Effects of a Novel Arginine Methyltransferase Inhibitor on T Helper Cell Cytokine Production (original) (raw)
. Author manuscript; available in PMC: 2011 May 1.
Abstract
The protein arginine methyltransferase (PRMT) family of enzymes catalyzes the transfer of methyl groups from S-adenosylmethionine to the guanidino nitrogen atom of peptidylarginine to form monomethylarginine or dimethylarginine. We created several less polar analogues of the specific PRMT inhibitor AMI-1, and one such compound was found to have improved PRMT inhibitory activity over the parent molecule. The newly identified PRMT inhibitor modulated T helper cell function and thus may serve as a lead for further inhibitors useful for immune-mediated disease treatment.
Keywords: Cytokines, Inhibitors, Protein Arginine Methyltransferase, T helper cell, NIP45
Introduction
Though methylation of arginine residues has been recognized for more than four decades, the first mammalian protein arginine methyltransferase (PRMT) was cloned just over ten years ago [1]. Since then, PRMTs have been shown to regulate transcription, protein and RNA subcellular localization, RNA splicing, DNA damage repair, and signal transduction [2]. Nine PRMT family members have been cloned and characterized to date with putative tenth and eleventh family members identified by homology searches[3]. Two types of PRMTs have been subclassified based on the symmetry of their reaction products. Using S-adenosylmethionine (SAM) as the methyl donor, Type I PRMTs (1,3,4,6,8) catalyze asymmetric modification of arginine residues, depositing two methyl groups on a single guanidine nitrogen atom, and Type II PRMTs (5,7,9) perform symmetric transfer, placing one methyl group per terminal nitrogen of the arginine side chain. Both Type I and Type II PRMTs catalyze monomethylation as a reaction intermediate[4].
Posttranslational modifications within T cell receptor signaling cascades allow T lymphocytes to initiate a rapid and appropriate immune response to pathogens. Indeed, co-engagement of the CD28 costimulatory receptor with the T cell receptor increases PRMT activity and Vav1 methylation[5]. Perturbation of PRMT activity through the use of methylation inhibitors leads to diminished Vav1 methylation as well as downstream IL-2 production[5]. PRMT5 promotes Nuclear Factor of Activated T cells (NFAT) driven promoter activity and IL-2 secretion[6]. Additionally, arginine methylation regulates cytokine gene transcription in T helper (Th) cells through arginine methylation of the NFAT cofactor NIP45[7]. These results demonstrate a role for arginine methylation in T cell function suggesting that PRMT inhibitors may be valuable for treatment of autoimmune diseases.
Since SAM is the methylation donor in the PRMT reaction, the use of SAM analogues is a logical strategy for the direct inhibition of PRMTs. As a SAM analogue, sinefungin can compete for SAM binding and inhibit the activity of all SAM-dependent methyltransferases including PRMTs[8]. Removal of the methyl group from SAM yields S-adenosylhomocysteine (SAH), which is broken down by SAH hydrolase[3]. SAH also acts as a methyltransferase inhibitor. Compounds, like (Z)-5′-fluoro-4′5′-didehydro-5′-deoxyadenosine (MDL 28,842) and adenosine dialdehyde (Adox), that hinder SAH hydrolase activity cause an increase in SAH and, thereby, inhibit methylation[9]. Though methylthioadenosine (MTA) was reported to inhibit methyltransferase activity directly, it is an inefficient direct inhibitor of PRMT activity and more likely acts via SAM catabolism[9–11]. Chemicals like MDL 28,842, Adox, sinefugin, and MTA are not specific to the PRMT pathways as they inhibit other SAM-dependent enzymes. Nonetheless, these inhibitors and similar molecules have been used widely in arginine methylation studies for lack of better reagents.
A non-nucleoside specific small molecule inhibitor of PRMTs, AMI-1, was recently identified by Bedford and coworkers by screening of a commercial chemical library[8]. Other inhibitors have been discovered using virtual screening methods or by creating analogues to molecules in the original AMI-1 study, identifying a variety of potential PRMT inhibitory structures [12–15]. Our goal was to generate a less polar version of AMI-1 while maintaining PRMT inhibitor properties hypothesizing that such modification may enhance biological activity. We describe here the identification of one such compound and the characterization its inhibitory properties, focusing on its modulation of T helper cell function.
Results
Chemistry
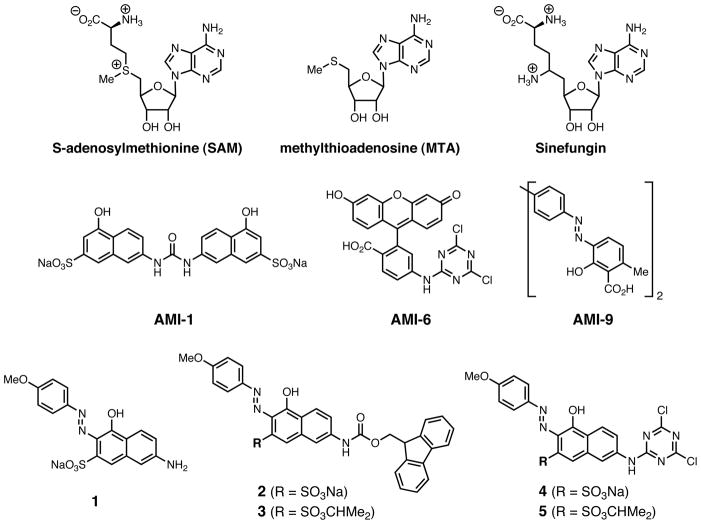
In the report disclosing the activity of AMI-1, Bedford and coworkers also identified fluorescein triazine derivative AMI-6 as a selective arginine methyltransferase inhibitor, and azo compound AMI-9 as a significantly more potent, but unselective, inhibitor of both lysine and arginine methyltransferases (Figure 1)[8]. In an effort to develop a more potent selective inhibitor, we melded elements of these two compounds. Accordingly, nonpolar functionality to the aminonaphthol sulfonate core of AMI-1 by appending the azo moiety of AMI-9 to one side, giving compound 1, and the dichlorotriazine group of AMI-6 to the other side, giving compound 4. The Fmoc intermediate compound 2 was also tested, as were the isopropyl sulfonate esters compounds 3 and 5, to assay the importance of the sulfonate negative charge and the possibility of providing a nonpolar prodrug form of the active charged species (Figure 1).
Figure 1. Known PRMT inhibitors and compounds synthesized in this study.
Chemical structures of SAM, MTA, Sinefungin, AMI-1, AMI-6, AMI-9, and compounds 1–5.
