Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex (original) (raw)
To satisfy the mitotic checkpoint and drive chromosome congression, the Mps1 kinase lets go of kinetochores by phosphorylating itself in trans (see also related papers by Maciejowski et al. and Santaguida et al. in this issue).
Abstract
Mps1 is an essential component of the spindle assembly checkpoint. In this study, we describe a novel Mps1 inhibitor, AZ3146, and use it to probe the role of Mps1’s catalytic activity during mitosis. When Mps1 is inhibited before mitotic entry, subsequent recruitment of Mad1 and Mad2 to kinetochores is abolished. However, if Mps1 is inhibited after mitotic entry, the Mad1–C-Mad2 core complex remains kinetochore bound, but O-Mad2 is not recruited to the core. Although inhibiting Mps1 also interferes with chromosome alignment, we see no obvious effect on aurora B activity. In contrast, kinetochore recruitment of centromere protein E (CENP-E), a kinesin-related motor protein, is severely impaired. Strikingly, inhibition of Mps1 significantly increases its own abundance at kinetochores. Furthermore, we show that Mps1 can dimerize and transphosphorylate in cells. We propose a model whereby Mps1 transphosphorylation results in its release from kinetochores, thus facilitating recruitment of O-Mad2 and CENP-E and thereby simultaneously promoting checkpoint signaling and chromosome congression.
Introduction
The Mps1 protein kinase plays an essential role in the spindle assembly checkpoint (SAC; Weiss and Winey, 1996), an inhibitory network that restrains anaphase until all kinetochores are stably attached to spindle microtubules (Musacchio and Salmon, 2007). Recent efforts have revealed that its catalytic activity is required for SAC function and chromosome alignment (Jelluma et al., 2008a,b; Tighe et al., 2008; Kwiatkowski et al., 2010; Sliedrecht et al., 2010; see Santaguida et al. in this issue). Exactly how Mps1 does this is unclear; although its SAC function is linked to kinetochore recruitment of Mad1 and Mad2 (Martin-Lluesma et al., 2002; Liu et al., 2003), Mps1 has also been implicated in aurora B regulation (Jelluma et al., 2008b), possibly explaining its role in chromosome alignment.
Previously, when we used an RNAi complementation assay to inhibit Mps1’s catalytic activity, Mad2 was largely undetectable at kinetochores (Tighe et al., 2008). However, Mad1 recruitment was only partially affected, suggesting two possibilities: either Mps1 recruits Mad1 via a noncatalytic role or only low level Mps1 activity is required for Mad1 recruitment. Either way, because Mad1 directly recruits Mad2 to the kinetochore (Chung and Chen, 2002), the simplest explanation was that Mps1 activity promotes the Mad1–Mad2 interaction.
This notion is at odds with current views of Mad2 function (Mapelli et al., 2007). The Mad2 template model posits that a Mad1 dimer acts as the kinetochore receptor for Mad2, which can adopt two conformations, open (O-Mad2) and closed (C-Mad2). When Mad2 binds Mad1, it adopts the closed conformation, forming the Mad1–C-Mad2 core complex. Because Mad2 can dimerize, this core complex in turn recruits two O-Mad2 molecules. These are handed over to Cdc20, and in doing so, Mad2 closes, generating two Cdc20–C-Mad2 complexes. Because the Mad1–C-Mad2 complex is very stable (Howell et al., 2004; Shah et al., 2004; Vink et al., 2006) and probably present in interphase where it binds the nuclear envelope (Campbell et al., 2001), it seems unlikely that Mps1 promotes binding of Mad2 to Mad1.
An alternative explanation comes from the fact that our observations were derived using an antibody against Mad2, called SM2.2 (Tighe et al., 2008). Because kinetochore-bound Mad2 effectively represents two different species, we reasoned that the effect observed in Mps1-deficient cells was not necessarily caused by a complete inability to recruit Mad2, but may reflect an inability to detect it using SM2.2 either because the antibody is conformation specific (i.e., it cannot bind C-Mad2) or possibly because C-Mad2 is masked. If either possibility is true, an attractive hypothesis emerges; if SM2.2 only detects O-Mad2 at kinetochores for whatever reason, perhaps Mps1 kinase activity is required to recruit O-Mad2 to the Mad1–C-Mad2 core complex. In this study, we test this hypothesis using a novel small molecule Mps1 inhibitor.
Results and discussion
AZ3146, a novel Mps1 inhibitor
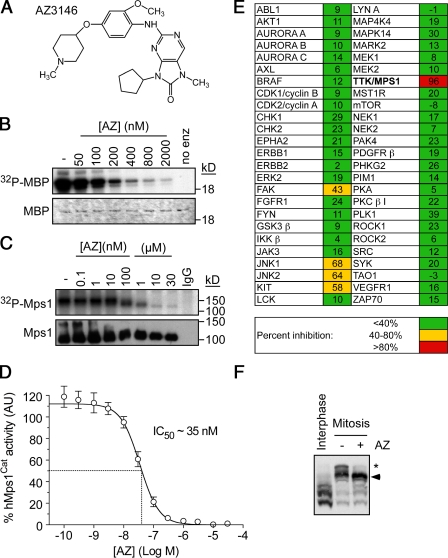
To identify Mps1 inhibitors, we used a high throughput in vitro kinase assay to screen a compound collection, yielding an inhibitor that was modified to enhance potency and selectivity, resulting in AZ3146 (Fig. 1 A). In in vitro kinase assays, AZ3146 inhibited human Mps1Cat with an IC50 (50% inhibitory concentration) of ∼35 nM (Fig. 1, B and D). AZ3146 also efficiently inhibited autophosphorylation of full-length Mps1 immunoprecipitated from human cells (Fig. 1 C). Screening a panel of 50 other kinases demonstrated minimal activity against 46 enzymes (Fig. 1 E). Only four kinases were inhibited by >40%, namely FAK, JNK1, JNK2, and KIT. Together, these data suggest that AZ3146 is a reasonably potent and selective Mps1 inhibitor. To measure the effect on Mps1 in cells, we used Phos tag gels to resolve phosphorylated isoforms (Kinoshita et al., 2006). When isolated from mitotic cells, Mps1 appeared as a major band and a minor, slower-migrating band (Fig. 1 F). Upon exposure to AZ3146, the minor band largely disappeared, demonstrating that it clearly inhibits phosphorylation of Mps1 in cells (Fig. 1 F and Fig. S1 B). Mitotic-specific phospho forms of aurora B and BubR1 were not affected by AZ3146 (Fig. S1 C), and AZ3146 did not inhibit Cdk1 or aurora B in mitotic cells (Fig. S1, A and D).
