J Domain Co-chaperone Specificity Defines the Role of BiP during Protein Translocation (original) (raw)
Abstract
Hsp70 chaperones can potentially interact with one of several J domain-containing Hsp40 co-chaperones to regulate distinct cellular processes. However, features within Hsp70s that determine Hsp40 specificity are undefined. To investigate this question, we introduced mutations into the ER-lumenal Hsp70, BiP/Kar2p, and found that an R217A substitution in the J domain-interacting surface of BiP compromised the physical and functional interaction with Sec63p, an Hsp40 required for ER translocation. In contrast, interaction with Jem1p, an Hsp40 required for ER-associated degradation, was unaffected. Moreover, yeast expressing R217A BiP exhibited defects in translocation but not in ER-associated degradation. Finally, the genetic interactions of the R217A BiP mutant were found to correlate with those of known translocation mutants. Together, our results indicate that residues within the Hsp70 J domain-interacting surface help confer Hsp40 specificity, in turn influencing distinct chaperone-mediated cellular activities.
Keywords: Chaperone Chaperonin, Endoplasmic Reticulum (ER), Genetics, Heat Shock Protein, Protein Degradation, Protein Translocation, Yeast
Introduction
Most newly synthesized proteins require the assistance of molecular chaperones to fold into their native conformations. In addition, chaperones facilitate the assembly of macromolecular structures. To perform these functions, chaperones non-covalently bind to surface-exposed hydrophobic patches, thus preventing protein aggregation and, in some cases, providing a secure folding environment (1, 2). One family of chaperones, heat shock proteins of 70 kDa (Hsp70s),3 forms a major component of the cellular folding and stress response machinery (3, 4). Hsp70s are quite abundant and can be found in nearly every cellular compartment in eukaryotes and in most prokaryotes. Not surprisingly, they catalyze a multitude of functions, including protein folding, transport, and degradation.
Hsp70s contain a conserved N-terminal ATPase domain, followed by a less conserved substrate binding domain (SBD) and a variable C-terminal “lid.” The lid is flexible and helps confine the substrate within the SBD. The Hsp70 ATP hydrolytic cycle correlates with substrate binding and release, such that the ADP-bound state of Hsp70s has a higher affinity for substrates than the ATP-bound state. Because the ATPase activity of Hsp70s is weak, Hsp40 co-chaperones are needed to accelerate ATP hydrolysis and promote maximal chaperone activity (5–7). Select Hsp40 family members can also deliver substrates to Hsp70s. The interaction between Hsp70s and Hsp40s is mediated by the Hsp40 J domain, an ∼70-amino acid motif that forms a four-helix bundle. Notably, an invariant HPD motif between helices 2 and 3 of the J domain plays a critical role in facilitating Hsp70-Hsp40 interaction.
In recent years, it has become clear that Hsp40s are more diverse than Hsp70s (8). For example, Escherichia coli encode three Hsp70s and six Hsp40s, whereas the budding yeast Saccharomyces cerevisiae expresses 14 Hsp70s and 22 Hsp40s. The diversity increases in higher eukaryotes, with humans expressing 20 Hsp70s and >50 Hsp40s. Moreover, a single Hsp70 can interact with multiple Hsp40s to drive distinct cellular processes. For example, the Hsp70 found within the endoplasmic reticulum (ER) of yeast, BiP (encoded by the KAR2 gene), interacts with three Hsp40 co-chaperones: Sec63p, which spans the ER membrane three times and presents its J domain in the ER lumen; Jem1p, which is ER membrane-associated; and Scj1p, which is a soluble ER-lumenal protein (9–14). The interaction between BiP and Sec63p is indispensable for the co- and post-translational translocation of nascent proteins into the ER (15–17). In contrast, neither Jem1p nor Scj1p are required for protein translocation. Instead, these Hsp40s interact with BiP to maintain the solubility of aberrant proteins (18), which are then retrotranslocated from the ER and degraded by the cytoplasmic 26 S proteasome via a process termed ER-associated degradation (ERAD) (19, 20). Unlike Jem1p and Scj1p, mutations in Sec63p have little effect on ERAD. Therefore, BiP function appears to be dictated by its interaction with Hsp40 partners. Nevertheless, it is impossible to predict which of the many possible Hsp70-Hsp40 pairs will function coordinately to effect specific cellular processes.
The features within Hsp70s that determine Hsp40 specificity are also poorly defined. Studies on the bacterial Hsp70, DnaK, showed that the J domain-interacting surface mapped to a charged cleft on the underside of the DnaK ATPase domain (21–23). In particular, an invariant Arg at position 167 was found to interact with the Asp in the HPD motif of DnaJ, a bacterial Hsp40. Indeed, when the analogous residue (Arg197) was mutated in mammalian BiP/GRP78, a reduced interaction with two ER-lumenal Hsp40s, ERdj2/SEC63 (24) and ERdj3 (25, 26), was observed. Whether this mutation equally affects interaction with the four other Hsp40 co-chaperones of BiP/GRP78 and whether Arg197 contributes to the ability of BiP/GRP78 to distinguish between Hsp40s is unclear. In addition, other putative J domain contacts have been identified in the SBD of DnaK (22, 27) and BiP/GRP78 (25), although their exact roles in Hsp70-Hsp40 interaction remain undefined.
To better understand the rules that govern the formation of functional Hsp70-Hsp40 pairs, we focused on the yeast ER, wherein the association between BiP and each of its cognate Hsp40 partners and BiP-mediated function are well defined. Using genetic, biochemical, and genomic tools, we discovered that an R217A mutant form of BiP interacts poorly with Sec63p, yet Jem1p interaction remains robust. Accordingly, yeast expressing R217A BiP exhibit translocation but not ERAD defects as well as genetic interactions that are diagnostic for defects in translocation. By creating new mutations in the BiP SBD, we also established the importance of substrate binding for both protein translocation and ERAD. These data indicate that additional residues within the J domain-interacting surface of Hsp70s help confer specificity for an Hsp40 partner and link a unique Hsp70-Hsp40 pair to a distinct chaperone-catalyzed process.
EXPERIMENTAL PROCEDURES
Plasmids and Yeast Strains
For the heterologous expression of mutant BiP proteins in E. coli, the KAR2 coding sequence cloned into plasmid pMR2623 (28) was mutagenized using the QuikChange site-directed mutagenesis kit (Stratagene) with the following primer pairs (underlined letters represent the altered sequence): (i) R217A, 5′ primer (GCTGGTTTGAACGTTTGGCAATTGTTAATGAACCAACCGC) and 3′ primer (GCGGTTGTTCATTAACAATTGCCAAAACGTTCAAACCAGC; (ii) K584X, 5′ primer (GGCCAAGGTGAATCTAGAAACTAATTAGAAAACTACGCTCAC) and 3′ primer (GTGAGCGTAGTTTTCTAATTAGTTTCTAGATTCAACCTTGGCC); (iii) S493F, 5′ primer (CGAGGTGAAAGAGCCATGTTTAAGGACAACAATCTATTAGG) and 3′ primer (CCTAATAGATTGTTGTCCTTAAACATGGCTCTTTCACCTCG). The resulting plasmids were transformed into E. coli RR1 cells for large scale protein purification (28).
To generate yeast strains constitutively expressing either the wild-type or mutant BiP proteins, the KAR2 coding sequence was first inserted into the pYES2 vector (2μ, URA3, PGAL1/10; Invitrogen); this strategy was essential to create constructs for the expression of the mutants. The KAR2 coding sequence was amplified from plasmid pMR713 (CEN4/ARS, LEU2, PKAR2-KAR2; a kind gift from M. D. Rose, Princeton University) with the following primer pair (italicized letters indicate the BamHI recognition site on the 5′ primer and the XhoI recognition site on the 3′ primer): 5′ primer, GTA_GGATCC_CCAGAGTAGTCTCAA; 3′ primer, TAC_CTCGAG_CTACAATTCGTCGTGTTC.
