Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis (original) (raw)
Abstract
Members of the GATA and RUNX families of genes appear to have conserved functions during hematopoiesis from Drosophila to mammals. In Drosophila, the GATA factor Serpent (Srp) is required in blood cell progenitors for the formation of the two populations of blood cells (plasmatocytes and crystal cells), while the RUNX factor Lozenge (Lz) is specifically required for crystal cell development. Here we investigate the function and the mechanisms of action of Lz during hematopoiesis. Our results indicate that Lz can trigger crystal cell development. Interestingly, we show that Lz function is strictly dependent on the presence of functional Srp and that Srp and Lz cooperate to induce crystal cell differentiation in vivo. Furthermore, we show that Srp and Lz directly interact in vitro and that this interaction is conserved between Drosophila and mammals. Moreover, both Srp and mouse GATA1 synergize with mouse RUNX1 to activate transcription. We propose that interaction and cooperation between GATA and RUNX factors may play an important role in regulating blood cell formation from Drosophila to mammals.
Keywords: Drosophila/GATA/hematopoiesis/RUNX
Introduction
Hematopoietic development provides an excellent paradigm to address how multipotent cells generate a spectrum of cell types through the combinatorial action of transcription factors (Orkin and Zon, 2002). Recent investigations suggest that Drosophila will prove to be a valuable model to study the mechanisms of hematopoietic cell fate choice. Indeed, various aspects of hematopoietic development have been conserved during evolution (Evans and Banerjee, 2003). As in vertebrates, Drosophila hematopoiesis occurs in two waves from mesodermally derived progenitors. First, during early embryogenesis, blood cells (hemocytes) originate from the head mesoderm (Tepass et al., 1994). A second wave of hematopoiesis occurs during the larval stages in a specialized organ formed from the dorsal mesoderm, the lymph gland (Rizki, 1978). Hemocytes differentiate into two major classes: plasmatocytes and crystal cells (Lebestky et al., 2000). Plasmatocytes, which represent 95% of the hemocytes, function as macrophages and play a crucial role in host defense and development (Tepass et al., 1994; Franc et al., 1996). Crystal cells participate in melanization, an insect-specific process involved in the encapsulation of foreign bodies and in wound healing (Rizki et al., 1980). Functionally and morphologically, Drosophila blood cells most closely resemble vertebrate monocytes/ macrophages and granulocytes.
Strikingly, several transcription factors that are important for hematopoiesis and immunity in vertebrates have been shown to play similar roles in Drosophila. First, members of the GATA family are recurrently used during hematopoiesis. For instance, in vertebrates, GATA2 plays a critical role in the proliferation and/or survival of hematopoietic stem cells and controls the production of mast cells (Tsai and Orkin, 1997). Moreover, while GATA2 is downregulated to permit erythrocytic differentiation (Persons et al., 1999), GATA1 is necessary for the differentiation of erythrocytes and megakaryocytes (Pevny et al., 1991; Fujiwara et al., 1996). In Drosophila, the only GATA factor known to participate in hematopoiesis is encoded by the gene serpent (srp) (Rehorn et al., 1996) that is alternatively spliced to give rise to different isoforms containing either one (SrpC) or two (SrpNC) GATA zinc fingers (Waltzer et al., 2002). Similarly to GATA2, srp is expressed in the blood cell precursors and is required for the proliferation and maintenance of this population (Rehorn et al., 1996; Sam et al., 1996). However, srp may also have a function later in hematopoiesis as Srp is still detected in mature plasmatocytes and crystal cells (Lebestky et al., 2000). Secondly, members of the friend of GATA (FOG) family are implicated in hematopoietic development in both mammals and Drosophila. FOG proteins interact with the N-terminal zinc finger of GATA factors and modulate their activity (Haenlin et al., 1997; Tsang et al., 1997). In mouse, the interaction between FOG1 and GATA1 is necessary for normal erythroid and megakaryocytic differentiation (Tsang et al., 1998). In Drosophila, the FOG factor U-shaped (Ush) represses crystal cell fate (Fossett et al., 2001) and inhibits the expression of the apoptotic body receptor croquemort (crq) in plasmatocytes, most likely by interacting with SrpNC (Waltzer et al., 2002). Thirdly, factors containing a Runt domain (hereafter collectively called RUNX factors) participate in hematopoiesis in both vertebrates and Drosophila. In mouse, RUNX1, also known as acute myeloid leukemia factor 1 (AML1), is required for the emergence of definitive hematopoietic stem cells (Okuda et al., 1996; Wang et al., 1996). In addition, RUNX1 may promote myeloid progenitor differentiation, as suggested by in vitro assays and the frequent chromosomal translocations affecting RUNX1 that are associated with human acute myeloid leukemia (Speck and Gilliland, 2002). In Drosophila, Lozenge (Lz), which exhibits 71% identity with RUNX1 in the Runt domain, also plays a role in hematopoiesis (Lebestky et al., 2000). Indeed, lz expression initiates in a small fraction of _srp_-expressing hemocytes in response to Notch signaling and is necessary for crystal cell production (Lebestky et al., 2000, 2003). Taken together, the functional conservation of the GATA, FOG and RUNX proteins in Drosophila and vertebrate hematopoiesis suggests that aspects of the molecular circuitry that control blood cell development might also be conserved.
Recent genetic studies suggest that Drosophila hematopoiesis is organized in a hierarchical cascade of transcription factors controlling progenitor differentiation into the two major cell types: plasmatocytes and crystal cells. Thus srp expression in prohemocytes is essential for the expression of the transcription factors glial cell missing (gcm) and gcm2 (Bernardoni et al., 1997; Alfonso and Jones, 2002), lz (Lebestky et al., 2000) and ush (Fossett et al., 2001). Subsequently, while gcm and gcm2 are required for hemocyte differentiation into mature plasmatocytes, lz is required for crystal cell determination. Interestingly, this hierarchical cascade is not absolutely linear, as the conserved GATA/FOG interaction provides a feedback loop by which Ush impinges on srp function by interacting with SrpNC (Waltzer et al., 2002). Whether other players in this cascade interact with each other is not known.
Here we investigate the function and mechanism of action of the RUNX factor Lz during blood cell formation in Drosophila. We show that Lz cooperates with Srp to induce crystal cell differentiation in vivo. Our data suggest that this cooperation is mediated by the formation of an Srp/Lz complex that synergistically activates transcription. Consistent with this, we find that Srp and Lz interact through their conserved GATA zinc finger and Runt domain, respectively, and that this interaction is conserved through evolution as mouse GATA1 and RUNX1 can interact with each other and with their Drosophila counterparts. Finally, we show that GATA and RUNX factors can synergize to activate transcription in cell culture. Together, these results suggest that the interaction and cooperation between GATA and RUNX factors may play an important role during hematopoiesis from Drosophila to vertebrates.
