Harnessing Traction-Mediated Manipulation of the Cell-Matrix Interface to Control Stem Cell Fate (original) (raw)
. Author manuscript; available in PMC: 2010 Dec 1.
Published in final edited form as: Nat Mater. 2010 Apr 25;9(6):518–526. doi: 10.1038/nmat2732
Abstract
Stem cells sense and respond to the mechanical properties of the extracellular matrix. However, both the extent to which extracellular matrix mechanics affect stem cell fate in 3D micro-environments and the underlying biophysical mechanisms are unclear. We demonstrate that the commitment of mesenchymal stem cell (MSC) populations changes in response to the rigidity of 3D micro-environments, with osteogenesis occurring predominantly at 11–30 kPa. In contrast to previous 2D work, however, cell fate was not correlated with morphology. Instead, matrix stiffness regulated integrin binding as well as reorganization of adhesion ligands on the nanoscale, both of which were traction-dependent and correlated with osteogenic commitment of MSC populations. These findings suggest that cells interpret changes in the physical properties of adhesion substrates as changes in adhesion ligand presentation, and that cells themselves can be harnessed as tools to mechanically process materials into structures that feedback to manipulate their fate.
Keywords: Hydrogel, Extracellular matrix (ECM), Mechanotransduction, Förster Resonance Energy Transfer (FRET), Mesenchymal Stem Cell (MSC)
Cell therapies hold great clinical promise, but control of transplanted cell fate remains a significant challenge1. Material-based transplantation systems offer a promising means to control cell fate for regeneration of functional tissue or controlled disease ablation2,3. As cell-matrix interactions are central to eukaryote biology4,5, these materials systems are often designed to exploit those interactions to manipulate cell fate, typically via presentation of integrin binding ligands (e.g. RGD) derived from natural extracellular matrix (ECM) proteins6–9. Interestingly, in-vitro studies with such synthetic ECM analogs have demonstrated that in 2D cultures, cell fate can also be manipulated by changing the rigidity of the substrate without altering ligand presentation10,11. However, although this previous work has identified important correlations between matrix rigidity and cell phenotype, the biophysical mechanisms that allow cells to sense matrix compliance remain unclear. Moreover, since most work linking substrate mechanics to cell fate has been done in 2D model systems, the extent to which ECM rigidity affects cell phenotype in more physiologically relevant 3D culture systems is also uncertain12–14.
We first tested the effects of 3D matrix rigidity on commitment of a clonally-derived murine mesenchymal stem cell (mMSC) line, D115,16, as clonally-derived cell lines provide more definitive information regarding fate decisions than heterogeneous primary cell populations. The relative homogeneity of mMSC compared to primary human MSC (hMSC) was confirmed via histology, as significant numbers of cells in the naïve hMSC population (10 ± 2%) exhibited Alkaline Phosphatase (ALP) Activity (a marker of osteogenesis), compared to <1% in the naïve mMSC population (Fig. S1). Naïve D1 cells were encapsulated into 3D hydrogel synthetic ECM formed by alginate polymers that present integrin-binding RGD peptides17. The elastic modulus (E), a quantitative metric of matrix rigidity, was varied between 2.5 and 110 kPa (Fig. S2), and the density of RGD was varied in parallel. The broad range of peptide density used in our studies is comparable to the RGD density of commonly used biological matrices, including tumor-derived ECM (140µM)13. As the matrices used in these studies are not susceptible to enzymatic degradation by mammalian cells, their mechanical properties are expected to remain constant over the course of differentiation studies. Thus, any observed effects of E on cell fate can be distinguished from indirect effects stemming from matrix degradation, which in-and-of-itself has been linked to cell fate in previous work18,19.
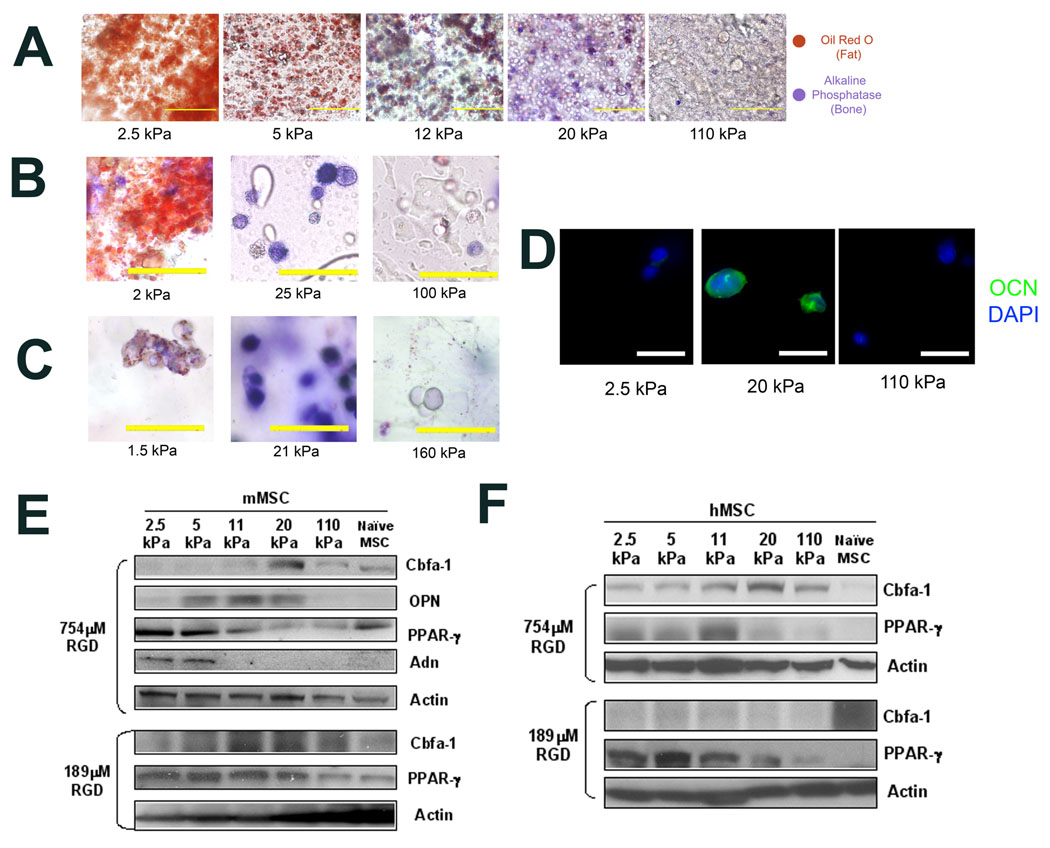
Cell phenotype was analyzed 1 week after encapsulation, and this revealed that matrix rigidity had significant effects on clonally derived mMSC phenotype, with osteogenic commitment occurring primarily at intermediate E (11–30 kPa) and adipogenic lineage predominating in softer (2.5–5 kPa) micro-environments (Fig. 1A). The generality of the relationship between matrix elasticity and stem cell fate was confirmed by studies in which mMSC were encapsulated into RGD-modified agarose20 or poly(ethylene glycol dimethacrylate) (PEGDM)21 hydrogels of varying rigidity (Fig. S2). Consistent with results obtained in alginate-based synthetic ECM, mMSC underwent optimal osteogenic commitment within matrices of intermediate stiffness (Fig. 1B–C). Histological results were verified by analysis of the expression of osteogenic biomarkers Core Binding Factor α1 (Cbfa-1), Osteopontin (OPN) and Osteocalcin (OCN), and adipogenic biomarkers Peroxisome Proliferator-Activated Receptor Gamma (PPAR-γ) and Adiponectin (Adn) (Fig. 1D–E, S1). Similar results were found at two different RGD densities (754µM and 189µM; Fig. 1E).