The resulting PCR product was digested with BamHI and XhoI and ligated into pYES2 to generate the p_GAL1-KAR2_ plasmid construct. The primer pairs described above were then used in site-directed mutagenesis experiments to introduce the R217A, K584X, and S493F mutations into p_GAL1-KAR2_, thus generating the vectors p_GAL1-kar2-R217A_, p_GAL1-kar2-K584X_, and p_GAL1-kar2-S493F_, respectively. These constructs were used to establish that mutant protein overexpression does not lead to a dominant negative effect on cell growth (see “Results”).4 Next, to constitutively express BiP from the PTEF1 promoter present in the p414TEF1 vector (CEN4/ARS, TRP1, PTEF1) (29), the KAR2 gene cloned into p_GAL1-KAR2_ was removed using BamHI and XhoI and ligated into p414TEF1; p414TEF1 was digested with the restriction enzymes BamHI and SalI. However, transformation of the ligation mixture into E. coli failed to yield clones of the desired genotype. Therefore, the ligation mixture was transformed directly into the yeast strain MMY8-2 (_MAT_α, _his3-Δ_200, _leu2-Δ_1, ura3-52, _trp1-Δ_63, kar2::HIS3, pMR397 (2μ, URA3, KAR2)) (30), and transformants were selected on synthetic complete medium lacking both uracil and tryptophan and containing 2% glucose. Viable clones were then plated onto selective synthetic complete medium lacking tryptophan and containing 2% glucose and 1 mg/ml 5-fluoroorotic acid. Clones that survived in the presence of 5-fluoroorotic acid had the genotype _MAT_α, his3-Δ_200, leu2-Δ_1, ura3-52, trp1-Δ_63, kar2::HIS3, pTEF1-KAR2 (CEN4/ARS, TRP1, PTEF1-KAR2) and were called TEF1-KAR2. The TEF1-R217A, TEF1-K584X, and TEF1-S493F mutant strains were created in a similar manner using the p_GAL1-kar2-R217A, p_GAL1-kar2-K584X, and p_GAL1-kar2-S493F vectors, respectively, which were described above.
To generate wild-type and mutant kar2 strains amenable to manipulation in the UPR-based genetic screen (see below), we first inserted the KAR2 or kar2-R217A coding sequence upstream of the NATMX6 antibiotic resistance cassette in the pFA6a-NATMX6 vector (31). Accordingly, the KAR2 gene containing the 3′-UTR (henceforth referred to as KAR2-UTR) was amplified with the following primer pair (the underlined sequence in the 5′ primer represents the PvuII recognition site and in the 3′ primer represents the BamHI recognition site): 5′ primer, GTCCCCAAGAGCAGCTGCAAGGGAAA; and 3′ primer, CAATAGTGATGGGATCCGATGAGATGA.
The resulting PCR product was digested with the indicated restriction enzymes and inserted into the vector pFA6a-NATMX6 to generate the plasmid construct p_KAR2-UTR-NAT_. Site-directed mutagenesis was then used to insert the R217A mutation into p_KAR2-UTR-NAT_ with the primer pair described above, creating p_kar2-R217A-UTR-NAT_. Next, to carry out targeted gene replacement, the KAR2-UTR-NAT (or kar2-R217A-UTR-NAT) cassette cloned into p_KAR2-UTR-NAT_ (or p_kar2-R217A-UTR-NAT_) was amplified with the primer pair: 5′ primer (AAAGATTAACGTGTTACTGTTTTACTTTTTTAAAGTCCCCAAGAGTAGTCTCAAGGGAAAAAGCGTATC), and 3′ primer (CCATTTCAGTATTAGGTTCTCGAGCCTTTCAACTCTCTCTGTTATAATGTGAATTCGAGCTCGTTTAAAC), and the resulting PCR product was transformed into the BY4743 strain (MATa/α, _his3-_Δ_0/his3-Δ_0, _leu2-_Δ_0/leu2-Δ_0, _ura3-_Δ_0/ura3-Δ_0, TRP1/TRP1, _LYS2/lys2-Δ_0, _MET15/met15-Δ_0, KAR2/KAR2; Invitrogen). Transformants were selected on yeast extract-peptone-dextrose medium supplemented with 100 μg/ml nourseothricin. To identify strains in which one copy of the chromosomal KAR2 gene was replaced with KAR2-UTR-NAT (or kar2-R217A-UTR-NAT), PCR amplification was performed with the following primer pair: 5′ primer, GGCTATGTAATTCTAAAGATTAACGT; 3′ primer, GTTATCTTAGGGTCATACTCATCAATT. Strains in which homologous recombination had taken place at a single KAR2 chromosomal locus yielded PCR products that contained two bands, corresponding to endogenous KAR2 (2.1 kb) and KAR2-UTR-NAT (or kar2-R217A-UTR-NAT) (3.9 kb). Next, the confirmed diploid strains were subjected to sporulation and tetrad analysis, and haploid strains of the genotype MATa, his3_-Δ_1, leu2_-Δ_0, ura3_-Δ_0, met15_-Δ_0, KAR2-UTR-NAT (or kar2-R217A-UTR-NAT) were selected for further analysis. The _NAT_-marked wild-type strain was called KAR2::NAT and is isogenic to BY4741. The _NAT_-marked kar2-R217A mutant strain was called kar2-R217A::NAT.
To generate a strain containing a hypomorphic allele of KAR2, the KAR2 gene lacking the 3′-UTR (henceforth referred to as kar2-DamP) was amplified from yeast genomic DNA with the following primer pair (the underlined sequence in the 5′ primer represents the PvuII recognition site and in the 3′ primer represents the BamHI recognition site): 5′ primer, GTCCCCAAGAGCAGCTGCAAGGGAAA; 3′ primer, CAACCTTGAAGGATCCAGCAGCAAAA. The resulting PCR product was digested with PvuII and BamHI and inserted upstream of the NATMX6 antibiotic resistance cassette in pFA6a-NATMX6 to generate the p_kar2-DamP-NAT_ plasmid. The strategy described above was then used to create the kar2-DamP::NAT strain.
Protein Purification and ATP Hydrolysis Assays
Hexahistidine-tagged wild-type and mutant BiP proteins expressed from pMR2623 were purified from E. coli RR1 cells using a previously optimized protocol (28). GST-tagged J domains from Sec63p and Jem1p were purified from E. coli BL21(DE3) and E. coli TG1 cells, respectively, according to established protocols (30, 32).
Steady state ATPase assays were performed at 30 °C as previously described (28, 33). The amount of BiP present in each reaction and the ratios of the J domain-containing cofactors or peptide p5 (CLLLSAPRR; a kind gift from L. Gierasch, University of Massachusetts, Amherst, MA) to BiP are indicated in the legends for Figs. 1 and 2. The J domain-containing cofactors and peptide were preincubated with BiP on ice for 10 min in the absence of radiolabeled ATP before initiating the assay upon the addition of [α-32P]ATP (PerkinElmer Life Sciences).
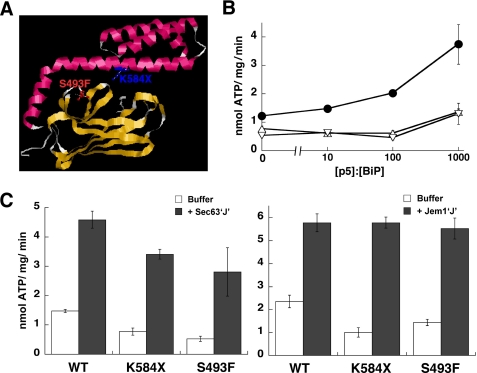
FIGURE 1.