Results
lozenge can induce crystal-cell-specific gene expression
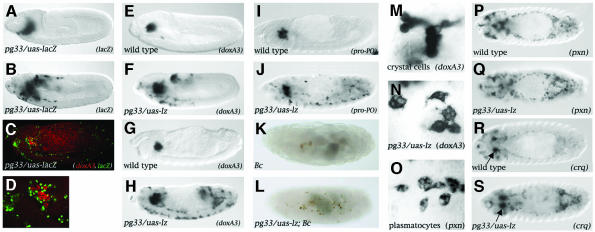
The choice between crystal cell and plasmatocyte differentiation appears to be regulated mainly by the antagonistic action of Gcm on Lz. Notably, it was shown that forced expression of gcm in crystal cell precursors redirects their fate toward a plasmatocyte destiny (Lebestky et al. 2000). To gain further insight into the mechanisms controlling the choice between plasmatocyte and crystal cell formation, we asked whether lz is sufficient to impose the crystal cell fate. Normally, Lz expression is activated in a small subset of hemocytes that will give rise to crystal cells but is absent from the vast majority of plasmatocyte-forming hemocytes (Lebestky et al., 2000). We used the UAS/GAL4 system to express Lz ectopically in plasmatocytes to see whether they would be converted into crystal cells. The Gal4 driver line pg33 contains a P(Gal4) insertion upstream of silver, a gene with no known function in hematopoiesis (Bourbon et al., 2002). This line drives specific expression of a uas-lacZ reporter gene in plasmatocytes from stage 9 onward (Figure 1 A–D and Supplementary figure 1 available at The EMBO Journal Online). Note that no LacZ expression is detected in crystal cells (Figure 1C and D). To monitor crystal cell formation, we analyzed the expression of an early crystal-cell-specific marker, the monophenol mono-oxygenase DoxA3 that is expressed as early as stage 9 (see Supplementary figure 1), as well as that of the mature crystal-cell-specific marker prophenoloxidase (Pro-PO), an enzyme of the melanization cascade that can be detected from stage 11 onward (Waltzer et al., 2002). Upon _pg33_-driven expression of Lz in plasmatocytes, we observed ectopic expression of doxA3 in cells that migrated along the normal plasmatocyte migration paths (compare Figure 1F with E and B). At the end of embryogenesis, _doxA3_-expressing cells were scattered throughout the embryo (compare Figure 1H with G). _pg33/uas-l_z induced ectopic pro-PO expressing cells along a similar pattern (compare Figure 1J with I). Crystal cells can also be visualized by using the Black cell (Bc) mutation, which causes premature melanization within the cell. When this mutation was introduced into a pg33/uas-lz context, we observed ectopic melanized cells dispersed throughout the embryo by the end of embryogenesis (compare Figure 1L with K). However, hemocytes ectopically expressing Lz morphologically resembled mature plasmatocytes rather than crystal cells (compare Figure 1N with O and M, respectively). Consistent with their morphology, hemocytes ectopically expressing Lz expressed the plasmatocyte markers peroxidasin (pxn) (Nelson et al. 1994) and croquemort (crq) in a normal pattern (Franc et al., 1996) (compare Figure 1Q and S with P and R, respectively). Thus Lz does not seem to repress plasmatocyte differentiation. Note that the plasmatocyte cell fate was not overridden by Lz even when Lz was misexpressed precociously throughout the mesoderm using the twist-gal4 driver (Supplementary figure 2).
Fig. 1. Lz can induce the crystal-cell-specific genetic program in plasmatocytes. (A–D) The Gal4 line pg33 drives expression in the plasmatocytes. Side views of (A) pg33/uas-lacZ stage 9, (B) pg33/uas-lacZ stage 11 and (C) pg33/uas-lacZ stage 14 embryos processed to reveal lacZ expression (A and B) or doxA3 mRNA (red) and nuclear-LacZ (green) (C). (D) Higher magnification of the head region in (C). (E–L) _pg33_-driven expression of Lz induces the ectopic expression of the crystal cell marker doxA3 and pro-PO in plasmatocytes as well as their melanization in a Bc mutant context. (E–H) Side views of wild-type (E, G and I) or pg33/uas-lz (F, H and J) embryos; doxA3 expression at stage 11 (E and F) or stage 14 (G and H); pro-PO expression at stage 14 (I and J). Side views of Bc (K) or pg33/uas-lz; Bc (L) stage 17 embryos. (M–S) Lz does not repress plasmatocyte cell fate. (M) High magnification of the head region around the proventriculus showing wild-type stage 15 crystal cells expressing doxA3. (N) High magnification of cells ectopically expressing doxA3 in a pg33-gal4/uas-lz stage 15 embryo (localized ventrally in the trunk). (O) High magnification of wild-type stage 15 plasmatocytes expressing pxn (localized ventrally in the trunk). (P–S) Dorsal views of wild-type (P and R) or pg33/uas-lz (Q and S) stage 14 embryos: (P and Q) pxn expression; (R and S) crq expression. The arrows in R and S indicate crystal cells expressing crq.
Taken together, these results indicate that Lz can induce in plasmatocytes features specific of crystal cells, namely the expression of genes specific to this lineage as well as the enzymatic cascade required for melanization.
srp is required for lz-mediated activation of crystal-cell-specific genes
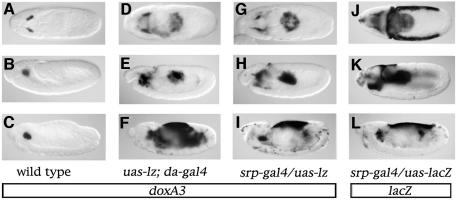
In order to test whether Lz could also induce expression of crystal-cell-specific genes in non-hemogenic territories, we misexpressed Lz throughout the embryo using the da-gal4 driver and analyzed pro-PO and doxA3 expression. As early as stage 9, we detected ectopic expression of doxA3 in the hematopoietic primordium as well as in the posterior midgut (compare Figure 2D with A). At later stages, doxA3 could still be detected in hemocytes and in the posterior midgut as well as in the fat body (a lateral derivative of the mesoderm) and in the amnioserosa (compare Figure 2E and F with B and C). Similar results were obtained when we analyzed pro-PO expression (Supplementary figure 3). Thus, the activation of crystal cell markers expression in response to Lz is spatially restricted.
Fig. 2. Lz–mediated activation of crystal-cell-specific genes is restricted to _srp_-expressing cells. (A–I) Side views of doxA3 expression in stage 9 (A, D and G), stage 11 (B, E and H) or stage 14 (C, F and I) embryos: (A–C) wild-type; (D–F) uas-lz; da-gal4; (G–I) srp-gal4/uas-lz. (J–L) Side views of lacZ expression in srp-gal4/uas-lacZ stage 9 (J), stage 11 (K) or stage 14 (L) embryos.
