Innate immune natural killer cells and their role in HIV and SIV infection (original) (raw)
. Author manuscript; available in PMC: 2011 May 1.
Published in final edited form as: HIV Ther. 2010 Jul 1;4(4):483–504. doi: 10.2217/HIV.10.28
Abstract
The findings that early events during HIV-1 and SIV infection of Asian rhesus macaques dictate the levels of viremia and rate of disease progression prior to the establishment of mature and effective adaptive immune responses strongly suggest an important role for innate immune mechanisms. In addition, the fact that the major target of HIV and SIV during this period of acute infection is the gastrointestinal tissue suggests that whatever role the innate immune system plays must either directly and/or indirectly focus on the GI tract. The object of this article is to provide a general overview of the innate immune system with a focus on natural killer (NK) cells and their role in the pathogenesis of lentivirus infection. The studies summarized include our current understanding of the phenotypic heterogeneity, the putative functions ascribed to the subsets, the maturation/differentiation of NK cells, the mechanisms by which their function is mediated and regulated, the studies of these NK-cell subsets, with a focus on killer cell immunoglobulin-like receptors (KIRs) in nonhuman primates and humans, and finally, how HIV and SIV infection affects these NK cells in vivo. Clearly much has yet to be learnt on how the innate immune system influences the interaction between lentiviruses and the host within the GI tract, knowledge of which is reasoned to be critical for the formulation of effective vaccines against HIV-1.
Keywords: differentiation, HIV, innate immunity, KIRs, NK cells, regulation, SIV
The response of the host to viral infections involves the presence of naturally occurring antiviral agents at the site of infection (tissue-specific antiviral factors) and the co-ordinated sequential orchestration of cells belonging to the innate immune system, followed by the adaptive immune system. The mechanisms by which such host responses are induced, co-ordinated and regulated dictate the outcome of viral infection [1]. During the acute phase of infection, the rapidity by which the signals are communicated between cells at the site of infection and the innate immune effector cells, which are required to contain the infection, clearly play an important role in the outcome of the infection. In some cases, such innate immune effector cells are present locally at the site of infection, facilitating the rapid control of infection. However, in most other cases the innate immune effector cells have to be recruited to the site of infection from the circulation or from remote cellular depots in efforts to contain such infections. This recruitment is mediated by the generation of chemical signals in the form of chemokine gradients from cells at the site of infection, which radiate like sonar waves until they reach their cognate receptors expressed by the innate immune effector cells. Response to such signals thus requires the conversion of the innate immune cells that exist at a normal physiologically steady state to a ‘migratory’ state. The migration of these cells has been shown to be tightly regulated. While some efforts to study the trafficking of cells of the innate immune system and its subsets have been made, the detailed mechanisms that are involved in such regulation of trafficking, particularly in humans and nonhuman primates (NHPs), are at present poorly understood [2]. It is generally thought that the innate immune system basically controls the level of viremia until the adaptive immune system can be effectively generated. One of the major cell lineages comprising the innate immune system that responds to such signals are the natural killer (NK) cells, which have been calculated to consist of 2 billion cells in a healthy adult [3]. NK cells were thought to be a primitive cell lineage [4,5], which evolved prior to the development of the adaptive immune system based on a number of reasons. Thus, while cells of the adaptive immune system utilize clonal sets of somatically rearranged receptors for antigen recognition, the cells of the innate immune system, including NK cells, utilize nonclonal germ line-encoded receptors. The major general differences between adaptive and innate immune cells are summarized in Table 1. The more recent findings of a large number of cell-surface molecules expressed by NK cells, which upon ligation exert either inhibitory or activating signals (a function unique to mammalian species), coupled with the fact that their cytolytic function is mediated by a mechanism that is quite similar to conventional CD8+ effector cytotoxic T cells suggests that these two cell lineages may in fact have coevolved from a common ancestral effector cell lineage [6].
Table 1.