The K584X and S493F BiP mutants are defective for peptide-stimulated ATPase activity. A, the Lys584 and Ser493 residues were mapped onto the crystal structure of the SBD and lid domain of the bacterial Hsp70 DnaK (68) after aligning the BiP and DnaK protein sequences. Although Lys584 is conserved between BiP and DnaK, Ser493 is Ala448 in DnaK. B, ATPase assays were performed in the presence of increasing molar ratios of peptide p5 (CLLLSAPRR) (69). Reactions contained wild-type (WT) (●), K584X (▵), or S493F (▿) BiP at a final concentration of 0.7 μm. Data represent the means of a minimum of three independent experiments ± S.E. C, ATPase assays were performed in the absence (white bars) or presence (gray bars) of the J domains from Sec63p (left panel) or Jem1p (right panel), as described (30, 32). The wild-type or mutant BiP proteins (2.1 μm) and the J domains were present in equimolar amounts. Data represent the means of a minimum of three independent experiments ± S.E. (error bars).
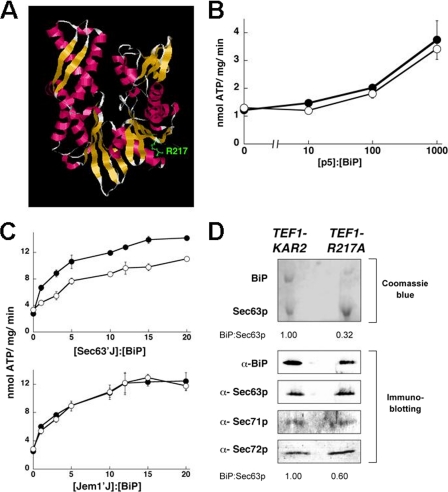
FIGURE 2.
R217A BiP exhibits J domain specificity. A, residue R217 was mapped onto the crystal structure of the ATPase domain of bovine Hsc70 (44) after aligning the protein sequences of BiP and Hsc70. This residue is conserved. B, the ATPase activity of wild-type (●) and R217A BiP (○) at a final concentration of 0.7 μm was measured in the presence of increasing molar ratios of peptide p5. C, the ATPase activity of wild-type or R217A BiP at a final concentration of 0.7 μm was measured in the absence or presence of increasing amounts of the J domains from Sec63p or Jem1p. Data represent the means of a minimum of three independent experiments ± S.E. (error bars). D, the Sec63p complex, which contains BiP, Sec63p, Sec71p, and Sec72p, was purified from microsomes derived from TEF1-KAR2 or TEF1-R217A yeast, resolved by SDS-PAGE, and subjected to Coomassie Brilliant Blue staining or immunoblot analysis. The ratio of BiP to Sec63p from each analysis is indicated below the corresponding set of panels.
Purification of the Sec63 Complex
The Sec63 complex containing BiP, Sec63p, Sec71p, and Sec72p was purified from microsomes derived from TEF1-KAR2 and TEF1-R217A yeast under non-denaturing conditions using a three-step chromatographic protocol (34). The only modification involved the substitution of the Superose-6 resin that was recommended for size exclusion chromatography with a 32-ml Sephacryl S-300 column (Amersham Biosciences) that was run under gravity. After purification, the components of the complex were detected by Coomassie Brilliant Blue staining and immunoblotting. Images of Coomassie Brilliant Blue-stained gels were captured using Image Station 440CF (Eastman Kodak Co.), and the BiP/Sec63p ratio was quantified using the Kodak 1D software. For immunoblotting, the following antibodies were used: anti-BiP (34); anti-Sec63, which recognizes the J domain of Sec63p (a kind gift from R. Schekman, University of California, Berkeley, CA); anti-Sec71p (35); and anti-Sec72p (34). Primary, bound antibodies were decorated with the appropriate horseradish peroxidase-conjugated secondary antibody, and signals were detected using the SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific).
Serial Dilutions
The TEF1-KAR2 and TEF1-R217A strains were grown to logarithmic phase (_A_600 = 0.6–0.8) in selective synthetic complete medium containing 2% glucose for ∼16 h. The TEF1-K584X and TEF1-S493F strains were slow growing and required longer incubation periods to reach the logarithmic phase of growth. 10-fold serial dilutions of equivalent OD units of each strain were spotted onto solid medium and cultured at the indicated temperatures for 2 days. Where indicated, the growth medium was supplemented with 8 mm dithiothreitol (DTT).
Pulse Labeling of Cells and Immunoprecipitation
To measure the stability of BiP in wild-type and mutant TEF1-KAR2 strains, cells were radioactively labeled and chased as previously described (36). Briefly, 20 OD units of logarithmic phase cells were harvested, washed, and resuspended in synthetic complete medium lacking methionine to 10 OD units/ml. After recovery at 30 °C for 30 min, the cells were labeled with 20 μl of Express 35S labeling mix (PerkinElmer Life Sciences) for 10 min. Next, cycloheximide was added to a final concentration of 200 μg/ml to stop protein translation, and samples were collected at 0, 20, 40, and 60 min. At each time point, 4 OD units of cells were harvested, 0.1 m sodium azide was added, and cell extracts were prepared using glass bead lysis in the presence of protease inhibitors. The radioactivity in each sample was measured using a scintillation counter (Beckman), and for each sample, a total volume corresponding to ∼5 × 106 cpm was treated with polyclonal anti-BiP antiserum (see above) for >12 h at 4 °C followed by incubation with protein A-Sepharose for 2 h at 23 °C. Immunoprecipitated proteins were resolved by SDS-PAGE, and the radiolabeled proteins were detected by autoradiography. Relative amounts of radioactivity, which corresponded to relative protein levels, were quantified using Image Gauge software (FujiFilm).
To analyze prepro-α-factor (ppαF) translocation in vivo, the TEF1-KAR2 and TEF1-R217A strains were transformed with plasmid pSM36-ΔgppαF-HA, which expresses an HA-tagged version of ppαF lacking the core consensus glycosylation sequences (37). Transformants were grown to logarithmic phase, labeled, and chased as described above, except that samples were collected at 0 and 10 min. Immunoprecipitation of a sample volume corresponding to ∼10 × 106 cpm was performed with an anti-HA monoclonal antibody (Roche Applied Science) and analyzed as above.
To evaluate the effects of the KAR2 mutations on CPY* degradation, a plasmid encoding an HA-tagged version of CPY*, pDN431 (38), was transformed into wild-type and mutant TEF1-KAR2 strains. Transformants were grown to logarithmic phase, labeled, and chased for the indicated times, and immunoprecipitates were analyzed as above.
β-Galactosidase Assays to Measure the Induction of the UPR
Wild-type and mutant TEF1-KAR2 strains were transformed with the UPR reporter plasmid, pJC104 (a kind gift from P. Walter, University of California, San Francisco) (39), which contains the lacZ gene encoding β-galactosidase downstream from four copies of the unfolded protein response element. The readout for UPR induction (i.e. β-galactosidase expression) was assayed using a standard protocol (40). Prior to harvesting, cells were grown to logarithmic phase at 30 °C, shifted to 37 °C for 1 h, or treated with 8 mm DTT for 1 h at 30 °C.
In Vitro Translocation and ERAD Assays
To measure the translocation and ERAD efficiencies of wild-type and mutant forms of ppαF, respectively, ER-derived microsomes were prepared from the TEF1-KAR2, TEF1-R217A, TEF1-K584X, and TEF1-S493F strains as described (41). In brief, 2000–3000 OD units of logarithmic phase cells were harvested, washed, and subjected to lyticase treatment in order to digest the cell wall. After digestion, spheroplasts lacking the cell wall were centrifuged through 0.8 m sucrose, 1.5% Ficoll 400, 20 mm HEPES-NaOH, pH 7.4. Next, the spheroplasts were homogenized in 0.1 m sorbitol, 50 mm KOAc, 2 mm EDTA, 20 mm HEPES-NaOH, pH 7.4, supplemented with protease inhibitors, and microsomes were collected as a floating fraction at the top of 1.0 m sucrose, 50 mm KOAc, 20 mm HEPES-NaOH, pH 7.4, 1 mm DTT. Finally, the microsomes were washed and resuspended in 20 mm HEPES-NaOH, pH 6.8, 150 mm KOAc, 5 mm MgOAc, 250 mm sorbitol at a protein concentration of 10–12 mg/ml (_A_280 ∼40/ml). Single-use aliquots were stored at −80 °C.