Interestingly, all the tissues that responded to Lz also express the GATA factor Srp (Sam et al., 1996). To substantiate this observation, we ectopically expressed Lz under the control of an _srp_-gal4 driver that recapitulates most of the embryonic expression of srp except that in the fat body. As shown in Figure 2G–L, there was a striking correlation between the pattern of expression of a lacZ reporter gene placed under the control of srp-gal4 and the doxA3 expression pattern when Lz was ectopically expressed with the same driver or with da-gal4 (compare Figure 2J, K and L with G, H and I or D, E and F, respectively). Note that ectopic induction of doxA3 expression was delayed compared with lacZ, particularly in the amnioserosa. These results show that Lz-induced activation of the crystal cell program is restricted to _srp_-expressing cells and that all the _srp_-expressing cells can activate this program in the presence of Lz.
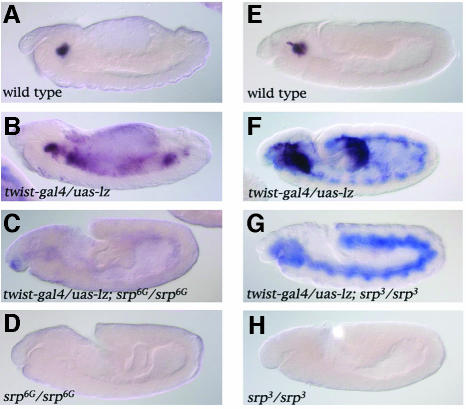
The above results suggest that srp might be required for Lz-induced gene expression. In order to test this hypothesis, we expressed Lz in an _srp_-mutant background. Expression of the Lz transgene was visualized by in situ hybridization with a probe against lz and we monitored crystal cell differentiation with a probe against doxA3. As expected, expression of neither doxA3 nor lz was detected in an srp null mutant (_srp_6G) (compare Figure 3D with A). Consistent with the above results, when Lz was expressed throughout the mesoderm using the twist-gal4 driver in a wild-type embryo, ectopic expression of doxA3 was detected in the anterior mesoderm and posterior midgut by stage 11 (Figure 3F) as well as in the fat body by stage 14 (Figure 3B). In contrast, no doxA3 expression was observed in an _srp_6G mutant when Lz was expressed in the mesoderm (Figure 3C). To investigate further the need for srp, we then made use of the _srp_3 allele in which hemocyte precursors appear to form (Sam et al., 1996; Fossett et al., 2001). In this mutant, neither doxA3 nor lz were expressed (compare Figure 3H with E). As for _srp_6G, _twist_-driven expression of Lz was unable to induce doxA3 expression in an _srp_3 mutant (compare Figure 3G with F). These results indicate that srp is required for _lz_-induced crystal-cell-specific gene expression.
Fig. 3. srp is required for _lz_-mediated activation of crystal-cell-specific genes. Side views of doxA3 (dark purple) and lz (blue staining) expression in stage 14 (A–D) or stage 11 (E–H) embryos: (A and E) wild-type; (B and F) twist-gal4/uas-lz; (C) twist-gal4/uas-lz; srp_6G/srp_6G; (D) srp_6G/srp_6G; (G) twist-gal4/uas-lz; srp_3/ srp_3; (H) srp_3/srp_3.
Lz and Srp interact through a conserved domain
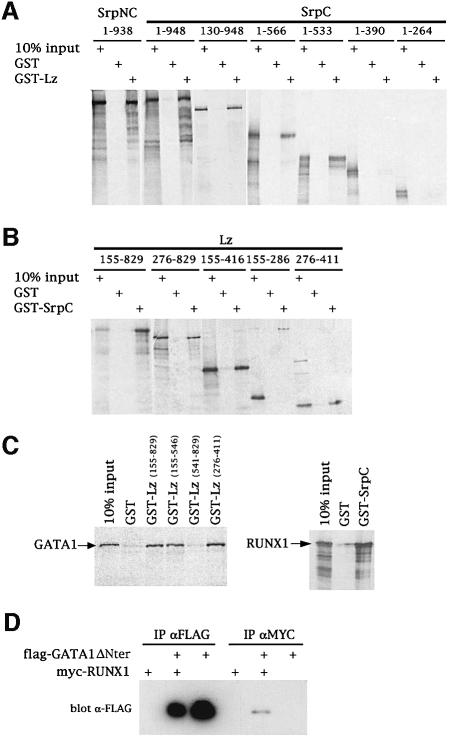
Although it is possible that Lz requires a cofactor induced by srp expression, we favored the hypothesis that Lz directly interacts with Srp. Accordingly, we looked for physical interaction between Srp and Lz using a pulldown assay in vitro. We found that in vitro translated [35S]methionine-labeled SrpC and SrpNC bound to GST-Lz but not to GST alone (Figure 4A). Reciprocally, we observed that in vitro translated Lz bound to GST-SrpC but not to GST alone (Figure 4B). To characterize the mutual binding domains within Lz and Srp, we expressed various truncated versions of these proteins either as GST fusions or as in vitro translated proteins. As shown in Figure 4A, the GATA zinc finger of SrpC (amino acids 478–533) is required for the interaction with GST-Lz. Moreover, the Runt domain of Lz (amino acids 276–407) is required and sufficient for the interaction with GST-SrpC (Figure 4B). These experiments demonstrate that Lz and Srp bind directly to each other through the RUNT domain and the GATA zinc finger, respectively. Given that these domains are highly conserved in their vertebrate counterparts, we tested the binding of mouse GATA1 to Lz and of mouse RUNX-1 to Srp. As shown in Figure 4C (left panel), in vitro translated GATA1 specifically interacted with the Lz Runt domain. Similarly, RUNX1 bound to GST-SrpC (Figure 4C, right panel). Thus the GATA–RUNX interaction is conserved between species.
Fig. 4. GATA and RUNX factors interact through their conserved Runt and GATA zinc finger domains. (A and B) Characterization of the interaction between Srp and Lz by pulldown assays. Equivalent molar amounts of the GST fusion proteins were tested for their interaction with the various in vitro translated [35S]methionine-labeled protein fragments as indicated. (C) The GATA–RUNX interaction is conserved across species. Pulldown assays between in vitro translated mouse GATA1 and various GST-Lz domains (left panel) or between in vitro translated mouse RUNX1 and GST-SrpC (right panel). (D) RUNX1 and GATA1 deleted of its N-terminal domain interact in vivo. COS-7 cells were transfected with pCDNA-mycRUNX1 and pCDNA-flagGATA1ΔNter as indicated in the upper part of the panel. Cell extracts were immunoprecipitated (IP) using either anti-flag or anti-myc and immunoblotted with anti-flag.
While this article was in preparation, Elagib et al. (2003) reported that RUNX1 interacts with GATA1. Using immunoprecipitations, these authors showed that the N-terminal domain of GATA-1, which is not conserved between species, was required for the interaction. On the contrary, our results show that GATA factors interact with RUNX proteins through their conserved zinc fingers. In order to clarify this point, we performed immunoprecipitation with GATA1 and RUNX1. As shown in Figure 4D, a flag-tagged version of N-terminally deleted GATA1 expressed in COS-7 cells was specifically co-immunoprecipitated with myc-tagged RUNX1. This result lends further support to our conclusion that GATA and RUNX proteins interact together and that the interaction is mediated though their conserved GATA zinc finger and Runt domain, respectively.
