The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells (original) (raw)
Abstract
Members of the Tc1_/mariner_ superfamily of transposable elements isolated from vertebrates are transpositionally inactive due to the accumulation of mutations in their transposase genes. A novel open reading frame-trapping method was used to isolate uninterrupted transposase coding regions from the genome of the frog species Rana pipiens. The isolated clones were ∼90% identical to a predicted transposase gene sequence from Xenopus laevis, but contained an unpredicted, ∼180 bp region encoding the N-terminus of the putative transposase. None of these native genes was found to be active. Therefore, a consensus sequence of the transposase gene was derived. This engineered transposase and the transposon inverted repeats together constitute the components of a novel transposon system that we named Frog Prince (FP). FP has only ∼50% sequence similarity to Sleeping Beauty (SB), and catalyzes efficient cut-and-paste transposition in fish, amphibian and mammalian cell lines. We demonstrate high-efficiency gene trapping in human cells using FP transposition. FP is the most efficient DNA-based transposon from vertebrates described to date, and shows ∼70% higher activity in zebrafish cells than SB. Frog Prince can greatly extend our possibilities for genetic analyses in vertebrates.
INTRODUCTION
Genome sequencing projects of different model organisms allow scientists to take a global view of genomes. However, the task of functional genomics is even more challenging: assigning functions to the known sequences. Phenotype-driven genetic screens in vertebrate model systems have been valuable in the identification of mutations affecting embryonic as well as post-embryonic development, thereby providing important insight into vertebrate development and presenting animal models for human diseases.
Mutagenesis screens using ethylnitrosourea (ENU) have identified a large number of loci important in embryonic development of the zebrafish (Danio rerio) (1,2) and the mouse (3). Although ENU is highly mutagenic, it introduces base-pair changes into DNA; thus, identification of mutant genes by positional cloning is a difficult and slow process. Insertional mutagenesis by integrating viruses is a powerful alternative to chemical mutagenesis (4,5). However, genetic design of virus vectors is restricted due to the constraints of the virus in terms of size, structure and regulation of expression. Yet another possible strategy for transgenesis and insertional mutagenesis, one that has been extremely valuable in exploring gene function in Drosophila (6) and Caenorhabditis elegans (7), is the use of transposon-based vectors.
The Tc1/mariner superfamily of transposable elements is widespread in nature (8,9). These elements are framed by terminal inverted repeats (IR), and contain a single gene encoding a transposase. Both mariner_- (10–14), and Tc1-like elements (15–18) are able to transpose in species other than their hosts, and are therefore emerging tools for functional genomics in several organisms (9,19–21). However, all Tc1/mariner_ transposons isolated to date from vertebrates are transpositionally inactive. The general inactivity of these elements is the result of accumulation of mutations: a process called ‘vertical inactivation’ (22).
To address the above problem, an ancestral Tc1-like element called Sleeping Beauty (SB) was reactivated by deducing a consensus sequence of an active transposase based on sequence data collected from different fish species (23). SB shows efficient transposition in a variety of vertebrate cell lines in tissue culture (15) and in the mouse in vivo, both in somatic tissues (24) and in the germline (25,26).
SB shows no host-restrictions in vertebrates, but the efficiency of transposition in cell lines derived from different species is variable (15). Therefore, having a palette of different, vertebrate-derived transposons with different host preference widens the potential of transposons as genomic tools in vertebrates. Here, we describe the reconstruction of an active Tc1-like transposable element from the Northern Leopard Frog (Rana pipiens). The R.pipiens genome was estimated to contain about 8000 copies of a transposable element most closely related to Txr elements in Xenopus laevis. We devised an open reading frame (ORF)-trapping method for the selection of uninterrupted transposase ORFs. One of the transposase genes isolated by this method contained only two mutations compared to a consensus sequence. These mutations were corrected, and the resulting transposase gene together with the inverted repeats isolated from the R.pipiens genome constitute the components of a novel transposon system that we named Frog Prince (FP), after the fairy tale by the Grimm brothers. FP exhibits efficient and precise cut-and-paste transposition in cell lines of major vertebrate taxa, and shows an ∼70% higher activity than SB in zebrafish cells. We demonstrate the functional usefulness of FP for high-efficiency gene trapping in human cells. Frog Prince can be a complementary, transposon-based tool for genetic analyses in vertebrates.
MATERIALS AND METHODS
Open reading frame trapping and plasmids
Transposase coding regions from R.pipiens genomic DNA were amplified by using the primer 5′-GACTGCGGCCGCAAATCTACATGG GCCTGTGTGAAAAAGTG specific for the start of the presumptive Txr transposase gene predicted by Lam et al. (27), and 5′-ATAACTGGTTGGGCCACCCTTAGC specific for the end but lacking a translational stop codon. The PCR products were digested with NotI (underlined) and cloned into the NotI/SmaI sites in pCMVβFUSa (28). The plasmids containing ORFs served as templates for another round of PCR amplification, using primers Txr-start (5′-CTGACCGCGGCCGCATCATGCCGAGACCCAAAGAAATTCAGG) and Txr-stop (5′-CTGAGCGGCCGCTAATAACTGGTTGGGCCACCCTTAGC). The PCR products were digested with restriction enzymes SacII and NotI (underlined), and cloned into the respective sites of the carp β-actin promoter-containing expression plasmid pFV4a (29). Additional transposase coding sequences were PCR-amplified from R.pipiens genomic DNA with primers Txr-start and Txr-stop. The PCR products were digested with SacII and NotI, and cloned into pFV4a.
Site-specific mutagenesis with ligase chain reaction was done to obtain the consensus _Rana_-type transposase. Txr-start and Txr-stop served as end-primers, whereas Txr-S/T (5′-CTGTGGACCGATGAGACAAAAGTGGAAC) and Txr-C/R (5′-CCAGGACGCTGTAAAAGCCTCATTGCACG) were the mutagenic primers to obtain the desired Ser→Thr(152) and Cys→Arg(315) coding changes, respectively. The PCR product was cloned into the SacII/NotI sites of pFV4a, resulting in pFV-FP.