Some distinguishing characteristics between cells of the innate and adaptive immune systems.
| Innate | Adaptive | |
|---|---|---|
| Encoding of receptors | Germline | Somatically |
| Distribution of receptors | Nonclonal | Clonal |
| Repertoire of receptors | Limited | Very large |
| Nature of targets | Invariant | Variable |
| Recognition of targets | Perfect | Imperfect |
| Kinetics of response | Fast | Slow |
| Generation of memory | No | Yes |
In addition, a series of more recent findings has made us re-evaluate our views on previously ascribed differences between innate and adaptive immune cells (Box 1). Unlike T or B cells, NK cells were thought to exert their effector function without prior ‘education’. However, subsequent discoveries in recent years have shown that these cells share functions previously only ascribed to cells involved in adaptive immune responses, such as mature T and B cells, and have led us to rethink the role that this cell lineage plays in response to foreign insults. Thus, NK cells have been shown to undergo a process similar to T cells in terms of negative/positive selection and self/nonself discrimination, and are now known to require ‘licensing’ and ‘arming’ [7]. Prior to acquiring full maturation, the NK cells within the bone marrow undergo ‘education’, which is conferred by the interaction of select inhibitory receptors expressed by this cell lineage with MHC class I molecules expressed by other cell lineages [8]. The NK cells have also been shown to possess an ‘editing’ function by their ability to eliminate immature dendritic cells (DCs) [9,10], execute ‘immunoregulatory’ function [11,12] leading to the coinage of the term ‘NKregs’, and lastly, acquiring ‘immunological memory’ [13,14]. Some of these functions ascribed for NK cells continue to be a subject of debate and generate controversy, and are reasoned to probably be secondary to the discovery of the heterogeneity that exists amongst this cell lineage, discussed in more detail later. Clearly, the advancement of technologies, such as polychromatic analysis of cell-surface markers and microarray analysis of mRNA coding for gene products that serve to distinguish subsets within a given cell lineage, have provided powerful new tools that have allowed for a more sophisticated analysis of the potential existence of sub-lineages within the NK-cell lineage. In addition, the ability to generate knock-out, knock-in and transgenic strains of mice have greatly facilitated the discovery of new molecules and assays involving NK cells, which has prompted studies to identify their homologs in humans and NHPs. These phenotypic and functional advances have led to the findings that such select sublineages not only play distinct roles but also appear to interact with other cell lineages (i.e., cross talk), such as cells of the DC lineage, and these early innate immune interactions perhaps dictate the quality and quantity of the adaptive immune response against pathogenic insults. The important role of the innate immune system in contributing to disease outcome is highlighted recently by the discovery of the role that the NK-cell lineage potentially plays in dictating the outcome of HIV-1 infection [15–17]. This article will summarize our current views of NK-cell heterogeneity both in terms of phenotype and function, potential mechanisms of regulation and cytolytic function together with our current views on their potential role in HIV infection of humans and, as a model, SIV infection of NHPs.
Box 1. Evidence for the hypothesis that natural killer cells play a role in the orchestration of immune responses.
- ▪
Natural killer (NK) cells appear to undergo a process similar to negative/positive selection by T cells such as ‘licensing’ and ‘arming’ - ▪
In the bone marrow NK cells have been shown to undergo ‘education’ via the interaction of select inhibitory receptors with MHC-class I to acquire full functional maturation - ▪
NK cells have been shown to possess an ‘editing’ function by their ability to delete immature dendritic cells - ▪