GATA1 and Srp synergize with RUNX1 to activate transcription in vitro
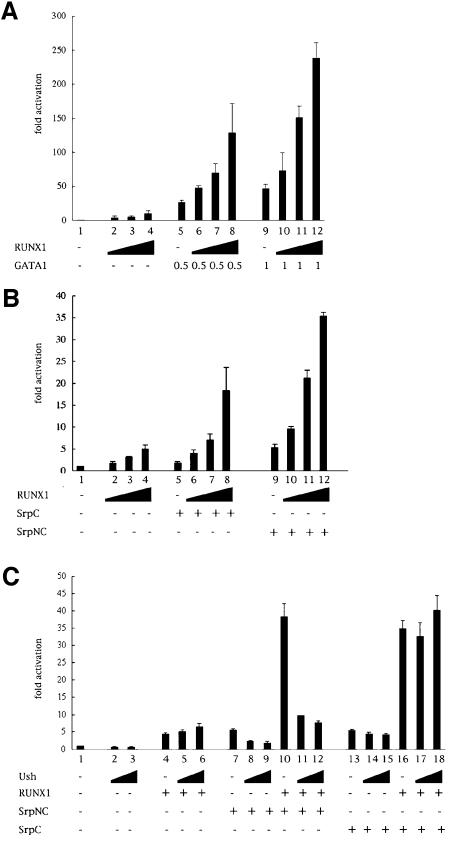
To determine the functional consequences of the GATA–RUNX interaction, we first analyzed the effect of RUNX1 on GATA1-mediated transactivation. We transfected COS-7 cells with a reporter plasmid containing the luciferase reporter gene placed under the control of nine GATA binding sites linked to a minimal promoter (Newton et al., 2001). Upon expression of GATA1, we observed a strong activation of the reporter gene (Figure 5A, lanes 5 and 9). Unexpectedly, RUNX1 weakly activated this reporter gene (Figure 5A, lanes 2–4). Interestingly, when we coexpressed GATA1 with increasing amounts of RUNX1, we observed a dose-dependent synergistic transactivation of the reporter gene (Figure 5A, lanes 6–8 and 10–12). This cooperation was not observed in the absence of the GATA binding site upstream of the reporter gene (data not shown). We then assessed Srp transactivation potential in this heterologous assay. Expression of SrpC or SrpNC modestly, but reproducibly, activated the expression of the reporter gene (Figure 5B, lanes 5 and 9). This activation was dependent on the presence of GATA binding sites upstream of the reporter gene (data not shown). Remarkably, in the presence of increasing amounts of RUNX1, SrpNC and SrpC induced a robust transactivation (Figure 5B, lanes 6–8 and 10–12) that was up to 2-fold greater than the sum of the activation induced by each factor on its own. Thus, like GATA1, Srp can interact and synergize with RUNX1 to induce transcription in vitro. On the contrary, despite Lz interacted with Srp and GATA1, it could not substitute for RUNX1 in this transactivation assay (dat not shown). It is possible that Lz does not function normally in this heterologous system. Notably the temperature of 37°C may be detrimental to Lz function as suggested by the existence of a temperature-sensitive allele of lz.
Fig. 5. GATA and RUNX synergize to induce transcription from a GATA-responsive promoter. Luciferase activities of the GATA reporter pGATA-luc in transfected COS-7 cells in the presence of different combination of expression vectors for GATA1, RUNX1, SrpC, SrpNC and Ush. Results are expressed as fold activation relative to empty expression vector. (A) RUNX1 synergizes with GATA1 to activate the GATA reporter gene. (B) RUNX1 synergizes with SrpC and SrpNC to activate the GATA reporter gene (C) Ush antagonizes SrpNC- but not SrpC-mediated transactivation and cooperation with RUNX1.
Members of the FOG family interact with the GATA N-terminal zinc finger and can either repress or enhance GATA-mediated activation (Haenlin et al., 1997; Tsang et al., 1997). In Drosophila, the FOG factor Ush interacts with SrpNC and represses crystal cell formation as well as crq expression (Fossett et al., 2001; Waltzer et al., 2002). We decided to test the effect of Ush on Srp- and on Srp/RUNX1-mediated activation. As shown in Figure 5C, Ush repressed SrpNC-mediated transactivation (lanes 8 and 9 compared with lane 7). Moreover, the transactivation observed in the presence of both RUNX1 and SrpNC was strongly repressed by Ush (lanes 11 and 12 compared with lane 10). This repression is specific to the interaction between Ush and SrpNC since it is not observed in the case of SrpC- or SrpC/RUNX1-mediated transactivation (compare lanes 14 and 15 with lane 13, and lanes 17 and 18 with lane 16, respectively). Taken together, these data show that the interaction of SrpNC with Ush inhibits SrpNC-induced activation and impedes the cooperation between SrpNC and RUNX1. These results are consistent with previous genetic studies showing that Ush inhibits crystal cell formation (Fossett et al., 2001) and SrpNC-induced activation of crq (Waltzer et al., 2002).
lz and srp synergize to induce crystal-cell-specific gene activation in vivo
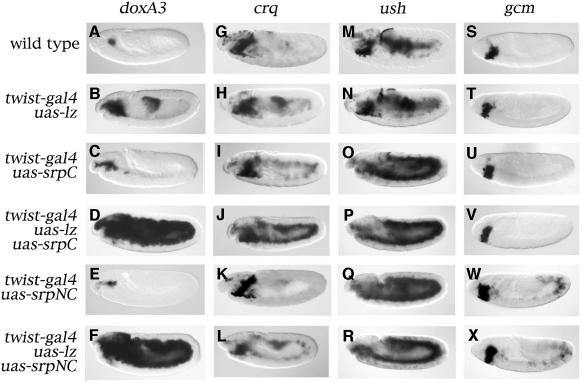
Next we evaluated whether GATA and RUNX factors can cooperate in vivo during Drosophila blood cell development. We have shown previously that _twist_-driven misexpression of srp can induce the expression of plasmatocyte-specific genes in most of the mesoderm. In contrast, ectopic pro-PO activation was restricted to the head mesoderm (Waltzer et al., 2002). Our present results indicate that Lz can induce the crystal-cell-specific program only in _srp_-expressing cells. We surmised that Lz and Srp might cooperate to induce crystal-cell-specific gene expression. In order to test this hypothesis, we coexpressed Lz with SrpC or SrpNC in the mesoderm and we monitored the expression of crystal-cell-specific genes. Whereas Lz, SrpC or SrpNC alone induced restricted ectopic activation of doxA3 (Figure 6B, C and E compared with A, respectively), the coexpression of SrpC or SrpNC with Lz had dramatic consequences: not only did we observe expression of doxA3 throughout the mesoderm, but its expression was also much stronger (Figure 6D and F). We observed similar results when we assessed pro-PO expression (Supplementary figure 3) or the expression of two other crystal cell markers, CG1633 and CG8193 (L.Waltzer, unpublished results). These data indicate that Srp cooperates with Lz to induce the activation of the crystal-cell-specific program.