Left and right IRs were amplified from R.pipiens genomic DNA using the primer Txr-1R (5′-TACAGTGGTGTGAAAAACTATTTGCCC) specific for the ends of the Txr transposon in X.laevis at GeneBank locus XLRIBSIG, and either Txr-F3 (5′-AAGACTTTGGAGTGGCCTAG) or 5′-GGAACTCTGCCATGCAGGCC, pointing towards the start or the stop codons of the R.pipiens transposase genes, respectively. The PCR products were cloned into the HincII site of pUC19 resulting in pE5 containing the left IR, and pV3 containing the right IR, respectively. pE5-neo was constructed by cloning the EcoRI/BamHI fragment of pRc/CMV (Invitrogen) containing an SV40 promoter/enhancer, the neomycin phosphotransferase (neo) gene and the SV40 poly-A signal into the MunI site of pE5. The EcoRI/HindIII fragment of pE5-neo was subsequently cloned into the HindIII site of pV3 resulting in pTxr-neo.
To obtain the _R.pipiens_-specific IR sequences, splinkerette-PCR (30) was done on Sau3AI digested genomic DNA. The primers for the nested PCR specific for the transposase gene were Txr-F3 and Txr-C/R. The consensus _R.pipiens_-specific IR sequences were derived by PCR, using primer 5′-TACAGTGGTGTGAAAAAGTGTTTGCCCCCTTCCTCATTTCCTGTTCC on pTxr-neo as template. The product was cloned into the SmaI site of pUC19 and the construct was named pFP-neo. The consensus sequence of a predicted, active, full-length transposon was deposited in GenBank (accession nos AY261370–AY261372 and TPA BK001476).
The gene trap plasmid pFP/GT-neo was generated by subcloning the first splice acceptor site of the mouse engrailed-2 fused to an ATG-less neo gene followed by the zeo gene equipped with both a bacterial and a CMV promoter (Invitrogen) between the FP IRs. A Klenow-filled HindIII–SpeI fragment of pMiLRgeo (17) was cloned into FP resulting in pFP/GT-geo.
Copy number determination and phylogenetic analysis
Genomic copy number was estimated by dot-blotting as described (31). Known amounts of pFV-FP were blotted alongside with known quantities of R.pipiens genomic DNA, and probed with a 32P-labeled, full-length transposase gene. Radioactivity of the dots was quantified with a Storm PhosphorImager (Molecular Dynamics) using the Imagequant program, and a linear range of data was used to estimate the copy number. Consensus amino acid sequences of transposases were aligned in ClustalX. The tree was generated with neighbor-joining method, with 1000 replication of resampling. The tree was displayed in Phylip version 3.6.
Cell culture and transfection
HeLa and CHO-K1 cells were maintained in DMEM; FHM, A6 and PAC2 cells were cultured in L-15 medium containing 10 or 15% (PAC2) FCS. Transposition assays were done as described (23). Briefly, 3 × 105 cells were transfected with 100 ng of each the transposon donor plasmid and the transposase-expressing helper plasmid using Fugene6 transfection reagent (Roche). Two days post-transfection, the cells were re-plated and selected in 400 µg/ml (PAC2) 1 mg/ml (CHO-K1) and 1.4 mg/ml G418 (HeLa, FHM and A6) G418 (Gibco). HeLa cells transfected with pFP/GT-neo were selected with 1 mg/ml G418 and/or 200 µg/ml zeocin (Invitrogen). After 2 weeks of selection, the resistant colonies were either stained and counted, or picked and expanded to individual cultures.
Analyses of excision footprints and genomic junction fragments
Low molecular weight DNA was isolated from HeLa cells transfected with pFP-neo and pFV-FP 2 days post-transfection using a modified protocol of QIAprep Spin (Qiagen). About 2 µg template DNA was used for a series of PCR reactions using pUC19 backbone-specific primers: 5′-CCTCTGACACATGCAGCTCCCGG and 5′-CAGTAAGAGAATTATGCAGTGCTGCC in PCR1, 5′-TCACAGCTTGTCTGTAAGCGG and 5′-TCTTTCCTGCGTTATCCCCTGATTC in PCR2, and 5′-TTCGCCATTCAGGCTGCGCAACTG and 5′-CAGCTGGCACGACAGGTTTCCCG in PCR3. PCR products were cloned with pGEM-T Vector SystemI (Promega) and sequenced.
To obtain genomic flanks of integrants, splinkerette PCR (30) was performed on Sau3AI-digested DNA isolated from individual G418-resistant HeLa clones. The transposon- specific primers for the nested PCR were 5′-AATTGAACTCAGGTGTGGACAACC and 5′-AGGTGTGGCAATAATCAGGCCTGGGTGTG for the left, and Txr-C/R and 5′-ACAATTCTGCAAAGTTGAGTGGGC for the right side of the transposon.
RT–PCR and rescue of gene trap transposons
cRACE was performed as described by Maruyama et al. (32). Poly-A RNA from an expanded, individual, G418-resistant HeLa clone was reverse transcribed with Superscript-II (Gibco) using the primer 5′-TTCTGCTTCATCAGCAGGATATCC. The _geo_-specific primers used for the inverse PCR were 5′-CATCGCAGGCTTCTGCTTCAATC and 5′-AACGGCAAGCCGTTGCTGATTCG for the first round, 5′-ACCACGCTCATGGATAATTTCACC and LacZF2 5′-CATGGTCAGGTCATGGATGAGCAG for the second round and LacZF2 and 5′-CTTCGCTATTACGCCAGCTGG for the third round of PCR.
For plasmid rescue, genomic DNA was isolated from either pooled, or expanded individual G418/zeocin double-resistant HeLa clones. Approximately 10 µg DNA was digested with SpeI, which does not cut within the gene trap transposon in pFP/GT-neo. The fragments were ethanol precipitated and self-ligated using T4 DNA ligase, under dilute conditions and electroporated into DH10B bacteria, which were than selected on plates containing 50 µg/ml zeocin.