Next, radiolabeled ppαF and ΔgppαF were synthesized using the plasmid templates pDJ100 (42) and pGem2α36–3Q (43), respectively, with Promega's TnT® coupled reticulocyte lysate system. Each plasmid template was mixed on ice with Express 35S labeling mix, ribonuclease inhibitor, SP6 RNA polymerase, an amino acid mixture lacking methionine, and rabbit reticulocyte lysate, according to the manufacturer's instructions. The reaction was incubated at 30 °C for 90 min, and the resulting radiolabeled translation product was aliquoted and stored at −80 °C.
For in vitro translocation assays, microsomes derived from wild-type or mutant TEF1-KAR2 strains were mixed with radiolabeled ppαF and an ATP-regenerating system, and the assay was performed as described previously (41). For in vitro ERAD assays (43), translocation of ΔgppαF was initially carried out by incubating radiolabeled ΔgppαF with microsomes prepared from wild-type or mutant TEF1-KAR2 strains and an ATP-regenerating system for 1 h at 20 °C. Next, to measure ERAD efficiency, microsomes containing translocated ΔgpαF were harvested, washed, and either mixed with 0.5 mg/ml of cytosol derived from RSY607 yeast (_MAT_α, leu2-3, 112, ura3-52, PEP4::URA3) and an ATP-regenerating system or directly resuspended in reaction buffer (20 mm HEPES, pH 6.8, 150 mm KOAc, 250 mm sorbitol, and 5 mm MgOAc). The reaction mixture was incubated at 30 °C for 20 min, after which proteins were trichloroacetic acid-precipitated, resolved by SDS-PAGE, and analyzed by autoradiography. The data were quantified using Image Gauge software (FujiFilm).
UPR-based Genetic Analysis
The UPR-based genetic interactions of wild-type and mutant kar2 strains were assessed as described (45). Briefly, double mutant strains containing a _NATMX6_-marked kar2 allele and a single _KANMX6_-marked gene deletion that perturbs steady-state UPR levels were generated using the systematic genetic analysis method (46). Next, high throughput flow cytometry was used to measure the fluorescence of a GFP reporter of UPR activity and a constitutive RFP control in each double mutant. The genetic interaction exhibited by each double mutant was quantified using a π-score, which represents the difference between the observed reporter levels in the double mutant strain and those expected if there was no genetic interaction. Correlations between the patterns of π-scores of each kar2 mutant and the π-scores of every other mutant in the data set described by Jonikas et al. (45) were calculated using uncentered Pearson correlation.
RESULTS
The Functional Interaction with a J Domain Is Unaffected by Mutations in the BiP Substrate Binding Domain
We previously reported that mutations in the substrate binding pocket of yeast BiP compromised substrate affinity and ERAD efficiency but did not affect J domain-stimulated ATPase activity (30). To explore this phenomenon in greater detail, we created the following mutants (Fig. 1A). First, an S493F mutation was introduced into the BiP SBD. This mutation is predicted to disrupt a salt bridge between the SBD and lid and thus may alter the “hinge” that entraps substrates (47). Moreover, the loss of the hinge may affect J domain-mediated allosteric regulation of Hsp70s. Second, we introduced a translational stop site at position 584, which generates the K584X mutant. The resulting protein lacks part of the SBD and the entire lid. Surface plasmon resonance studies demonstrated that the K584X mutant binds to the same range of peptide substrates as wild-type BiP, albeit with a higher off-rate in the presence of ADP (48). This C-terminal deletion also lacks residue Leu585, which may contribute to allosteric regulation by Hsp40 partners (22).
To begin, hexahistidine-tagged wild-type and mutant BiP proteins were purified from E. coli, and steady-state ATPase activities were measured in the presence of increasing amounts of a peptide substrate previously shown to accelerate ATP hydrolysis of BiP (30). Over a 3-log range of peptide concentrations, the K584X and S493F BiP mutants were found to be defective for peptide-stimulated ATPase activity as compared with the wild-type protein (Fig. 1B). This was anticipated based on the predicted effects of these mutations on the lid domain. Interestingly, even in the absence of peptide, the K584X and S493F mutants exhibited reduced ATP hydrolytic activities (Fig. 1B). These reduced rates may result from an altered conformation of the SBD, which might in turn affect communication between this domain and the ATPase domain (49).
Next, we analyzed the steady-state rates of ATP hydrolysis upon the addition of the J domains from two BiP Hsp40 co-chaperones, Sec63p and Jem1p (30, 32). As above, the K584X and S493F mutants showed a decreased rate of endogenous activity as compared with the wild-type protein; however, their ATPase activities were proficiently stimulated by either J domain (Fig. 1C). Together, these data indicate that BiP mutants can be obtained that are proficient for Hsp40 co-chaperone stimulation but are unable to exhibit functional peptide interaction.
The R217A BiP Mutant Displays J Domain Specificity
To complement our studies on BiP mutations that specifically compromise peptide binding, we wished to examine mutations that exclusively alter J domain interaction. To this end, several point mutations were introduced into the putative J domain-interacting surface on BiP. In this report, we will focus on the R217A mutation (Fig. 2A). Based on published studies (22, 24, 25), the alteration of the conserved Arg residue at position 217 should impinge upon the ability of J domains to interact with BiP.
First, the steady state ATPase activity of a hexahistidine-tagged form of R217A BiP purified from E. coli (1.5 ± 0.09 nmol of ATP hydrolyzed/mg/min) was found to be identical to that of the wild-type protein (1.5 ± 0.06 nmol of ATP hydrolyzed/mg/min). Consistent with these data, mutations in the analogous residue in mammalian BiP had no effect on ATP hydrolysis or on ATP binding (25). Furthermore, the peptide-stimulated ATPase activities of wild-type and R217A BiP were identical (Fig. 2B). This discovery suggested that J domain interaction might be compromised without affecting peptide binding. This result was also intriguing because earlier work with DnaK showed that mutating a conserved Glu that is 3 residues C-terminal to this site disrupts the coupling of ATP hydrolysis to substrate binding (50).
Second, to examine whether R217A BiP functionally interacts with the J domains from Sec63p and Jem1p, reactions were assembled that contained wild-type or R217A BiP and increasing amounts of the Sec63p or Jem1p J domains. To our surprise, R217A BiP exhibited a decreased extent of ATPase stimulation by the J domain from Sec63p but was proficiently stimulated to wild-type levels by the Jem1p J domain (Fig. 2C). For example, at a 3-fold molar excess of the Sec63p J domain to BiP, the ATPase activity of the wild-type chaperone was enhanced by almost 300%, whereas the R217A mutant was stimulated only by ∼65%. Even at the highest concentrations examined, there was a significant difference between the ability of Sec63′J′ to stimulate the ATPase activities of wild-type versus R217A BiP.
To confirm these findings, we also examined the interaction between wild-type or R217A BiP and tagged forms of the Sec63p and Jem1p J domains in vitro. These assays were also performed in the presence of ADP or ATP, in order to ensure that any observed interactions reflected bona fide chaperone-co-chaperone associations. We found that there was an ∼3-fold difference in the ATP- and ADP-dependent association between wild-type BiP and both Sec63′J′ and Jem1′J′ (supplemental Fig. S1). In contrast, whereas the R217A BiP and Jem1′J′ interaction was within 9% of wild-type efficiency with the R217A mutant, there was a 65% decrease in the association of this mutant protein with Sec63′J′.
