-[carbonylbis[imino-3,1-phenylenecarbonylimino{4-methyl-3,1-phenylene}carbonylimino]]bis-1,3,5-naphthalenetrisulfonic acid); NF023 (8,8′-[carbonylbis[imino-3,1-phenylenecarbonylimino]]bis-1,3,5-naphthalenetrisulfonic acid); NF110 (4,4′,4″,4‴-[carbonylbis[imino-5,1,3-benzenetriylbis(carbonylimino)]]tetrakisbenzenesulfonic acid); NF157 (8,8′-[carbonylbis[imino-3,1-phenylenecarbonylimino(4-fluoro-3,1-phenylene)carbonylimino]]bis-1,3,5-naphthalene trisulfonic acid); NF279 (8,8′-[carbonylbis(imino-4,1-phenylenecarbonylimino-4,1-phenylenecarbonylimino)]bis-1,3,5-naphthalenetrisulfonic acid); and NF449 (4,4′,4″,4″′-[carbonyl-bis[imino-5,1,3-benzenetriyl bis-{carbonylimino}]]tetrakis-1,3-benzenedisulfonic acid). C, NF007 (8-[3-nitrobenzamido]-1,3,5-naphthalenetrisulfonic acid).
FIGURE 8.
Effect of suramin analogs on FGFR3 signaling. A, RCS chondrocytes were treated with FGF2 (5 ng/ml) alone or in the presence of an inhibitor (suramin (40 μm); NF449 or NF023 (25 μm); NF007, NF110, NF157, or NF279 (20 μm)), grown for 72 h, and counted. Note the differential inhibitor abilities to rescue the FGF2-mediated RCS growth arrest. Data represent an average from four wells with the indicated S.D. (error bars). Statistical differences are indicated (Student's t test; *, p < 0.01). B, the FGFR3 kinase assay was carried out as described under “Experimental Procedures” with the recombinant TK domain of FGFR3 as a kinase, recombinant STAT1 as a substrate, and a suramin analog added to the kinase reaction. FGFR3-mediated phosphorylation of STAT1 was detected by WB with a phospho-STAT1(Tyr701) antibody. Note the differential effects of inhibitors on the FGFR3-mediated phosphorylation of STAT1, which corresponds to their activity in the RCS growth arrest assay (A). Data are representative of five experiments.
DISCUSSION
Activating mutations in the FGFR3 receptor tyrosine kinase account for more then 10 human conditions, including syndromes affecting the skeleton (hypochondroplasia, achondroplasia, thanatophoric dysplasia, SADDAN, Muenke syndrome), skin (epidermal nevi, seborrhaeic keratosis, acanthosis nigricans), and cancer development (multiple myeloma, prostate, bladder cancer, seminoma) (1, 20–24). Although its role in human disease makes FGFR3 an attractive candidate for therapy, there is no treatment for FGFR3 disorders available to date, thus warranting development of novel approaches to target FGFR3 signaling (25–27). Here, we took advantage of RCS chondrocytes, which represent the most extensively studied cell model for FGFR3-related skeletal dysplasia (8–12). We used the well characterized growth-inhibitory response of RCS chondrocytes to FGFR3 activation as a reporter for compound library screening aimed at identification of novel inhibitors of FGFR3 signaling (7). A molecular library consisting of 1120 molecules was screened, leading to identification of several potential antagonists of FGFR3 signaling.3 In this article, we characterized the inhibitory effect of a compound named NF449 on FGFR3 signaling.