RESULTS
Isolation of transposase genes from Rana pipiens with ORF-trapping
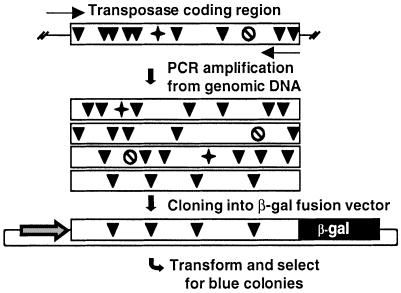
Relatively high copy number of inactive transposable elements in genomes practically prohibits the isolation of functional transposase genes using non-selective methods. In search for potentially active transposase genes in vertebrates, we devised an open reading frame (ORF)-trapping method. The procedure is based on generating a pool of PCR products from genomic DNA using primers flanking the transposase gene sequences (Fig. 1). The 5′-primer contains the predicted translational initiation signal, and the 3′-primer lacks the stop codon. The PCR products are then cloned into an expression vector to generate fusion genes with lacZ. The recombinant plasmids are transformed into Escherichia coli, and plated on X-gal-containing plates. Blue colonies can only arise if the cloned sequences are in frame with the lacZ gene, and do not contain a stop codon.
Figure 1.
Strategy for trapping transposase ORFs from the R.pipiens genome. Transposase genes are PCR-amplified from genomic DNA (arrows show primers). A collection of transposase coding regions (boxes) can be amplified. The vast majority of these genes are defective due to point mutations (black arrowhead), frameshifts (star) and premature translational stop codons (crossed circle). ORFs can be selected by cloning the PCR products in fusion with the lacZ gene driven by the CMV promoter, transformation into E.coli, and plating on X-gal-containing plates.
We applied the ORF-trap on genomic DNA from R.pipiens, using PCR primers designed to the consensus sequence of Txr elements in X.laevis (27). Three resultant blue bacterial colonies indicated the presence of transposase-coding sequences that did not contain premature stop codons. All three sequences were longer than the predicted consensus Txr transposase gene (27), and contained part of the 5′-terminal inverted repeat followed by sequences that were missing in the Txr copies. Apparently, Txr elements contain a conserved deletion of 180 bp covering the N-terminal part of the transposase gene. Similarly, a conserved deletion has been described in Tdr1 elements in zebrafish (31). The rest of the R.pipiens sequences and Txr coding regions showed ∼90% similarity (data not shown).
The genomic copy number of the R.pipiens transposon was estimated by dot blotting. Assuming that the size of the R.pipiens haploid genome is 6.6 × 109 bp (33), we estimated that the transposase gene is represented about 8000 times per haploid genome. To assess what fraction of these elements contains intact ORFs, additional transposase coding regions were PCR-amplified from the R.pipiens genome, and cloned without selecting for ORFs. Seven transposase genes were sequenced and, to our surprise, we found that three of them contained ORFs (Supplementary Material, Fig. 1A). These results suggest that this transposon family is a relatively young component of the R.pipiens genome.
The two components of the Frog Prince transposon system
The 10 transposase genes isolated above were aligned to generate a consensus sequence (Supplementary Material, Fig. 1A). The consensus R.pipiens transposase gene (Fig. 2) encodes a typical Tc1-like transposase containing an N-terminal DNA-binding domain composed of two predicted helix–turn–helix motifs (9), a bipartite nuclear localization signal (34), an AT-hook motif (35) and a catalytic domain with the DD(34)E signature (36) (Fig. 2). The 10 transposase genes were ∼99% identical to the consensus sequence, and one of them differed only in two nucleotides from the consensus, resulting in two amino acid substitutions in its ORF (clone Rana10 in Supplementary Material Fig. 1A). One of these mutations was a T152S exchange in the first part of the catalytic domain of the transposase, and the other was an R315C substitution close to the C-terminus of the protein. Site-specific PCR mutagenesis was used to derive the sequence of the consensus R.pipiens transposase gene (Fig. 2).
Figure 2.
Consensus sequence of the full-length Frog Prince transposable element. The IRs are displayed in black background. The DRs are indicated in white boxes. The encoded amino acid sequence of the transposase is displayed below the DNA sequence. The amino acids that are predicted by PredictProtein to form the α-helices within the two helix–turn–helix motifs in the N-terminal DNA-binding domain are underlined. The AT-hook motif is boxed, the NLS sequence is typed bold and the DDE residues are typed against a black background. The asterisks indicate base pair positions where sequences between the R.pipiens and X.laevis elements are different within the transposase binding sites, and where replacements were introduced in the transposase gene.
In order to derive the binding sites for the _R.pipiens_-type transposase, linker-mediated PCR was applied on genomic DNA to amplify the complete inverted repeats together with genomic flanking sequences. Alignments of five different clones revealed 214 bp long, perfect inverted repeats flanking the transposase genes (Fig. 2). The R.pipiens transposons are typical IR/DR-type elements, i.e. each IR contains two putative transposase binding sites, which are represented as directly repeated sequences (DRs) at its ends (9). The DRs are 21 nucleotides long, and differ in one nucleotide between the outer and internal sites. The IR sequences together with the consensus transposase gene constitute the components of a novel transposable element system that we named Frog Prince (FP) (Fig. 2).
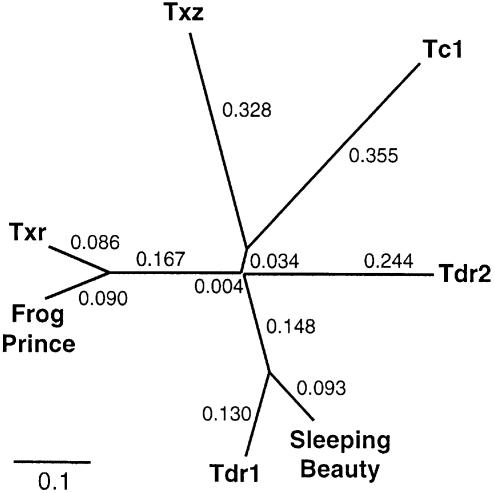
The presumptive transposase binding sites of FP differ in two nucleotides from those of a Txr insertion in the X.laevis ribosomal protein S1 gene (GenBank XLRIBS1G). Thus, these two subfamilies of transposons have likely diverged relatively recently. To determine the phylogenetic position of Frog Prince among other Tc1-like transposase genes, amino acid sequences of Tc1 from C.elegans (GenBank NP-493808), Txr and Txz from X.laevis (27), Tdr1 (31) and Tdr2 (Tzf) from zebrafish (34,37), SB which represents the salmonid subfamily of fish elements (23) and the putative FP transposase were used to generate a phylogenetic tree (Fig. 3). The topology of the unrooted tree shows significant phylogenetic distance between the SB and the FP transposases, and displays that Frog Prince is most closely related to the Txr elements.