Together, these data indicate that a single amino acid on the surface of an Hsp70 helps distinguish between different J domains. The data also indicate that Hsp70 alleles can be obtained that exhibit J domain specificity. However, although the structures of the Jem1p and Sec63p J domains are predicted to be similar, we note that their sequences are only ∼40% identical, which might provide additional BiP-interacting residues in the case of Jem1p.
Yeast Expressing BiP Substrate Binding Mutants Are Sensitive to ER Stress
Our in vitro analyses indicated that K584X and S493F BiP are defective for substrate binding, whereas R217A BiP exhibits a defect for Sec63p J domain interaction. To examine the in vivo consequences of expressing these mutant BiP proteins, we engineered yeast to produce each protein as the only copy of BiP in the cell; preliminary work established that none of the mutants exhibit a dominant negative effect.4 Because mutations in KAR2 that affect its function can activate the UPR and enhance its transcription (30, 51), the genes encoding wild-type and mutant BiP were transcribed from the constitutive TEF1 promoter (29); the level of the wild-type protein expressed from PTEF1 was identical to that from the endogenous promoter of BiP at 30 °C.4 The resulting strains were denoted TEF1-KAR2, TEF1-R217A, TEF1-K584X, and TEF1-S493F.
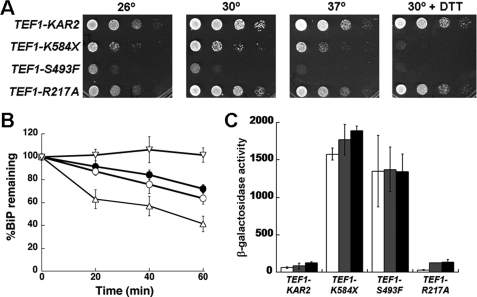
We observed that, similar to the wild-type TEF1-KAR2 strain, TEF1-R217A yeast grew robustly at all temperatures and in the absence or presence of DTT, a reducing agent that causes ER stress (Fig. 3A). In contrast, the TEF1-K584X and TEF1-S493F strains showed modest and strong DTT-sensitive growth defects, respectively (Fig. 3A). To examine whether these phenotypes resulted from protein instability, BiP levels were examined in the TEF1-KAR2, TEF1-R217A, TEF1-K584X, and TEF1-S493F strains by pulse-chase analysis. We found that the K584X mutant was turned over more rapidly than wild-type or R217A BiP (Fig. 3B), suggesting that the lid domain confers stability and that DTT sensitivity of the TEF1-K584X strain (Fig. 3A) may arise from reduced protein levels. In contrast, S493F BiP appeared to be hyperstable.
FIGURE 3.
Characterization of yeast strains expressing wild-type, K584X, S493F, or R217A BiP from the PTEF1 promoter. A, 10-fold serial dilutions of yeast expressing wild-type or mutant BiP were plated onto selective medium. The plates were incubated at 26, 30, or 37 °C for 2 days. Where indicated, growth was also tested in the presence of 8 mm DTT. B, to measure the stability of the wild-type and mutant BiP proteins, pulse-chase immunoprecipitation assays were performed with the TEF1-KAR2 (●), TEF1-R217A (○), TEF1-K584X (▵), and TEF1-S493F (▿) strains grown at 30 °C. At each time point, data were standardized to the amount of protein at the start of the chase and represent the means of three or more independent experiments ± S.E. C, UPR induction in the wild-type and indicated mutant strains was analyzed using a β-galactosidase reporter assay. Cells were incubated either at 30 °C (white bars), shifted to 37 °C for 1 h (gray bars), or treated with 8 mm DTT for 1 h at 30 °C (black bars). Data represent the means of two independent experiments ± range of the observed values. The DTT-stimulated UPR induction in the TEF1-KAR2 strain was 2-fold higher than the control, whereas in the TEF1-R217A strain it was 4.5-fold higher than the corresponding control.
The growth defects of TEF1-K584X and TEF1-S493F yeast could also be attributed to a constitutive induction of the unfolded protein response (UPR) by the BiP mutants (30, 51, 52). Because the addition of DTT leads to an accumulation of unfolded proteins and, consequently, UPR induction, this may cause a more severe growth defect. To measure the UPR, the wild-type and mutant strains were transformed with a reporter plasmid, and UPR levels were assessed in the absence or presence of DTT or at elevated temperatures. The TEF1-K584X and TEF1-S493F strains showed a greater than 10-fold higher induction of the UPR at 30 °C when compared with the TEF1-KAR2 strain, but UPR levels in TEF1-R217A and wild-type cells were low and quite similar to one another under each condition (Fig. 3C). Moreover, stressors such as elevated temperature and DTT did not further exaggerate the already high level of UPR induction in the TEF1-K584X and TEF1-S493F strains. These data demonstrate that the expression of K584X and S493F BiP results in maximal UPR induction and provide an explanation for the growth defects of the respective mutant strains. The data also indicate that TEF1-R217A yeast do not exhibit a basal ER stress response, although the strains exhibit a defective BiP-Sec63p interaction. We therefore propose that this strain can be used to monitor how a deficiency in Hsp40 interaction affects Hsp70-mediated cellular processes. Hence, we primarily focused next on characterizing the R217A mutant form of yeast BiP.
Sec63p Complex Formation Is Reduced in R217A BiP-expressing Yeast
BiP can be isolated in a complex with Sec63p, Sec71p, and Sec72p from detergent-solubilized ER membranes (34). The stoichiometric interaction between BiP and Sec63p, which is mediated by the J domain of Sec63p, localizes BiP at the ER membrane, where it facilitates protein translocation (14, 16). Because we observed a reduced interaction of R217A BiP with the J domain of Sec63p in vitro (Fig. 2C), we asked whether the BiP-Sec63p interaction was maintained in TEF1-R217A yeast. Accordingly, microsomes were prepared from TEF1-KAR2 or TEF1-R217A yeast, solubilized in octylglucopyranoside, and subjected to three chromatographic steps. Individual components of the complex were identified using Coomassie Brilliant Blue staining and by immunoblotting after SDS-PAGE; the immunoblot analysis confirmed that each component resided in the isolated complex (Fig. 2D, Immunoblotting). As previously reported (34), wild-type BiP and Sec63p were present in equal amounts in the complex purified from the TEF1-KAR2 wild-type strain (Fig. 2D, Coomassie Blue). However, there was an ∼70% reduction in the amount of R217A BiP that co-purified with Sec63p when the complex was isolated from the TEF1-R217A strain. Therefore, the in vitro defect in R217A BiP-Sec63p interaction was also evident in vivo and in fact was magnified in vivo, as observed in other studies (28). As a control, we also examined the interaction between wild-type or the R217A mutant and an epitope-tagged version of Jem1p. We discovered that the wild-type and mutant BiP bound with equal efficiencies to Jem1p when an anti-BiP pull-down was performed using yeast lysates (supplemental Fig. S2). This result cements our conclusion that the R217A mutant interacts proficiently with Jem1p.
Yeast Expressing R217A BiP Exhibit Translocation but Not ERAD Defects
Protein translocation across the ER membrane is the first commitment step in the secretory pathway. Given that BiP, along with Sec63p, plays a vital role during protein translocation (15–17), we examined translocation efficiency in the TEF1-KAR2 and TEF1-R217A strains. Each strain was transformed with a plasmid encoding an HA-tagged version of a prepro-α-factor mutant that is unable to acquire _N_-linked core glycans (ΔGppαF) (37). The appearance of a signal sequence-processed form of the substrate (i.e. pαF) after radiolabeling and immunoprecipitation is indicative of translocation. We discovered that the untranslocated product, denoted ppαF in Fig. 4A, was present in the TEF1-R217A strain but absent from wild-type cells, suggesting that R217A BiP expression results in translocation defects (Fig. 4A). The relatively subtle translocation defect in this experiment most probably arises from proteasome-mediated degradation of the untranslocated species (see below).4
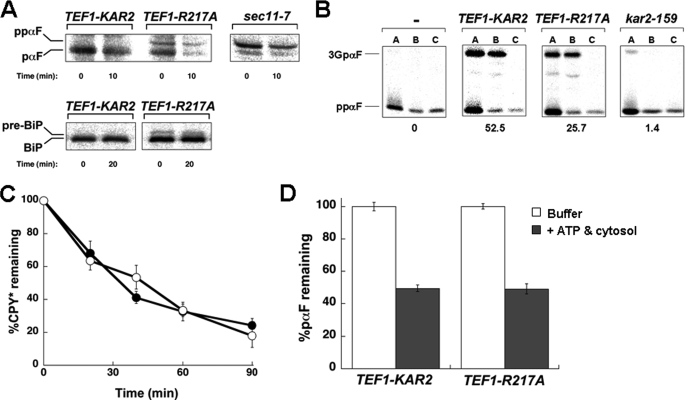
FIGURE 4.