NK cells execute ‘immunoregulatory’ functions much like Tregs with the coining of the term NKregs - ▪
In select models they have been shown to display a ‘memory’ response
NK cells: phenotypic heterogeneity & differentiation
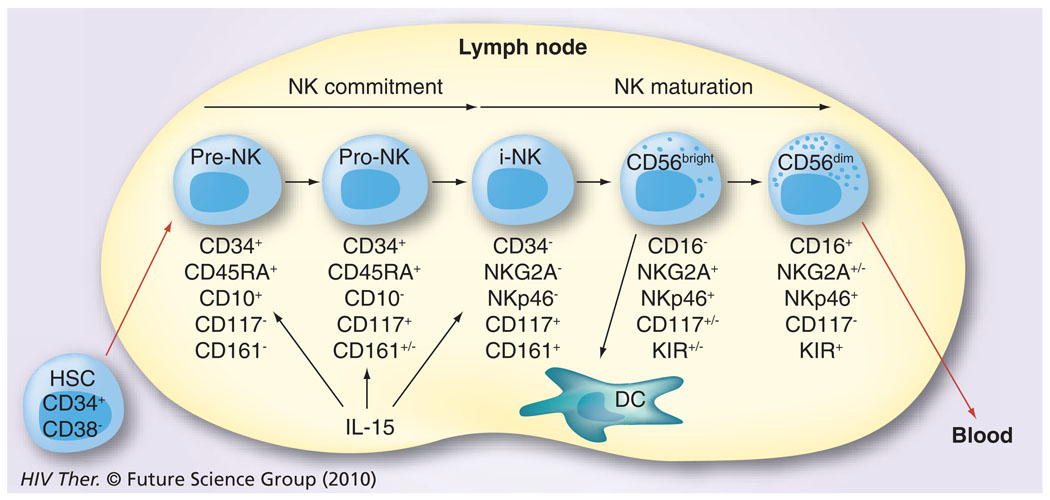
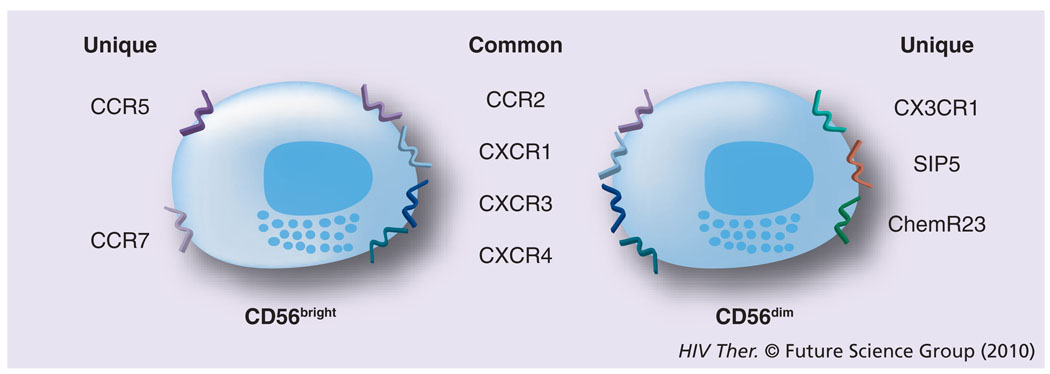
Initially, the difficulties in defining appropriate cell-surface markers to distinguish NK-cell lineage led to the belief that there was only a single NK-cell population, which was distinguished in humans by the absence of the CD3 molecules and the expression of the CD56 receptor [18,19]. Additional problems stemmed from the fact that in mice the surface markers of the NK cells were quite different and exhibited limited homology to their human counterparts. Further studies have demonstrated that several subsets of NK cells exist in humans. Thus, more than 95% of NK cells are CD3−cells that express CD8α-chain homodimers (α/α). This cell population can be further differentiated by the relative density of expression of CD56 (i.e., neural cell adhesion molecule [NCAM], a homophilic binding glycoprotein, expressed on the surface of neurons, glia, skeletal muscle and NK cells) and CD16 (i.e., the Fcγ-RIII receptor) cell-surface markers. Two major subpopulations were characterized by the combination of these markers – the CD3−CD56brightCD16− and CD3−CD56dimCD16+ population [20]. The smaller population of CD3−CD56brightCD16+ is generally considered together with other CD56bright NK cells. The major NK-cell subsets have also been shown to differentially express CD27 [21]. Thus, while the CD3−CD56dimCD16+ population fails to express CD27, the CD3−CD56brightCD16− population expressed readily detectable levels of CD27. While the NK cells constitute a relatively minor fraction of mononuclear cells in peripheral blood (~5%), in the lymph nodes (LNs) their numbers are much higher (up to 50%) and LNs are currently considered as a major site for NK-cell education, similar to the role that the thymus plays for the education of T cells. The relative abundance of NK-cell subtypes in the blood and the LN is quite distinct. Thus, whereas approximately 90% of NK cells are of the CD16−. CD27+CD56bright phenotype within the LN, the CD16+CD27−CD56dim phenotype is the predominant NK-cell phenotype in the peripheral blood [22,23]. Originally, it was not clear whether these two phenotypically distinct populations both represent terminally differentiated sublineages or if one is a developmental precursor of another. The current view supported by data from a series of recent studies is that the CD56bright NK cells in the LN eventually develop into CD56dim NK cells (Figure 1) [24,25] by a not as yet fully understood process involving cross-talk with other cell types, such as DCs and fibroblasts. These two cell lineages are also functionally quite different. The CD56brightCD16− NK cells express homing receptors, such as CCR7 and CD62L, and have the capacity to secrete cytokines, such as IFN-γ and TNF-α, while the CD56dimCD16+ NK cells are the major contributors of MHC unrestricted cytotoxicity and are rich in granzyme and perforin. In addition, since CD16 is a molecule that represents the Fcγ-RIII receptor capable of binding specific antibodies, the NK cells that express CD16 function as mediators of the antibody-dependent cell cytotoxicity (ADCC), a function originally attributed only to cytotoxic T cells [26]. These two major subsets of NK cells have also been shown to express both shared and distinct sets of chemokine receptors [2], which may play a role in their trafficking based on the chemokines being synthesized at the site of infection. Thus, as seen in Figure 2, the CD56bright and the CD56dim NK cells share the expression of CCR2, CXCR1, CXCR3 and CXCR4, but whereas only the CD56bright express CCR5 and CCR7, the CD56dim are distinguished by their expression of CXCR1, SIP5 and chem23. The role of chemokines in influencing human NK-cell function has recently been highlighted [27].