Figure 3.
Phylogenetic position of Frog Prince among Tc1-like transposons. Consensus transposase sequences used for the unrooted cladogram were the following: for C.elegans Tc1, GenBank NP-493808; for Danio rerio Tdr1, sequence published in (31); for D.rerio Tdr2 (Tzf), sequence published in (37); for X.laevis TXr and TXz sequences published in (27); for Sleeping Beauty, sequence published in (23); and for Frog Prince, sequence in Figure 2. Numbers at the branches indicate the phylogenetic distances calculated by ClustalX.
Transposition of Frog Prince
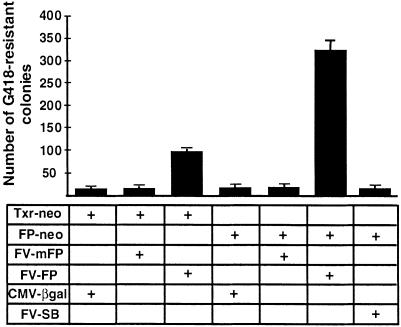
Sleeping Beauty shows high transpositional activity in human cells (23). Therefore, the initial tests for transpositional activity of the Frog Prince element were done in cultured HeLa cells, using a transposition assay established for SB (23). The assay is based on cotransfection of a helper plasmid expressing the transposase and a donor construct containing the transposon with a neomycin-resistance (neo) gene between the terminal IRs. Cells containing transposon insertions can be selected by the antibiotic G418 due to chromosomal integration and expression of the neo gene. The efficiency of transposition is assessed from an increase in the number of G418-resistant colonies in the presence of transposase. The reconstructed consensus R.pipiens transposase ORF (in pFV-FP), and its predecessor gene containing the two amino acid changes (in pFV-mFP) were transfected together with either the Txr-type (pTxr-neo) or with the _Frog Prince_-type (pFP-neo) substrate constructs (Fig. 4). A 17-fold increase in colony number was detected when pFV-FP was cotransfected with its own substrate, pFP-neo (Fig. 4). The mFP transposase was completely inactive, indicating that replacement of either or both of the amino acids T(152) and R(315) were deleterious to transposase activity. The significant sequence similarity between the Xenopus and Rana elements could still allow cross-mobilization between them, as it is the case among the hAT-superfamily elements hobo and Hermes (38). Indeed, we observed a 5-fold increase in the number of G418-resistant cell colonies when pFV-FP was cotransfected with pTxr-neo (Fig. 4). Thus, the R.pipiens transposase can cross-mobilize a X.laevis transposon, indicating that the two transposon families in these species diverged recently. In contrast, no cross-mobilization was observed between FP and Sleeping Beauty (Fig. 4). Taken together, the data demonstrate that we successfully derived an active transposon system from the R.pipens genome, and that Frog Prince can significantly increase the efficiency of transgene integration from plasmid-based vectors to the human genome.
Figure 4.
Transposition and substrate recognition of Frog Prince in human HeLa cells. Different combinations of donor and helper plasmids indicated in the table were cotransfected into HeLa cells. Transfection of pCMV-βgal with the donor plasmids served as control. The efficiency of transgene integration was estimated by counting G418-resistant colonies. The numbers on the left represent the mean values of the numbers of colonies per 105 cells plated after three independent transfections. The error bars indicate SEM.
Cut-and-paste transposition of Frog Prince into the human genome
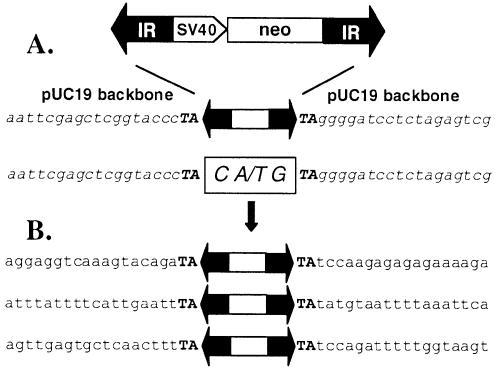
Tc1/mariner elements transpose via a cut-and-paste mechanism (39). During the first step of this process, the element is excised by a pair of staggered double-strand DNA breaks. The host DNA repair machinery seals the gap and, according to the number of the protruding nucleotides, a small insertion indicates the former presence of a transposon. Tc1/mariner elements generate footprints in the range of 2–4 bp (39–42). Primers flanking the transposons were used in a series of nested PCR to identify the footprints left behind by FP transposition in the donor plasmids. Sequencing of the PCR products revealed that FP transposition leaves a CTG or CAG triplet at the excision site (Fig. 5A), indicating that excision of FP generates 3-nucleotide-long overhangs.
Figure 5.
_Frog Prince_-mediated cut-and-paste transposition into human chromosomes. (A) Excision. On top, a schematic of the _FP_-neo element is shown. IRs are represented by black arrows, the SV40 promoter and the neomycin-resistance marker gene (neo) are indicated. pUC19 vector backbone sequences that flank the element in the donor construct are shown in italics. The transposon footprints are depicted in the white box. (B) Integration. Three regions of human genomic sequences that served as target sites for the transposase are illustrated below. Flanking TA target site duplications are typed in bold.
Tc1/mariner elements transpose into TA dinucleotides, which are duplicated and flank the integrated transposon (9). Flanking sequences of three integrated FP transposons were isolated from individual G418-resistant HeLa clones. All three FP insertions were flanked by the expected TA dinucleotides, followed by different human genomic sequences (Fig. 5B). In sum, these data show that Frog Prince follows precise cut-and-paste transposition into various locations in the human genome.