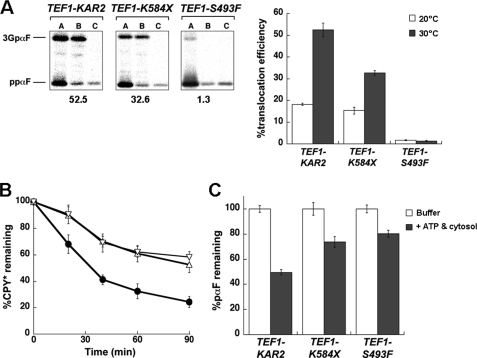
Yeast expressing R217A BiP exhibit a defect in protein translocation. A, the ER translocation of ΔGppαF and pre-BiP was analyzed at the indicated time points during a pulse-chase immunoprecipitation experiment using TEF1-KAR2 and TEF1-R217A yeast. The signal peptidase-deficient strain, sec11-7, served as a positive control. B, translocation assays with wild-type ppαF were performed using microsomes derived from TEF1-KAR2 or TEF1-R217A yeast. As controls, reactions either lacked microsomes (−) or contained microsomes derived from the kar2–159 mutant strain. After 60 min, each reaction was aliquoted and treated with buffer (A), trypsin (B), or trypsin and Triton X-100 (C). The percentage of translocation efficiency is indicated below each panel and corresponds to the means from a minimum of four independent experiments. C, the degradation of a soluble ERAD substrate, CPY*, was measured by pulse-chase analysis in the TEF1-KAR2 (●) and TEF1-R217A (○) strains. At each time point, data were standardized to the amount of protein at the start of the chase and represent the means of a minimum of three independent experiments ± S.E. D, ERAD assays were performed to assess the degradation of pαF using microsomes from TEF1-KAR2 or TEF1-R217A yeast. Reactions were performed in the absence (white bars) or presence (gray bars) of an ATP regeneration system and 0.5 mg/ml yeast cytosol. Data were standardized to the amount of protein remaining in the absence of cytosol and ATP regeneration system and represent the means of a minimum of four independent experiments ± S.E. (error bars).
To demonstrate that the slower migrating protein represented the signal sequence-containing species, we also performed this analysis in sec11-7 yeast, which carry a mutation in a signal peptidase subunit (53). The major product in this mutant was ppαF, which co-migrated with the untranslocated species observed only in the TEF1-R217A strain (Fig. 4A). To confirm that TEF1-R217A yeast exhibit a translocation defect, we examined whether a signal sequence-containing form of BiP (i.e. pre-BiP) was present in TEF1-R217A but not in wild-type yeast. Consistent with data demonstrating a ΔGppαF translocation defect, a fraction of BiP accumulated in the uncleaved form in TEF1-R217A cells (Fig. 4A).
To further establish that the R217A BiP allele confers a translocation defect, we used an in vitro assay in which the translocation of wild-type ppαF can be monitored in ER-derived microsomes (34). Upon translocation into the ER, the signal sequence of ppαF is cleaved, and the protein becomes triply glycosylated to form 3GpαF. When microsomes derived from TEF1-KAR2 wild-type yeast were examined in this assay, a translocation efficiency of ∼50% was obtained (Fig. 4B). However, microsomes containing R217A BiP translocated ppαF only half as efficiently. Of note, these experiments were performed with saturating levels of microsomes so that the observed effect does not reflect a limiting microsome concentration. Moreover, as a control for this experiment, we noted that translocation was virtually absent in microsomes prepared from kar2–159 yeast, as observed previously (41, 54). Together, these data demonstrate that R217A BiP is unable to support maximal levels of translocation, an effect that most likely arises from reduced interaction between R217A BiP and Sec63p.
Because the R217A mutant interacted with Jem1p at wild-type levels (Fig. 2C), we anticipated that yeast expressing this mutant would be ERAD-proficient (18). Therefore, the degradation of a model ERAD substrate, CPY* (55), was assessed in TEF1-KAR2 and TEF1-R217A yeast. We observed that CPY* was turned over in the TEF1-R217A strain as efficiently as in the wild-type strain (Fig. 4C). To substantiate these data, we evaluated the degradation of pαF in an in vitro system. pαF is the translocation product of ΔGppαF, and due to its inability to acquire _N_-linked glycans, it becomes an ERAD substrate. In yeast microsomes, the retrotranslocation and proteasome-mediated degradation of pαF occur in a cytosol- and ATP-dependent manner (43, 56). Consistent with our in vivo results, we observed that microsomes prepared from the TEF1-R217A strain degraded pαF identically to microsomes derived from the wild-type strain (Fig. 4D), confirming that R217A BiP expression does not affect ERAD.
To provide controls for these experiments and to examine the impact of compromised substrate binding on protein translocation and ERAD, the same assays were performed with the TEF1-K584X and TEF1-S493F mutants described above. We discovered that, in both cases, translocation and ERAD were inhibited in vitro and in vivo (Fig. 5). For example, the translocation efficiency of ppαF into microsomes harboring the S493F mutant was ∼2%, whereas the K584X mutant supported ppαF translocation at about half the efficiency of wild-type BiP (Fig. 5A). Also, in ERAD assays, there was ∼3-fold more CPY* remaining in both mutant strains after a 90-min chase, as compared with wild-type cells (Fig. 5B). These data show that the expression of substrate binding mutants of BiP affects multiple functions and emphasize the essential nature of substrate binding for BiP activity and Hsp70 activity in general.
FIGURE 5.
Yeast expressing K584X and S493F BiP are translocation- and ERAD-defective. A, the translocation of wild-type ppαF was assessed at 30 °C using microsomes derived from TEF1-KAR2, TEF1-K584X, or TEF1-S493F yeast. After 60 min, each reaction was split and treated either with buffer (A), trypsin (B), or trypsin and Triton X-100 (C). The percentage of translocation efficiency is indicated below each panel. The graph on the right compares the percentage of translocation efficiency of the microsomes when the assay was performed at 20 °C (white bars) or 30 °C (gray bars). Data represent the means of a minimum of four independent experiments ± S.E. (error bars). B, the degradation of CPY*, a soluble ERAD substrate, was measured at 30 °C in the TEF1-KAR2 (●), TEF1-K584X (▵), and TEF1-S493F (▿) strains by pulse-chase analysis. At each time point, data were standardized to the amount of protein at the start of the chase and represent the means of a minimum of three independent experiments ± S.E. C, ERAD assays were performed at 30 °C to assess the degradation of pαF using microsomes from TEF1-KAR2, TEF1-K584X, or TEF1-S493F yeast. Reactions were carried out in the absence (white bars) or presence (gray bars) of an ATP regeneration system and 0.5 mg/ml yeast cytosol. Data were standardized to the amount of protein remaining in the absence of cytosol and the ATP regeneration system and represent the means of a minimum of three independent experiments ± S.E.