Characterization of PRMT inhibitors
To test the inhibitory activity of the compounds, we compared the ability of recombinant rat GST-PRMT1 or recombinant human GST-PRMT4 to methylate histone 4 or histone 3, respectively, giving the IC50 values shown in Table 1. AMI-1 was a more effective inhibitor of PRMT1 than PRMT4 (Table 1). Compounds 1, 2, 4, and 5 inhibited both PRMT1 and PRMT4, with the latter enzyme being more sensitive to the added compounds. Comparisons between compounds 2 vs. 3 and compounds 4 vs. 5 demonstrated that the charged sulfonate group is advantageous. The triazine derivative compound 4 was most potent with an IC50 of 4.15 μM for PRMT1 and 2.65 μM for PRMT4 (Table 1) similar to the reported 8.8 μM IC50 value of the parent compound AMI-1 for human PRMT1. Further investigations focused on compound 4.
Table 1.
Inhibition of histone methylation by PRMT1 and PRMT4 in the presence of compounds depicted in Figure 1.
| Compound | PRMT1 IC50 | PRMT4 IC50 |
|---|---|---|
| AMI-1 | 8.8 μM[a] | 169.8 ± 10.7 μM |
| 1 | 397.3 ± 44.2 μM | 196 ± 19.8 μM |
| 2 | 332.5 ± 79 μM | 56.1 ± 14.6 μM |
| 3 | no inhibition | no inhibition |
| 4 | 4.15 ± 1.6μM | 2.65 ± 0.6 μM |
| 5 | 56.7 ± 10.9 μM | 52.4 ± 6.6 μM |
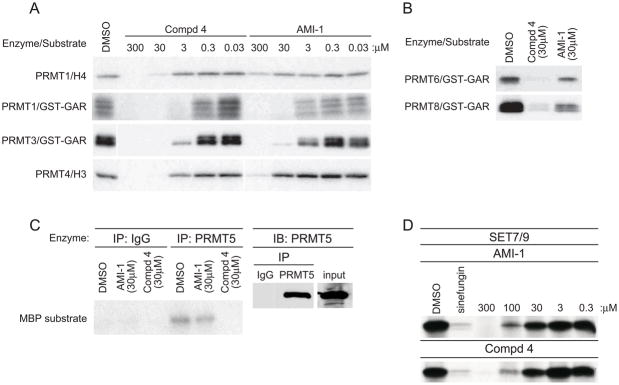
We determined the specificity of compound 4 by evaluating its effects on a panel of catalytically active recombinant Type I PRMTs using AMI-1 for comparison (Figure 2A and 2B). Using in vitro methylation assays with increasing concentrations of inhibitory compounds, compound 4 proved effective against PRMT1, PRMT3 and PRMT4. The substrate identity influenced the inhibitory activity of AMI-1, which most potently inhibited PRMT1 methylation of GST-GAR (GST fused to the glycine and arginine rich region of fibrillarin) compared to histone 4 (Figure 2A, top two panels). The published AMI-1 IC50 value for PRMT1 was determined using the glycine and arginine rich GST-Npl3 substrate[8]. Compound 4 prevented GST-GAR methylation by PRMT6 and PRMT8 while AMI-1 was less effective against these enzymes (Figure 2B). Next, we examined the potency of compound 4 on Type II PRMTs. Since the activity of recombinant PRMT5 is several hundredfold lower than PRMT5 isolated from mammalian cells, we performed methyltransferase assays using PRMT5 immunoprecipitated from 293T cells. [16]. While compound 4 inhibited the activity of PRMT5, AMI-1 was ineffective as a PRMT5 inhibitor (Figure 2C). In addition, compound 4 was selective for arginine methyltransferases over the SET domain-containing H3K4 lysine methyltransferase SET7/9, requiring at least 30-fold higher concentrations to inhibit recombinant SET7/9 activity relative to compound 4 inhibition of PRMT1 (Figure 2A and 2C).
Figure 2. Comparison of AMI-1 and compound 4 inhibitory activity.
a) in vitro methylation reactions with recombinant GST-PRMT1, GST-PRMT3, and GST-PRMT4 with indicated substrate and [3H]SAM in the presence of increasing concentrations of AMI-1 or 4. b) in vitro methylation reactions with recombinant GST-PRMT6 or GST-PRMT8 together with GST-GAR and [3H]SAM in the presence of 30μM AMI-1 or 4. c) Immunoprecipitated PRMT5 or isotype control from 293 cell extracts was subjected to in vitro methylation reactions using the indicated concentrations of AMI-1 or 4 and MBP as substrate (left panel). Reaction inputs were determined by immunoblotting with PRMT5 antisera (right panel). d) in vitro methylation reactions with recombinant Set7/9 with calf thymus histones as substrate and [3H]SAM in the presence of increasing concentrations of AMI-1 or 4. Data are representative of at least three independent experiments.
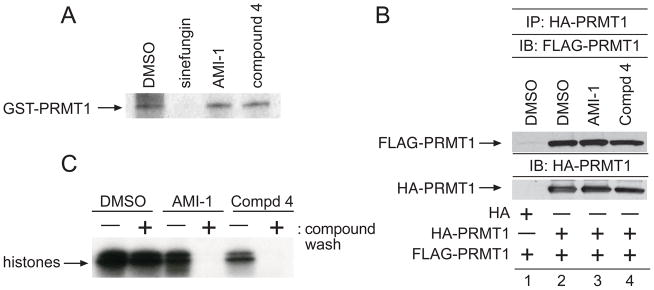
Since SAM serves as the methyl donor in PRMT-dependent methylation reactions, we examined whether compound 4 inhibits PRMT activity by competing for SAM binding. Recombinant PRMT1 was incubated in the presence of radiolabeled SAM and a 50-fold molar excess of sinefungin, AMI-1, or compound 4, followed by UV irradiation to crosslink the bound SAM to the protein. As previously published, the SAM analogue sinefungin was competitive with SAM for binding while AMI-1 was not [8]. Analysis by SDS-PAGE and visualization by fluorography (Figure 3A) revealed that compound 4 did not block SAM binding to PRMT1.
Figure 3. Characterization of Compound 4 inhibitory activity.
a) GST-PRMT1 was UV-crosslinked to [3H]SAM in the presence of DMSO, sinefugin (100μM), AMI-1 (100μM), or 4 (100μM), separated by SDS-PAGE, and visualized by fluorography b) 293T cells were transfected with HA-PRMT1 or FLAG-PRMT1. Lysates from the FLAG-PRMT1 transfection were incubated with HA-PRMT1 immunoprecipitates in the presence of DMSO (lane 2), AMI-1 (100μM, lane 3), or 4 (100μM, lane 4), resolved by SDS-PAGE, and the immunoblot was incubated with an antibody to FLAG. Reprobing the immunoblot with an antibody to HA demonstrated equal loading. Specificity of the HA-PRMT1/FLAG-PRMT interaction was determined by incubating immunoprecipitates from vector only transfected cells with FLAG-NIP45 lysates c) Incubations of GST-PRMT1 glutathione beads with DMSO, AMI-1, or 4 were divided into two aliquots. Bead aliquots were washed either in the presence (+) or absence (−) of indicated compounds. Washed aliquots were immediately subjected to in vitro methylation assays using calf thymus histones. Data are representative of three independent experiments.