Fig. 1. Matrix Compliance Alters Mesenchymal Stem Cell Fate in 3D Matrix Culture.
(A–C). In-situ staining of encapsulated clonally derived mMSC (D1) for Alkaline Phosphatase (ALP) activity (Fast blue; osteogenic biomarker, blue) and neutral lipid accumulation (Oil Red O; adipogenic biomarker, red) after 1 week of culture in the presence of combined osteogenic and adipogenic chemical supplements within encapsulating matrices comprised of (A) RGD-modified alginate, (B) RGD-modified agarose, or (C) RGD-modified PEGDM hydrogels presenting 754µM RGD with varying E. (D). Immunofluorescence staining for OCN (green) and the nuclear counterstain DAPI (blue) in cryosectioned alginate matrices of varying E containing mMSC. (E). Western analysis of osteogenic (Cbfa-1, OPN) and adipogenic (PPAR-γ, Adn) protein expression in mMSC cultured in RGD-alginate hydrogels for 1 week. (F). Western analysis of Cbfa-1 and PPAR-γ protein expression in primary hMSC after 1 week of 3D culture within alginate matrices in which both E and RGD density were varied in parallel. E values shown are for hydrogels after 1 day in culture, after which point no changes in E occur. Scale bars: (A); 100µm, (B,C); 50µm, (D); 20µm.
These studies were next repeated with primary hMSC to confirm their physiologic relevance. Population trends with hMSC were consistent with trends observed using the clonally derived mMSC line, with osteogenesis occurring most prevalently within matrices of intermediate rigidity and high RGD density (Fig. 1F, S1). However, hMSC exhibited more heterogeneous lineage commitment in response to ECM mechanical properties (Fig. S1), consistent with the baseline heterogeneity of the naïve cell population (Fig. S1). Together, these results from clonally derived and primary stem cell populations are consistent with the hypothesis that matrix elasticity modulates stem cell lineage specification in 3D culture. Further studies would determine if this same phenomenon holds for other lineages and stem cell types. Our findings also agree with previous reports detailing the effects of specific values of E on osteogenic specification in 2D culture10,11, and a recent study that reported hMSC differentiation was influenced by matrix elasticity in 3D culture22. However, the specific moduli found optimal for osteogenesis in the recent 3D cell culture report was distinct from our findings, and the previous 2D culture studies, perhaps due to the different metrics used to examine differentiation in that study, or the shear-thinning nature of the materials used in that work22.
Having establishing a role for matrix elasticity in regulating MSC phenotype in 3D culture, we investigated potential underlying mechanisms. In 3D matrices, MSC lineage commitment may be affected by small molecules (e.g. calcium ions) used to control E. This possibility was addressed by performing studies in which the concentration of available calcium ions was decoupled from the rigidity of the matrix encapsulating cells. In those studies, mMSC fate correlated with matrix stiffness, not the amount of soluble calcium (Fig. S3). A second possibility is that altering matrix mechanics regulates diffusion of instructive morphogens23. However, direct measurements of the relative diffusion coefficient (Deff) for a model protein, revealed no statistically significant changes within the E range of alginate matrices used in this work (Fig. S3). Hence, changes in morphogen diffusion alone are unlikely to underlie the observed dependence of cell fate on E.
The relation between cell morphology and fate in 3D culture was next examined, as it has often been observed in 2D culture studies that matrix rigidity affects cell morphology 24,25, and morphology in and of itself has been correlated to hMSC fate26. Strikingly, though, E had no significant affects on gross mMSC morphology in 3D matrices either a short time (2 hr) (Fig. 2A, S4), or at later (24 hr and 1 week; data not shown) time-points, and cells appeared grossly spherical. This finding is consistent with recent work utilizing other hydrogels with nm-scale pore sizes to encapsulate cells27. Phalloidin-staining revealed µm-sized cortical protrusions of the cells into the surrounding matrix (Fig. S4, Fig. 2B). These protrusions were only present in RGD-modified alginate, and were eliminated by treating mMSC with a cell-traction inhibiting agent, (2,3)-butanedione-monoxime (BDM; Fig S4). However, no strong correlations were observed between these protrusions and matrix elasticity. Likewise, although there was a subtle decrease in nuclear sphericity (ψ) and volume (VN) as E increased, there was no statistically significant relationship between E and either parameter (Fig. 2C, S4). These findings contrast to reports correlating nuclear morphology to osteogenesis in 2D culture28, though different metrics were used to quantify nuclear morphology in this 3D study as compared to the aforementioned 2D work. Importantly, these data relating matrix elasticity to cell morphology contrast with previous 2D studies that suggested gross cell morphology changes are required to alter MSC fate in response to substrate rigidity10,24. However, our data is consistent with reports of in vivo versus in vitro morphology of other cell types. For example, fibroblasts exhibit marked changes in spreading as the rigidity of 2D culture substrates is modulated25, but exhibit a characteristic, spindle-shaped morphology in 3D matrix culture and in vivo, even though the tissues where they reside exhibit large disparities in compliance and adhesion ligand presentation12.
Fig. 2. Cell and Nuclear Morphology are not Strongly Correlated to Mechanics of 3D Matrices.
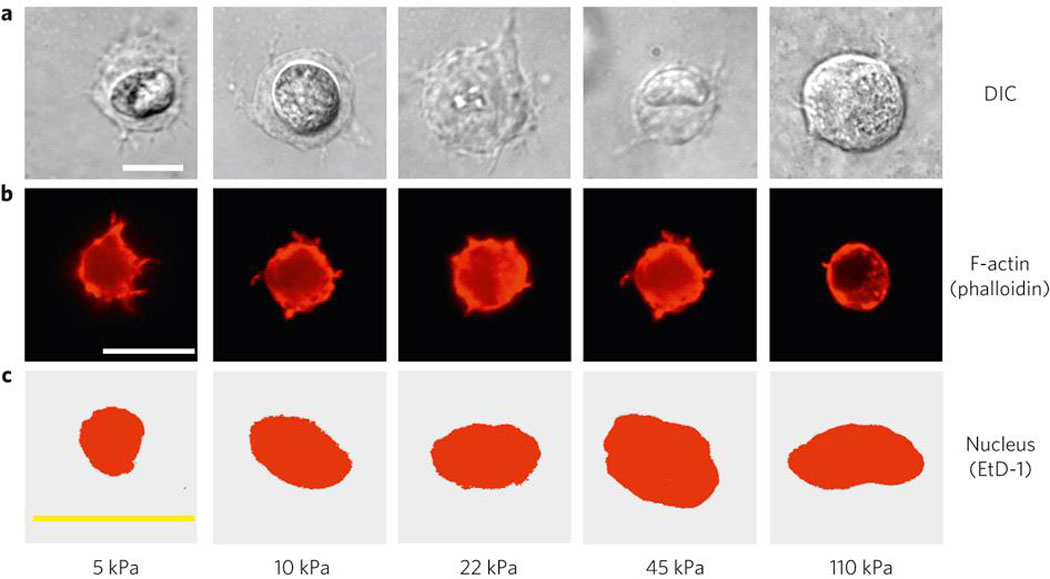
(A–C). Representative micrographs showing cross-sections of mMSC 2 hr after encapsulation into 3D alginate matrices with varying E and constant (754 µM) RGD density, visualized by (A) Differential-Interference Contrast (DIC), (B) F-Actin staining (Alexa Fluor 568-Phalloidin), or (C) Nuclear staining (Ethidium Homodimer). E values shown are for hydrogels at the time of cell encapsulation. Scale bars: 10µm.