Figure 1.
AZ3146, a novel Mps1 inhibitor. (A) AZ3146 (AZ) is shown. (B) In vitro kinase assay measuring the ability of Mps1Cat to phosphorylate myelin basic protein (MBP). (C) In vitro kinase assays measuring the activity of Mps1 immunoprecipitated from nocodazole-arrested HeLa cells. (D) Dose–response curve showing inhibition of Mps1Cat by AZ3146. (E) Activity of AZ3146 against a kinase panel. (F) Phos tag immunoblot of Mps1 isolated from interphase cells or nocodazole-arrested cells treated with MG132 ± 2 µM AZ3146 for 2 h. Arrowhead, major band; asterisk, minor band. Error bars indicate SEM derived from three experiments.
AZ3146 overrides the spindle checkpoint
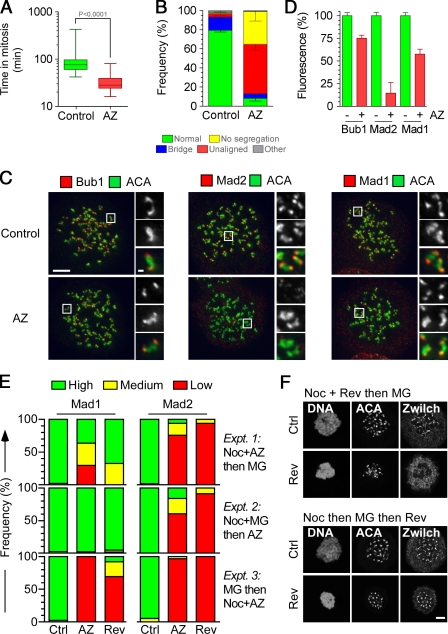
Cell cycle analysis showed that HeLa cells treated with nocodazole and 2 µM AZ3146 only delayed mitosis briefly and then rereplicated their genomes (Fig. S2, A–C), demonstrating that the drug overrides the SAC. AZ3146 also silenced an already established SAC signal; after release from a nocodazole block, AZ3146 dramatically accelerated mitotic exit (Fig. S2 D). This was blocked by MG132, a proteasome inhibitor (Fig. S2 E), ruling out direct inhibition of Cdk1 activity. Time-lapse analysis of HeLa cells expressing a GFP-tagged histone showed that during an otherwise unperturbed mitosis, AZ3146 reduced the time to complete mitosis from ∼90 min in controls to ∼32 min (Fig. 2 A). Strikingly, ∼90% of AZ3146-treated cells underwent abnormal mitoses; although ∼50% entered anaphase without aligning all of their chromosomes, ∼30% exited mitosis without undergoing obvious chromosome segregation (Fig. 2 B). Although AZ3146 had a dramatic effect on kinetochore localization of Mad2, reducing its levels to ∼15%, its effect on Mad1 was less pronounced, with levels remaining at ∼60% (Fig. 2, C and D). These data are entirely consistent with our prior observations, namely a dramatic inhibition of Mad2 and a partial effect on Mad1 (Tighe et al., 2008). Thus, taking together the enzyme data, the SAC override phenotype, and the effect on Mad1 and Mad2 localization, these data show that AZ3146 behaves as one would predict for an Mps1 inhibitor, strongly arguing that it is a useful tool for probing Mps1 function.
Figure 2.
AZ3146 compromises SAC function and inhibits Mad2 localization. (A) Box and whisker plot of HeLa GFP-H2B cells treated with or without AZ3146 (AZ) and analyzed by time-lapse microscopy, showing the time from nuclear envelope breakdown to chromosome decondensation. Values were derived from ≥75 cells. (B) Bar graph quantifying mitotic phenotypes. Values were derived from ≥115 cells. (C) Images of HeLa cells treated with nocodazole and MG132 for 2 h with or without AZ3146, fixed, and stained to detect centromeres (ACA) and either Bub1, Mad1, or Mad2. Insets show higher magnification views of individual kinetochore pairs (boxed regions). (D) Bar graph quantifying pixel intensities at kinetochores normalized to ACA in mitotic HeLa cells treated with or without AZ3146. Values were derived from ≥228 kinetochores. (E) Bar graphs scoring kinetochore staining of Mad1/2 as high, medium, or low after exposure to nocodazole (Noc) or MG132 (MG) plus either AZ3146 or reversine (Rev). Data were derived from a two experiments in which at least 50 cells were counted per condition. (F) Images of HeLa cells stained to detect Zwilch and centromeres after exposure to nocodazole, reversine, and MG132 in the sequences indicated. AZ3146 was used at 2 µM and reversine at 500 nM. Ctrl, control. Error bars indicate SEM. Bars: (C and F) 5 µm; (C, insets) 0.5 µm.