PRMT1 has been shown to form dimers in crystal structure studies, and mutations within the dimerization interface reduce methyltransferase activity[4, 17]. To test the possibility that compound 4 inhibits PRMT1 activity by preventing oligomerization, we performed coimmunoprecipitation experiments (Figure 3B). Equal volumes of HA-PRMT1 and FLAG-PRMT1 transfected 293T cell lysates were mixed and incubated with DMSO (lane 2), AMI-1 (100μM) (lane 3) or compound 4 (100μM) (lane 4) during the coimmunoprecipitation. Specificity of the HA-PRMT1/FLAG-PRMT1 interaction was determined using an empty HA vector (Figure 3B, lane 1). The presence of either compound did not interfere with the interaction between HA-PRMT1 and FLAG-PRMT1, indicating that compound 4 does not interfere with PRMT1 oligomerization.
To examine whether compound 4 is a reversible inhibitor, we performed washout experiments. Recombinant GST-PRMT1 bound to glutathionine beads was preincubated with compound 4 (100μM) or AMI-1 (100μM). The beads were then washed with methylation buffer only (Figure 3C, indicated by “−“) or with methylation buffer containing indicated compound (Figure 3C, indicated by “+”) prior to methylation reactions using calf thymus histones as a source of the PRMT1 substrates histone 4 and histone 2A [18]. Inhibition by both compound 4 and AMI-1 was relieved by the washout, demonstrating that both are reversible PRMT inhibitors.
Biological activity
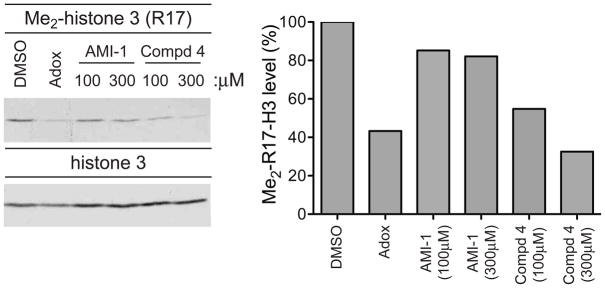
To determine whether compound 4 is cell permeable, we examined the effect of compound 4 on cellular PRMT activity. 293T cells were incubated with DMSO, compound 4, or the general methylation inhibitor adenosine dialdehyde (Adox)[8]. Cell extracts were immunoblotted and incubated with an antibody recognizing H3R17 methylation (Figure 4). Over this period no cellular toxicity with these treatments was observed (data not shown). At 100μM, compound 4 induced more than 40% reduction in H3R17 methylation, a significant increase in inhibitory activity relative to AMI-1.
Figure 4. Compound 4 is cell permeable.
293T cells were treated with DMSO, AMI-1 (100μM or 300μM), 4 (100μM or 300μM), or Adox (20μM) for 24 hrs. Histone extracts were immunoblotted for H3R17 methylation (left panel). Quantification of the methylation levels of compound-treated samples relative to vehicle-treated samples is depicted in the right panel. Data are representative of three independent experiments.
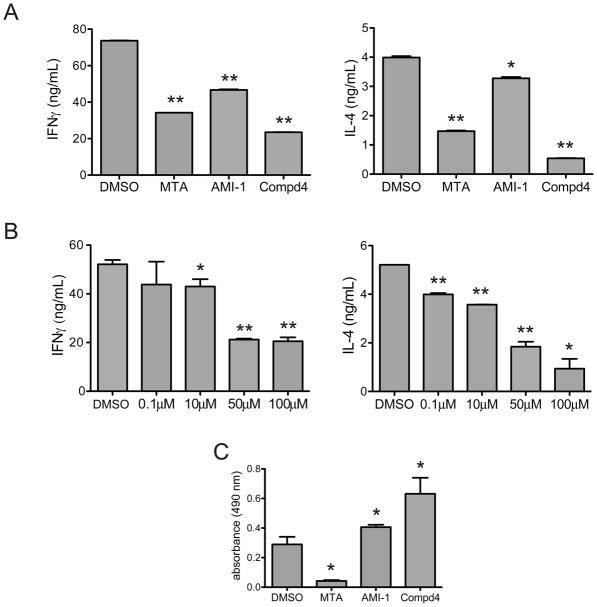
Since compound 4 interferes with cellular PRMT activity, we examined its effects on PRMT-dependent gene regulation. Type 1 T helper (Th1) cells modulate the immune response largely by the secretion of interferon γ (IFNγ), while type 2 T helper (Th2) cells secrete interleukin 4 (IL-4)[19]. PRMTs have been shown to regulate T helper cell activation and cytokine secretion [5, 7, 20]. Indeed, PRMT1 augments both IFNγ and IL-4 promoter activity, and general methylation inhibitors decrease IFNγ and IL-4 transcript levels [7]. We examined the effect of compound 4 on Th1 and Th2 cells cytokine expression (Figure 5). As shown previously, MTA diminished both IFNγ and IL-4 production (Figure 5A) [7]. Treatment with compound 4 reduced Th1 IFNγ production more than 60% and Th2 IL-4 levels more than 75%, while incubation with AMI-1 reduced T helper cytokine expression by less than 40%. Compound 4 inhibited IFNγ secretion by Th1 cells and IL-4 secretion by Th2 cells in a dose dependent manner with significant effects seen at 10 μM for IFNγ secretion and at 0.1 μM for IL-4 secretion (Figure 5B). Thus, IL-4 production is more sensitive than IFNγ to treatment with compound 4. Importantly, compound 4 did not affect T helper cell viability (Table 2). Indeed, both AMI-1 and compound 4 enhanced T helper cell proliferation, the latter doing so to a greater degree, suggesting a possible correlation between PRMT inhibition and T cell proliferation, while MTA treatment inhibited T cell proliferation (Figure 5C). Thus, the reduced T helper cell cytokine expression following AMI-1 or compound 4 treatment is not a result of increased cell death or reduced cell numbers.
Figure 5. Effects of Compound 4 on T helper cell function and proliferation.
a) Th1 or Th2 were stimulated with plate-bound anti-CD3 in the presence of DMSO, MTA (100μM), AMI-1 (100μM), and 4 (100μM). Supernatants were analyzed by ELISA to determine Th1 production of IFNγ (left panel) or Th2 production of IL-4 (right panel). b) Th1 or Th2 cells were stimulated with plate-bound anti-CD3 in the presence of DMSO or varying concentrations of 4, and IFNγ (left panel, Th1 cells) or IL-4 (right panel, Th2 cells) levels were determined by ELISA. c) T helper cells were stimulated with plate-bound anti-CD3 in the presence of DMSO, MTA, AMI-1, or 4. Cellular proliferation was determined by the MTS assay. * p < 0.05, ** p < 0.01. Data are representative of at least three independent experiments.