As neither cell nor nuclear morphology appeared to be required for MSC fate changes in response to the rigidity of 3D micro-environments – in particular, the biphasic relationship between E and osteogenic commitment of MSC - alternative means for cellular mechanosensing were next investigated. Previous work identifying molecular sensors of ECM rigidity has concentrated on deformation of extracellular adhesion molecules or of proteins that comprise the focal adhesion complex within the cell5,29–33. However, there has been little focus on the bimolecular interactions between the matrix and integrins, despite previous work34,35 suggesting that matrix rigidity and adhesion ligand density act in concert to regulate fate. We thus hypothesized that changes in substrate stiffness alter cell fate, at least partially, by directly modulating integrin-adhesion ligand bond formation. This possibility is supported by the finding that mechanical forces can directly affect the lifetime of bimolecular bonds36, and models that predict a biphasic relationship between applied force and bond lifetime37–39. It is also possible that cells may use different integrin receptors as stiffness and dimensionality of the matrix are varied12. In 2D culture, clonally derived mMSC utilized αV-integrins to adhere to monolayers of either fibronectin (FN) or vitronectin (VN; lacking the PHSRN ‘synergy sequence’ of FN purported to be essential for α5β1-integrin ligation40), but only used α5-integrins when adherent to FN (Fig. 3A). However, in 3D, α5-integrins localized to the cell-matrix interface in an RGD-dependent manner (Fig. 3B), suggesting they were being utilized for adhesion. An ELISA technique was developed to directly address the possibility of α5-integrin mediated adhesion in 3D, and to quantitatively compare specific integrin ligation by cells within or on compliant RGD-modified hydrogels (Fig. S5). ELISA quantification revealed that in 2D culture, αV-integrins acted as RGD receptors, but α5-integrins did not (Fig. 3C). However, mMSC encapsulated within 3D matrices with the same E and RGD density did use α5-integrins as RGD receptors (Fig. 3C). The maximum number of αV-RGD bonds did not change significantly between 2D and 3D matrices presenting high densities of ligand, implying that α5-RGD binding in 3D culture is not simply a consequence of αV-integrin saturation. Strikingly, analysis of integrin ligation in 3D matrices with varying E revealed a biphasic relationship between α5-RGD bonds and E that peaked at 22 kPa, correlating with osteogenic commitment (Fig. 3D). αV-integrins exhibited a similar biphasic relationship between matrix stiffness and RGD-binding and peaked at 22 kPa in 3D (data not shown). In contrast to the biphasic relationship between E and integrin-RGD bond formation observed in 3D, when mMSC were cultured in 2D on alginate substrates, the number of RGD-αV-integrin bonds increased in a hyperbolic fashion as matrix stiffness was raised, with no statistically significant difference in bond formation between 22 kPa and stiffer substrates (Fig. S5). Adhesion assays were performed at an early time-point (2 hr) after encapsulation or seeding of cells, to minimize the influence of changes in cellular gene expression on bond formation.
Figure 3. Mechanically-Controlled α5-integrin-RGD bond Formation Correlates with Stem Cell Osteogenic Lineage in 3D.
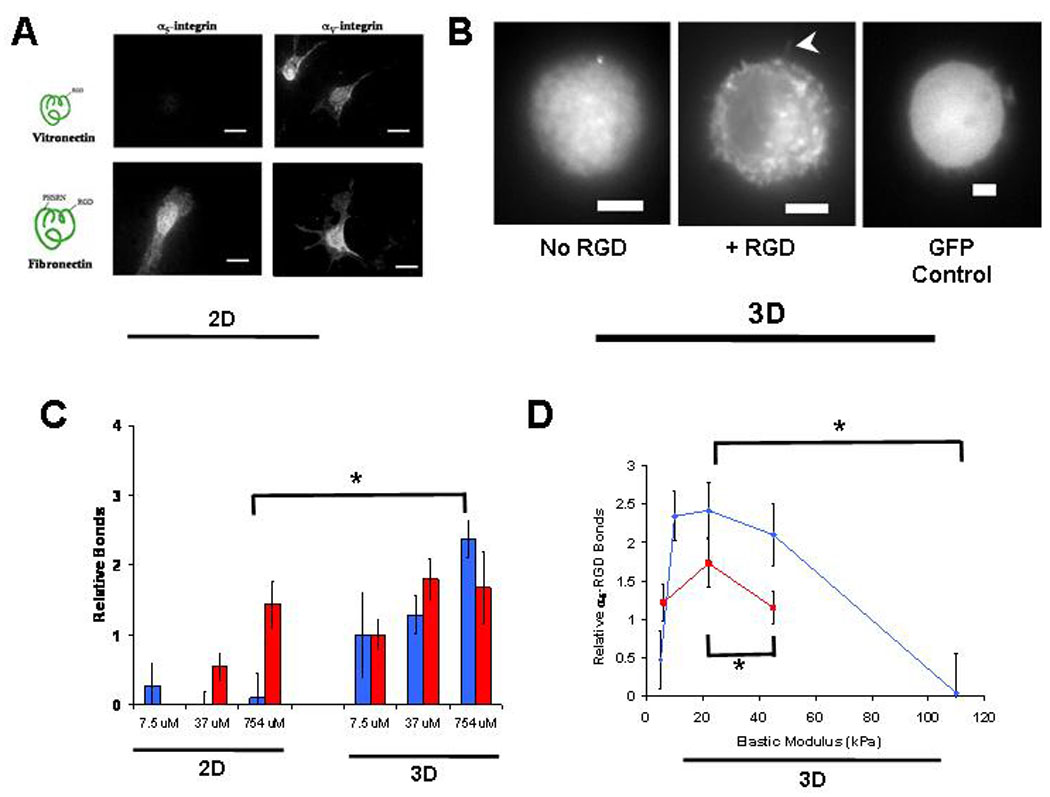
(A). Immunofluorescence analysis of α5 and αV integrins bound to ECM in mMSC adherent to FN or VN coated glass (2D culture). (B). Localization of EGFP-α5-integrins or naked EGFP in mMSC encapsulated into 3D alginate matrices with or without RGD. Note, α5-integrins appear clustered within cells and localized to the cell-matrix interface at the periphery of confocal cross-sections in RGD-modified matrices (arrowhead). (C). ELISA quantification of α5 ( ) and αV (
) and αV ( ) integrin binding to RGD-biotin presented at varying density by either 2D or 3D alginate matrices (* p < 0.01, _t_-test). (D). α5-integrin-RGD bond formation in matrices with varying stiffness presenting either 37 µM (
) integrin binding to RGD-biotin presented at varying density by either 2D or 3D alginate matrices (* p < 0.01, _t_-test). (D). α5-integrin-RGD bond formation in matrices with varying stiffness presenting either 37 µM ( ) or 754 µM (
) or 754 µM ( ) RGD-biotin (* p < 0.01, _t_-test). α5-integrin binding to matrices presenting 754µM RGE-biotin was negligible. Error bars are SEM (n = 4–5). Scale bars: (A), 20µm; (B), 5µm.