Figure 1. The maturation/differentiation of natural killer cells in lymph nodes based on progressive expression of cell surface markers.
HSCs migrate from the bone marrow to lymph nodes. They first undergo commitment to the NK cell lineage (pre-NK and pro-NK) and then develop through to the immature NK cell stage. These NK cells (CD16−CD56bright) stay within the lymph node and interact with DCs or further mature into cytotoxic NK cells characterized by the expression of CD16 and CD56dim. A few of the former and the majority of the latter are released for recirculation in the peripheral blood.
DC: Dendritic cell; HSC: Hematopoietic stem cell precursor; NK: Natural killer.
Figure 2. The two major subsets of natural killer cells also express a variety of chemokine receptors.
Some of these are common to both and others are unique to the two subsets.
Other NK-cell lineages
Besides these two major NK-cell lineages there are other subsets of NK cells, which perform highly specialized functions. These include the uterine NK cells and the gastrointestinal (GI) NK cells.
Uterine NK cells
The uterine NK cells have received considerable attention for their potential role at the fetal– maternal interface [28], in pregnancy loss [29] and pre-eclampsia [30]. It is important to note that even at this specific tissue site there appears to be heterogeneity among the NK cells. Thus, the NK cells residing in the endometrial tissues (eNK) are distinguished both phenotypically and functionally from the NK cells that reside within the decidual tissues (dNK). These cells possess some unique markers and perform functions associated not only with their cytotoxicity, but are also involved in other processes including, for example, attracting the trophoblast and correct remodeling of spinal arteries [31].
New NKid on the block
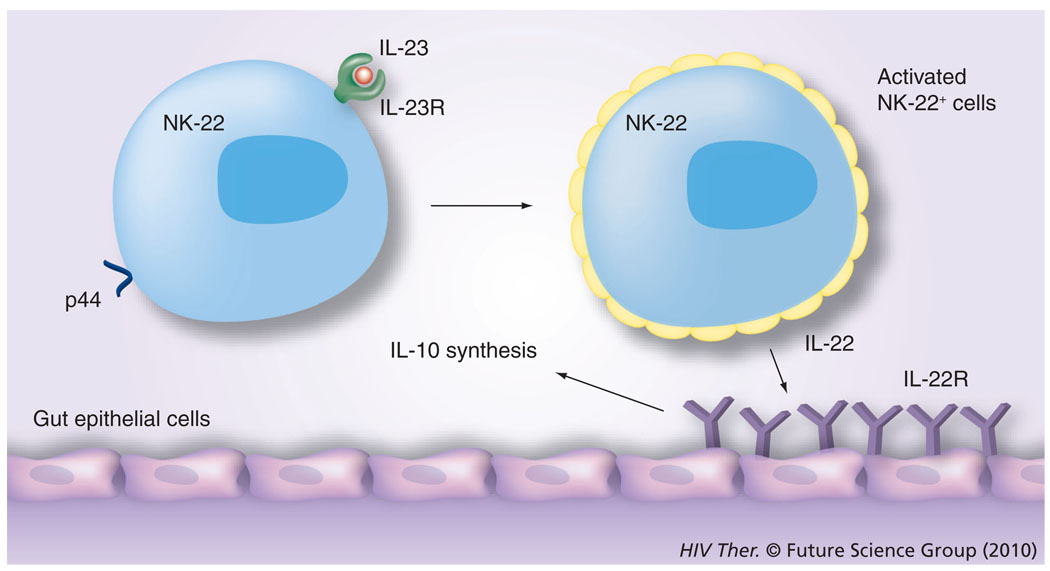
The discovery of GI NK cells has attracted considerable attention because the GI tract is the major target of both HIV and SIV infection, and while the role that these cells play, particularly during the acute infection period, has yet to be defined, it is currently thought that these gut-associated NK cells are perhaps a distinct NK-cell lineage from those in the peripheral circulation. They are characterized by the expression of NKp44 in humans and NKp46 in mice and by the synthesis of IL-22 and are also termed the NK-22 subset [32]. They also express the IL-23 receptor and have been shown to be activated in the presence of IL-23, which leads to the synthesis of IL-22, and which in turn binds to the IL-22 receptor