In RCS chondrocytes, NF449 rescued two major cellular phenotypes of pathological FGFR3 signaling in skeletal dysplasias (i.e. the growth arrest and extracellular matrix loss), as evidenced by cell counting experiments, matrix visualization by Alcian blue staining, and matrix proteoglycan labeling by [35S]sulfate (Figs. 1 and 2, A and B). Importantly, NF449 also rescued the inhibitory effect of FGFR3 activation on murine limb development in organ culture (Fig. 2, C–F, and supplemental Fig. S1), thus demonstrating its capacity to inhibit FGFR3 signaling in chondrocytes both in vitro and in vivo. To test whether this is also the case in the multiple myeloma-cellular environment, we determined the effect of NF449 on FGFR3 signaling in OPM2 and KMS11 cells lines, which represent well characterized models of pathological FGFR3 signaling in multiple myeloma. OPM2 and KMS11 cells were established from patients suffering from multiple myeloma and up-regulate FGFR3 as a result of t(4;14)(p16.3;q32) translocation. In addition, OPM2 and KMS11 harbor highly activating FGFR3 mutations (K650E and Y373C, respectively), common in severe forms of FGFR3-related skeletal dysplasia and cancer (1, 2). In agreement with previous studies, FGFR3 activation in OPM2 and KMS11 cell lead to activation of ERK MAPK and transcriptional induction of CCL3 and CCL4 chemokines (16, 17),3 with both phenotypes being vulnerable to NF449 (Fig. 3). Our data demonstrate that NF449 is not only capable of inhibiting FGFR3 signaling in the multiple myeloma cellular environment but also capable of inhibiting the signaling of FGFR3 carrying highly activating mutations.
How does the NF449 inhibit FGFR3 signaling? Little is known about the biological activities of NF449, which was originally described as an antagonist of both G-proteins (selective to the Gαs subunit) and P2X adenine receptors (28, 29). Although these findings open the attractive possibility of involvement of G-proteins or P2X receptors in FGFR3 signaling, the lowest NF449 concentration active in the RCS growth arrest assay (∼6 μm; Fig. 1A) was significantly higher than that needed to inhibit Gαs or P2X receptors (∼0.1 μm for Gαs, subnanomolar for P2X) (28, 29). In addition, NF023 and NF110, which are both potent inhibitors of P2X receptors (30), showed no activity in the RCS growth arrest assay (Fig. 8A). Because a treatment with three P2X receptor agonists (i.e. ATP, MethyleneATP, and BzATP) did not inhibit growth or sensitize RCS chondrocytes to FGF2 (data not shown), we concluded that NF449 inhibits FGFR3 signaling independent of P2X receptors. Interestingly, the same conclusion was reached by Guzmán-Arángues et al. (31), who recently reported the inhibitory effect of another P2X receptor antagonist, pyridoxalphosphate-6-azophenyl-2′,4′-disulfonicacid tetrasodium salt, on the signaling of FGFR3 with an achondroplasia mutation (G380R) in chondrogenic RCJ3.1 cells.
Because NF449 inhibited the FGF2-mediated phosphorylation of PLCγ, a direct substrate of the FGFR3 kinase (Fig. 4A), we hypothesized that NF449 targets FGFR3 itself, either extracellularly or at the intracellular kinase domain. NF449 exhibits close structural similarity with suramin, a polysulfonylated naphtyl urea derivative (Fig. 7). Suramin is known to inhibit FGFR activation via formation of suramin-FGF multimers that are incapable of binding FGFR (32). In addition, several studies using smaller structural units of suramin (such as sulfonylated naphthalene) indicated that the suramin can bind to the heparin binding pocket in FGF (32–36). Because formation of binary FGF·FGFR complex requires participation of proteoglycans, such as heparin (37–39), the competitive binding of suramin to the heparin binding site would interfere with the FGF-FGFR interaction, thus preventing subsequent FGFR activation (40). In line with these findings, we observed reversal of the FGFR3-mediated RCS growth arrest after suramin treatment (Fig. 8A). Considering the close structural similarity of NF449 and suramin, one of the possible modes of NF449 action on FGFR3 signaling observed here might be extracellular (i.e. via interference with assembly of the heparin·FGF2·FGFR3 complexes essential for proper FGFR3 activation).