The Genetic Interaction Profile of R217A BiP Provides Additional Support for a Translocation-specific Defect
The kar2-R217A allele is the first BiP mutant identified that is translocation-defective but ERAD-proficient. The absence of UPR induction in response to R217A BiP expression also suggests that this mutation has no effect on protein folding in the ER. In accordance with this hypothesis, we found that the transport of CPY, which is sensitive to kar2 alleles that alter folding efficiency (57, 58), was unaffected in R217A BiP-expressing yeast.4
We next used an unbiased genetic approach to measure the physiological impact of various BiP mutant alleles. In this approach, the pattern of genetic interactions of a given mutant allele provides an in depth measure of its phenotypic impact. Thus, by comparing the genetic interaction patterns of different mutants, one can, in a systematic manner, identify alleles that function in closely related biological pathways because these alleles will have similar genetic interaction patterns (46, 59). To perform this analysis, we first created an integrated version of the kar2-R217A allele and confirmed that there was a translocation defect (data not shown). Next, we systematically measured genetic interactions between the wild-type or the R217A allele and deletions in a set of 350 genes linked to ER physiology (45). These 350 genes were selected in a screen for deletions that altered the ER folding environment as evidenced by perturbations in the UPR resulting from their deletion (45). We measured the genetic interactions of the R217A BiP allele by looking for genes whose deletions in combination with kar2-R217A produced unusually high or low levels of UPR activation, as assessed through the use of a fluorescent UPR reporter assay (supplemental Fig. S1 and supplemental Table S1) (45). As a control, we also measured UPR levels in a wild-type KAR2 background and in the background of a hypomorphic (DAmP) allele of BiP in which the 3′-UTR has been deleted. The DAmP allele results in a ∼2-fold decreased expression of the wild-type BiP protein (59).4
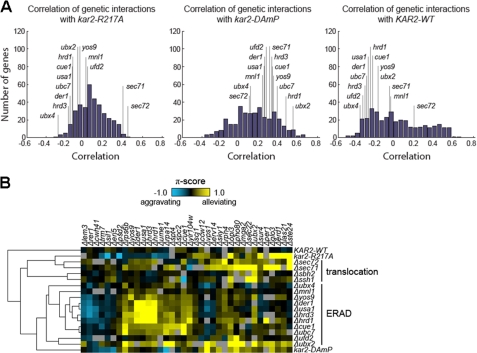
Strikingly, we found that the pattern of genetic interactions of the kar2-R217A allele was highly correlated with the pattern seen for the deletion of the two nonessential components of the Sec63 complex, _sec71_Δ and _sec72_Δ (Fig. 6, A and B), and poorly correlated with the pattern seen for deletion of ERAD components. This suggests that the R217A mutation perturbs ER physiology in a manner very similar to the perturbation in _sec71_Δ and _sec72_Δ yeast and is different from the perturbation in ERAD mutants. These data are consistent with a specific translocation defect in kar2-R217A. In contrast, the genetic interactions of the hypomorphic kar2-DAmP allele showed very poor correlations with _sec71_Δ and _sec72_Δ (Fig. 6A). These data suggest that the similarity in the perturbation of ER homeostasis in kar2-R217A yeast to _sec71_Δ and _sec72_Δ yeast is due to the R217A mutation and not a general defect in BiP function.
FIGURE 6.
The genetic interactions exhibited by kar2-R217A resemble those exhibited by the _sec71_Δ and _sec72_Δ translocation-deficient strains. Genetic interactions were quantified by using a π-score, which represents the difference between the observed double mutant UPR reporter level and that expected in the absence of a genetic interaction (45). A, correlations between the patterns of π-scores of the kar2-R217A, kar2-DAmP, or KAR2 strains and the π-scores of every other mutant in the data set of Jonikas et al. (45) obtained in the UPR-based genetic analysis are depicted using histograms. Select correlations, including those to _sec71_Δ, _sec72_Δ, and known components of the ERAD pathway are highlighted. B, a “heat map” comparing select genetic interactions exhibited by the wild-type and mutant kar2 strains with known translocation and ERAD mutants.
Interestingly, the high correlation values between the genetic interaction patterns of kar2-R217A, _sec71_Δ, and _sec72_Δ mutants are driven by a strong alleviating interaction with ste24_Δ, which is shared by all three mutants (Fig. 6B). Ste24p belongs to a conserved family of zinc metalloproteases and proteolyzes CAA_X sequences in C-terminal prenylation acceptor motifs (60). These results raise the possibility of a previously unappreciated role for Ste24p in the translocation machinery or in the translocation of a subset of secreted proteins.
DISCUSSION
Since their discovery over 30 years ago as components of the cellular heat shock response machinery (61), we have learned much about the various essential roles performed by Hsp70s and their J domain-containing Hsp40 cofactors. Moreover, given the availability of whole genome sequences, there is a greater appreciation for the potential number of Hsp70-Hsp40 pairs that can form in different organisms and within distinct cellular compartments. Because the Hsp40s are more abundant and diverse than the Hsp70s (8), it is hypothesized that specific Hsp40s engineer their cognate Hsp70s to carry out unique functions. In keeping with this hypothesis, a yeast cytosolic Hsp70, Ssa1p, interacts with the Hsp40 cofactor Ydj1p to catalyze ER translocation and folding, whereas interaction with Ydj1p and Hlj1p, a second Hsp40 cofactor, is required for Ssa1p to target aberrant membrane proteins for ERAD (33, 62–65). However, the molecular determinants underlying functional bias remain undefined. Is Hsp70-Hsp40 coupling dependent primarily on the J domain of the Hsp40s, or have Hsp70s evolved mechanisms/unique sequences to distinguish between multiple Hsp40s?
In this study, we came to the surprising conclusion that a single amino acid change in the ER-lumenal Hsp70, BiP, alters Hsp40 specificity and, consequently, BiP-mediated functions in yeast. The substitution, R217A, is within the predicted J domain binding surface of BiP and somewhat unexpectedly compromised the ability of BiP to interact with Sec63p but not Jem1p. Because Sec63p is required for protein translocation but is dispensable for ERAD and because Jem1p contributes to ERAD, we conjectured that this mutant would be translocation-defective but ERAD-proficient. Indeed, by using a variety of in vivo and in vitro studies, we obtained evidence to support our hypothesis. We also discovered that defects in the ability of BiP to bind substrates resulted in both reduced ERAD and translocation efficiency. This result indicates that substrate recognition strongly contributes to several BiP functions.
Our isolation of an allele-specific mutation in the gene encoding BiP, as reported here, complements a previous effort in the laboratory. Specifically, the current study indicates that a unique translocation-defective mutant of BiP can be obtained (i.e. R217A); in contrast, the prior publication (30) noted the generation of an ERAD-specific BiP mutant (i.e. P515L). To begin to define the broader impact of these BiP alleles on ER homeostasis, we employed a recently developed screen in which UPR induction levels in double mutant strains are used to assess genetic interactions between the respective genes (45). In agreement with our biochemical data, we found that R217A BiP, but not a hypomorphic allele resulting from decreased expression of the wild-type BiP protein, exhibited genetic interactions that were highly correlated with two mutants in components of the Sec63 complex, _sec71_Δ and _sec72_Δ (Fig. 6). More broadly, the analysis of the genetic interactions of the kar2-R217A allele argues that the genetic interaction approach based on quantitative measures of UPR induction, which initially focused on deletions of nonessential alleles, can be extended to analyze mutant alleles of essential genes. Therefore, through future efforts with other essential genes, it should be possible to isolate novel genetic interactions that contribute predominantly to the ERAD or translocation pathways. Given the fact that additional players in these pathways are still being identified (15, 19, 66), we believe that this strategy will prove worthwhile.