expressed by gut epithelial cells, leading to the synthesis of IL-10 by these cells (Figure 3) and this cascade of interactions is reasoned to be the normal physiological function of this cell lineage [32]. While the detailed mechanisms by which this unique subset of IL-22-synthesizing NKp46 cells localized to the gut tissue continue to unravel, it appears that this lineage is also characterized by the synthesis of antimicrobial peptides [33] and the intracellular expression of unique transcription factors, such as RORγt [34], although its ability to mediate its NK function is not linked to the expression of the p46 molecule [35].
Figure 3. The most recently described natural killer cell subset is the natural killer subset that is present in mucosal lymphoid tissues, such as the tonsil and mesenteric lymph nodes.
These NK cells express p44 in the case of humans and the IL-23R. It is reasoned that dendritic cells, upon activation following exposure to harmful infectious agents, synthesize IL-23, which binds to the IL-23R on NK cells. These activated NK cells synthesize IL-22. The gastrointestinal epithelial cells express the receptor for IL-22 and, upon ligation, synthesize IL-10, which supposedly downregulates the degree of inflammation and homeostasis is maintained.
NK: Natural killer; R: Receptor.
Differentiation & maturation of NK cells
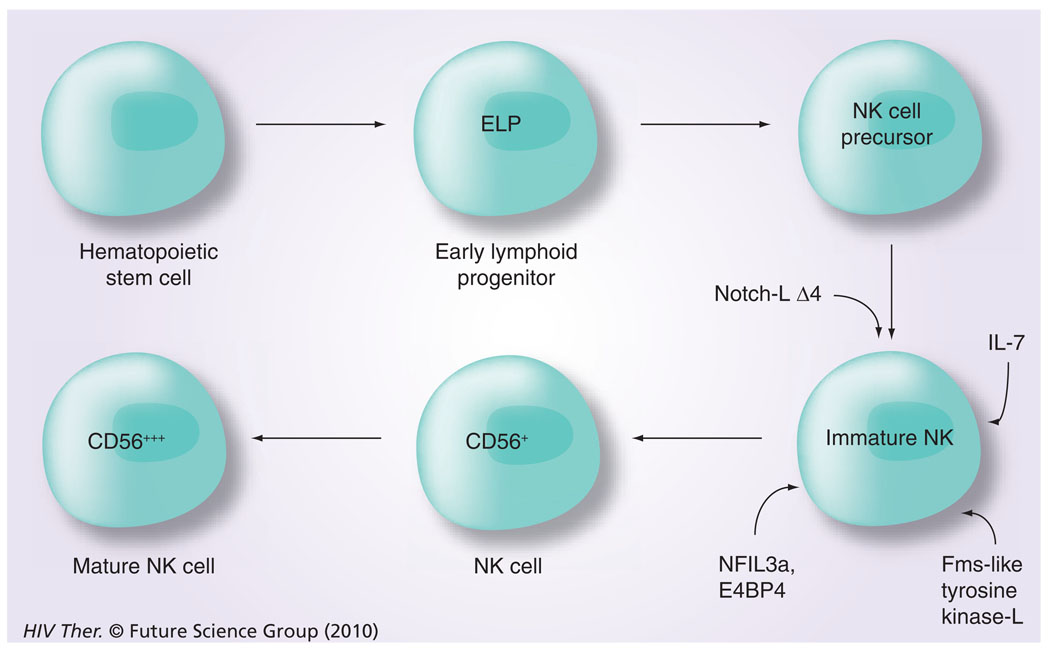
The differentiation and maturation of NK cells during ontogeny is a complex and an incompletely understood process, which seems to progress at various paces in different organs (i.e., spleen, liver, gut tissues and the bone marrow) and studies in mice have shown the involvement of RAG [36]. With regard to differentiation, a role for a number of transcription factors along with the presence of select cytokines has been documented (Figure 4). These include a role for Notch-LΔ4, IL-7 and the Fms-like tyrosine kinase 3 ligand [37], a requirement for NFil3A [38] and an exposure to membrane-bound IL-15 for complete maturation from hematopoietic progenitor cells to mature NK cells progressively from CD56brightCD16− killer cell immunoglobulin-like receptor (KIR)− cells to CD56dimCD16+KIR− and finally to CD56dimCD16+KIR+ cells [39]. The role of IL-7 and IL-15 in the differentiation process has been further studied and it appears that a balance between levels of these two cytokines may in fact lead to the differentiation of particular NK-cell subpopulations [40], which are mediated by various intracellular pathways, including the p38 MAPK pathway [41]. There have also been reports on the