) RGD-biotin (* p < 0.01, _t_-test). α5-integrin binding to matrices presenting 754µM RGE-biotin was negligible. Error bars are SEM (n = 4–5). Scale bars: (A), 20µm; (B), 5µm.
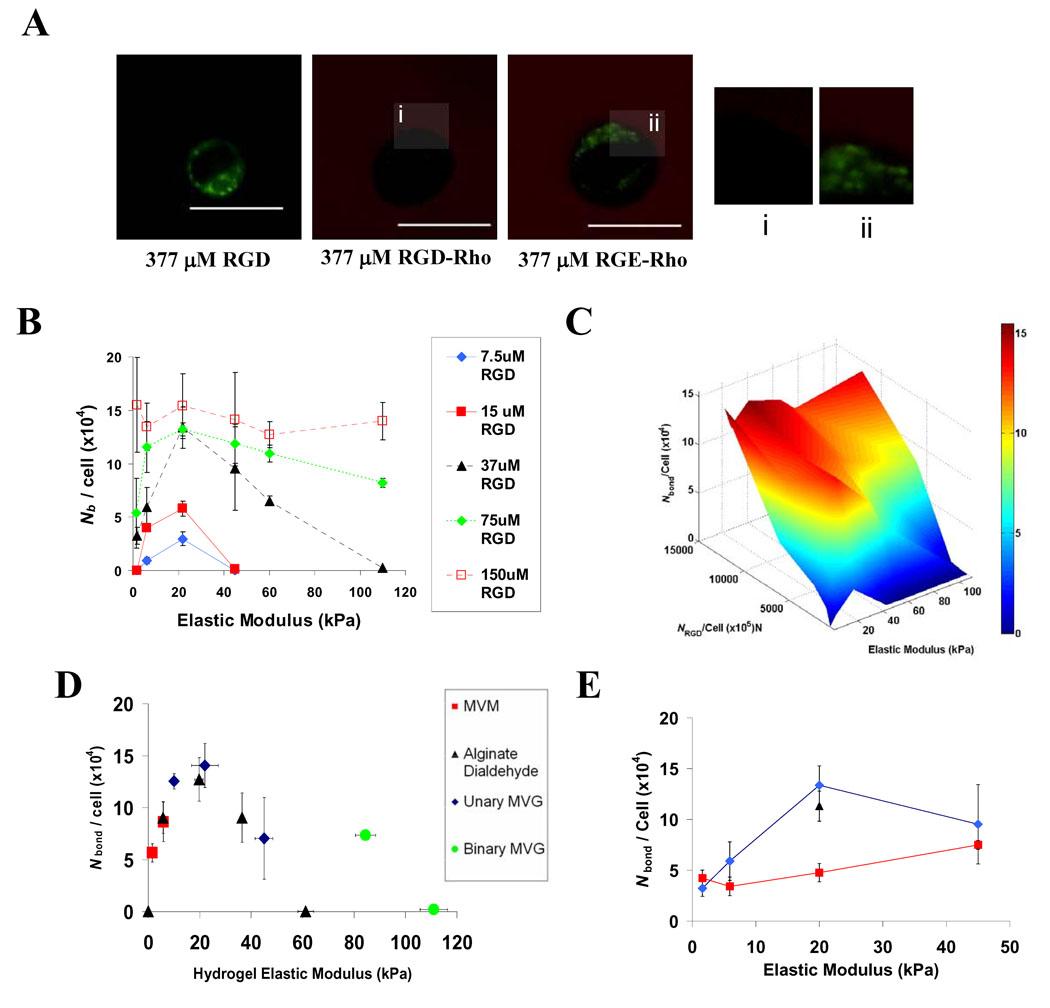
To provide an absolute measure of bond formation between integrins and RGD presented by 3D hydrogels, to complement the relative values obtained biochemically, and to allow a broader examination of the variable space modulating RGD-integrin ligation, we used a non-invasive technique based on Förster Resonance Energy Transfer (FRET41–43) between a membrane-intercalating fluorescein dye and rhodamine labeled peptides (Fig. 4A, S6). The intensity of fluorescein emission at the cell-matrix interface was diminished in the presence of integrin-binding RGD adhesion peptides due to FRET, but not with control RGE peptides (Fig. 4A), as in previous work41, confirming this technique as a specific measure of cell-RGD bonds. Likewise, emission spectra of entire hydrogels (~ 106 cells) indicated FRET between cells and RGD, with the magnitude of energy transfer depending on matrix rigidity (Fig. S6). The number of mMSC-RGD bonds per cell (Nb) calculated from FRET measurements of cell-encapsulating hydrogels indicated that bond formation was regulated both by the density of available RGD and matrix rigidity (Fig. 4B–C). As in ELISA studies of relative integrin-RGD ligation, the relationship between Nb and E, was biphasic and correlated with osteogenesis, with optimal RGD ligation at 22 kPa. Even when high densities of peptide (150µM RGD) were presented, a similar, albeit diminished, relationship was noted (Fig. 4B–C). The decreased dependence of Nb on E observed for Nb exceeding 1.4 × 105 bonds/cell contrasts with the biochemical analysis (Fig. 3D), suggesting saturation of the FRET signal at very high RGD densities. The biphasic relationship between Nb and matrix rigidity was maintained as E was varied over a wide range using a variety of different polymers and crosslinking molecules (Fig. 4D). In contrast, there was virtually no correlation between Nb and the concentration of calcium used to crosslink certain hydrogels (Fig. S7), suggesting that changes in Nb and correlated osteogenic specification reported here are not secondary to changes in matrix-associated calcium. Importantly, the impact of matrix compliance on bond formation was lost when cells were treated with BDM (Fig. 4E), indicating that cell contractility is necessary for matrix mechanics to regulate integrin binding. These findings are consistent with reports indicating that mechanosensing is an active process that requires cell-traction to be exerted on the matrix via motor proteins such as Myosin-II10,24,29–33. As in other adhesion studies, FRET measurements of bond formation were performed at an early time-point (2 hr) after encapsulating cells into hydrogels. Control experiments performed with cyclohexamide treated cells confirmed that protein synthesis did not affect cell-RGD bond formation (Fig. 4E).
Figure 4. Cell-RGD Bond Formation Exhibits a Biphasic Dependence on Matrix Stiffness.