However, our findings reveal an alternative mechanism of FGFR3 inhibition by both NF449 and suramin. As demonstrated by cell-free kinase assays utilizing both recombinant and immunopurified FGFR3 as a kinase and recombinant STAT1 as a substrate (18), both NF449 and suramin inhibited FGFR3 kinase activity (Figs. 5, 6, and 8 and supplemental Figs. S2 and S3). It is important to note that in the majority of these experiments, both FGFR3 kinase and STAT1 substrate components were recombinant proteins, and, in addition, the FGFR3 kinase was a truncated form that lacked the extracellular, FGF, and heparin-binding domains. Our data thus indicate that NF449 inhibits FGFR3 via directly targeting its intracellular tyrosine kinase domain. Although the exact mechanism of this inhibition is not known and is a subject of ongoing investigation, our data suggest that NF449 targets the kinase domain of FGFR3 at a site other than the ATP binding site (Fig. 6A).
Fig. 1B demonstrates that NF449-mediated rescue of FGF2-mediated growth arrest in RCS chondrocytes significantly exceeds the similar activity of C-natriuretic peptide. C-natriuretic peptide is a recently discovered potentially therapeutic antagonist of FGFR3 signaling that suppresses the pathological FGFR3 signaling in cartilage via inhibition of one of its intermediates, the ERK MAP kinase pathway (12, 13). Because NF449 and C-natriuretic peptide target FGFR3 signaling at different levels (FGFR3 kinase versus activation of the ERK pathway) (12), their combination could prove useful to achieve stable therapeutic inhibition of FGFR3 signaling. Similarly, NF449 might be combined with SU5402, which represents a well established inhibitor of FGFR kinases that acts via binding the ATP pocket of FGFR (19). When compared directly with NF449, SU5402 proved to be a more potent inhibitor of FGFR3 kinase activity, achieving nearly complete inhibition at the 10 μm scale (supplemental Fig. S3_A_). A similar effect was achieved in a growth arrest experiment, where SU5402 rescued the FGFR3-mediated RCS growth arrest more potently then NF449 (supplemental Fig. S3_B_). Unlike NF449, however, SU5402 showed significant cellular toxicity throughout its active range, potentially limiting its therapeutic use. Because both inhibitors target FGFR3 differently (i.e. via the ATP-binding pocket (SU5402) and independent of ATP (NF449)), we asked whether NF449 and SU5402 can synergistically inhibit FGFR3. Fig. 6 demonstrates that potent FGFR3 inhibition can be achieved by both inhibitors used together at low concentration, thus overcoming the toxic effects evident at higher SU5402 concentrations.
As a suramin analog, NF449 belongs to a large group of chemicals defined as symmetric polysulfonates with a central urea bridge (Fig. 7) (41, 42). Using both the RCS growth arrest assay and cell-free FGFR3 kinase assay, we determined the activity of several such compounds, including NF007, NF023, NF110, NF157, and NF279. We found differential activities among the tested compounds, with NF157 and NF279 inhibiting FGFR3 signaling similarly to NF449 or suramin, whereas NF007, NF023, and NF110 were virtually inactive (Fig. 8). Based on their structure, the studied compounds can be divided into three groups. First, NF007, the “one half” of the suramin molecule, corresponds essentially to the active moiety (i.e. 3,1-phenylenecarbonylimino-1,3,5-naphtalenetrisulfonic acid of suramin). The fact that NF007 does not possess any inhibitory activity toward FGFR3 (Fig. 8) implies that the isolated active moiety of suramin has insufficient selectivity with respect to FGFR3 binding site.
In the second group, consisting of suramin, NF157, and NF279, the basic phenylcarbonyl urea core is symmetrically substituted with either two 3,1-phenylenecarbonylimino-1,3,5-naphthalenetrisulfonic acid groups (NF279) or two 3,1-phenylenecarbonylimino-1,3,5-naphthalenetrisulfonic acid groups (suramin, NF157). The only difference between suramin and NF157 is a CH3 or F substituent in the 3,1-phenylenecarbonylimino moiety at position 4 (Fig. 7). Because these two compounds display similar activity toward FGFR3 (Fig. 8), it is unlikely that the nature or position of substituents contributes to the activity and/or specificity of suramin, NF157, and NF279 toward FGFR3.