Mps1 plays two roles when recruiting Mad1 and Mad2 to kinetochores
We were puzzled why inhibiting Mps1 had a dramatic effect on kinetochore localization of Mad2 but only partially effected Mad1. Indeed, other studies suggest that Mps1 inhibition does dramatically affect Mad1 (Jelluma et al., 2008b; Kwiatkowski et al., 2010; Sliedrecht et al., 2010); e.g., inhibition of Mps1 using another small molecule, reversine, blocked recruitment of Mad1 and its kinetochore receptor, the Rod–Zw10–Zwilch (RZZ) complex (Santaguida et al., 2010). We reasoned that these differences may be a result of when Mps1 was first inhibited either before or during mitosis. To test this, we designed three experiments, exposing cells to nocodazole, MG132, and AZ3146, but in different orders. After treatment, cells were fixed, stained, and the intensity of Mad1 at kinetochores was scored as high, medium, or low (Fig. S3 A).
First, we treated cells with nocodazole and AZ3146 for 2 h, adding MG132 for the last hour. Like our previous experiments, in this regimen, some cells should enter mitosis in the presence of AZ3146, whereas others would already be in mitosis. This yielded a mixed effect on Mad1: although some cells still had high levels of Mad1 at kinetochores, others had low levels (Fig. 2 E, Expt. 1). Thus, the aforementioned partial effect on Mad1 (Fig. 2 D) appears to arise from the fact that when we quantitate pixel intensities in a small population of cells, some have relatively normal levels of Mad1 at kinetochores, whereas in others, it is markedly reduced (Fig. S3 A). Next, we treated cells with nocodazole and MG132 for 2 h, adding AZ3146 for the last hour. Because MG132 blocks entry into and exit out of mitosis, in this regimen, all mitotic cells should have entered mitosis before AZ3146 exposure. Under these conditions, Mad1 was now largely unaffected (Fig. 2 E, Expt. 2). Finally, we treated cells with MG132 for 2 h, adding nocodazole and AZ3146 for the last hour. In this regimen, before AZ3146 treatment, all mitotic cells should have aligned chromosomes with kinetochores stripped of Mad1/2 (DeLuca et al., 2003). Therefore, adding nocodazole to destabilize microtubules should lead to rerecruitment of Mad1. Significantly, although Mad1 was present at kinetochores in controls, kinetochores in cells treated with nocodazole plus AZ3146 were devoid of Mad1 (Fig. 2 E, Expt. 3). Note that AZ3146 inhibited kinetochore localization of Mad2 regardless of the experimental regimen (Fig. 2 E).
The simplest explanation for these data is that Mps1 is involved in two distinct steps. First, it is required upon mitotic entry to get Mad1 to kinetochores (or, in the case of experiment 3, it is required in mitosis to rerecruit Mad1 when microtubules are removed from attached kinetochores). Although Mps1 activity is no longer needed to maintain Mad1 localization, it is continuously required during mitosis to promote Mad2 recruitment. Importantly, reversine yielded virtually identical phenotypes (Fig. 2 E). Furthermore, when cells were treated with reversine before mitotic entry, the RZZ component Zwilch was largely absent from kinetochores (Fig. 2 F). However, when cells accumulated in mitosis before exposure to reversine, Zwilch localization was normal.
Mps1 activity is required to recruit O-Mad2 to the Mad1–C-Mad2 core
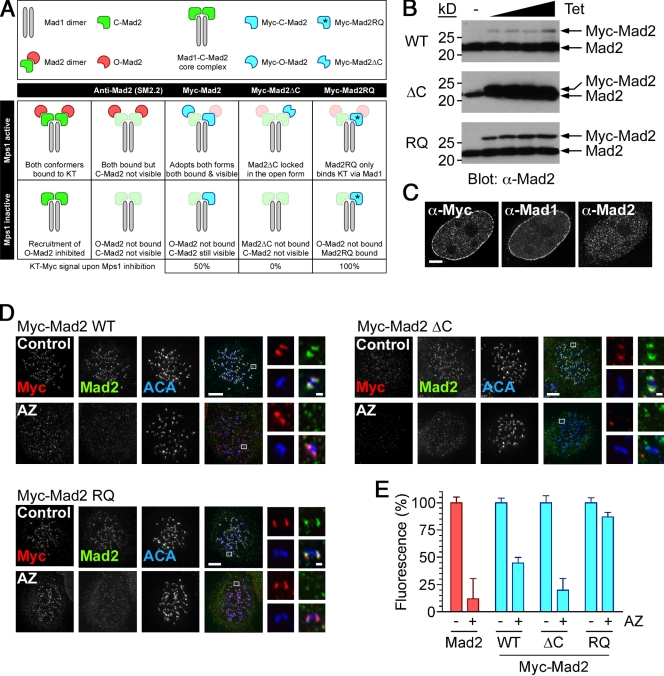
The aforementioned data suggest that during mitosis, Mps1 activity is required continuously to recruit Mad2 to kinetochore-bound Mad1 (Fig. 2 E, Expt. 2). However, this notion contradicts data showing that the Mad1–C-Mad2 interaction is very stable (Howell et al., 2004; Shah et al., 2004; Vink et al., 2006). Because a key step in checkpoint activation is recruitment of O-Mad2 to kinetochore-bound Mad1–C-Mad2 (Musacchio and Salmon, 2007), we hypothesized that perhaps Mps1 activity is required to recruit O-Mad2 to Mad1–C-Mad2 complexes already at kinetochores. In that case, upon Mps1 inhibition, kinetochore-bound Mad2 should only fall to ∼50% (Fig. 3 A), yet AZ3146 abolishes Mad2 localization (Fig. 2, C–E). Therefore, we speculated that the Mad2 antibody SM2.2 does not efficiently detect C-Mad2 at the kinetochore either because the antibody cannot bind C-Mad2 or because the relevant epitopes are masked.
Figure 3.