(A). Representative confocal images of fluorescein (green) and TAMRA-RGD emission from mMSC encapsulated into hydrogels presenting 377 µM RGD, RGD-TAMRA or RGE-TAMRA. The cell-matrix interface is shown at high resolution in insets i and ii. (B). Calculated Nb for mMSC in matrices where available RGD density and elastic modulus were varied in parallel. (C). Response surface depicting Nb as a function of E and NRGD / cell revealed significant effects of both RGD density and the interaction between RGD density and elastic modulus (2-way ANOVA; p < 0.01). (D). Curve of Nb versus E generated from FRET studies using matrices formed from various alginate polymers and crosslinking agents presenting a constant density (37 µM) of RGD. (E). Curve of Nb versus E for untreated cells ( ), or cells treated with either 20mM BDM (
), or cells treated with either 20mM BDM ( ) or 20µg/mL cyclohexamide (▲) encapsulated into matrices presenting 37µM RGD. FRET analyses of cell-RGD bonds were performed 2 hr after encapsulating cells, and E values shown are for hydrogels at the time of cell encapsulation. Scale bars: (B); 10µm.
) or 20µg/mL cyclohexamide (▲) encapsulated into matrices presenting 37µM RGD. FRET analyses of cell-RGD bonds were performed 2 hr after encapsulating cells, and E values shown are for hydrogels at the time of cell encapsulation. Scale bars: (B); 10µm.
An additional role for cell contractility in controlling bond number was next revealed by studying peptide clustering by the matrix-encapsulated cells. With nanoporous hydrogel substrates, it is typical to assume that cells are capable of binding any adhesion molecules that lie within 5–10nm of the plasma membrane44. However, a direct comparison of the number of bound integrins measured by FRET to the density of RGD within the matrix surrounding mMSC revealed that if the RGD distribution within the alginate matrix remained homogenous, cells would have to probe nearly 50nm into the surrounding material to access the needed number of peptides (Fig. 5A–B). Thus, it seemed likely that cells utilized traction forces to mechanically reorganize the RGD peptides presented by these hydrogel matrices on the nanometer scale, clustering RGD near integrins while the peptides remain bound to the material (Fig. 5C) – a process somewhat analogous to traction-mediated unfolding of fibronectin which subsequently reveals cryptic peptide epitopes32. To directly address this possibility, a second FRET technique11 was used to measure traction mediated clustering of RGD by encapsulated mMSC (Fig. 5D; Fig. S8). Traction mediated matrix reorganization peaked at 22 kPa, correlating strongly with the peak in Nb and osteogenic commitment (Fig. 5E). The finding that RGD clustering was maximized in matrices of intermediate rigidity is consistent with previous studies indicating that cells cultured on very compliant substrates cannot assemble the cytoskeleton-associated adhesion complexes required to exert significant traction forces45, whereas on very rigid substrates the cells cannot generate enough force to deform the matrix11. Matrix mechanics and dimensionality may change integrin ligation by a number of different mechanisms, including altered integrin-RGD bond life-time, and the observed changes in the nanoscale organization of the ECM. It is also possible that intracellular changes in activation and transport of-integrins, as well as their binding to focal adhesion components, and the resistance of these intracellular bonds to rupture46–50, contribute to mechanically dependent integrin ligation in 3D matrix culture. This mechanism may work independently, or more likely, in concert with previously identified cell-contractility dependent intracellular signaling pathways29–33 to influence cell fate. Because of the physical continuity of integrins with both matrix and focal adhesions, manipulations intended to selectively perturb one of these components (e.g., over-expression of focal adhesion proteins or pharmacologic inhibition of cell traction) are likely to perturb others. This highlights the importance of technologies capable of probing cellular mechanotransduction without mechanically perturbing the system32,33,43, including the FRET assays used in these studies.
Figure 5. Cell-Traction Mediated Reorganization of Ligands Presented by Synthetic ECM.
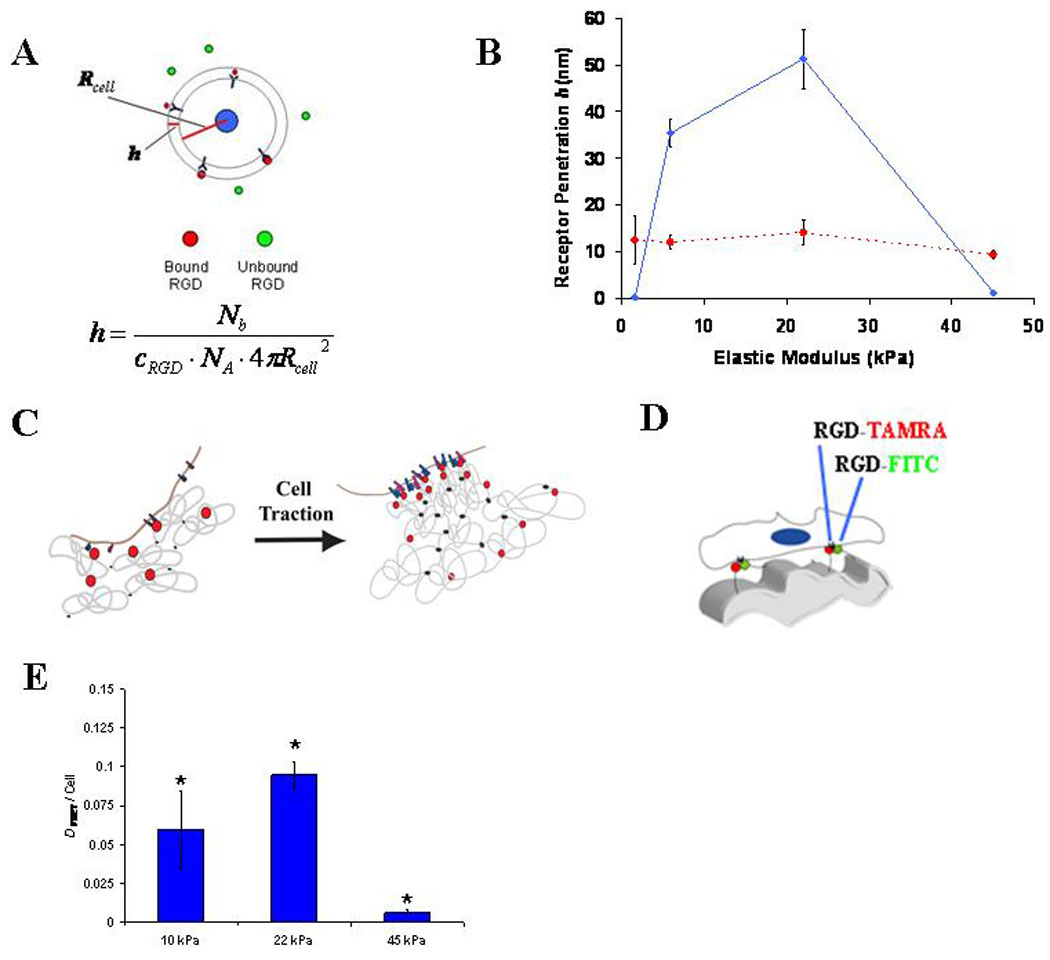
(A). Schematic of the method used to calculate the minimum depth of protrusion of integrin receptors into RGD-modified alginate. As shown, cells were assumed to be spherical with Rcell equal to 10µm. The equation used to calculate the minimum receptor penetration depth, h, is shown. (B). Measurements of mMSC-RGD bond number (Nb) in 3D matrices presenting either 15 µM ( ) or 150 µM (
) or 150 µM ( ) RGD were used to calculate h. (C). Schematic depicting enhanced cell-RGD bond formation due to nanoscale RGD clustering mediated by cell traction forces. (D). Schematic of FRET assay to monitor cell-traction mediated nanoscale RGD-clustering of RGD-CFsC and RGD-TAMRA attached to different alginate chains. (E). FRET measurements of nanoscale RGD-clustering by encapsulated mMSC (* p < 0.01 compared to other conditions, Holm-Bonferonni test). FRET analyses of integrin ligation and nanoscale-RGD reorganization were performed 2 hr after encapsulating cells, and E values shown are for hydrogels at the time of cell encapsulation. Schematic drawings are not meant to be to scale. Error bars are SEM for calculated protrusion depth calculations (n = 3–5) and SD (n = 3) for clustering measurements.