The third group studied consists of NF023, NF110, and NF449, characterized by symmetrical substitution of the basic phenylcarbonyl urea core directly with mono- and disulfonic acids of imino-benzene (NF110 and NF449) or trisulfonic acids of imino-naphthalene (NF023). The most obvious difference when compared with the first group is the absence of 4,1-phenylenecarbonylimino linker, typical for suramin, NF157, and NF279 (Fig. 7). Although NF023 and NF110 showed no FGFR3-inhibitory activity, the NF449 activity was comparable with the activities of suramin, NF157, and NF279 (Fig. 8). As demonstrated by Hausmann et al. (43), selectivity of suramin analogs to P2X receptors is primarily determined by the location of sulfonic acid groups. Three-dimensional structural modeling suggests that the location of sulfonic acid groups is also responsible for the different activities toward FGFR3. The NF449 model indicates that four of its eight sulfonic acid groups have a spatial arrangement virtually identical to those in suramin, NF157, and NF279 (supplemental Figs. S4 and S6). In contrast, the inactive suramin analogs have either an insufficient number (NF110) or inappropriate spatial orientation of the sulfonic acid groups (NF023) (supplemental Figs. S5 and S6). Correspondingly, modeling indicates that the binding of suramin, NF157, NF279, and NF449 to FGFR3 is mediated by four sulfonic groups having a rectangular arrangement separated roughly by 5 and 10 Å (supplemental Fig. S6). However, not only the number and spatial separation of sulfonic acid groups but also their mutual orientation appears critical for selectivity toward FGFR3, as illustrated by supplemental Fig. S6.
Taken together, our data suggest that both the number and exact location of negatively charged sulfonic groups are important for FGFR3-inhibitory activity. Symmetrical suramin analogs based on 4,4′-carbonylbis(imino-3,1-phenylenecarbonylimino)bis-1,3-benzenedisulfonic acid might thus represent attractive, novel lead compounds in the development of FGFR3 inhibitors.
Supplementary Material
Supplemental Data
Acknowledgments
We thank K. Pejchalova and P. Mekikian for primer design and excellent technical assistance and P. Jelinkova for statistical analysis of the data.
*
This work was supported, in whole or in part, by National Institutes of Health Grants 5P01HD022657-21A and 1R01AR055556-01A209 (to S. M.). This work was also supported by the Multiple Myeloma Research Foundation; Ministry of Education, Youth, and Sports of the Czech Republic Grant MSM0021622430; Grant Agency of the Czech Republic Grants 301/09/0587 and 204/09/H058; Academy of Sciences of the Czech Republic Grants AVOZ50040507 and AVOZ50040702; a Winnick Family Research Scholars Award (to W. R. W.); and a European Molecular Biology Organization Installation Grant (to V. B.).
3
K. Pejchalova and W. Wilcox, unpublished results.
2
The abbreviations used are:
FGF
fibroblast growth factor
FGFR
FGF receptor
MethyleneATP
α,β-methyleneadenosine 5′-triphosphate
BzATP
2′(3′)-_O_-(4-benzoylbenzoyl)adenosine-5′-triphosphate
RCS
rat chondrosarcoma
CHO
Chinese hamster ovary
WB
Western blot
ERK
extracellular signal-regulated kinase
MAPK
mitogen-activated protein kinase
MEK
mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
PLC
phospholipase C.