AZ3146 inhibits kinetochore recruitment of O-Mad2. (A) A scheme is shown predicting the behavior of endogenous Mad2 and Myc-tagged mutants in cells after inhibition of Mps1 activity. (B) Immunoblots of Tet-induced HeLa lines probed to detect Mad2. (C) Images of an interphase HeLa cell expressing Myc-Mad2 wild type stained to detect Mad1, Mad2 using SM2.2, and an anti-Myc antibody. (D) Images of nocodazole-treated cells stained to detect the Myc tag (red), Mad2 (green), and centromeres (ACA; blue) after a 2-h exposure to 2 µM AZ3146 (AZ) and MG132. Insets show higher magnification views of individual kinetochore pairs (boxed regions). (E) Bar graph quantitating either Mad2 (red) or Myc tag (blue) pixel intensities at kinetochores normalized to ACA. WT, wild type; RQ, Mad2RQ. Values indicate SEM derived from ≥245 kinetochores. Bars: (C and D) 5 µm; (C, insets) 0.5 µm.
To test this, we generated cells expressing Myc-tagged Mad2 (Fig. 3 B and Fig. S3 B), reasoning that anti-Myc antibodies might detect it regardless of conformation or cellular context. Interestingly, Mad1 and Mad2 localize to the nuclear envelope in interphase (Campbell et al., 2001; Shah et al., 2004), and although we could readily detect Mad2 there using anti-Myc antibodies, SM2.2 did not decorate the envelope (Fig. 3 C), suggesting that it does not recognize C-Mad2 when bound to Mad1. In mitosis, both anti-Myc antibodies and SM2.2 decorated kinetochores (Fig. 3 D). Significantly, although AZ3146 reduced SM2.2 signal to background levels, anti-Myc antibodies still gave obvious kinetochore staining (Fig. 3, D and E). Importantly, however, quantitation showed that anti-Myc staining was reduced to 45 ± 5% (Fig. 3 E), i.e., approaching the 50% value predicted by our hypothesis.
To test the hypothesis further, we analyzed two Mad2 mutants. Mad2ΔC cannot adopt the closed conformation (Mapelli et al., 2007) or bind Mad1 and, thus, can only bind kinetochores via dimerizing with endogenous C-Mad2 (Fig. 3 A). Mad2RQ can bind Mad1 but cannot dimerize (Mapelli et al., 2007) and, thus, can only target kinetochores by binding Mad1 (Fig. 3 A). If Mps1 is required to recruit O-Mad2 to the core, upon exposure to AZ3146, Mad2ΔC should be absent from kinetochores, whereas Mad2RQ should be unaffected (Fig. 3 A). We generated stable cell lines expressing Myc-Mad2ΔC and Myc-Mad2RQ (Fig. 3 B and Fig. S3 B). Although both mutants localized to kinetochores in mitosis (Fig. 3 D), Myc-Mad2ΔC did not bind the nuclear envelope (Fig. S3 C). However, consistent with our hypothesis in response to AZ3146, Mad2ΔC was no longer abundant at kinetochores, whereas Mad2RQ appeared unaffected (Fig. 3 D). Indeed, quantitation showed that Myc-Mad2ΔC was reduced to a background level, whereas Myc-Mad2RQ remained at ∼90% (Fig. 3 E). Thus, these observations strongly support the notion that during mitosis, Mps1 kinase activity is required to recruit O-Mad2 to kinetochore-bound Mad1–C-Mad2.
Mps1 is required for chromosome alignment
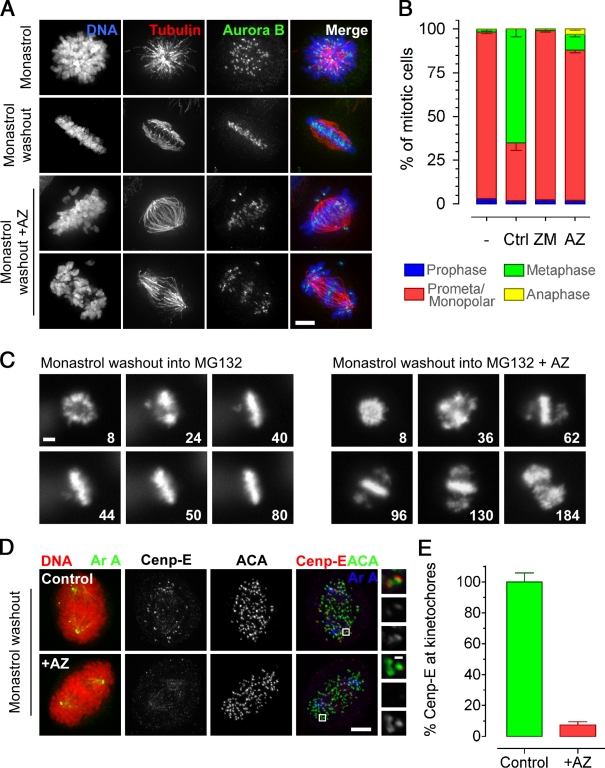
To confirm and further probe the role of Mps1 in chromosome alignment (Maure et al., 2007; Jelluma et al., 2008b), we analyzed the effect of AZ3146 after a monastrol washout (Mayer et al., 1999; Kapoor et al., 2006). Note that cells were also treated with MG132 to prevent mitotic exit caused by SAC override. Under these conditions, AZ3146 inhibited chromosome alignment in >85% of cells (Fig. 4, A and B), yielding either a mild phenotype with few unaligned chromosomes or a severe phenotype with most chromosomes near the poles (Fig. 4 A). Time-lapse microscopy showed that AZ3146-treated cells typically aligned all but a few chromosomes (Fig. 4 C, 96 min). However, sometime later, sister chromatids appeared to separate followed by rather chaotic attempts to realign (Fig. 4 C, 184 min; and Fig. S2 F). Thus, the mild phenotype appears to reflect Mps1’s role in efficient chromosome alignment, whereas the severe phenotype, at least in some cells, may be a consequence of the prolonged mitotic arrest.
Figure 4.