Trapping human genes with Frog Prince
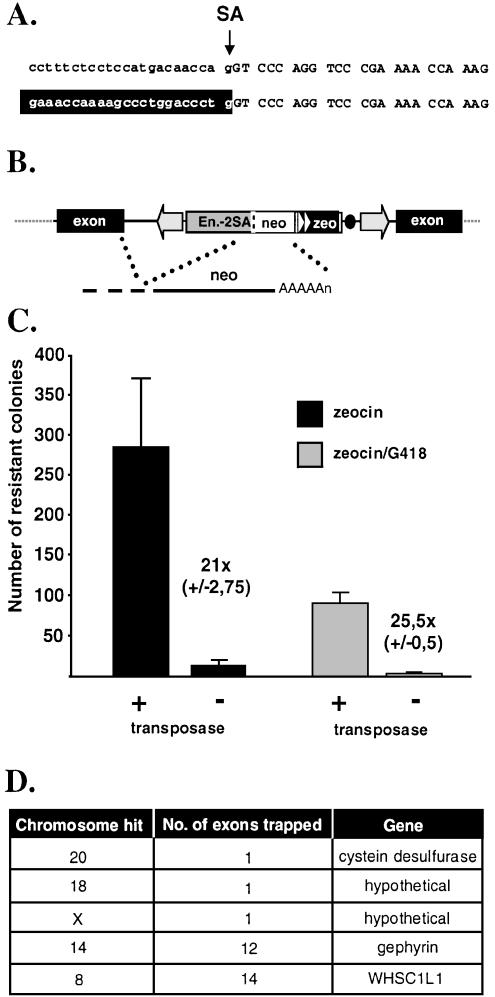
High frequency, precise transposition into different genomic loci suggests that genome-wide gene trapping is feasible with FP. A prerequisite of successful transposon-based gene trapping is that the terminal IRs do not contain potential splice sites. To examine this possibility, an _FP_-based gene trap vector (pFP/GT-geo) containing a splice acceptor (SA) sequence of the mouse engrailed-2 gene followed by the lacZ-neo (geo) fusion was constructed and used for transposition in HeLa cells. Twenty-six out of 27 individual _neo_-resistant colonies were positive for β-galactosidase activity, indicating at least one successful gene trap event per clone (data not shown). LacZ fusion transcripts were identified from G418-resistant, β-galactosidase-positive cells with cRACE (32). We identified a transcript in which splicing generated a fusion between an endogenous RNA and the marker exactly at the engrailed-2 SA (Fig. 6A). These data indicate that the inverted repeats of FP do not interfere with the desired splicing event between a splice donor sequence of an endogenous transcript and the engrailed-2 SA within the transposon.
Figure 6.
Gene trapping with Frog Prince in human HeLa cells. (A) Fusion transcript. On top, nucleotide sequences of the engrailed-2/lacZ junction in pFP/GT-geo are shown. Intron sequences are typed in lowercase, exon sequences are in uppercase. The arrow indicates the splice acceptor site (SA). Human transcript sequences (depicted against black background) are fused to the engrailed-2 exon due to correct splicing at the SA. (B) Gene trapping with pFP/GT-neo. Engrailed-2 sequences are depicted by the gray box. Triangles indicate the eukaryotic and prokaryotic promoters. The black dot represents the bacterial origin of replication. _A_s stand for polyadenylation signal. (C) Efficiency of gene trapping with FP. Numbers of antibiotic-resistant colonies are indicated on the _y_-axis. Zeocin selection was used to deduce the transpositional efficiency in the presence of the helper (pFV-FP) versus the control (pCMV-β) plasmid (black columns). Gene trapping efficiencies were determined by using zeocin/G418 double selection (gray columns). Numbers next to the columns indicate the fold difference in numbers of colonies obtained in the presence versus absence of the transposase. (D) Gene trapping events identified by transposon rescue.
Next, we wanted to determine the efficiency of gene trapping and to identify the tagged genes. For this purpose, an _FP_-based donor plasmid (pFP/GT-neo) was constructed which contains engrailed-2 intron sequences with the SA, a glycine bridge to allow proper folding of the marker in protein fusions (43), an ATG-less neo gene, a zeocin resistance gene (zeo) driven by dual eukaryotic/bacterial promoters and a plasmid origin of replication (Fig. 6B). All chromosomal transposition events give rise to zeocin-resistant cells. A subset of transformant cells will be G418-resistant, if the transposon inserted into an intron of an expressed gene in the proper orientation, and if splicing occurred in-frame with neo. The plasmid origin of replication within the element can be used to isolate the integrated transposon from genomic DNA by plasmid rescue.
Based on the numbers of zeocin-resistant cell colonies, transposition efficiency of FP/GT-neo was comparable to that of FP-neo (Fig. 6C). The number of zeocin/G418 double-resistant colonies was about one-third of those resistant to zeocin alone, indicating that ∼30% of all transposition events occurred in introns of expressed genes and in-frame splicing took place (Fig. 6C). Five insertion sites of the FP gene trap transposons were identified. All of them mapped to introns of genes in different chromosomes, in the correct orientation (Fig. 6D). Our results suggest that FP can potentially target a large fraction of genes in the human genome.
Frog Prince is active in various vertebrate cell lines
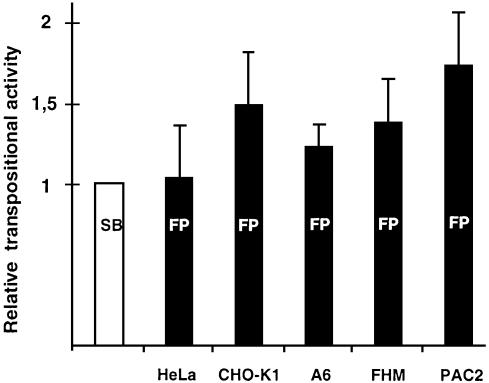
SB has varying transpositional activity in different vertebrate cell lines (15). However, SB is a synthetic element of fish origin and Frog Prince was reconstructed from the genome of an amphibian. Thus, the same set of cell lines can provide different permissive environments to the two transposon systems. To test this hypothesis, we compared the activities of the two systems in cultured cell lines derived from two mammalian, an amphibian and two fish species with the standard transposition assay (Fig. 7). The plasmids used for cotransfections were pFV-SB (15) and pT/neo (23) in the case of Sleeping Beauty, and pFV-FP and pFP-neo in the case of the Frog Prince system. The vector backbones, the promoters, the poly-A signals and the transposon marker genes were identical in the constructs of the two systems. FP proved to be more active than SB in some of the cell lines tested (Fig. 7). Most importantly, FP appeared ∼70% more active than SB in the PAC2 zebrafish cell line, (P = 0.02). These data demonstrate that transposition of Frog Prince is not restricted to phylogenetically close taxa, and that it is the most active transposable element in vertebrate species described to date.