REFERENCES
- 1.Passos-Bueno M. R., Wilcox W. R., Jabs E. W., Sertié A. L., Alonso L. G., Kitoh H. (1999) Hum. Mutat. 14, 115–125 [DOI] [PubMed] [Google Scholar]
- 2.Chesi M., Nardini E., Brents L. A., Schröck E., Ried T., Kuehl W. M., Bergsagel P. L. (1997) Nat. Genet. 16, 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Intini D., Baldini L., Fabris S., Lombardi L., Ciceri G., Maiolo A. T., Neri A. (2001) Br. J. Haematol. 114, 362–364 [DOI] [PubMed] [Google Scholar]
- 4.Cappellen D., De Oliveira C., Ricol D., de Medina S., Bourdin J., Sastre-Garau X., Chopin D., Thiery J. P., Radvanyi F. (1999) Nat. Genet. 23, 18–20 [DOI] [PubMed] [Google Scholar]
- 5.L'Hôte C. G., Knowles M. A. (2005) Exp. Cell Res. 304, 417–431 [DOI] [PubMed] [Google Scholar]
- 6.Plowright E. E., Li Z., Bergsagel P. L., Chesi M., Barber D. L., Branch D. R., Hawley R. G., Stewart A. K. (2000) Blood 95, 992–998 [PubMed] [Google Scholar]
- 7.Krejci P., Pejchalova K., Wilcox W. R. (2007) Invest. New Drugs 25, 391–395 [DOI] [PubMed] [Google Scholar]
- 8.Krejci P., Masri B., Salazar L., Farrington-Rock C., Prats H., Thompson L. M., Wilcox W. R. (2007) J. Biol. Chem. 282, 2929–2936 [DOI] [PubMed] [Google Scholar]
- 9.Krejci P., Bryja V., Pachernik J., Hampl A., Pogue R., Mekikian P., Wilcox W. R. (2004) Exp. Cell Res. 297, 152–164 [DOI] [PubMed] [Google Scholar]
- 10.Dailey L., Laplantine E., Priore R., Basilico C. (2003) J. Cell Biol. 161, 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aikawa T., Segre G. V., Lee K. (2001) J. Biol. Chem. 276, 29347–29352 [DOI] [PubMed] [Google Scholar]
- 12.Krejci P., Masri B., Fontaine V., Mekikian P. B., Weis M., Prats H., Wilcox W. R. (2005) J. Cell Sci. 118, 5089–5100 [DOI] [PubMed] [Google Scholar]
- 13.Yasoda A., Komatsu Y., Chusho H., Miyazawa T., Ozasa A., Miura M., Kurihara T., Rogi T., Tanaka S., Suda M., Tamura N., Ogawa Y., Nakao K. (2004) Nat. Med. 10, 80–86 [DOI] [PubMed] [Google Scholar]
- 14.Sahni M., Ambrosetti D. C., Mansukhani A., Gertner R., Levy D., Basilico C. (1999) Genes Dev. 13, 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronchetti D., Greco A., Compasso S., Colombo G., Dell'Era P., Otsuki T., Lombardi L., Neri A. (2001) Oncogene 20, 3553–3562 [DOI] [PubMed] [Google Scholar]
- 16.Krejci P., Mekikian P. B., Wilcox W. R. (2006) Leukemia 20, 1165–1168 [DOI] [PubMed] [Google Scholar]
- 17.Masih-Khan E., Trudel S., Heise C., Li Z., Paterson J., Nadeem V., Wei E., Roodman D., Claudio J. O., Bergsagel P. L., Stewart A. K. (2006) Blood 108, 3465–3471 [DOI] [PubMed] [Google Scholar]
- 18.Krejci P., Salazar L., Goodridge H. S., Kashiwada T. A., Schibler M. J., Jelinkova P., Thompson L. M., Wilcox W. R. (2008) J. Cell Sci. 121, 272–281 [DOI] [PubMed] [Google Scholar]
- 19.Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997) Science 276, 955–960 [DOI] [PubMed] [Google Scholar]
- 20.Hernández S., de Muga S., Agell L., Juanpere N., Esgueva R., Lorente J. A., Mojal S., Serrano S., Lloreta J. (2009) Mod. Pathol. 22, 848–856 [DOI] [PubMed] [Google Scholar]