existence of differences in the frequencies of CD34+, CD45RA+ pre-NK cells in the blood versus secondary lymphoid tissues. Thus, while the blood and bone marrow appear to contain low frequencies (<1 to <10% of CD34+ cells), the secondary lymphoid tissues are highly enriched for the pre-NK cells [42]. In addition, similar pre-NK cells have been documented to exist in the gut-associated lymphoid tissues [43,44]. Whether these are similar to the Lin−CD127+ cells that express RORγC in mesenteric LNs, as reported by Cupedo et al. [45], remains unclear. The Lin−CD127+ cells appear to share characteristics with lymphoid tissue inducer and their progeny were shown to be the CD56+ CD127+ NK cells, which synthesize both IL-17 and IL-22.
Figure 4. Intracellular transcription factors that play a role in the differentiation of natural killer cells and the requirement for exposure to membrane bound IL-15 for full natural killer cell maturation.
ELP: Early lymphoid progenitor; L: Ligand; NK: Natural killer.
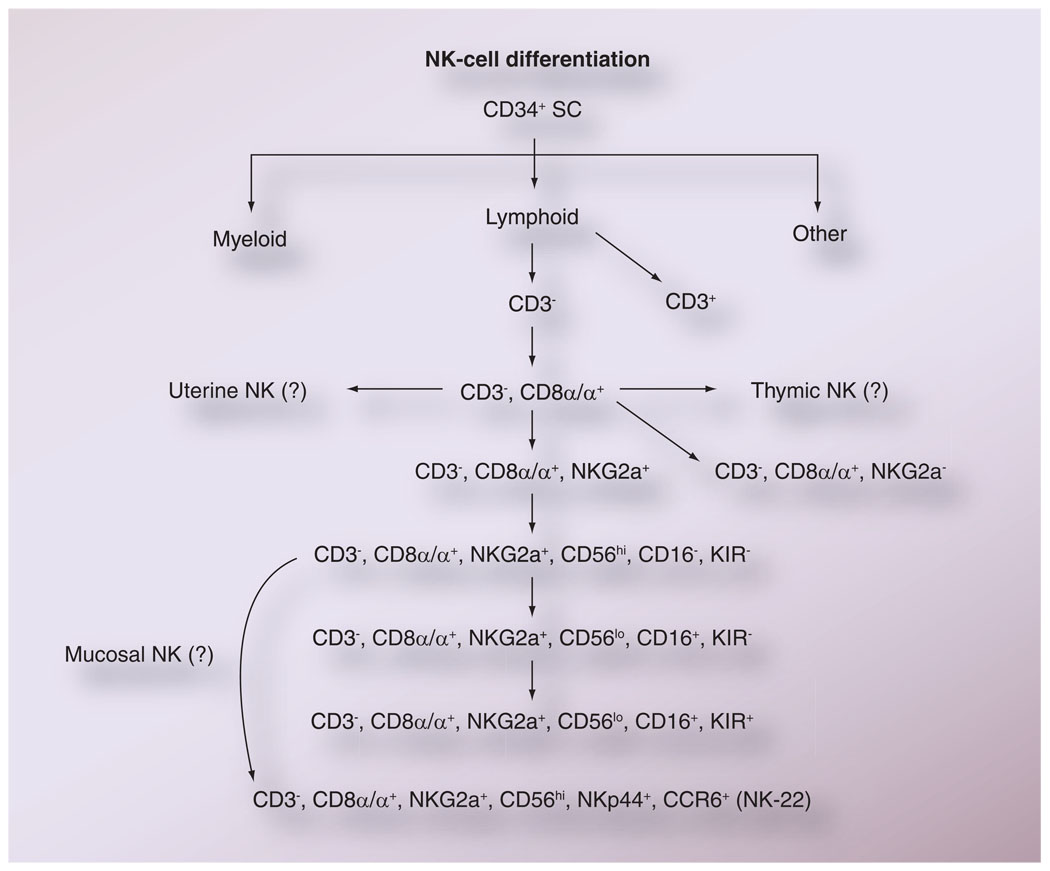
With regard to maturation, while controversies continue to exist, there have been numerous attempts to utilize data on the sequential appearance of cell-surface markers by NK cells as indicators of the maturation of NK cells (Figure 5). It is the general opinion that the majority of NK cells do not express CD3 but do express CD8α homodimers, and based on phenotypic analysis of cells from patients undergoing bone marrow transplantation, an ordered expression of cell-surface markers seems to occur as depicted in Figure 5. Of interest is the finding that the KIR molecules are only expressed by the most differentiated NK cells, according to this scheme, and suggests that while the cytokine-generating property of NK cells is independent of KIR-mediated regulation, the cytolytic function is probably influenced by KIR and KIR-ligand interactions. It is important to note that at present it is not clear whether the uterine and gutlocalized NK-22 cells are subsets of progenitor NK cells or a cell lineage that differentiates from a common NK-cell progenitor cell.
Figure 5. The presumed differentiation of natural killer cells based on sequential expression of cell surface markers.
As indicated, it is not as yet clear whether the NK cells found in uterine tissues, within the thymus and the mucosal tissues, originate from the same common precursor NK cells as those found in the blood and lymph nodes.
NK: Natural killer; SC: Stem cell.
NK-cell regulation