AZ3146 inhibits chromosome alignment. (A) Images of HeLa cells stained to detect the chromosomes (blue), tubulin (red), and aurora B (green) after monastrol washout into 2 µM AZ3146 (AZ) and MG132 for 90 min. (B) Bar graph quantitating the mitotic phenotypes observed after monastrol washout into AZ3146 or ZM447439 (ZM). Values were derived from ≥115 cells in three independent experiments. Ctrl, control. (C) Time-lapse sequences of GFP-H2B HeLa cells after monastrol washout into AZ3146 and MG132. Numbers represent minutes after monastrol washout. (D) Images of HeLa cells stained to detect the chromosomes (red), aurora A (green/blue), CENP-E (red), and centromeres (ACA; green) after monastrol washout into AZ3146 and MG132 for 60 min. Insets show higher magnification views of individual kinetochore pairs (boxed regions). (E) Bar graph quantitating CENP-E staining normalized to ACA at polar kinetochores in AZ3146-treated cells. The mean control value is normalized to 100%. At least 300 polar kinetochores were analyzed in ≥23 cells. Error bars indicate SEM. Bars: (A, C, and D) 5 µm; (D, insets) 0.5 µm.
Mps1 activity is required for centromere protein E (CENP-E) localization
In light of Mps1 regulating aurora B via borealin phosphorylation (Jelluma et al., 2008b), we were surprised that the alignment defect was not typical of that observed after aurora B inhibition (Ditchfield et al., 2003). Indeed, AZ3146 had no obvious effect on several bona fide aurora B substrates (Fig. S1, D–F). Therefore, we turned our attention to other factors to explain the chromosome alignment defect. The phenotype observed in AZ3146-treated cells is quite exquisite; most chromosomes align, K-fibers are evident (Fig. 4 A), and only a few chromosomes remain near the spindle poles (Fig. 4, A–C). This reminded us of the phenotype typical of CENP-E inhibition (Wood et al., 1997; Weaver et al., 2003; Kapoor et al., 2006). Strikingly, 1 h after monastrol washout, the level of CENP-E at kinetochores was reduced to ∼10% (Fig. 4, D and E). Consistent with this being an on-target effect of AZ3146, when Mps1-RNAi cells were reconstituted with the Mps1M602A allele, 1NM-PP1 inhibited kinetochore localization of CENP-E (Fig. S3 D). Thus, CENP-E appears to be an important downstream effector of Mps1 kinase activity. Note that this is entirely consistent with a previous study analyzing Xenopus laevis egg extracts (Abrieu et al., 2001).
Inhibition of Mps1 increases its abundance at kinetochores
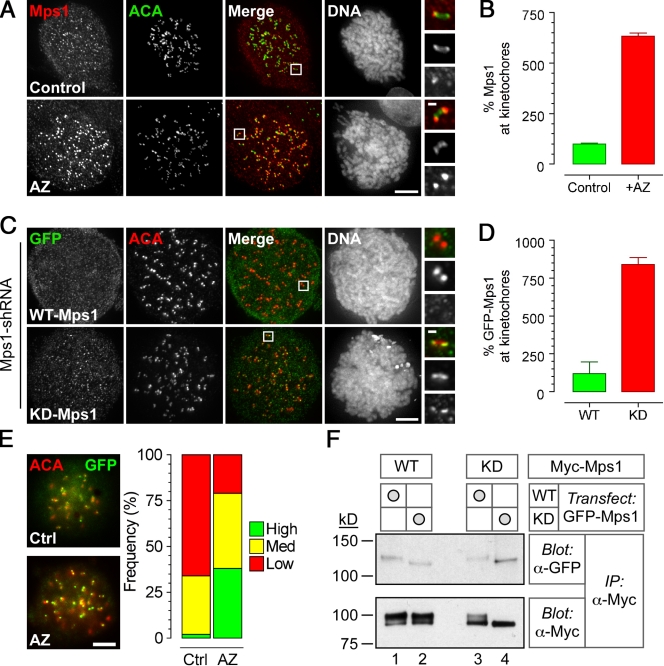
These data indicate that Mps1 contributes to mitotic fidelity by promoting kinetochore recruitment of O-Mad2 and Cenp-E. Although detailing the exact molecular mechanisms will require further experimentation, one observation may prove informative. In the presence of AZ3146, Mps1 staining at kinetochores increased by about sixfold (Fig. 5, A and B). This is not simply caused by increasing epitope accessibility, as direct fluorescence from a GFP-Mps1 fusion was also more evident at kinetochores in AZ3146-treated cells (Fig. 5 E).
Figure 5.
AZ3146 enhances kinetochore localization of Mps1. (A) Images of HeLa stained to detect Mps1 (red), centromeres (ACA; green), and the chromosomes after treatment with nocodazole and MG132 for 2 h ± 2 µM AZ3146 (AZ). (B) Bar graph quantitating Mps1 at kinetochores normalized to ACA. Values were derived from ≥345 kinetochores in ≥18 cells with the mean in control cells normalized to 100%. (C) Images of HeLa cells expressing GFP-Mps1, either wild type (WT) or kinase dead (KD), stained to detect GFP-Mps1 (green), centromeres (ACA; red), and chromosomes. Note that endogenous Mps1 was first repressed by RNAi. Before fixation, cells were treated with nocodazole and MG132 for 2 h with or without AZ3146. (D) Bar graph quantitating GFP pixel intensities at kinetochores normalized to ACA. Values were derived from ≥240 kinetochores in ≥17 cells with the mean in controls cells normalized to 100%. (E) Images of HeLa cells expressing GFP-Mps1 treated with or without AZ3146 and a bar graph scoring direct GFP fluorescence from kinetochores as high, medium, or low are shown. Data are representative of two independent experiments in which ≥50 cells were counted. Ctrl, control. (F) Immunoblots of anti-Myc immune complexes isolated from HEK293 cells stably expressing Myc-tagged Mps1 transgenes and transiently transfected with vectors encoding GFP-tagged Mps1 transgenes, either wild type or kinase dead. Membranes were blotted as indicated. IP, immunoprecipitation. (A and C) Insets show higher magnification views of individual kinetochore pairs (boxed regions). Error bars indicate SEM. Bars: (A, C, and E) 5 µm; (A and C, insets) 0.5 µm.