- 21.Muenke M., Gripp K. W., McDonald-McGinn D. M., Gaudenz K., Whitaker L. A., Bartlett S. P., Markowitz R. I., Robin N. H., Nwokoro N., Mulvihill J. J., Losken H. W., Mulliken J. B., Guttmacher A. E., Wilroy R. S., Clarke L. A., Hollway G., Adès L. C., Haan E. A., Mulley J. C., Cohen M. M., Jr., Bellus G. A., Francomano C. A., Moloney D. M., Wall S. A., Wilkie A. O. (1997) Am. J. Hum. Genet. 60, 555–564 [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner C., van Oers J. M., Vogt T., Landthaler M., Stoehr R., Blaszyk H., Hofstaedter F., Zwarthoff E. C., Hartmann A. (2006) J. Clin. Invest. 116, 2201–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafner C., Landthaler M., Mentzel T., Vogt T. (2010) Br. J. Dermatol. 162, 508–512 [DOI] [PubMed] [Google Scholar]
- 24.Goriely A., Hansen R. M., Taylor I. B., Olesen I. A., Jacobsen G. K., McGowan S. J., Pfeifer S. P., McVean G. A., Meyts E. R., Wilkie A. O. (2009) Nat. Genet. 41, 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aviezer D., Golembo M., Yayon A. (2003) Curr. Drug Targets 4, 353–365 [DOI] [PubMed] [Google Scholar]
- 26.Meyer A. N., McAndrew C. W., Donoghue D. J. (2008) Cancer Res. 68, 7362–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qing J., Du X., Chen Y., Chan P., Li H., Wu P., Marsters S., Stawicki S., Tien J., Totpal K., Ross S., Stinson S., Dornan D., French D., Wang Q. R., Stephan J. P., Wu Y., Wiesmann C., Ashkenazi A. (2009) J. Clin. Invest. 119, 1216–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun K., Rettinger J., Ganso M., Kassack M., Hildebrandt C., Ullmann H., Nickel P., Schmalzing G., Lambrecht G. (2001) Naunyn Schmiedebergs Arch. Pharmacol. 364, 285–290 [DOI] [PubMed] [Google Scholar]
- 29.Hohenegger M., Waldhoer M., Beindl W., Böing B., Kreimeyer A., Nickel P., Nanoff C., Freissmuth M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soto F., Lambrecht G., Nickel P., Stühmer W., Busch A. E. (1999) Neuropharmacology 38, 141–149 [DOI] [PubMed] [Google Scholar]
- 31.Guzmán-Aránguez A., Crooke A., Yayon A., Pintor J. (2008) Eur. J. Pharmacol. 584, 72–77 [DOI] [PubMed] [Google Scholar]
- 32.Kathir K. M., Kumar T. K., Yu C. (2006) Biochemistry 45, 899–906 [DOI] [PubMed] [Google Scholar]
- 33.Botta M., Manetti F., Corelli F. (2000) Curr. Pharm. Des. 6, 1897–1924 [DOI] [PubMed] [Google Scholar]
- 34.Sola F., Capolongo L., Moneta D., Ubezio P., Grandi M. (1999) Cancer Chemother. Pharmacol. 43, 241–246 [DOI] [PubMed] [Google Scholar]
- 35.Zamai M., Hariharan C., Pines D., Safran M., Yayon A., Caiolfa V. R., Cohen-Luria R., Pines E., Parola A. H. (2002) Biophys. J. 82, 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozano R. M., Jiménez M., Santoro J., Rico M., Giménez-Gallego G. (1998) J. Mol. Biol. 281, 899–915 [DOI] [PubMed] [Google Scholar]
- 37.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 38.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 39.Yeh B. K., Eliseenkova A. V., Plotnikov A. N., Green D., Pinnell J., Polat T., Gritli-Linde A., Linhardt R. J., Mohammadi M. (2002) Mol. Cell Biol. 22, 7184–7192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammadi M., Olsen S. K., Goetz R. (2005) Curr. Opin. Struct. Biol. 15, 506–516 [DOI] [PubMed] [Google Scholar]
- 41.Firsching A., Nickel P., Mora P., Allolio B. (1995) Cancer Res. 55, 4957–4961 [PubMed] [Google Scholar]
- 42.Kassack M. U., Braun K., Ganso M., Ullmann H., Nickel P., Böing B., Müller G., Lambrecht G. (2004) Eur. J. Med. Chem. 39, 345–357 [DOI] [PubMed] [Google Scholar]
- 43.Hausmann R., Rettinger J., Gerevich Z., Meis S., Kassack M. U., Illes P., Lambrecht G., Schmalzing G. (2006) Mol. Pharmacol. 69, 2058–2067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data