Overall, our results indicate that J domain selection is mediated at least in part by a single position in yeast BiP. This discovery, in turn, suggests that the diversification and complexity of J domain selection evolved from a primordial BiP-J protein pair. Over time, gene duplication and other genome-wide events, combined with amino acid drift, might have led to the origin of novel J domain proteins, thus resulting in the development of new chaperone-co-chaperone pairs with distinct functions. In effect, the studies described in this report have turned back the clock such that an Hsp70, BiP, has lost its ability to choose a partner. In the future, it will be exciting to probe whether this is a general phenomenon and whether a similar mechanism of action exists when Hsp70s must choose among a range of available nucleotide exchange factors (67).
Supplementary Material
Supplemental Data
Acknowledgments
We thank S. Nishikawa, R. Schekman, M. D. Rose, and L. Gierasch for reagents and discussions.
*
This work was supported, in whole or in part, by National Institutes of Health Grants GM75061 (to J. L. B.) and GM54068 (to L. M. H.). This work was also supported by the Cancer Center CORE Grant CA21765 and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital (to L. M. H.).
4
S. S. Vembar and J. L. Brodsky, unpublished data.
3
The abbreviations used are:
Hsp70
heat shock protein of 70 kDa
Hsp40
heat shock protein of 40 kDa
SBD
substrate binding domain
ER
endoplasmic reticulum
ERAD
ER-associated degradation
UPR
unfolded protein response
DTT
dithiothreitol
ppαF
prepro-α-factor
UTR
untranslated region.
REFERENCES
- 1.Saibil H. (2000) Curr. Opin. Struct. Biol. 10, 251–258 [DOI] [PubMed] [Google Scholar]
- 2.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 3.Mayer M. P., Bukau B. (2005) Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daugaard M., Rohde M., Jäättelä M. (2007) FEBS Lett. 581, 3702–3710 [DOI] [PubMed] [Google Scholar]
- 5.Hennessy F., Nicoll W. S., Zimmermann R., Cheetham M. E., Blatch G. L. (2005) Protein Sci. 14, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh P., Bursać D., Law Y. C., Cyr D., Lithgow T. (2004) EMBO Rep. 5, 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig E. A., Huang P., Aron R., Andrew A. (2006) Rev. Physiol. Biochem. Pharmacol. 156, 1–21 [DOI] [PubMed] [Google Scholar]
- 8.Qiu X. B., Shao Y. M., Miao S., Wang L. (2006) Cell Mol. Life Sci. 63, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlenstedt G., Harris S., Risse B., Lill R., Silver P. A. (1995) J. Cell Biol. 129, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silberstein S., Schlenstedt G., Silver P. A., Gilmore R. (1998) J. Cell Biol. 143, 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikawa S., Endo T. (1997) J. Biol. Chem. 272, 12889–12892 [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa S., Endo T. (1998) Biochem. Biophys. Res. Commun. 244, 785–789 [DOI] [PubMed] [Google Scholar]
- 13.Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. (1989) J. Cell Biol. 109, 2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldheim D., Rothblatt J., Schekman R. (1992) Mol. Cell Biol. 12, 3288–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapoport T. A. (2007) Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 16.Brodsky J. L. (1996) Trends Biochem. Sci. 21, 122–126 [PubMed] [Google Scholar]
- 17.Johnson A. E. (1997) Trends Cell Biol. 7, 90–95 [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 21.Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P., Lafer E. M., Sousa R. (2007) Mol. Cell 28, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suh W. C., Burkholder W. F., Lu C. Z., Zhao X., Gottesman M. E., Gross C. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gässler C. S., Buchberger A., Laufen T., Mayer M. P., Schröder H., Valencia A., Bukau B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. (2005) J. Cell Biol. 168, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awad W., Estrada I., Shen Y., Hendershot L. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y., Awad W., Petrova K., Hendershot L. M. (2008) EMBO J. 27, 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh W. C., Lu C. Z., Gross C. A. (1999) J. Biol. Chem. 274, 30534–30539 [DOI] [PubMed] [Google Scholar]
- 28.McClellan A. J., Endres J. B., Vogel J. P., Palazzi D., Rose M. D., Brodsky J. L. (1998) Mol. Biol. Cell 9, 3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumberg D., Müller R., Funk M. (1995) Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 30.Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. (2003) Mol. Biol. Cell 14, 3437–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Driessche B., Tafforeau L., Hentges P., Carr A. M., Vandenhaute J. (2005) Yeast 22, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 32.Corsi A. K., Schekman R. (1997) J. Cell Biol. 137, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cyr D. M., Lu X., Douglas M. G. (1992) J. Biol. Chem. 267, 20927–20931 [PubMed] [Google Scholar]
- 34.Brodsky J. L., Schekman R. (1993) J. Cell Biol. 123, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldheim D., Yoshimura K., Admon A., Schekman R. (1993) Mol. Biol. Cell 4, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrow M. W., Brodsky J. L. (2001) Traffic 2, 705–716 [DOI] [PubMed] [Google Scholar]
- 37.Kim W., Spear E. D., Ng D. T. (2005) Mol. Cell 19, 753–764 [DOI] [PubMed] [Google Scholar]
- 38.Ng D. T., Spear E. D., Walter P. (2000) J. Cell Biol. 150, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox J. S., Walter P. (1996) Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 40.Rose M., Botstein D. (1983) Methods Enzymol. 101, 167–180 [DOI] [PubMed] [Google Scholar]
- 41.Brodsky J. L., Hamamoto S., Feldheim D., Schekman R. (1993) J. Cell Biol. 120, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen W., Garcia P. D., Walter P. (1986) Cell 45, 397–406 [DOI] [PubMed] [Google Scholar]
- 43.McCracken A. A., Brodsky J. L. (1996) J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 45.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., Schuldiner M. (2009) Science 323, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 47.Fernández-Sáiz V., Moro F., Arizmendi J. M., Acebrón S. P., Muga A. (2006) J. Biol. Chem. 281, 7479–7488 [DOI] [PubMed] [Google Scholar]
- 48.Misselwitz B., Staeck O., Rapoport T. A. (1998) Mol. Cell 2, 593–603 [DOI] [PubMed] [Google Scholar]
- 49.Moro F., Fernández-Sáiz V., Muga A. (2004) J. Biol. Chem. 279, 19600–19606 [DOI] [PubMed] [Google Scholar]
- 50.Buchberger A., Valencia A., McMacken R., Sander C., Bukau B. (1994) EMBO J. 13, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimata Y., Kimata Y. I., Shimizu Y., Abe H., Farcasanu I. C., Takeuchi M., Rose M. D., Kohno K. (2003) Mol. Biol. Cell 14, 2559–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 53.Böhni P. C., Deshaies R. J., Schekman R. W. (1988) J. Cell Biol. 106, 1035–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brodsky J. L., Goeckeler J., Schekman R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 56.Werner E. D., Brodsky J. L., McCracken A. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13797–13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons J. F., Ferro-Novick S., Rose M. D., Helenius A. (1995) J. Cell Biol. 130, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.te Heesen S., Aebi M. (1994) Eur. J. Biochem. 222, 631–637 [DOI] [PubMed] [Google Scholar]
- 59.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. (2005) Cell 123, 507–519 [DOI] [PubMed] [Google Scholar]
- 60.Tam A., Schmidt W. K., Michaelis S. (2001) J. Biol. Chem. 276, 46798–46806 [DOI] [PubMed] [Google Scholar]
- 61.Ang D., Liberek K., Skowyra D., Zylicz M., Georgopoulos C. (1991) J. Biol. Chem. 266, 24233–24236 [PubMed] [Google Scholar]
- 62.Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 63.Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. (2004) Mol. Biol. Cell 15, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caplan A. J., Cyr D. M., Douglas M. G. (1992) Cell 71, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 65.Cyr D. M. (1995) FEBS Lett. 359, 129–132 [DOI] [PubMed] [Google Scholar]
- 66.Johnson A. E. (2009) J. Cell Biol. 185, 765–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaner L., Morano K. A. (2007) Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pierpaoli E. V., Gisler S. M., Christen P. (1998) Biochemistry 37, 16741–16748 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data