Fig. 6. Srp and Lz cooperate to induce crystal-cell-specific genes in vivo. Side views of stage 11 (A–R) or stage 8 (S–X) embryos processed to reveal the expression of doxA3 (A–F), crq (G–L), ush (M–R) or gcm (S–X). (A, G, M and S) wild-type. (B, H, N and T) twist-gal4/uas-lz. (C, I, O and U) twist-gal4; uas-srpC. (D, J, P and V) twist-gal4/uas-lz; uas-srpC. (E, K, Q and W) twist-gal4; uas-srpNC. (F, L, R and X) twist-gal4/uas-lz; uas-srpNC.
As mentioned above, srp can induce ectopic activation of various genes implicated in hematopoiesis. Among them, crq can be activated ectopically by SrpC but not by SrpNC, owing to the inhibitory action of Ush (Waltzer et al., 2002). Moreover, although crq was thought to be specifically expressed in plasmatocytes, we have observed that it is also expressed in crystal cells. Indeed, the main site of expression of crq in stage 14/15 embryos is a bilateral cluster of cells next to the proventriculus (Figure 1R, arrow) that also express doxA3 and are absent in an lz mutant (Supplementary figure 1). Thus, at least at the RNA level, crq is expressed in both plasmatocytes and crystal cells during late embryogenesis. This suggests that SrpC and Lz may also cooperate to induce crq expression as they do to induce crystal-cell-specific genes. Accordingly, when Lz was coexpressed with SrpC, we observed a strong increase in the level of crq expression throughout the mesoderm compared with the activation induced by Srp or Lz alone (compare Figure 6J with I and H). However, when Lz was coexpressed with SrpNC, a modest ectopic activation of crq throughout the mesoderm was observed (Figure 6L). Since we have shown previously that SrpNC alone does not induce ectopic expression of crq owing to the inhibitory effect of Ush (Waltzer et al., 2002), these results, as well as those of our transactivation assays, suggest that the balance between Ush and Lz can modulate the transcriptional response to SrpNC in vivo.
We also assessed the expression of ush and gcm, two genes whose expression is initiated in the hematopoietic primordium and is maintained specifically in plasmatocytes (Lebestky et al., 2000; Fossett et al., 2001). ush can be activated throughout the mesoderm in response to SrpC or SrpNC, whereas gcm is only activated by SrpNC (Waltzer et al., 2002). Interestingly, when Lz was coexpressed with SrpC or SrpNC, Srp-induced activation of ush did not seem to be either enhanced or repressed (compare Figure 6P and R with O and Q). Similarly, SrpNC-induced ectopic activation of gcm was not affected by the coexpression of Lz, and SrpC could not ectopically activate gcm expression even in the presence of Lz (compare Figure 6W with X and U with V). Thus Lz does not cooperate with Srp to induce ush or gcm, two known antagonists of crystal cell development.
In conclusion, Srp and Lz seem to cooperate only in the activation of genes expressed in crystal cells. This suggests that the Srp/Lz complex provides the molecular selectivity to trigger crystal cell differentiation during hematopoiesis.
srp and lz cooperate to induce crystal cell formation
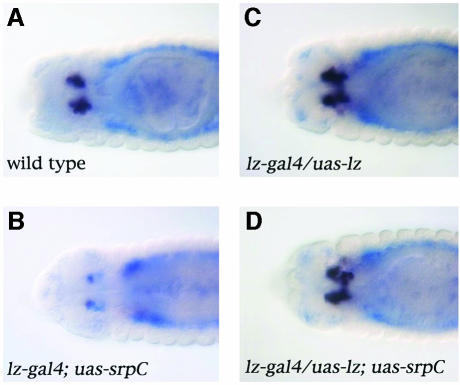
All the results shown here support the hypothesis that Lz and Srp act in concert to induce crystal cell development. In apparent contradiction, we have previously shown that _lz_-driven expression of Srp in crystal cell precursors inhibits their differentiation (Waltzer et al., 2002). Intriguingly, Srp expression in crystal cells is lower than in plasmatocytes (Lebestky et al., 2000), suggesting that the downregulation of Srp in these cells is important for their differentiation. Moreover, by overexpressing srp in crystal cell precursors, we altered the stoichiometry of Srp versus Lz. We hypothesized that increasing the dose of Lz might revert the srp gain-of-function phenotype. Accordingly, we used the lz-gal4 driver to express uas-srpC and/or uas-lz in crystal cell progenitors and we monitored crystal cell differentiation by assessing doxA3 expression. As shown in Figure 7, overexpression of Lz did not seem to affect crystal cell determination (compare Figure 7C with A). On the contrary, SrpC reduced the number of crystal cells and repressed their differentiation, judging by the repression of doxA3 expression (compare Figure 7B with A). However, when Lz was coexpressed with SrpC, formation and differentiation of the crystal cells were restored (Figure 7D). Similar results were obtained with SrpNC or when we assessed pro-PO expression (not shown). Thus the relative levels of Srp and Lz are important for crystal cell development and these two factors cooperate to induce their formation in vivo.
Fig. 7. The balance between Srp and Lz is critical for crystal cell formation. Dorsal view of stage 14 embryos processed to reveal doxA3 expression (black staining) and srp expression (blue staining): (A) wild-type; (B) _lz_-gal4; uas-srpC; (C) _lz_-gal4/uas-lz; (D) _lz_-gal4/uas-lz; uas-srpC.
Discussion
In Drosophila, as in vertebrates, members of the GATA and RUNX families play critical roles during hematopoiesis. In this study, we have investigated the mechanisms by which the RUNX factor Lz controls the formation of crystal cells, one of the two embryonic types of blood cell in Drosophila. We provide evidence that Lz interacts and cooperates with the GATA factor Srp during this process and that this interaction is conserved through evolution.
Lz induces the crystal cell genetic program
In the embryo, Drosophila blood cell progenitors can differentiate into two cell types: plasmatocytes or crystal cells. Crystal cell precursors are first detectable as a small fraction of hemocytes in which Lz is expressed. Whereas Lz function is continuously required for crystal cell development, its mode of action in this process was unknown (Lebestky et al., 2000). Our results provide strong evidence that Lz can trigger the genetic program necessary for crystal cell function as its misexpression in plasmatocytes induced the ectopic expression of crystal-cell-specific genes in these cells and their melanization in a Bc mutant context. Contrary to our findings, Lebestky and coworkers failed to observe induction of the crystal cell fate when Lz was overexpressed under the control of a heat-shock promoter (Lebestky et al., 2000). This difference may be due to the marker they analyzed and/or the method of misexpression used.