To confirm that this reflected an on-target effect of AZ3146, we compared kinetochore localization of wild-type and kinase-dead GFP-Mps1 fusions after repression of the endogenous kinase by RNAi (Tighe et al., 2008). Strikingly, the kinase-dead mutant was about eightfold more abundant at kinetochores compared with wild-type (Fig. 5, C and D), supporting the notion that inhibiting Mps1’s activity does indeed increase its levels at kinetochores. Because FRAP experiments have shown that GFP-Mps1 has a relatively short residency time at kinetochores (Howell et al., 2004), one explanation is that inhibiting Mps1 activity reduces its turnover at kinetochores.
Mps1 dimerizes and transphosphorylates
Importantly, the GFP-Mps1 kinase-dead fusion only increased at kinetochores when we repressed the endogenous kinase (Fig. 5 C). Therefore, we wondered whether Mps1 functions as a dimer that requires transphosphorylation to be released from the kinetochore. Interestingly, Mps1 transphosphorylation has been demonstrated in vitro, but there is no evidence to support dimerization in cells (Kang et al., 2007). Therefore, we transiently transfected GFP-tagged Mps1 into cells stably expressing Myc-tagged Mps1. Myc-tagged proteins were immunoprecipitated and blotted to detect GFP. When we immunoprecipitated Myc-Mps1, GFP-Mps1 was readily detectable (Fig. 5 F and Fig. S3 E), which is consistent with dimerization. Interestingly, when either fusion was catalytically active, a portion of the Myc-tagged protein was phosphorylated (Fig. 5 F). In contrast, when both fusions were catalytically inactive, the Myc-Mps1 band shift was absent (Fig. 5 F, lane 4). Together, these observations support the notion that Mps1 does dimerize and transphosphorylate in mitotic cells. Whether this regulates its localization requires further investigation.
Coupling SAC signaling with chromosome congression
Our observations suggest a two-step role for Mps1. Around the onset of mitosis, Mps1 activity is required to first recruit the Mad1–C-Mad2 core to kinetochores, possibly indirectly by promoting RZZ recruitment. During mitosis, Mps1 activity is continuously required to recruit O-Mad2 to the Mad1–C-Mad2 core. Although Mps1 may achieve this directly, e.g., by phosphorylating the core or O-Mad2, this seems unlikely because the Mad2 template mechanism can be recapitulated in vitro without Mps1 activity (Vink et al., 2006). Therefore, perhaps Mps1 kinase activity counters a cellular inhibitor not present in these in vitro assays. Interestingly, p31comet may act as an inhibitory “cap” on the Mad1–C-Mad2 core (Musacchio and Salmon, 2007). Thus, an attractive hypothesis is that Mps1 phosphorylates and inactivates p31comet, removing the inhibitory cap to allow recruitment of O-Mad2 and thereby kick starting the Mad2 template reaction. However, we presently have no evidence linking Mps1 and p31comet. As Mps1 levels at kinetochores increase when its activity is blocked, perhaps Mps1 is itself an inhibitory cap that must be released for checkpoint signaling. Having first recruited the Mad1–C-Mad2 core, perhaps Mps1 transphosphorylation promotes an autorelease mechanism that allows recruitment of O-Mad2, kick starting the Mad2 template mechanism.
We confirm a role for Mps1 activity in chromosome alignment. Although we observe no obvious effect on aurora B function, Mps1 activity is clearly required for CENP-E recruitment after monastrol washout. Interestingly, however, Mps1-RNAi did not prevent CENP-E recruitment (Jelluma et al., 2008b; Tighe et al., 2008). Because inactive Mps1 accumulates at kinetochores, perhaps Mps1 and CENP-E occupy mutually exclusive binding sites. In this scenario, Mps1 autorelease would normally expose the CENP-E–binding site. However, when its catalytic activity is inhibited, Mps1 blocks CENP-E recruitment by remaining bound to the kinetochore. In contrast, when the entire Mps1 protein is depleted by RNAi, the CENP-E–binding site is vacant, allowing its recruitment. Consistent with this notion, the levels of kinetochore-bound CENP-E have been observed to increase after Mps1 depletion (Jelluma et al., 2008b). Although speculative, these ideas warrant further exploration and should provide interesting insights into how Mps1 coordinates spindle checkpoint signaling and chromosome congression.
Materials and methods
Inhibitor screen
N-terminal GST-tagged full-length human Mps1 kinase (GenBank accession no. NM_003318) was expressed in insect cells and purified via the GST epitope tag. A 12 µl mixture of recombinant enzyme, an FITC-labeled peptide substrate (FITC-DHTGFLTEYVATR-CONH2), 12 µM ATP, and a buffer solution (50 mM Hepes, pH 7.5, 0.015% vol/vol Brij-35, 1 mM DTT, and 10 mM MgCl2) were incubated with test compounds at room temperature for 25 min. Test compounds were prepared and further diluted in DMSO and placed into 384-well low volume white polystyrene plates (Greiner) using an acoustic liquid handler (Echo; Labcyte, Inc.). Reactions were stopped by the addition of 40 mM, 0.1% coating reagent (Caliper Life Sciences), 100 mM Hepes, pH 7.5, 0.015% vol/vol Brij-35, and 5% DMSO. Inhibitors of Mps1 activity were identified using a microfluidic chip and laser-induced fluorescence (LabChip LC3000; Caliper Life Sciences) to measure the conversion of the fluorescent-labeled peptide to a phosphorylated product.