Figure 7.
Activity of Frog Prince in comparison with Sleeping Beauty in diverse vertebrate species. The donor and helper plasmids of FP and SB were cotransfected in HeLa (human), CHO-K1 (hamster), A6 (X.laevis), FHM (fathead minnow) and PAC2 (zebrafish) cell lines. Transposition efficiencies were calculated by deriving ratios between the numbers of G418-resistant cell clones obtained in the presence versus in the absence of the transposases. Activities of FP (indicated by black columns) were compared to those of SB (white column). Mean values of relative efficiencies are derived from at least three independent transfections, and are indicated on the _y_-axis. Transpositional efficiency of SB was normalized to the value 1 for each cell line. The error bars show SEM.
DISCUSSION
We have reconstructed an active Tc1-like transposable element from the frog species R.pipiens, and named this transposon Frog Prince. Two approaches were used to derive the two components of the transposon system. First, a consensus transposase gene was derived by a combination of a novel ORF-trapping method (Fig. 1) with comparative phylogenetic analysis and site-directed PCR mutagenesis. A particular advantage of the ORF-trap method is that those transposase coding regions that are recovered by this procedure were likely functional recently, and thus have the smallest number of amino acid substitutions affecting activity. We applied the ORF-trapping assay to search for Tdr1 (31) and Txr (27) transposase ORFs in the zebrafish and X.laevis genomes, respectively. Neither of the cloned PCR pools of amplified transposase genes led to colonies with β-galactosidase activity, consistent with very old age and severe mutational damage of these elements (data not shown). Our attempt to find active transposase genes in the R.pipiens genome was based on the assumption that a genome from a genus other than Xenopus might contain copies of a Txr-like transposon in a better conserved state. Our success in identifying transposase ORFs from the R.pipiens genome suggests that our ORF-trap method should be useful for the isolation of uninterrupted coding regions of any gene, for which inactive copies exist in the same genome. The second component of the transposon system, the terminal inverted repeats, were isolated from the R.pipiens genome by unselective PCR amplification, generation of sequence alignments and selection of sequences devoid of mutations.
The reconstructed transposase gene and its specific IRs make up the two functional components of a putative, complete transposable element (Fig. 2). FP is probably identical, or very similar to the transposon that originally colonized the R.pipiens genome. Vertical inactivation leads to time-proportional accumulation of mutations in the elements after the invasion of a genome (22). Thus, the more a transposon resembles its once active ancestor, the younger a component it is of the colonized genome. The well-conserved state of _FP_-like elements (Supplementary Material, Fig. 1A) therefore suggests that germline invasion of this species by a founder FP element must have taken place relatively recently in evolutionary terms. Our success in reconstructing an active transposon from frogs underscores the importance of comparative phylogenetic analysis. Deriving a consensus transposon sequence from a single species can lead to a false prediction of the active transposon sequence. This is because some of the inactive elements can be amplified to an extent that the majority of transposons in a genome are derived from a single particular mutant. This might have happened in both the zebrafish (31) and the X.laevis genomes (27), where only elements with a conserved deletion can be detected. Thus, sequence comparisons between closely related elements in the X.laevis and R.pipiens genomes (Fig. 3) were of key importance in predicting the sequence of an active transposase.
Transpositional activity of FP was established by devising a two-component, in vivo transposition assay based on _trans_-complementation of a genetically marked transposon by the transposase. The FP system mediates precise, cut-and-paste transposition into the chromosomes of human somatic cells (Fig. 5). Since mapping of a large number of Sleeping Beauty insertions showed a random pattern of distribution in the human genome (44), we expect that a substantial number of TA dinucleotides can also serve as integration sites for FP.
FP is active in cell lines of representatives of major vertebrate taxa, and in some cell lines it has higher transpositional activity than SB (Fig. 7). In considering possible explanations for the significantly more efficient transposition of FP in zebrafish cells, the higher intrinsic transpositional activity of FP can presumably be ruled out, as the two systems were about equally active in human HeLa cells (Fig. 7). More likely, the zebrafish cellular environment is more favorable for the FP system because of the absence of repressing activities that interfere with SB. Since transposases can mobilize inactive elements in trans, the ratio of inactive to active elements in eukaryotic genomes increases (22). Some of these inactive copies might function as repressors either by dominant-negative complementation (22), or by competition with transposase in substrate binding. Similarly, thousands of dispersed endogenous elements might inhibit the activity of an exogenously supplied transposase by transposase titration (45). Sleeping Beauty is a fish transposon, and the zebrafish genome contains about 1000 copies of a Tc1-like transposon, Tdr1 (31). It is likely that the endogenous Tdr1 elements can interfere with SB transposition, since they share over 80% sequence identity with SB, both on the DNA and protein sequence levels (31). In contrast, Frog Prince is a phylogenetically distant element (Fig. 3) with only ∼50% transposase sequence identity to either SB (Supplementary Material, Fig. 1B) or Tdr1 (not shown). Furthermore, _FP_-like transposons are not present in the zebrafish genome (data not shown). Thus, the newly introduced FP system is perhaps immune or at least less vulnerable to the above inhibitory mechanisms in zebrafish cells.
The ability of FP to precisely integrate single copies of foreign DNA into various chromosomal loci in a variety of vertebrate genomes allows us to propose the usefulness of the Frog Prince system in transgenesis and insertional mutagenesis. We have tested the ability of FP to efficiently trap expressed genes in a simple cotransfection assay in human cells. Klinakis et al. (2000) showed that the Tc1/mariner element Minos could potentially tag all human genes in HeLa cells. However, only ∼1–10% of all Minos insertions was estimated to represent actual gene trap events. We estimate that ∼30% of all selectable Frog Prince transposon insertions occur in genes (Fig. 6). To our knowledge, such high gene trapping frequencies have not been seen with other vectors. It is yet to be determined whether the higher gene trapping efficiency of FP reflects a different insertion site preference as compared to the Minos element. In comparison with retroviruses that preferentially integrate into the 5′-regions of genes (46), the integration pattern of Tc1-like transposons is more random (44). Therefore, transposon insertions are expected to produce a different mutational spectrum than retroviruses.