) RGD were used to calculate h. (C). Schematic depicting enhanced cell-RGD bond formation due to nanoscale RGD clustering mediated by cell traction forces. (D). Schematic of FRET assay to monitor cell-traction mediated nanoscale RGD-clustering of RGD-CFsC and RGD-TAMRA attached to different alginate chains. (E). FRET measurements of nanoscale RGD-clustering by encapsulated mMSC (* p < 0.01 compared to other conditions, Holm-Bonferonni test). FRET analyses of integrin ligation and nanoscale-RGD reorganization were performed 2 hr after encapsulating cells, and E values shown are for hydrogels at the time of cell encapsulation. Schematic drawings are not meant to be to scale. Error bars are SEM for calculated protrusion depth calculations (n = 3–5) and SD (n = 3) for clustering measurements.
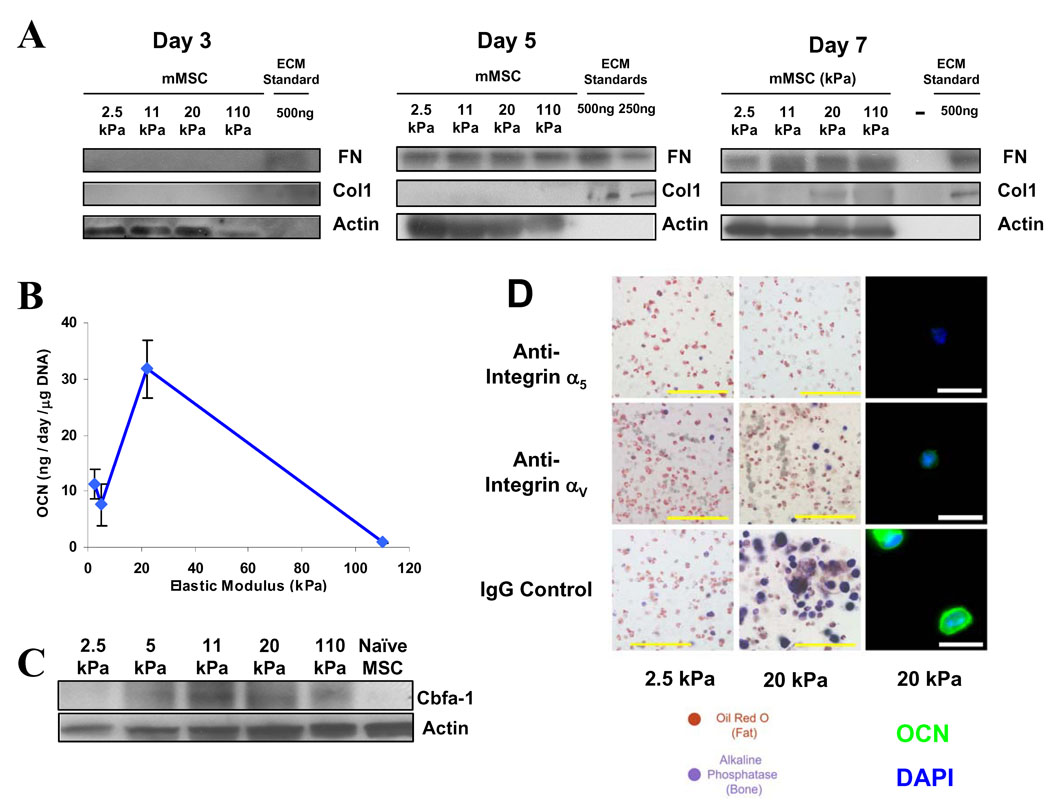
In 3D culture, endogenous ECM assembly is likely to occur, and this may diminish cells’ sensitivity to both the ligand presentation and elasticity of synthetic ECM analogs. FN synthesis became significant after 5 days of culture, but did not appear to change as a function of matrix elasticity (Fig. 6A). In contrast, collagen I (Col I) expression only became significant after one week, and occurred predominantly in matrices that promoted osteogenesis (Fig. 6A). Furthermore, even after 21 days of culture in 3D, secretion of osteocalcin (a hallmark of late osteoblastic differentiation) by mMSC exhibited a marked biphasic dependence on ECM rigidity, (Fig. 6B), while Cbfa-1 expression followed a similar, albeit slightly diminished, trend (Fig. 6C). Together, these data suggest that bonds between RGD and integrins act to trigger lineage-specific protein expression, which persists even in the face of endogenous ECM synthesis. Alternatively, bonds between integrins and RGD coupled to the materials used in these studies may persist, even when there is significant endogenous ECM deposition, and thus provide continuous signaling to cells.
Figure 6. Long Term Regulation of Osteogenic Commitment and Role of Specific Integrins in Stem Cell Fate in 3D Matrices.
(A). Western analysis of matrix synthesis over the time-course of mMSC culture in matrices with varied rigidity which present 754µM RGD. (B). Normalized osteocalcin (OCN) secretion by mMSC after 3 weeks of 3D matrix culture. (C). Western analysis of Cbfa-1 expression in mMSC after 3 weeks of matrix culture. (D). Histologic analysis of encapsulated mMSC cultured for 1 week in matrices of varying stiffness but constant RGD density (754µM) in which α5-RGD bonds or αV-RGD bonds were inhibited with 50µg/mL function blocking antibodies: In-situ staining for Alkaline Phosphatase activity (Fast blue) and Neutral Lipid accumulation (Oil Red O) (left) or OCN immunofluorescence (green) and DAPI nuclear counterstain (blue) of cells in cryosectioned matrices (right). E values shown are for hydrogels after 1 day in culture. Error bars are SD (n = 3). Scale bars: ALP/ORO stain: 100µm; immunofluorescence: 20µm.