Table 2.
Viability of T helper cells in the presence of MTA or compound 4.
| % viable | % apoptotic | % necrotic | |
|---|---|---|---|
| DMSO | 95.9 | 2.2 | 2.0 |
| MTA (100 μM) | 90.9 | 4.9 | 4.2 |
| cpd 4 (100 μM) | 94.2 | 1.7 | 3.0 |
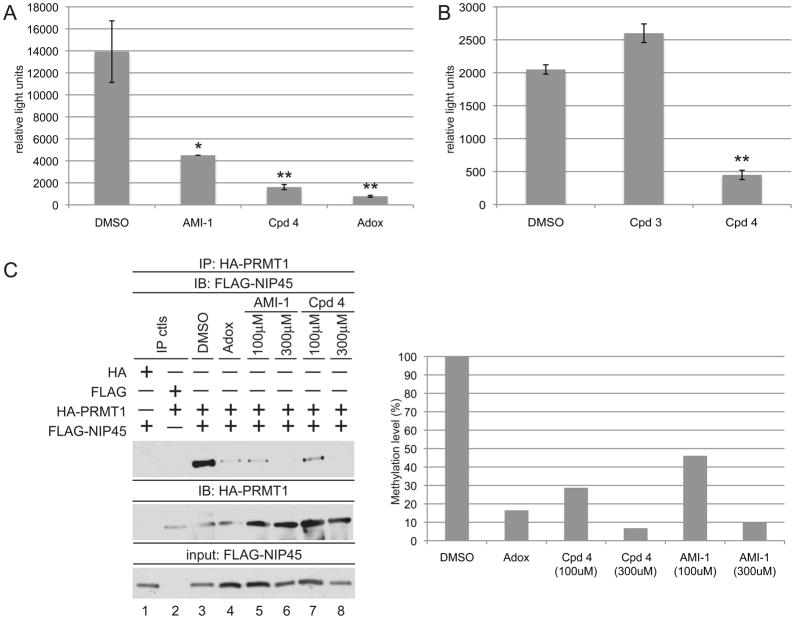
As compound 4 is a cell permeable PRMT inhibitor capable of modulating T helper cell cytokine secretion, it could suppress IL-4 levels by altering promoter activation or affecting RNA stability. Accordingly, we tested the effect of compound 4 on the activity of a Th2 selective region of the IL-4 promoter (−760 to +68), which is responsive to transactivation by NFATc2 and its binding partner NIP45 [21]. We transfected Jurkat cells, a human T cell line that contains endogenous IL-4 promoter transactivating factors, with an IL-4 luciferase reporter. The transfected cells were incubated with DMSO, compound 4, AMI-1, and Adox (Figure 6A). As expected, Adox greatly diminished IL-4 promoter reporter activity [7]. Incubation with AMI-1 decreased promoter activity, while compound 4 diminished IL-4 promoter activity even further. Incubation with 3, which did not exhibit in vitro PRMT inhibitory activity (Table 1), also did not interfere with IL-4 promoter activity (Figure 6B). These data support the notion that the decrease in Th2 IL-4 production occurred at least in part at the transcriptional level.
Figure 6. Compound 4 inhibits IL-4 promoter activity and the interaction between NIP45 and PRMT1.
a) Jurkat cells were transfected with the IL-4 luciferase reporter (3μg) along with TK-Renilla luciferase vector (10ng) as an internal control. Transfectants were pretreated with DMSO, AMI-1 (100μM), 4 (100μM), or Adox (20μM) for 18hrs prior to 6hr stimulation with PMA/Ionomycin. Luciferase values were calculated relative to TK-Renilla luciferase internal controls. Similar results were obtained in at least three independent experiments. * p < 0.05, ** p < 0.01. b) same as in a) except cells were treated with 3 (100μM) or 4 (100μM). ** p < 0.01 c) 293T cells transfected with HA-PRMT1 and FLAG-NIP45 expression vectors were treated with DMSO (lane3), Adox (20μM, lane 4), AMI-1 (100μM, lanes 5–6), or 4 (100μM, lanes 7–8). Lysates were immunoprecipitated with anti-HA agarose, and immunoprecipitates were probed for FLAG-NIP45 using an antibody recognizing the FLAG epitope (top panel). HA-PRMT immunoprecipitate levels were evaluated by reblotting with an antibody to HA (middle panel). Bottom panel demonstrates FLAG-NIP45 input levels. Quantification of FLAG-NIP45/HA-PRMT association levels are depicted relative to the DMSO treated sample (right panel). Data are representative of three independent experiments.
The nuclear protein NIP45 (NFAT Interacting Protein 45kDa) was isolated by virtue of its ability to interact with NFATc2 in a yeast two-hybrid screen [21]. NIP45 activity has been reported to combine with two critical Th2 transcription factors, c-Maf and NFATc2, to induce the expression of IL-4 in a normally non-IL-4 producing cell line [21]. NIP45 contains an arginine-rich amino terminus, which is a substrate of PRMT1. Arginine methylation facilitates the interaction between NIP45 and NFAT, thereby augmenting cytokine expression [7]. As a transcriptional coactivor, PRMT1 is recruited to the NFAT complex via NIP45, forming a tripartite complex that likely serves to enhance NFAT-driven transcriptional activity [7, 22]. Inhibition of methyltransferase activity by MTA treatment diminished the PRMT1/NIP45 interaction [7]. To determine whether inhibition of PRMT activity by compound 4 would also disrupt PRMT1/NIP45 association, 293T cells were transfected with HA-PRMT1 and FLAG-NIP45, and coimmunoprecipitation assays were performed. As predicted, Adox treatment reduced the association between PRMT1 and NIP45 compared to incubation with the compound vehicle DMSO (Figure 6C, compare lanes 3 and 4). Additionally, both AMI-1 and compound 4 interfered with the NIP45 and PRMT1 interaction, supporting the notion that both are bona fide arginine methyltransferase inhibitors (Figure 6C, lanes 5–8). These data suggest that compound 4 may diminish Th2 IL-4 production, at least in part, by interfering with the function of NIP45.