It is worth noting that the plasmatocyte cell fate does not seem to be repressed upon ectopic Lz expression. Several factors expressed in plasmatocytes may prevent Lz from repressing their differentiation along this path. For instance srp, gcm and ush are expressed at high levels in pro-hemocytes and in plasmatocytes, while they are normally downregulated (srp) or not expressed (gcm and ush) in crystal cells (Lebestky et al., 2000; Fossett et al., 2001). Upon ectopic expression of lz in plasmatocytes, the expression of these genes was not repressed and thus they might still promote plasmatocyte differentiation. In addition, contrary to crystal cells, plasmatocytes are migratory cells that will be exposed to changing environmental cues; thus their cellular identity must be maintained cell autonomously and may be locked in place early on. Hence the concomitant upregulation of Lz and repression of ush and gcm in crystal cell progenitors is probably a critical step for normal blood cell differentiation.
Srp and Lz cooperate to induce crystal cell fate
Our results indicate that Srp and Lz act in concert to induce crystal cell development. We demonstrate that the induction of crystal-cell-specific genes by Lz is strictly dependent on the presence of Srp. Moreover, we show that Lz and Srp bind to each other and that Srp synergized with RUNX1 to induce transcription in cell culture. Taken together, these lines of evidence suggest that Srp and Lz form a complex that activates the transcription of the genes required for crystal cell differentiation in vivo.
Interestingly, the transcriptional cooperation between Srp and Lz seems restricted to genes expressed in crystal cells, such as doxA3, pro-PO and crq. How this target gene specificity is achieved is not known. In cell culture, Srp can cooperate with RUNX1 to activate transcription from a synthetic promoter containing multimerized GATA binding sites. However, in vivo, while Srp alone activates the expression of crq, gcm and ush, cooperation between Srp and Lz was not observed in expression of gcm and ush, two genes that antagonize crystal cell formation. The Srp/Lz complex may only bind to promoters containing certain GATA binding sites. Alternatively, it is possible that Lz also participates in the tethering of the Srp/Lz complex to RUNX binding sites. In order to bind these sites, RUNX proteins have to form dimers with CBFβ. In Drosophila, the CBFβ homologs Brother and Big Brother are ubiquitously expressed during embryogenesis, but it is not known whether they play a role in hematopoiesis (Golling et al., 1996; Kaminker et al., 2001). Interestingly, doxA3 and pro-PO proximal promoter regions contain both GATA and RUNX consensus binding sites, which suggests that both partners may participate in the recruitment (L.Waltzer, unpublished observation). Defining the _cis_-responsive elements in crystal-cell- and plasmatocyte-specific genes, respectively, should help to define the molecular mechanisms of cooperation between Srp and Lz.
Our finding that Srp plays a direct part in crystal cell differentiation was unexpected as forced expression of Srp in these cells inhibited their development, suggesting that srp has to be downregulated to allow their differentiation (Waltzer et al., 2002; Evans and Banerjee, 2003). Instead, we propose that Srp is necessary early on for all hemocyte fate (including that of crystal cell precursors) and subsequently, in balance with Lz, for crystal cell differentiation. A similar situation has been described for GATA2 which is required at a high level for proliferation of blood cell progenitors and at a lower level in differentiated blood cells, such as mature erythrocytes and mast cells (Jippo et al., 1996; Harigae et al., 1998; Tsai and Orkin, 1997). Our study suggests that the balance between Srp and Lz, rather than the absolute level of Srp, may be critical for crystal cell differentiation. Indeed, the inhibition of crystal cell formation due to the overexpression of Srp was counterbalanced by increasing the levels of Lz. The fact that Srp is required even at stages when its overexpression can be inhibitory highlights that Srp function at a given moment is dependent on its precise level of expression. It also probably reflects the existence of cross-regulation between a combination of transcription factors whose equilibrium in the progenitors is critical for cell fate choice.
Cross-regulations between Ush, Lz and Srp
An important component of this equilibrium is the FOG factor Ush. Members of the FOG family interact specifically with GATA N-terminal zinc fingers and act either as coactivators or corepressors (Haenlin et al., 1997; Tsang et al.,1997; Wang et al., 2002). As such, Ush can only interact with SrpNC, and not with SrpC, and we have previously proposed that Ush controls hematopoiesis by repressing SrpNC-mediated activation (Waltzer et al., 2002). Our present data support this hypothesis since Ush directly repressed SrpNC-mediated activation in our transactivation assay. During hematopoiesis, Ush was shown to repress crystal cell formation (Fossett et al., 2001). Consistent with this role, we observed that Ush inhibits the transactivation mediated by the SrpNC/RUNX complex. Conversely, the inhibitory effect of Ush on SrpNC-induced activation of crq can be partially relieved by Lz in vivo. Since Ush is coexpressed with Srp and Lz in crystal cell precursors (Fossett et al., 2001), a competition between Ush and Lz to modulate SrpNC activity might influence cell fate choice. Indeed, Lebestky and coworkers observed that a small fraction of the Lz-expressing cells differentiate into plasmatocytes (Lebestky et al., 2000). We propose that the cross-interactions between Srp, Lz and Ush might constitute a mechanism whereby one cell type is chosen at the expense of the other. Similar regulation of GATA activity occurs during chicken eosinophil differentiation. Eosinophil progenitors express GATA1, FOG1 and C/EBP. FOG1 was shown to repress the expression of eosinophil-specific markers by blocking GATA-induced transactivation, while C/EBP interacts and cooperates with GATA1 to induce eosinophil differentiation (Yamaguchi et al., 1999; Querfurth et al., 2000). Thus dose-dependent antagonistic or cooperative interactions between GATA factors and their partners appear to be a conserved means of regulating lineage choice and differentiation.
Conserved interaction and cooperation between GATA and RUNX
We have shown here that GATA and RUNX factors cooperate in vivo to control Drosophila blood cell formation. Several lines of evidence suggest that the functional relationship between GATA and RUNX factors may be conserved in vertebrate hematopoiesis. First, we have shown that the interaction between Srp and Lz occurs through highly conserved domains. Secondly, we have found that GATA1 binds to Lz and that RUNX1 interacts with Srp. Thirdly, in cotransfection assays, we have demonstrated that RUNX1 and GATA1 bind to each other and synergistically activate transcription. Elagib and coworkers have also recently described RUNX1 binding to GATA1 (Elagib et al., 2003). Contrary to what we observed, these authors failed to detect an interaction with RUNX1 when GATA1 N-terminal transactivation domain was deleted. The reason for this discrepancy is unclear. Interestingly though, they showed that these two factors cooperate to induce the expression of megakaryocyte-specific integrin in vitro, suggesting that RUNX1 may participate with GATA1 to induce megakaryocytic differentiation. Moreover, haploinsufficient mutations in RUNX1 are associated with familial platelet disorder with predisposition to acute myelogenous leukemia (FPD/AML) (Song et al., 1999; Michaud et al., 2002), suggesting a role for RUNX1 in human megakaryopoiesis. While it is well established that GATA1 is required for normal megakaryopoiesis (Vyas et al., 1999), a role for RUNX1 in this process has not yet been demonstrated. Furthermore, acquired mutations in GATA1 are specifically associated with megakaryocytic leukemia and transient myeloproliferative disorder in children with Down syndrome (Wechsler et al., 2002; Groet et al., 2003). As RUNX1 is located on chromosome 21, it is tempting to speculate that the interaction between GATA1 and RUNX1 may participate in the development of these megakaryocytoses. Finally, RUNX1 is coexpressed with GATA1, GATA2 or GATA3 at different stages of hematopoiesis (Speck and Gilliland, 2002). Thus the GATA/RUNX complex may regulate various aspects of hematopoiesis from stem cell emergence and proliferation to lineage determination and terminal differentiation.