In the era of functional genomics, there is a sore need for developing efficient means to explore the roles of genes in different cellular functions. Both invertebrate and vertebrate transposon systems hold potentials for transgenesis and insertional mutagenesis in model organisms (6,9,11,12, 47,48). The availability of alternative transposon systems in the same species opens up new possibilities for genetic analyses. For example, piggyBac transposons can be mobilized in Drosophila in the presence of stably integrated P elements (48). Because P element- and _piggyBac_-based systems show different integration site preferences (48,49), the number of fly genes that can be insertionally inactivated by transposable elements can greatly be increased. P element vectors have also been used to introduce components of the mariner transposable element into the Drosophila melanogaster genome by stable germline transformation. In these transgenic flies, mariner transposition can be studied without accidental mobilization of P elements (50). We have shown that Frog Prince and Sleeping Beauty do not detectably interact in an in vivo transposition assay (Fig. 4). Thus, FP can be used as a genetic tool in the presence of SB, and vice versa, which considerably broadens the utility of these elements. As an alternative transposon system, significantly different from any other active transposon, Frog Prince can expand our possibilities for transposon-mediated genetic manipulations in vertebrates.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
[Supplementary Material]
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Fiedler and E. Stüwe for their technical assistance, and members of the Transposition Group at the MDC for critical reading of the manuscript. We also thank Akira Hikosaka for his help in the phylogenetic analysis. The PAC2 zebrafish cell line was a kind gift from W. Driever. The Minos-based gene trap vector was kindly provided by C. Savakis. This work was supported by EU grant QLG2-CT-2000-00821.
DDBJ/EMBL/GenBank accession nos AY261370–AY261372
REFERENCES
- 1.Driever W., Solnica-Krezel,L., Schier,A., Neuhauss,S., Maliki,J., Stemple,D., Stainier,D., Zwartkruis,F., Abdelilah,S., Rangini,Z., Belak,J. and Boggs,C. (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development, 123, 37–46. [DOI] [PubMed] [Google Scholar]
- 2.Haffter P., Granato,M., Brand,M., Mullins,M.C., Hammerschmidt,M., Kane,D.A., Odenthal,J., van Eeden,F.J., Jiang,Y.J., Heisenberg,C.P., Kelsh,R.N., Furutani-Seiki,M., Vogelsang,E., Beuchle,D., Schach,U., Fabian,C. and Nusslein-Volhard,C. (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development, 123, 1–36. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K.V. (2000) Finding the genes that direct mammalian development: ENU mutagenesis in the mouse. Trends Genet., 16, 99–102. [DOI] [PubMed] [Google Scholar]
- 4.Amsterdam A., Burgess,S., Golling,G., Chen,W., Sun,Z., Townsend,K., Farrington,S., Haldi,M. and Hopkins,N. (1999) A large-scale insertional mutagenesis screen in zebrafish. Genes Dev., 13, 2713–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zambrowicz B.P., Friedrich,G.A., Buxton,E.C., Lilleberg,S.L., Person,C. and Sands,A.T. (1998) Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature, 392, 608–611. [DOI] [PubMed] [Google Scholar]
- 6.Cooley L., Kelley,R. and Spradling,A. (1988) Insertional mutagenesis of the Drosophila genome with single P elements. Science, 239, 1121–1128. [DOI] [PubMed] [Google Scholar]
- 7.Zwaal R.R., Broeks,A., van Meurs,J., Groenen,J.T. and Plasterk,R.H. (1993) Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl Acad. Sci. USA, 90, 7431–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson H.M. (1995) The Tc1-mariner superfamily of transposons in animals. J. Insect Physiol., 41, 99–105. [Google Scholar]
- 9.Plasterk R.H., Izsvak,Z. and Ivics,Z. (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet., 15, 326–332. [DOI] [PubMed] [Google Scholar]
- 10.Coates C.J., Jasinskiene,N., Miyashiro,L. and James,A.A. (1998) Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc. Natl Acad. Sci. USA, 95, 3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadool J.M., Hartl,D.L. and Dowling,J.E. (1998) Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc. Natl Acad. Sci. USA, 95, 5182–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman A., Dawson,A., Mather,C., Gilhooley,H., Li,Y., Mitchell,R., Finnegan,D. and Sang,H. (1998) Transposition of the Drosophila element mariner into the chicken germ line. Nature Biotechnol., 16, 1050–1053. [DOI] [PubMed] [Google Scholar]
- 13.Schouten G.J., van Luenen,H.G., Verra,N.C., Valerio,D. and Plasterk,R.H. (1998) Transposon Tc1 of the nematode Caenorhabditis elegans jumps in human cells. Nucleic Acids Res., 26, 3013–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Sankar,U., Lampe,D.J., Robertson,H.M. and Graham,F.L. (1998) The Himar1 mariner transposase cloned in a recombinant adenovirus vector is functional in mammalian cells. Nucleic Acids Res., 26, 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izsvák Z., Ivics,Z. and Plasterk,R.H. (2000) Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol., 302, 93–102. [DOI] [PubMed] [Google Scholar]
- 16.Raz E., van Luenen,H.G., Schaerringer,B., Plasterk,R.H. and Driever,W. (1998) Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr. Biol., 8, 82–88. [DOI] [PubMed] [Google Scholar]
- 17.Klinakis A.G., Zagoraiou,L., Vassilatis,D.K. and Savakis,C. (2000) Genome-wide insertional mutagenesis in human cells by the Drosophila mobile element Minos. EMBO Rep., 1, 416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catteruccia F., Nolan,T., Loukeris,T.G., Blass,C., Savakis,C., Kafatos,F.C. and Crisanti,A. (2000) Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature, 405, 959–962. [DOI] [PubMed] [Google Scholar]