Finally, to confirm that within 3D culture, mMSC fate was mediated by integrin-RGD bonds, we examined cells encapsulated into RGD-modified matrices of varying rigidity in the presence of function-blocking antibodies against either α5-integrins or αV-integrins. Blocking RGD-integrin bond formation with either antibody did not affect cell viability or proliferation significantly (Fig S9). However, blocking RGD binding to α5-integrins significantly diminished osteogenesis, and enhanced adipogenesis in an antibody dose-dependent fashion (Fig. 6D, S9). Cells were still capable of undergoing osteogenic commitment, although to a greatly diminished extent, when αV-RGD interactions were blocked (Fig. 6D, S9). These results are consistent with previous work highlighting roles for both α5 and αV-integrin ligation in osteogenesis51,52.
Altogether, the work presented here implicates integrin-adhesion ligand bonds as morphology-independent sensors of both matrix elasticity and dimensionality. This finding is likely to be broadly important to the biology of a variety of processes, from embryogenesis to cancer4,5,13,14, and useful in designing materials to control cell fate for medical applications2,3. Further, differences in integrin ligation in standard tissue culture versus compliant 3D micro-environments are likely to partially explain discrepancies often observed between standard 2D tissue culture versus in-vivo, and highlight the importance of synthetic ECM approaches for both basic investigations and drug screening.
In vivo and in vitro, cells actively probe their micro-environment for adhesion ligands by deforming themselves and their matrix on multiple spatiotemporal scales5,29–33. Here, we demonstrate that one can exploit this process to mechanically tune the molecular interface between cells and the matrix, which in turn plays a significant role in determining MSC fate. Our findings suggest that cells interpret mechanically disparate substrates as having different adhesion ligand presentation, even if their chemical compositions are identical. From an engineering standpoint, cell traction forces may be harnessed as tools to mechanically tune synthetic materials in-situ, generating a new cell-matrix interface which subsequently contributes to cell fate decisions. This may present a general strategy to create functionally complex materials from structurally simple templates, as materials processed in this manner would gain functional characteristics inherent to the cells with which they interact.
Methods Summary
3D Mesenchymal Stem Cell Culture and Lineage Analysis
Alginates (FMC Biopolymer) and agarose were covalently coupled with the integrin binding peptide (Gly)4-Arg-Gly-Asp-Ala-Ser-Ser-Lys-Tyr (Peptides International)17,20. PEGDM polymers were photo-crosslinked in the presence of acryloyl-PEG-GRGDS21. For cell encapsulation studies, mixtures of varying wt % polymer were mixed with stem cells (20 million clonally derived mMSC (D1) per mL, or 15 million hMSC/mL) and crosslinked to form hydrogels. The elastic modulus E of hydrogel matrices was measured using an Instron 3342 mechanical apparatus at a compression rate of 1mm/min. Cell-encapsulating hydrogels were transferred to FBS-supplemented Dulbecco’s Modified Eagle Media (DMEM, Invitrogen) containing a combination of osteogenic and adipogenic chemical supplements. After 1 week in culture, lineage specification was assessed by in-situ staining for Alkaline Phosphatase Activity (Fast Blue) and Neutral Lipids (Oil Red O) in the same samples, by OCN staining in cryosectioned matrices, or by biochemical analysis of cell lysates obtained by recovering cells from alginate matrices with 50mM ethylenediaminetetraacetic acid (EDTA) in PBS. Western analysis of cell phenotype was performed to assess population-level expression levels of FN and ColI, as well as adipogenic (PPAR-γ, Adn) and osteogenic (Cbfa-1, Osteopontin) biomarkers.
In certain experiments, cell, encapsulating, calcium-crosslinked alginate matrices were combined with cell-free alginate matrices in the same media so that the concentration of calcium ions available to cells could be controlled independent from the rigidity of cell-encapsulating matrices (Fig. S3). The relative diffusion coefficient for a model protein, bovine serum albumin (BSA; 67 kDa) was measured via release of rhodamine-labeled BSA from alginate matrices of varying rigidity (Fig. S3).
Biochemical Measurements of Integrin-Adhesion Ligand Bond Formation
Clonally derived mMSC (D1) were encapsulated into alginate matrices presenting G4RGDASSK(biotin)Y. G4RGEASSK(biotin)Y-OH presenting alginates were used as negative controls. After encapsulating cells, hydrogels were incubated in FBS-supplemented media for 2 hr. Hydrogels were dissolved into 250µg/mL alginate lyase, and cell lysates were prepared with RIPA buffer (Sigma). Equal amounts of protein were probed onto Neutravidin-coated stripwells (Thermo Scientific), and biotin-RGD-bound integrins were probed by ELISA using polyclonal antibodies against either α5 or αV integrin subunits, HRP-tagged secondary antibodies (Jackson Immunolabs), and QuantaBlue fluorogenic HRP substrate (Thermo Scientific). QuantaBlue fluorescence was read in a Biotek plate reader and the relative number of bonds between integrins and RGD was calculated according to Equation 1:
| Nb,αX=RFIsample−RFIblankRFIcontrol−RFIblank | Equation 1 |
|---|
where RFI refers to the relative fluorescence intensity of samples, controls or blank (lysate from cells that were not encapsulated into alginate-RGD-biotin). The control was a 3D, 22 kPa matrix modified with 15 µM RGD-biotin (Fig. 3C,D).
FRET Measurements of Integrin-Adhesion Ligand Bond Formation
FRET analysis of integrin-adhesion ligand bonds was performed by monitoring energy transfer between a membrane-intercalating fluorescein dye and rhodamine labeled peptides as previously described by Kong et al41. Clonally derived mMSC (D1) were labeled with 5-hexadecanoylaminofluorescein (Invitrogen) for 24 hr, and encapsulated into alginate matrices presenting G4RGDASSK(tetramethylrhodamine)Y-OH. G4RGEASSK(tetramethylrhodamine)Y-OH peptides were used as negative controls. Hydrogels were formed by mixing cells with various alginate polymers and crosslinking either with CaSO4 (calcium-alginate) or poly(acrylamide-co-hydrazide; alginate dialdehyde). After encapsulating cells, hydrogels were incubated in FBS-supplemented media for 2 hr. The emission spectrum from hydrogels was collected using a Fluorimeter (Jobin Horiba) with 488nm excitation, and normalized to cell density by dissolving hydrogels into PBS containing 50mmol/L EDTA (calcium alginate) or 250µg/mL alginate lyase (alginate dialdehyde). The degree of energy transfer (DFRET) was calculated according to Equation 2:
where I refers to emission intensity at 520nm, I0 to the 520nm emission intensity in a donor control sample with the same peptide density, elastic modulus and drug treatment but with RGD peptides rather than RGD-TAMRA peptides. DFRET was used to calculate the number of integrin-RGD bonds based on a linear calibration curve generated by performing parallel FRET-binding and 125I-RGD binding studies in solution41.
FRET between cells and RGD was visualized using a Laser Scanning Confocal Microscope (Zeiss LSM 510). Excitation was provided by an argon laser (488nm), and fluorescein (green, 500–530nm) and rhodamine (red, 565–615) emission were collected simultaneously. Images were prepared by overlaying the green and red emission channels.
Further details are provided in Supplementary Information.