Discussion
Here, we have identified compound 4 as a selective PRMT inhibitor that targets both Type I and Type II PRMTs. The effectiveness of compound 4 on PRMT1 inhibition was dependent upon the substrate used with methylation of GST-GAR being more responsive to inhibition than histone 4. A histone 4 and an arginine and glycine rich peptide were recently compared in kinetic studies with PRMT1. The Km of the histone 4 peptide was about ten-fold lower than the arginine and glycine rich peptide, suggesting that the inhibition threshold may differ between PRMT1 substrates [10]. Compound 4 is a reversible inhibitor, limiting its in vivo toxicity. Also, compound 4 was mostly inactive against the lysine methyltransferase Set7/9 in methylation assays. Importantly, compound 4 potently inhibited cellular H3R17 methylation supporting the notions that compound 4 is cell permeable and is capable of inhibiting endogenous PRMT activity. It is important to note that AMI-1 was first identified as a HIV-1 reverse transcriptase inhibitor [23]. Therefore, it will be important to determine whether compound 4, as a derivative of AMI-1, is a selective inhibitor of endogenous PRMT activity.
The hybrid structure of compound 4 incorporates features of the AMI-1, AMI-6, and AMI-9 compounds described by Bedford and colleagues[24]. AMI-1, AMI-6, and AMI-9 were all effective methyltransferase inhibitors. Only AMI-1 and AMI-6 demonstrated selectivity for the PRMTs, though AMI-6 was minimally active against a cellular PRMT substrate [24]. Computational modeling suggested AMI-1 spans the SAM and arginine binding pockets of PRMT1; however, AMI-1 did not compete for [3H]SAM binding to recombinant PRMT1 [24, 25]. Additionally, in a study using peptide-based fluorescent reporters, AMI-1 blocked PRMT1 binding to its substrate [26]. The mechanism of action of compound 4 is not completely clear, but it neither competed with SAM binding nor blocked PRMT1 dimerization.
In the first report of specific small molecule PRMT inhibitors, Bedford and coworkers used an antibody-based high throughput screening to identify several arginine methylation inhibitors (AMIs) (Table 2)[8]. Of these, AMI-1 showed interesting selectivity by not inhibiting the lysine methyltransferase Set7/9 but was only weakly cell permeable, limiting its use in vivo [8]. In a follow up studies, the bromo-moiety containing AMI-5 structure was used as a template to create several new inhibitors with similar potency to AMI-1 (low micromolar)[14, 15]. Using 26 AMI-analogues, one low micromolar PRMT1 inhibitor was identified, and cellular activity was not reported[25]. Virtual ligand screening using the published PRMT1 structure has resulted in several novel compounds with inhibitory activity (thyglycolic amide, allantodapsone)[12, 13]. Thompson and coworkers generated PRMT1 inhibitors using in situ bisubstrate generation (D2AAI), but none of these compounds was more potent than AMI-1 [27]. Recently, both Methylgene and Bristol-Myers Squibb have reported high potency (picomolar IC50) and selective PRMT4 inhibitors, though the Methylgene compound was not active in cellular assays and no cellular data was reported for the Bristol-Myers Squibb compounds [28–30]. Additionally, we find that while compound 4 inhibits both Type I and Type II PRMTs, AMI-1 distinguishes between the two PRMT subclasses. Thus, a variety of chemical structures can serve as PRMT inhibitors, and highly potent and selective PRMT structures are achievable.
The NFAT interacting protein NIP45 is also a PRMT1 substrate [7]. Arginine methylation of NIP45 promotes NFAT-driven transcription [7]. In previous studies, we used MTA treatment to support a role for PRMTs in T helper cell cytokine expression [7]. Using compound 4, we have extended these earlier studies showing that methyltransferase inhibition results in inhibition of IFNγ and IL-4 production, interference with IL-4 promoter activity, and impairment of the interaction between PRMT1 and NIP45.
Several lines of evidence strongly suggest that specific PRMT inhibitors may be valuable for the treatment of autoimmune diseases such as rheumatoid arthritis [5–7, 31–34]. PRMTs modify and regulate several critical immunomodulatory proteins. Post-translational modifications within T cell receptor signaling cascades allow T lymphocytes to initiate a rapid and appropriate immune response to pathogens. Co-engagement of the CD28 costimulatory receptor with the T cell receptor elevates PRMT activity and cellular protein arginine methylation, including methylation of the guanine nucleotide exchange factor Vav1 [5]. Incubation with MDL 28,842 diminished methylation of the guanine exchange factor Vav1 as well as IL-2 production. Similarly, MTA treatment and siRNA directed against PRMT5 both inhibited NFAT-driven promoter activity and IL-2 secretion[6]. We also demonstrated that arginine methylation of the NFAT cofactor NIP45 (NFAT Interacting Protein 45kD) within Th cells by PRMT1 promotes its association with NFAT, thereby driving NFAT-mediated cytokine gene expression[7]. In fibroblast cell lines, PRMT1 also cooperates with Carm1 to enhance NFkB p65-driven transcription and facilitate the transcription of p65 target genes like TNFα [33]. Symmetric dimethylation of Sm D1 and D3 forms an epitope for the production of anti-Sm autoantibodies, which are often found in lupus [31, 32, 34]. Taken together, these results demonstrate an important role for arginine methylation in inflammation, suggesting that PRMT inhibitors may be valuable for treatment of autoimmune diseases.
Inhibition of PRMT activity using AMI-1 or compound 4 augmented T helper cell proliferation. It will be important to determine whether compound 4 also enhances proliferation of transformed cells. The combination of compound 4 treatment along with a PRMT siRNA approach may provide some insight into the mechanism behind this phenomenon. Since PRMT activity promotes T helper cell cytokine production, compound 4 and more potent derivatives thereof may be useful for treating T helper cell-driven autoimmune diseases, such as multiple sclerosis. Further work to develop more potent derivatives is underway guided by docking studies of compound 4 to the PRMT1 crystal structure.
Materials and Methods
Mice and Cell Culture
Balb/c mice were obtained from The Scripps Research Institute breeding colony. All animal protocols were in accordance with The Scripps Research Institute Institutional Animal Care and Use Committee policy. T helper cells were isolated by magnetic bead selection (Miltenyi Biotech). T cells were cultured in RPMI and stimulated with plate-bound 1μg/mL anti-CD3 (2C11; BioXCell) plus 2μg/mL anti-CD28 (PV1; BioXCell), in the presence of IL-2 (NCI Biological Resources Branch). For Th1 skewing, 5ng/mL rIL-12 (14-8121; eBiosciences) and 10 μg/mL anti-IL-4 (11B11; NCI Biological Resources Branch) were added to the cell culture, and for Th2 skewing, 10 ng/mL rIL-4 (14-8041; eBiosciences) and 10 μg/mL anti-IFNγ (R4/GA2; BioXCell). For PMA/ionomycin stimulation, cells were incubated with 50 ng/mL PMA and 1mM ionomycin (EMD Biosciences). Compounds were dissolved in DMSO. AMI-1 (AK Scientific and EMD Biosciences), adenosine dialdehyde (Sigma), methylthioadenosine (Sigma), and sinefungin (Sigma) were solubilized in DMSO. Jurkat cells were grown in RPMI. 293T cells were grown in DMEM. Proliferation assays were performed using the CellTiter 96 Aqueous One Solution Proliferation Assay reagent (Promega).