In conclusion, our results show for the first time that the GATA and RUNX factors cooperate in vivo to control hematopoiesis. In Drosophila, the cooperation between Srp and Lz is central to the specification of crystal cell fate. We have also highlighted the fact that the GATA–RUNX interaction is conserved between Drosophila and mammals. This raises the possibility that a similar functional interaction between GATA and RUNX is important during hematopoietic development in mammals.
Materials and methods
Fly stocks
The uas-srpC and uas-srpNC transgenic lines were described previously (Waltzer et al., 2002). The plasmatocyte-specific driver pg33-gal4 was described previously (Bourbon et al., 2002). The uas-lz, lz-gal4, srp-gal4 and _srp_3 lines were kindly provided by P.Gergen, U.Banerjee, M.Meister and R.Reuter, respectively. Other stocks were provided by the Bloomington Drosophila Stock Center. All crosses and embryo collections were carried out at 25°C. Blue balancers or in situ hybridization against the transgenes were used to genotype the embryos.
In situ hybridizations and antibody stainings
In situ hybridizations were carried out as previously described (Peyrefitte et al., 2001). RNA probes for srp, crq, pxn, pro-PO, ush, gcm and lacZ have been described previously. To generate RNA probes for doxA3 exon 3 or lz cDNA, the corresponding DNA sequences were cloned by PCR in pGemTeasy. The corresponding antisense RNAs were transcribed in vitro using T7 or SP6 RNA polymerase.
Double-fluorescent immunostaining and in situ hybridization were carried out using fluorescein-UTP labeled doxA3 antisense probe, mouse antifluorescein antibody (1/500) (Roche), goat antimouse antibody conjugated to Alexa Fluor 546 (1/400) (Molecular Probe), rabbit anti-β-gal antibody (1/500) (Cappel Inc. Pharmaceutical) and goat antirabbit antibody conjugated to Alexa Fluor 488 (1/400) (Molecular Probe).
Pulldown assays
pBS-SrpC, pBS-SrpNC, pGEX2TK-SrpC and pGEX2TK-SrpNC were described previously (Waltzer et al., 2002). These plasmids and pET3c-Lz (Xu et al., 2000) were used as templates to subclone various domains of Srp or Lz into pGEX2TK or into pT7βlink by standard cloning techniques. pGEX2TK-derived expression plasmids were used to produce GST proteins in Escherichia coli BL21. pBS or pT7βlink-derived plasmids were used as a template to produce in vitro [35S]methionine-labeled proteins using a coupled transcription–translation system (Promega). Interaction assays were performed as described (Waltzer et al., 2002).
Reporter plasmids and mammalian expression vectors
The mouse RUNX1 expression plasmid pCMV-AML1b (McLarren et al., 2000) was provided by S.Stifani. The expression vector for myc-tagged RUNX1 (pCDNA-myc-PEBP2αB1) (Kim et al., 1999) was provided by Y.Ito. The mouse GATA-1 expression plasmid pCMV-GATA-1 (Newton et al., 2001) was provided by M.Crossley and was used as a template to generate pCDNA3-flag-GATA1ΔNter (81–413). Full-length SrpC and SpNC cDNA were subcloned from pBS-SrpC or pBS-SrpNC into the CMV-based expression plasmid pXJ42 to generate pXJ-SrpC and pXJ-SrpNC respectively. pXJ-Ush was described previously (Haenlin et al., 1997). The reporter plasmid pGATA-luc contains multimerized GATA binding sites upstream of the β-globin TATA box and the luciferase reporter gene (Newton et al., 2001).
Cells, transfection and reporter assay
COS-7 cells, grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% (v/v) fetal bovine serum (FBS), were seeded in 12-well culture plates at a density of 0.5 × 104 cells/well and transfected 24 h later using the calcium phosphate method. Fifty nanograms of the β-galactosidase reporter pCMV-βgal was used per transfection as an internal control. Empty pCMV expression vector was added as required to keep the amount of pCMV plasmid constant. pUC plasmid was added as carrier DNA to a total amount of 10 µg DNA per transfection. Cells were harvested 48 h after transfection, washed in PBS and lysed in reporter lysis buffer (Promega). Luciferase assays were performed using a Lumat LB 9501 luminometer. β-Galactosidase activity was measured with a Galacto-Light Plus kit (Applied Biosystems) according to the manufacturer’s instructions. Luciferase activities are representative of three independent experiments done in triplicate and have been normalized to β-galactosidase levels.
Immunoprecipitations and immunoblotting
COS-7 cells were plated in 100-mm diameter dishes and transfected using the DEAE/Dextran method. Forty-eight hours after transfection, cells were washed twice in PBS and lysed for 5 min at 4°C in lysis buffer (50 mM Tris–HCl pH 8, 300 mM NaCl, 10 mM MgCl2, 0.4% NP-40) supplemented with protease inhibitors (Roche). Soluble proteins were collected by centrifugation and diluted (v/v) with dilution buffer (50 mM Tris–HCl pH 8, 2.5 mM CaCl2, 0.4% NP-40) supplemented with DNase. Lysates were incubated for 3 h at 4°C with primary antibody-protein A Sepharose complexes. Immunoprecipitated proteins were washed with lysis buffer/dilution buffer and resolved on SDS–PAGE. Western blotting was performed using standard techniques. Monoclonal anti-Flag (M2) and anti-c-myc antibodies were purchased from Sigma and Roche, respectively.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to J.Smith, P.Blader and B.G.Monster for critically reading the manuscript. We thank B.Augé for expert technical assistance. We also thank U.Banerjee, M.Crossley, P.Gergen, K.Ito, M.Meister, R.Reuter and S.Stifani for plasmids and fly stocks. This work was supported by the Centre National de Recherche Scientifique (CNRS) and grants from the Association pour la Recherche sur le Cancer (ARC). G.F. is supported by a postdoctoral fellowship from the ARC.