In vitro kinase assays
His-tagged human Mps1Cat encoding amino acids 510–857 was generated as described previously (Chu et al., 2008). For kinase assays, 500 ng was added to buffer (25 mM Tris-HCl, pH 7.4, 100 mM NaCl, 50 µg/ml BSA, 0.1 mM EGTA, 0.1% β-mercaptoethanol, 10 mM MgCl2, and 0.5 µg/ml myelin basic protein), AZ3146, and 100 µM γ-[32P]ATP (2 µCi/assay). Reactions were incubated at 30°C for 20 min, spotted onto P81 paper, washed in 0.5% phosphoric acid, and immersed in acetone. Phosphate incorporation was determined by scintillation counting. For immunoprecipitation kinase assays, HeLa cells were treated with nocodazole for 14 h, mitotic cells isolated, washed in PBS, and lysed for 30 min in 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.5% NP-40, 5 mM EDTA, 5 mM EGTA, 40 mM β-glycerophosphate, 0.2 mM PMSF, 1 mM DTT, 1 mM sodium orthovanadate, 20 mM sodium fluoride, 1 µM okadaic acid (EMD), and complete EDTA-free protease inhibitor cocktail (Roche). Full-length Mps1 was immunoprecipitated as described previously (Holland et al., 2007). Purified complexes were washed with lysis buffer containing 100 mM NaCl and assayed as described for the recombinant protein. To quantify 32P incorporation, reactions were stopped with SDS sample buffer and separated by SDS-PAGE followed by phosphorimaging. The plate (Fujifilm) was analyzed using a phosphorimager (FLA-3000; Fujifilm) using AIDA software (Raytest Isotopenmessgerate GmbH). To assess the specificity of AZ3146, a single-point screen was carried using kinase profiling service (SelectScreen; Invitrogen). 50 kinases were selected and assayed with 1 µM AZ3146.
Cell lines and drugs
Parental HeLa cells, HeLa cells expressing GFP-tagged histone H2B, and Flp-in T-REx HeLa cells were used (Taylor and McKeon, 1997; Morrow et al., 2005; Tighe et al., 2008) and cultured as described previously (Taylor et al., 2001). Flp-in T-REx HEK293 cells were used as described previously (Tighe et al., 2004). A human Mad2 cDNA (Johnson et al., 2004) was cloned into pcDNA5/FRT/TO and mutagenized (QuickChange; Agilent Technologies) to create Mad2ΔC (Mad2 lacking amino acids 196–205) and Mad2 R133E, Q134A (Table S1). The resulting vectors were cotransfected into Flp-in T-REx HeLa cells with the Flp recombinase—encoding plasmid pOG44 (Tighe et al., 2004). Hygromycin-resistant colonies were pooled, expanded, and transgene expression was induced with 500 ng/ml tetracycline (Sigma-Aldrich). Nocodazole, taxol, monastrol, and MG132 (Sigma-Aldrich) were used at a final concentration of 0.2 µg/ml, 10 µM, 100 µM, and 20 µM, respectively. ZM447439 (Tocris) was used at 2 µM. The Mps1 inhibitor AZ3146 was used at a final concentration of 2 µM unless otherwise stated.
Cell biology and RNAi
DNA content, mitotic index measurement, and synchronization of HeLa cells at G1/S using a double-thymidine block were performed as described previously (Taylor and McKeon, 1997). In brief, cells were fixed in 70% ethanol, stained with MPM-2 antibodies (Millipore) followed by a FITC-conjugated donkey anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.), and stained with propidium iodide. Flow cytometric analysis was performed using a Cyan (Dako). For inhibitor washout experiments, cells grown on coverslips were treated with monastrol for 4 h. The cells were washed twice with media and once with media containing AZ3146 and MG132. 2 h later, the cells were processed for immunofluorescence. For analysis by time-lapse microscopy, HeLa cells stably expressing GFP–histone H2B were cultured on 30-mm glass-bottomed dishes (MatTek Co) and treated with monastrol for 2 h. The cells were washed before being imaged as described previously (Morrow et al., 2005). Repression of endogenous Mps1 was performed as described previously (Tighe et al., 2008). In brief, cells were seeded into a 24-well plate and transfected using Lipofectamine Plus (Invitrogen) with an Mps1 shRNA plasmid. The DNA–lipid complexes were added to the cells, and 24 h later, the cells were replated onto coverslips or 6-well plates and analyzed 24 h later. To analyze the effect of reversine on Zwilch localization, two regimens were used. To ensure that cells were in mitosis before addition of reversine, HeLa cells were first treated with 3.3 µM nocodazole for 12 h, and 10 µM MG132 was added for 30 min followed by 0.5 µM reversine for 1.5 h. To allow cells to enter mitosis in the presence of reversine, cells were first synchronized at G1/S using a double-thymidine arrest. 5 h after release, the cells were treated with 3.3 µM nocodazole and 0.5 µM reversine for 2 h followed by 10 µM MG132 for 1.5 h.
Antibody techniques and Phos tag gels
Immunoprecipitations, immunoblotting, and immunofluorescence were performed as described previously (Tighe et al., 2004; Morrow et al., 2005; Tighe et al., 2008) using the antibodies described in Table S2. In brief, cells were harvested and lysed on ice for 30 min. Lysates were cleared by centrifugation, precleared with protein G–Sepharose, and incubated for 2 h with antibody-coupled beads. After washing in lysis buffer, isolated proteins were resolved by SDS-PAGE, transferred onto a PVDF membrane (Millipore), blocked in 5% milk, and primary antibodies were added overnight. After washing and incubation with appropriate secondary antibodies, proteins were visualized using chemiluminescence (SuperSignal; Thermo Fisher Scientific) and imaged on film (Biomax MR; Kodak). For immunofluorescence, cells grown on coverslips were fixed in 1% formaldehyde, quenched in glycine, pH 8.5, permeabilized using PBS plus 0.1% Triton X-100 (PBST), and incubated with primary antibodies. After washes in PBST and incubation with appropriate secondary antibodies, cells were counterstained with 1 µg/ml Hoechst 33358 in PBST and mounting in 90% glycerol and 20 mM Tris-HCl, pH 8.0. To visualize microtubules, cells were preextracted in 100 mM Pipes, 1 mM MgCl2, 0.1 mM CaCl2, and 0.1% Triton X-100 for 90 s before fixation in 4% formaldehyde in the same buffer for 10 min. To stain for CENP-E, cells were preextracted with a buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% BSA, and 0.2% Triton X-100 for 2 min before fixation with 4% formaldehyde in PBS (Chan et al., 2000). Phos tag (Wako Chemicals USA, Inc.) was used according to the manufacturer’s instructions. In brief, 25 µM Phos tag and 50 µM MnCl2 were added to standard SDS-PAGE gels containing 6% acrylamide and visualized by immunoblotting.