Plasmids, Transfections, and Luciferase Assays
GST-PRMT1 and GST-CARM1 vectors were described previously[1, 35]. We thank Drs. H. Herschman, S. Richard, M. Bedford, and S. Clarke for GST-PRMT3, GST-PRMT6, GST-PRMT8, and GST-GAR vectors, respectively. Expression vectors for FLAG-PRMT1, HA-PRMT1, FLAG-NIP45, and IL-4 luciferase were described previously[7]. Transient transfection of 293 cells was performed using Fugene HD (Roche) according to manufacturer’s instructions. Jurkat cells were transfected using a BioRad electroporator (280V, 975μF). Thymidine kinase promoter driven Renilla luciferase was used as an internal control. Luciferase activity was determined using Promega’s Dual Luciferase Kit.
ELISA
Th1 or Th2 cells (1 × 106 cells/mL) from day 7 cultures were stimulated with plate-bound anti-CD3 (1μg/mL) for 24 hours. IL-4 or IFNγ protein levels in cell supernatants were measured by ELISA as described (eBiosciences)[7].
In Vitro Methylation Assays
Recombinant GST-PRMT1, GST-PRMT3, GST-PRMT4, GST-PRMT6, GST-PRMT8, and GST-GAR were prepared as described[21] and concentrated using a micron 10 filter device (Millipore). SET7/9 (ALX-201-178) was purchased from Enzo Life Sciences. Methylation reactions were performed as described using recombinant histone 3 (12-357; Millipore), histone 4 (12-347; Millipore), or calf thymus histones (H4380; Sigma)[7]. Reactions were quantified by densitometry (NIH ImageJ software) and IC50’s were calculated using linear regression.
Antibodies, Immunoprecipitations, and Immunoblots
Whole-cell lysates were prepared using 1% Triton x-100 lysis buffer. For coimmunoprecipitation experiments, cell lysates were prepared in a lysis buffer containing 100 mM NaCl, 50 mM TRIS (pH 7.5), 1 mM EDTA, 0.1% TX-100, 10 mM NaF, 1 mM PMSF, and 1 mM vanadate. Immunoprecipitations were performed using anti-HA agarose (Sigma). Primary antibodies used in these studies: anti HA-HRP (12CA5; Roche), anti-FLAG-HRP (M2; Sigma), anti-β-actin (ab8226; Abcam), and anti-histone 3 dimethylarginine 17 (07–214; Millipore).
Inhibitor Reversibility Assay
GST-PRMT1 bound to glutathione-agarose beads was incubated for 60 min on ice in the presence of 100μM of compounds. Samples were then washed three times with methylation reaction buffer (20mm Tris pH8.0, 200mM NaCl, 0.4mM EDTA) containing either 100μM inhibitor or DMSO. Samples were resuspended in methylation reaction buffer containing 100μM inhibitor or DMSO, 1μg Histones and 6μM S-adenosyl-[methyl-3H]methionine. Reactions were incubated for 90 min at room temperature and stopped with SDS sample buffer. Fluorography was performed as described[36].
Crosslinking
GST-PRMT1 (10 μg) was suspended in PBS containing 5mM DTT, 100μM inhibitor (Sinefungin, AMI-1 or compound 4) and 20μM S-adenosyl-[methyl-3H]methionine, and added to a 96-well plate pre-cooled on ice. Wells were exposed to short-wave UV light (UVP Inc. Model #UVGL-25) for 1 hour. Fluorography was performed as described[8].
Synthesis
All air- and moisture-sensitive reactions were performed under nitrogen in oven-dried or flame-dried glassware. Unless stated otherwise, reagents and solvents were purchased from VWR, Acros, TCI America, or Aldrich, and were used without further purification. All experiments were monitored by thin layer chromatography with visualization by exposure to ultraviolet light, iodine vapor, or a staining solution (5% phosphomolybdic acid in ethanol, anisaldehyde in EtOH, or aqueous KMnO4) followed by heating. Flash chromatography was performed using SINGLE StEP pre-packed MPLC columns (Thomson Instrument Company, Oceanside, CA). Melting points were determined using a Barnstead Electrothermal 9300 capillary melt apparatus and are uncorrected. Nuclear magnetic resonance (NMR) spectra were recorded with a Varian Inova-400 spectrometer (400 MHz for 1H, 100 MHz for 13C) in DMSO-_d_6 solvent. Routine mass spectra were obtained using an Agilent 1100 (G1946D) ESI MSD with mobile phase composed of 9:1 CD3CN:H2O containing 0.1% CF3CO2H. GC-MS analyses were performed on an HP GCD-II (Model 5810) instrument. Elemental analyses were performed by Midwest MicroLab, LLC. High-resolution mass spectra (HRMS) were recorded at the mass spectrometry facility at The Scripps Research Institute, La Jolla.
The synthetic steps are outlined in Fig. S1. In addition to _p_-methoxyaniline, the other four aromatic amines shown at the bottom of the figure were also tested and the resulting azo compounds elaborated into candidate structures, the last of which proved to be as effective as compounds 1–5, and will be the subject of further studies.
Compound 1
To a stirred solution of 4-methoxyaniline (665 mg, 5.40 mmol) in aqueous hydrochloric acid (30% v/v, 5.4 mL) was added dropwise a solution of sodium nitrite (401 mg, 5.83 mmol) in H2O (1 mL) at 0 °C. The reaction mixture was stirred for 15 min or until a positive test (deep blue color) for nitrous acid on potassium iodide-starch test paper was observed. The clear yellowish solution of diazonium salt was used immediately for the subsequent coupling reaction. Note: If the arylamine did not dissolve well in aqueous acid, the reaction mixture was sonicated or gradually heated to 70 °C until a clear solution was obtained. The solution was then cooled to 0–5 °C in an ice bath.
6-Amino-1-naphtol-3-sulfonic acid (1.3 g, 5.40 mmol) was dissolved first in 5 N aqueous sodium hydroxide (5 mL) and then diluted with water (20 mL). This solution was added dropwise to a solution of the above diazonium chloride salt at 0 °C with stirring. Aqueous sodium hydroxide was added to maintain the mixture at pH 7, and the solution stirred at 0 °C until a negative test on potassium iodide-starch test paper was observed. A fine brick red solid precipitated and was isolated by several cycles of settling, decanting, and dilution using pure water, followed by lyophilization, to give compound 1 in 70–80 % yield. 1H NMR (400 MHz, DMSO-d6) δ 16.3 (s, 1H), 7.92 (d, J = 8.4 Hz, 1H), 7.63 (d, J = 9.2 Hz, 2H), 7.21 (s, 1H), 7.01 (d, J = 9.2 Hz, 2H), 6.71–6.63 (m, 2H), 3.78 (s, 3H). 13C NMR (DMSO-d6, spectrum acquired at 40°C to aid solubility) δ 174.0, 157.1, 153.4, 143.4, 137.6, 136.9, 128.4, 127.6, 119.9, 119.5, 118.2, 114.6, 114.5, 109.7, 55.4. The compound appeared unchanged upon heating to 350 °C in a melting point capillary tube.