References
- Alfonso T.B. and Jones,B.W. (2002) gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev. Biol., 248, 369–383. [DOI] [PubMed] [Google Scholar]
- Bernardoni R., Vivancos,B. and Giangrande,A. (1997) glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol., 191, 118–130. [DOI] [PubMed] [Google Scholar]
- Bourbon H.M. et al. (2002) A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev., 110, 71–83. [DOI] [PubMed] [Google Scholar]
- Elagib K.E., Racke,F.K., Mogass,M., Khetawat,R., Delehanty,L.L. and Goldfarb,A.N. (2003) RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood, 101, 4333–4341. [DOI] [PubMed] [Google Scholar]
- Evans C.J. and Banerjee,U. (2003) Transcriptional regulation of hematopoiesis in Drosophila. Blood Cells Mol. Dis., 30, 223–228. [DOI] [PubMed] [Google Scholar]
- Fossett N., Tevosian,S.G., Gajewski,K., Zhang,Q., Orkin,S.H. and Schulz,R.A. (2001) The Friend of GATA proteins U-shaped, FOG-1 and FOG-2 function as negative regulators of blood, heart and eye development in Drosophila. Proc. Natl Acad. Sci. USA, 98, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc N.C., Dimarcq,J.L., Lagueux,M., Hoffmann,J. and Ezekowitz,R.A. (1996) Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity, 4, 431–443. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Browne,C.P., Cunniff,K., Goff,S.C. and Orkin,S.H. (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA, 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golling G., Li,L., Pepling,M., Stebbins,M. and Gergen,J.P. (1996) Drosophila homologs of the proto-oncogene product PEBP2/CBFβ regulate the DNA-binding properties of Runt. Mol. Cell. Biol., 16, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groet J. et al. (2003) Acquired mutations in GATA1 in neonates with Down’s syndrome with transient myeloid disorder. Lancet, 361, 1617–1620. [DOI] [PubMed] [Google Scholar]
- Haenlin M., Cubadda,Y., Blondeau,F., Heitzler,P., Lutz,Y., Simpson,P. and Ramain,P. (1997) Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev., 11, 3096–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigae H. et al. (1998) Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells, 3, 39–50. [DOI] [PubMed] [Google Scholar]
- Jippo T., Mizuno,H., Xu,Z., Nomura,S., Yamamoto,M. and Kitamura,Y. (1996) Abundant expression of transcription factor GATA-2 in proliferating but not in differentiated mast cells in tissues of mice: demonstration by in situ hybridization. Blood, 87, 993–998. [PubMed] [Google Scholar]
- Kaminker J.S., Singh,R., Lebestky,T., Yan,H. and Banerjee,U. (2001) Redundant function of Runt Domain binding partners, Big brother and Brother, during Drosophila development. Development, 128, 2639–2648. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke,M., Ogawa,E., Wee,H.J., Englmeier,U., Graf,T. and Ito,Y. (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J., 18, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T., Chang,T., Hartenstein,V. and Banerjee,U. (2000) Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science, 288, 146–149. [DOI] [PubMed] [Google Scholar]
- Lebestky T., Jung,S.H. and Banerjee,U. (2003) A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev., 17, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren K.W., Lo,R., Grbavec,D., Thirunavukkarasu,K., Karsenty,G. and Stifani,S. (2000) The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J. Biol. Chem., 275, 530–538. [DOI] [PubMed] [Google Scholar]
- Michaud J. et al. (2002) In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood, 99, 1364–1372. [DOI] [PubMed] [Google Scholar]
- Nelson R.E., Fessler,L.I., Takagi,Y., Blumberg,B., Keene,D.R., Olson,P.F., Parker,C.G. and Fessler,J.H. (1994). Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J., 13, 3438–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Mackay,J. and Crossley,M. (2001) The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem., 276, 35794–35801. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. and Zon,L.I. (2002) Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat. Immunol., 3, 323–328. [DOI] [PubMed] [Google Scholar]
- Persons D.A., Allay,J.A., Allay,E.R., Ashmun,R.A., Orlic,D., Jane,S.M., Cunningham,J.M. and Nienhuis,A.W. (1999) Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood, 93, 488–499. [PubMed] [Google Scholar]
- Pevny L., Simon,M.C., Robertson,E., Klein,W.H., Tsai,S.F., D’Agati,V., Orkin,S.H. and Costantini,F. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature, 349, 257–260. [DOI] [PubMed] [Google Scholar]
- Peyrefitte S., Kahn,D. and Haenlin,M. (2001) New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix–loop–helix genes. Mech. Dev., 104, 99–104. [DOI] [PubMed] [Google Scholar]
- Querfurth E., Schuster,M., Kulessa,H., Crispino,J.D., Doderlein,G., Orkin,S.H., Graf,T. and Nerlov,C. (2000) Antagonism between C/EBPβ and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev., 14, 2515–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehorn K.P., Thelen,H., Michelson,A.M. and Reuter,R. (1996) A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development, 122, 4023–4031. [DOI] [PubMed] [Google Scholar]
- Rizki T.M. (1978) The circulatory system and associated tissues. In Wright,T.R.F. (ed.) The genetics and biology of Drosophila. Academic Press, New York, pp. 229–335. [Google Scholar]
- Rizki T.M., Rizki,R.M. and Grell,E. (1980) A mutant affecting the crystal cells in Drosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol., 188, 91–99. [DOI] [PubMed] [Google Scholar]
- Sam S., Leise,W. and Hoshizaki,D.K. (1996) The serpent gene is necessary for progression through the early stages of fat-body development. Mech. Dev., 60, 197–205. [DOI] [PubMed] [Google Scholar]
- Song W.J. et al. (1999) Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet., 23, 166–175. [DOI] [PubMed] [Google Scholar]
- Speck N.A. and Gilliland,D.G. (2002) Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer, 2, 502–513. [DOI] [PubMed] [Google Scholar]
- Tepass U., Fessler,L.I., Aziz,A. and Hartenstein,V. (1994) Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development, 120, 1829–1837. [DOI] [PubMed] [Google Scholar]
- Tsai F.Y. and Orkin,S.H. (1997) Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood, 89, 3636–3643. [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara,Y., Hom,D.B. and Orkin,S.H. (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev., 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P., Ault,K., Jackson,C.W., Orkin,S.H. and Shivdasani,R.A. (1999) Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood, 93, 2867–2875. [PubMed] [Google Scholar]
- Waltzer L., Bataillé,L., Peyrefitte,S. and Haenlin,M. (2002) Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J., 21, 5477–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy,T., Binder,M., Marin-Padilla,M., Sharpe,A.H. and Speck,N.A. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA, 93, 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Crispino,J.D., Letting,D.L., Nakazawa,M., Poncz,M. and Blobel,G.A. (2002) Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J., 21, 5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J., Greene,M., McDevitt,M.A., Anastasi,J., Karp,J.E., Le Beau,M.M. and Crispino,J.D. (2002) Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet., 32, 148–152. [DOI] [PubMed] [Google Scholar]
- Xu C., Kauffmann,R.C., Zhang,J., Kladny,S. and Carthew,R.W. (2000) Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell, 103, 87–97. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Nishio,H., Kishi,K., Ackerman,S.J. and Suda,T. (1999) C/EBPβ and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: implication for C/EBPβ activity in eosinophil gene expression. Blood, 94, 1429–1439. [PubMed] [Google Scholar]