- 19.Akerley B.J., Rubin,E.J., Camilli,A., Lampe,D.J., Robertson,H.M. and Mekalanos,J.J. (1998) Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl Acad. Sci. USA, 95, 8927–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin E.J., Akerley,B.J., Novik,V.N., Lampe,D.J., Husson,R.N. and Mekalanos,J.J. (1999) In vivo transposition of _mariner_-based elements in enteric bacteria and mycobacteria. Proc. Natl Acad. Sci. USA, 96, 1645–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zagoraiou L., Drabek,D., Alexaki,S., Guy,J.A., Klinakis,A.G., Langeveld,A., Skavdis,G., Mamalaki,C., Grosveld,F. and Savakis,C. (2001) In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc. Natl Acad. Sci. USA, 98, 11474–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartl D.L., Lozovskaya,E.R., Nurminsky,D.I. and Lohe,A.R. (1997) What restricts the activity of _mariner_-like transposable elements? Trends Genet., 13, 197–201. [DOI] [PubMed] [Google Scholar]
- 23.Ivics Z., Hackett,P.B., Plasterk,R.H. and Izsvák,Z. (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish and its transposition in human cells. Cell, 91, 501–510. [DOI] [PubMed] [Google Scholar]
- 24.Yant S.R., Meuse,L., Chiu,W., Ivics,Z., Izsvák,Z. and Kay,M.A. (2000) Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nature Genet., 25, 35–41. [DOI] [PubMed] [Google Scholar]
- 25.Fischer S.E., Wienholds,E. and Plasterk,R.H. (2001) Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl Acad. Sci. USA, 98, 6759–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupuy A.J., Fritz,S. and Largaespada,D.A. (2001) Transposition and gene disruption in the male germline of the mouse. Genesis, 30, 82–88. [DOI] [PubMed] [Google Scholar]
- 27.Lam W.L., Seo,P., Robison,K., Virk,S. and Gilbert,W. (1996) Discovery of amphibian Tc1-like transposon families. J. Mol. Biol., 257, 359–366. [DOI] [PubMed] [Google Scholar]
- 28.Ivics Z. and Izsvák,Z. (1997) Family of plasmid vectors for the expression of β-galactosidase fusion proteins in eukaryotic cells. Biotechniques, 22, 254–256, 258. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z.J., Moav,B., Faras,A.J., Guise,K.S., Kapuscinski,A.R. and Hackett,P.B. (1990) Development of expression vectors for transgenic fish. Biotechnology, 8, 1268–1272. [DOI] [PubMed] [Google Scholar]
- 30.Devon R.S., Porteous,D.J. and Brookes,A.J. (1995) Splinkerettes–improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res., 23, 1644–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izsvák Z., Ivics,Z. and Hackett,P.B. (1995) Characterization of a Tc1-like transposable element in zebrafish (Danio rerio). Mol. Gen. Genet., 247, 312–322. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama I.N., Rakow,T.L. and Maruyama,H.I. (1995) cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Res., 23, 3796–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinogradov A.E. (1998) Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry, 31, 100–109. [DOI] [PubMed] [Google Scholar]
- 34.Ivics Z., Izsvák,Z., Minter,A. and Hackett,P.B. (1996) Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl Acad. Sci. USA, 93, 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izsvák Z., Khare,D., Behlke,J., Heinemann,U., Plasterk,R.H. and Ivics,Z. (2002) Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem., 277, 34581–34588. [DOI] [PubMed] [Google Scholar]
- 36.Doak T.G., Doerder,F.P., Jahn,C.L. and Herrick,G. (1994) A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common ”D35E” motif. Proc. Natl Acad. Sci. USA, 91, 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam W.L., Lee,T.S. and Gilbert,W. (1996) Active transposition in zebrafish. Proc. Natl Acad. Sci. USA, 93, 10870–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundararajan P., Atkinson,P.W. and O’Brochta,D.A. (1999) Transposable element interactions in insects: crossmobilization of hobo and Hermes. Insect Mol. Biol., 8, 359–368. [DOI] [PubMed] [Google Scholar]
- 39.van Luenen H.G., Colloms,S.D. and Plasterk,R.H. (1994) The mechanism of transposition of Tc3 in C. elegans. Cell, 79, 293–301. [DOI] [PubMed] [Google Scholar]
- 40.Lampe D.J., Churchill,M.E. and Robertson,H.M. (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J., 15, 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 41.Arca B., Zabalou,S., Loukeris,T.G. and Savakis,C. (1997) Mobilization of a Minos transposon in Drosophila melanogaster chromosomes and chromatid repair by heteroduplex formation. Genetics, 145, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo G., Ivics,Z., Izsvák,Z. and Bradley,A. (1998) Chromosomal transposition of a Tc1/_mariner_-like element in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA, 95, 10769–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronchain O.J., Hartley,K.O. and Amaya,E. (1999) A gene trap approach in Xenopus. Curr. Biol., 9, 1195–1198. [DOI] [PubMed] [Google Scholar]
- 44.Vigdal T.J., Kaufman,C.D., Izsvák,Z., Voytas,D.F. and Ivics,Z. (2002) Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol., 323, 441–452. [DOI] [PubMed] [Google Scholar]
- 45.Simmons M.J. and Bucholz,L.M. (1985) Transposase titration in Drosophila melanogaster: a model of cytotype in the P-M system of hybrid disgenesis. Proc. Natl Acad. Sci. USA, 82, 8119–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherdin U., Rhodes,K. and Breindl,M. (1990) Transcriptionally active genome regions are preferred targets for retrovirus integration. J. Virol., 64, 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawakami K., Shima,A. and Kawakami,N. (2000) Identification of a functional transposase of the Tol2 element, an _Ac_-like element from the japanese medaka fish and its transposition in the zebrafish germ lineage. Proc. Natl Acad. Sci. USA, 97, 11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hacker U., Nystedt,S., Barmchi,M.P., Horn,C. and Wimmer,E.A. (2003) _piggyBac_-based insertional mutagenesis in the presence of stably integrated P elements in Drosophila. Proc. Natl Acad. Sci. USA, 100, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spradling A.C., Stern,D.M., Kiss,I., Roote,J., Laverty,T. and Rubin,G.M. (1995) Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl Acad. Sci. USA, 92, 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lohe A.R. and Hartl,D.L. (2002) Efficient mobilization of mariner in vivo requires multiple internal sequences. Genetics, 160, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]