Compound 2
To a stirred suspension of compound 1 (3.36 g, 8.5 mmol) in methanol (72 mL) was added sodium carbonate (2.5 g, 24.5 mol) and 9-fluorenylmethyl chloroformate (Fmoc-Cl, 5.85 g, 22.7 mol) portionwise at room temperature. The reaction mixture was stirred for 24 h, after which 4 N HCl/dioxane (14 mL, 46 mol) was added and the suspension was stirred for 1h. The solvent was removed on a rotary evaporator and the crude product was triturated with diethyl ether, and compound 2 was isolated as a red solid. 1H NMR (200 MHz, DMSO-d6) δ 16.2 (s, 1H), 10.2 (s, 1H), 8.17 (d, J = 8.8 Hz, 1H), 7.95 (d, J = 8.8 Hz, 2H), 7.80 (m, 5H), 7.60 (m, 1H), 7.39 (m, 5H), 7.08 (d, J = 8.8 Hz, 2H), 4.56 (d, J = 3 Hz, 2H), 4.37 (t, J = 3 Hz, 1H), 3.83 (s, 3H). 13C (DMSO-d6) δ 170.4, 158.5, 153.3, 143.9, 143.6, 142.8, 140.8, 137.6, 136.2, 127.7, 127.2, 125.2, 124.5, 120.3, 119.9, 119.1, 117.9, 115.6, 114.8, 66.0, 55.5, 46.6. The compound appeared unchanged upon heating to 350 °C in a melting point capillary tube.
Compound 3
To a stirred suspension of compound 2 (6.0 g, 10.0 mol) in dioxane (80 mL) was added triisopropylorthoformate (9.2 mL, 48 mol) at room temperature. The reaction mixture was stirred at 60 °C overnight, cooled to room temperature, filtered, and the solid washed three times with CH2Cl2. The combined solutions were evaporated and the residue purified by column chromatography on silica gel (1:5 EtOAc:hexanes) to give compound 3 as a red solid (5.2 g, 83%). Rf: 0.8 (1:2 EtOAc:hexanes); 1H NMR (200 MHz, DMSO-d6) δ 16.2 (s, 1H), 10.2 (s, 1H), 8.26 (d, J = 8.8 Hz, 1H), 7.96 (m, 3H), 7.77 (m, 5H), 7.60 (m, 6H), 7.39 (m, 4H), 7.07 (d, J = 8.8 Hz, 2H), 4.81 (m, 1H), 4.56 (d, J = 3 Hz, 2H), 4.37 (t, J = 3 Hz, 1H), 3.83 (s, 3H), 1.26 (d, J =3Hz, 6H). 13C (DMSO-d6)δ 171.2, 159.2, 154.0, 144.6, 144.4, 143.5, 141.5, 138.3, 136.9, 128.4, 127.8, 125.8, 125.3, 120.9, 120.6, 119.9, 118.6, 116.3, 115.9, 66.7, 56.1, 55.5, 47.2, 27.7. The compound discolored at approximately 200 °C and partially sublimed at 250–255 °C, leaving a dark residue behind.
Compound 5
To a stirred solution of compound 3 (3.0 g, 4.7 mol) in DMF (8 mL) was added 4-methylpiperidine (722 μL, 6.11 mmol) at room temperature. After 2 h, the reaction mixture was partitioned between EtOAc and brine solution. The combined organic solution was washed with 1 N HCl to remove extra 4-methylpiperidine and then with water and brine, dried over anhydrous MgSO4, filtered, and concentrated. The crude product was purified by silica gel chromatography (1:5 EtOAc:hexanes) to give the intermediate amine (A in Fig. S1) as a red solid (1.67 g, 86%). Rf: 0.4 (1:1 EtOAc:hexanes); 1H NMR (200 MHz, CDCl3) δ 8.26 (d, J = 8.4 Hz, 1H), 7.64 (m, 3H), 6.91 (m, 4H), 4.88 (m, 1H), 3.85 (s, 3H), 1.33 (d, 6H). LCMS [M+H]+ 416.0
A stirred suspension of compound A (114 mg, 0.274 mmol) in acetone (1.5 mL) was treated with cyanuric chloride (75.8 mg, 0.411 mmol) and saturated NaHCO3 (34 μL) at room temperature. After 3h, the reaction mixture was filtered and the crude product was purified on silica gel, eluting with 1:5 EtOAc:hexanes to give 5 as a red solid.(127 mg, 83%) Rf: 0.8 (EtOAc/hexanes, 1/1); 1H NMR (200 MHz, DMSO-d6) δ 16.3 (s, 1H), 11.6 (s, 1H), 8.37 (d, J = 8 Hz, 1H), 8.08 (s, 1H), 7.96 (d, J = 8 Hz, 1H), 7.87 (s, 1H), 7.76 (d, J = 8 Hz, 2H), 7.15 (d, J = 8 Hz, 2H), 4.61 (m, 1H), 3.82 (s, 3H), 1.21 (d, J = 3 Hz, 6H); LCMS [M+H]+ 563.0.
Compound 4
To a stirred suspension of compound 5 (84.8 mg, 0.150 mmol) in 2-butanone (2 mL) was added sodium iodide (33.7 mg, 0.225 mmol) at room temperature. The reaction mixture was heated to 60°C and allowed to stir overnight. The reaction mixture was then cooled to room temperature, filtered, and washed with diethyl ether to provide compound 4 as a red solid (quant.). 1H NMR (400 MHz, DMSO-d6) δ 16.3 (s, 1H), 11.5 (s, 1H), 8.30 (d, J = 8 Hz, 1H), 8.07 (s, 1H), 7.81 (m, 2H), 7.52 (s, 1H), 7.11 (d, J = 7.4 Hz, 2H), 3.83 (s, 3H). The compound discolored when heated to 305–320 °C; melting was not evident up to 350 °C.
Supplementary Material
Supp Fig 1. Figure S1. Synthesis of new PRMT inhibitors.
Formation of a diazo linkage to 7-amino-4-hydroxynaphthalene-2-sulfonic acid was followed by elaboration of the amino group to give the candidate compounds.
Acknowledgments
We thank Dr. Sanja Arandjelovic for critical reading and review of the manuscript. This is manuscript # 20585 from TSRI. This work was partially supported by grants from NIAID AI067460-01 (K.A.M.), NIGMS GM085117 (K.A.M.), Skaggs Institute for Chemical Biology (M.G.F.), and the U.S. Department of Defense (W81XWH-05-1-0316) (M.G.F.). K.A.M is the recipient of an Arthritis Investigator Award and the Donald and Delia Baxter Foundation Young Career Scientist Award.