Image acquisition and processing
Immunofluorescence images were acquired at room temperature on a restoration microscope (Delta Vision RT; Applied Precision) using a 100× 1.40 NA Plan Apo objective and a filter set (Sedat Quad; Chroma Technology Corp.). The images were collected at room temperature using a charge-coupled device camera (CoolSNAP HQ; Photometrics) with a z-optical spacing of 0.2 µm. Raw images were deconvolved using the SoftWorx software (Applied Precision), and maximum intensity projections of these deconvolved images are described in Results and discussion. Time-lapse microscopy was performed on a manual microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with an automated stage (PZ-2000; Applied Biosystems) and an environmental control chamber (Solent Scientific), which maintained the cells at 37°C in a humidified stream of 5% CO2. Imaging was performed using either a 32× 0.40 NA long-distance A-Plan or a 40× 0.75 NA Neo Fluor objective. Shutters, filter wheels, and point visiting were driven by MetaMorph software (Universal Imaging). Images were taken using a camera (CoolSNAP HQ; Photometrics), and individual tagged image file format files were imported into PhotoShop (Adobe) for printing or into QuickTime (Apple) for videos.
Online supplemental material
Fig. S1 shows that AZ3146 does not inhibit Cdk1 or aurora B. Fig. S2 shows that AZ3146 overrides the spindle checkpoint. Fig. S3 shows that Mps1 activity is required for kinetochore localization of CENP-E. Table S1 lists the PCR primers used in this study. Table S2 lists the antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002133/DC1.
Acknowledgments
We are grateful to Bill Earnshaw, Don Cleveland, and Keith Gull for reagents and to members of the Taylor laboratory for comments on the manuscript.
L. Hewitt is supported by a studentship from the Biotechnology and Biological Sciences Research Council and the University of Manchester Alumni Funds. A. Tighe is funded by Cancer Research UK, S. Santaguida is supported by a fellowship of the Italian Foundation for Cancer Research, S. Green and C.D. Jones are employees of AstraZeneca UK Ltd, and S.S. Taylor is supported by a Senior Fellowship from Cancer Research UK.
Footnotes
Abbreviations used in this paper:
CENP-E
centromere protein E
RZZ
Rod–Zw10–Zwilch
SAC
spindle assembly checkpoint
References
- Abrieu A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., Labbé J.C. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93 10.1016/S0092-8674(01)00410-X [DOI] [PubMed] [Google Scholar]
- Campbell M.S., Chan G.K., Yen T.J. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., Yen T.J. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947 10.1038/35046598 [DOI] [PubMed] [Google Scholar]
- Chu M.L., Chavas L.M., Douglas K.T., Eyers P.A., Tabernero L. 2008. Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125. J. Biol. Chem. 283:21495–21500 10.1074/jbc.M803026200 [DOI] [PubMed] [Google Scholar]
- Chung E., Chen R.H. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 13:1501–1511 10.1091/mbc.02-01-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Howell B.J., Canman J.C., Hickey J.M., Fang G., Salmon E.D. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103–2109 10.1016/j.cub.2003.10.056 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Böttger F., Stemmann O., Taylor S.S. 2007. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J. Biol. Chem. 282:24623–24632 10.1074/jbc.M702545200 [DOI] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., McLeod I., Yates J.R., III, Cleveland D.W., Medema R.H., Kops G.J. 2008a. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS One. 3:e2415 10.1371/journal.pone.0002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., van den Broek N.J., Cruijsen C.W., van Osch M.H., Lens S.M., Medema R.H., Kops G.J. 2008b. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 132:233–246 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- Kang J., Chen Y., Zhao Y., Yu H. 2007. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc. Natl. Acad. Sci. USA. 104:20232–20237 10.1073/pnas.0710519105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 5:749–757 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski N., Jelluma N., Filippakopoulos P., Soundararajan M., Manak M.S., Kwon M., Choi H.G., Sim T., Deveraux Q.L., Rottmann S., et al. 2010. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 6:359–368 10.1038/nchembio.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Chan G.K., Hittle J.C., Fujii G., Lees E., Yen T.J. 2003. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 14:1638–1651 10.1091/mbc.02-05-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Massimiliano L., Santaguida S., Musacchio A. 2007. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 131:730–743 10.1016/j.cell.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., Nigg E.A. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- Maure J.F., Kitamura E., Tanaka T.U. 2007. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 17:2175–2182 10.1016/j.cub.2007.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Morrow C.J., Tighe A., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S. 2005. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 118:3639–3652 10.1242/jcs.02487 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952 [DOI] [PubMed] [Google Scholar]
- Sliedrecht T., Zhang C., Shokat K.M., Kops G.J. 2010. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PLoS One. 5:e10251 10.1371/journal.pone.0010251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 89:727–735 10.1016/S0092-8674(00)80255-X [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395 [DOI] [PubMed] [Google Scholar]
- Tighe A., Johnson V.L., Taylor S.S. 2004. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J. Cell Sci. 117:6339–6353 10.1242/jcs.01556 [DOI] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., Musacchio A. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Bonday Z.Q., Putkey F.R., Kops G.J., Silk A.D., Cleveland D.W. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E., Winey M. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111–123 10.1083/jcb.132.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S., Cleveland D.W. 1997. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 91:357–366 10.1016/S0092-8674(00)80419-5 [DOI] [PubMed] [Google Scholar]