Acknowledgements
We thank Dr. RF Horwitz for providing the human α5-integrin-EGFP plasmid, and Brian Tilton for performing FACs sorting of GFP-expressing mMSC at the Harvard FAS Center for Systems Biology. We also thank Evangelos Gatzogiannis for assisting with confocal microscopy at the Harvard University Center for Nanoscale Systems (CNS). The monoclonal antibody against Osteopontin (MPIIIB101) developed by Michael Solursh and Ahnders Franzen was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA. We acknowledge R Schmidt and KE Healy (University of California, Berkeley) for advice regarding nuclear morphology measurements, Drs. Andrew Putnam (University of Michigan), Hyun Joon Kong, Claudia Fischbach and Susan Hsiong for helpful discussions, and Drs. Herman Vandenburgh (Brown University) and Zhigang Suo for critical reading of the manuscript. We also acknowledge funding from NIH (R37 DE013033) and from the NIH “Systems-based Consortium for Organ Design and Engineering” training grant (JR-F, 1TL1EB008540–01, NIBIB), a Harvard Presidential Fellowship (PRA), an NSF Graduate Research Fellowship (NH), the Harvard College Research Program (ASM), an EMBO Long-Term Fellowship ALTF 42-2008 (DS) and the Wyss Institute for Biologically Inspired Engineering (SAB).
Footnotes
Supplementary Information is linked to the online version of the paper.
Author Contributions The experiments were designed by NH, PRA and DJM and carried out by NH, PRA, ASM, DS, and OAA. NH, SAB and JR-F provided new reagents/analytical tools. The manuscript was written by NH and DJM. The principal investigator is DJM.
References
- 1.Passier R, van Laake LW, Mummery CL. Stem-cell based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 2.Silva EA, Kim ES, Kong HJ, Mooney DJ. Material-based deployment enhances efficacy of endothelial progenitor cells. Proc. Natl. Acad. Sci. USA. 2008;105:14347–14352. doi: 10.1073/pnas.0803873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali OA, Huebsch N, Cao L, Dranoff G, Mooney DJ. Infection-mimicking materials to program dendritic cells in situ. Nat Mater. 2009;8:151–158. doi: 10.1038/nmat2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 5.Geiger B, Bershadsky A. Exploring the neighborhood: adhesion-coupled mechanosensors. Cell. 2002;110:139–142. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 6.Alsberg E, Anderson KW, Albeiruti A, Rowley JA, Mooney DJ. Engineering growing tissues. Proc. Natl. Acad Sci. USA. 2002;99:12025–12030. doi: 10.1073/pnas.192291499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;32:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 9.Klees RF, et al. Laminin-5 induces osteogenic gene expression in human mesenchymal stem cells through an ERK-dependent pathway. Mol Biol Cell. 2005;16:881–890. doi: 10.1091/mbc.E04-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeny HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc. Natl. Acad. Sci. USA. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 13.Fischbach C, et al. Cancer cell angiogenic capability is regulated by 3-D culture and integrin engagement. Proc. Natl. Acad. Sci. USA. 2009;106:399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 15.Hsiong SX, Boontheekul T, Huebsch N, Mooney DJ. Cyclic RGD Peptides Enhance 3D Stem Cell Osteogenic Differentiation. Tissue Eng A. 2009;15:263–272. doi: 10.1089/ten.tea.2007.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diduch DR, Coe MR, Joyner C, Owen ME, Balian G. Two cell lines from bone marrow that differ in terms of collagen synthesis, osteogenic characteristics, and matrix mineralization. J Bone Joint Surg Am. 1993;75:92–105. doi: 10.2106/00004623-199301000-00012. [DOI] [PubMed] [Google Scholar]
- 17.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 18.Alsberg E, et al. Regulating bone formation via controlled scaffold degradation. J Dent Res. 2001;80:2025–2029. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 19.Lutolf MP, et al. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 20.Connelly JT, Garcia AJ, Levenston ME. Interactions Between Integrin Ligand Density and Cytoskeletal Integrity Regulate BMSC Chondrogenesis. J. Cell. Physiol. 2008;217:145–154. doi: 10.1002/jcp.21484. [DOI] [PubMed] [Google Scholar]
- 21.Bencherif SA, et al. Influence of the degree of methacrylation on hyaluronic acid hydrogels properties. Biomaterials. 2008;29:1739–1749. doi: 10.1016/j.biomaterials.2007.11.047. [DOI] [PubMed] [Google Scholar]
- 22.Pek YS, Wan ACA, Ying JY. The effect of matrix stiffness on mesenchymal stem cell differentiation in a 3D thioxtropic gel. Biomaterials. 2010;31:385–391. doi: 10.1016/j.biomaterials.2009.09.057. [DOI] [PubMed] [Google Scholar]
- 23.Ghajar CM, et al. The effect of matrix density on the regulation of 3-D capillary morphogenesis. Biophys J. 2008;94:1930–1941. doi: 10.1529/biophysj.107.120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelham RJ, Jr, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity Sensing at the Leading Edge through αVβ3 Integrins and RPTPα. Biophys J. 2006;90:1804–1809. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 27.Benoit DS, Schwartz MP, Durney AR, Anseth KS. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater. 2008;7:816–823. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Discher DE, Janmey P, Wang YL. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 30.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 31.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc. Natl. Acad. Sci. USA. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung EH, et al. Biomimetic artificial ECMs stimulate bone regeneration. J Biomed Mater Res A. 2006;79:815–826. doi: 10.1002/jbm.a.30809. [DOI] [PubMed] [Google Scholar]
- 35.Engler AJ, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 37.Dembo M, Torney DC, Saxman K, Hammer D. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 38.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 39.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J Biol Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 41.Kong HJ, Boontheekul T, Mooney DJ. Quantifying the relation between ligand-receptor bond formation and cell phenotype. Proc. Natl. Acad. Sci. USA. 2006;103:18534–18539. doi: 10.1073/pnas.0605960103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakowicz JT. Principles of fluorescence spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
- 43.Huebsch ND, Mooney DJ. Fluorescent resonance energy transfer: A tool for probing molecular cell-biomaterial interactions in three dimensions. Biomaterials. 2007;28:2424–2437. doi: 10.1016/j.biomaterials.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hern DL, Hubbell JA. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J Biomed Mater Res. 1998;32:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 45.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danen EH, et al. Requirement for the synergy site for cell adhesion to fibronectin depends on the activation state of integrin alpha 5 beta 1. J. Biol. Chem. 1995;270:21612–21618. doi: 10.1074/jbc.270.37.21612. [DOI] [PubMed] [Google Scholar]
- 47.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 48.Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–995. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 49.Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J. Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 50.Ward MD, Dembo M, Hammer DA. Kinetics of cell detachment: peeling of discrete receptor clusters. Biophys J. 1994;67:2522–2534. doi: 10.1016/S0006-3495(94)80742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keselowsky BG, Collard DM, Garcia AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc. Natl. Acad. Sci. USA. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai CF, Cheng SL. Alphavbeta integrins play an essential role in BMP-2 induction of osteoblast differentiation. J. Bone Miner Res. 2005;20:330–340. doi: 10.1359/JBMR.041013. [DOI] [PubMed] [Google Scholar]