Footnotes
The following supplementary material is available:
Fig. S1. Synthesis of new PRMT inhibitors.
References
- 1.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 2.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Lee DY, Ianculescu I, Purcell D, Zhang X, Cheng X, Stallcup MR. Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function. Mol Endocrinol. 2007;21:1381–1393. doi: 10.1210/me.2006-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchet F, Cardona A, Letimier FA, Hershfield MS, Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. J Exp Med. 2005;202:371–377. doi: 10.1084/jem.20050176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mowen KA, Schurter BT, Fathman JW, David M, Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol Cell. 2004;15:559–571. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 8.Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 9.Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 10.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke SG. Inhibition of Mammalian Protein Methyltransferases by 5′-Methylthioadenosine (MTA): A Mechanism of Action of Dietary SAMe? In: Clarke SGaT., editor. The Enzymes: Protein Methyltransferases. Elsevier; London: 2006. pp. 467–486. [DOI] [PubMed] [Google Scholar]
- 12.Spannhoff A, Heinke R, Bauer I, Trojer P, Metzger E, Gust R, Schule R, Brosch G, Sippl W, Jung M. Target-Based Approach to Inhibitors of Histone Arginine Methyltransferases. J Med Chem. 2007;50:2319–2325. doi: 10.1021/jm061250e. [DOI] [PubMed] [Google Scholar]
- 13.Spannhoff A, Machmur R, Heinke R, Trojer P, Bauer I, Brosch G, Schule R, Hanefeld W, Sippl W, Jung M. A novel arginine methyltransferase inhibitor with cellular activity. Bioorg Med Chem Lett. 2007;17:4150–4153. doi: 10.1016/j.bmcl.2007.05.088. [DOI] [PubMed] [Google Scholar]
- 14.Mai A, Cheng D, Bedford MT, Valente S, Nebbioso A, Perrone A, Brosch G, Sbardella G, De Bellis F, Miceli M, et al. epigenetic multiple ligands: mixed histone/protein methyltransferase, acetyltransferase, and class III deacetylase (sirtuin) inhibitors. J Med Chem. 2008;51:2279–2290. doi: 10.1021/jm701595q. [DOI] [PubMed] [Google Scholar]
- 15.Mai A, Valente S, Cheng D, Perrone A, Ragno R, Simeoni S, Sbardella G, Brosch G, Nebbioso A, Conte M, et al. Synthesis and biological validation of novel synthetic histone/protein methyltransferase inhibitors. ChemMedChem. 2007;2:987–991. doi: 10.1002/cmdc.200700023. [DOI] [PubMed] [Google Scholar]
- 16.Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. M008660200 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. C700192200 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 20.Lawson BR, Manenkova Y, Ahamed J, Chen X, Zou JP, Baccala R, Theofilopoulos AN, Yuan C. Inhibition of transmethylation down-regulates CD4 T cell activation and curtails development of autoimmunity in a model system. J Immunol. 2007;178:5366–5374. doi: 10.4049/jimmunol.178.8.5366. [DOI] [PubMed] [Google Scholar]
- 21.Hodge MR, Chun HJ, Rengarajan J, Alt A, Lieberson R, Glimcher LH. NF-AT-Driven interleukin-4 transcription potentiated by NIP45. Science. 1996;274:1903–1905. doi: 10.1126/science.274.5294.1903. [DOI] [PubMed] [Google Scholar]
- 22.Bedford MT. Arginine methylation at a glance. Journal of cell science. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 23.Skillman AG, Maurer KW, Roe DC, Stauber MJ, Eargle D, Ewing TJ, Muscate A, Davioud-Charvet E, Medaglia MV, Fisher RJ, et al. A novel mechanism for inhibition of HIV-1 reverse transcriptase. Bioorg Chem. 2002;30:443–458. doi: 10.1016/s0045-2068(02)00502-3. S0045206802005023 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Cheng D, Yadav N, King R, Swanson M, Weinstein E, Bedford M. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 25.Ragno R, Simeoni S, Castellano S, Vicidomini C, Mai A, Caroli A, Tramontano A, Bonaccini C, Trojer P, Bauer I, et al. Small molecule inhibitors of histone arginine methyltransferases: homology modeling, molecular docking, binding mode analysis, and biological evaluations. J Med Chem. 2007;50:1241–1253. doi: 10.1021/jm061213n. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Xie N, Wu J, Yang C, Zheng YG. Inhibitory study of protein arginine methyltransferase 1 using a fluorescent approach. Biochem Biophys Res Commun. 2009;379:567–572. doi: 10.1016/j.bbrc.2008.12.119. S0006-291X(08)02522-9 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Osborne T, Roska RL, Rajski SR, Thompson PR. In situ generation of a bisubstrate analogue for protein arginine methyltransferase 1. J Am Chem Soc. 2008;130:4574–4575. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]
- 28.Allan M, Manku S, Therrien E, Nguyen N, Styhler S, Robert MF, Goulet AC, Petschner AJ, Rahil G, Robert Macleod A, et al. N-Benzyl-1-heteroaryl-3-(trifluoromethyl)-1H-pyrazole-5-carboxamides as inhibitors of co-activator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2009;19:1218–1223. doi: 10.1016/j.bmcl.2008.12.075. S0960-894X(08)01600-4 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Huynh T, Chen Z, Pang S, Geng J, Bandiera T, Bindi S, Vianello P, Roletto F, Thieffine S, Galvani A, et al. Optimization of pyrazole inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2009;19:2924–2927. doi: 10.1016/j.bmcl.2009.04.075. S0960-894X(09)00569-1 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Purandare AV, Chen Z, Huynh T, Pang S, Geng J, Vaccaro W, Poss MA, Oconnell J, Nowak K, Jayaraman L. Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett. 2008;18:4438–4441. doi: 10.1016/j.bmcl.2008.06.026. S0960-894X(08)00675-6 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Mahler M, Fritzler MJ, Bluthner M. Identification of a SmD3 epitope with a single symmetrical dimethylation of an arginine residue as a specific target of a subpopulation of anti-Sm antibodies. Arthritis Res Ther. 2005;7:R19–29. doi: 10.1186/ar1455. ar1455 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahler M, Raijmakers R, Dahnrich C, Bluthner M, Fritzler MJ. Clinical evaluation of autoantibodies to a novel PM/Scl peptide antigen. Arthritis Res Ther. 2005;7:R704–713. doi: 10.1186/ar1729. ar1729 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. Embo J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. M000300200 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 36.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig 1. Figure S1. Synthesis of new PRMT inhibitors.
Formation of a diazo linkage to 7-amino-4-hydroxynaphthalene-2-sulfonic acid was followed by elaboration of the amino group to give the candidate compounds.