The regulation of the function of NK cells has been a subject of study by several laboratories and has focused primarily on either cell-surface receptors (intrinsic) that are expressed by the NK-cell lineage(s) or on molecules synthesized by other cell lineages that influence NK-cell function (extrinsic).
Intrinsic regulation
Among the intrinsic molecules are the cell-surface receptors that have been categorized into those for which ligation results in inhibition of NK-cell function as compared with others that have been shown to augment NK-cell function [46] and have predominantly focused on the lytic function of NK cells. It is clear that the regulation of the non-cytolytic function of NK cells also needs study. Among the receptors that have been shown to be involved in the regulation of NK-cell function are the KIR family of both activating and inhibitory molecules (discussed more in detail later), the NKG2/CD94 heterodimeric series of molecules [47], the p30, p44 and p46 series of molecules [48–50], the inhibitory function of Siglec-7 [51] and the 2B4 molecule [52]. Evidence to date indicates that the net results of the interactions between these molecules and their cognate ligands dictate the quality and quantity of innate immune responses that are finally generated. As far as the KIR molecules are concerned, it is important to note that the relative affinity of the activating KIR molecules for their cognate classical and nonclassical MHC class I molecules is far lower than the affinity of the inhibitory KIRs and their cognate MHC class I ligands, suggesting a dominant role for the inhibitory KIRs [53]. Similar studies aimed at identifying the minimal requirements for the induction of activation versus inhibitory signals by receptors other than KIR have also been conducted [54].
Extrinsic regulation
Among the extrinsic molecules that have been shown to regulate NK-cell function are the cytokines that induce NK-cell activation and expansion, which include IL-2, IL-15, IL-21 and others that are involved in regulation of NK-cell function. Those involved in regulation of NK-cell function include the cytokines TGF-β [55] and IL-17 [56], activin-A (a member of the TGF-β superfamily) responsible for DC regulation of NK-cell function [57], and molecules synthesized by the prostate gland termed prostasomes [58]. Owing to the importance of the gut tissue in HIV/SIV infection, the role of regulation of NK function within the gut tissue has received attention. Thus, it is thought that the TH17−CD4+ T cells play a dual role by synthesizing IL-22 upon stimulation by IL-23 and by synthesizing IL-17, which contributes to the decreased function of NK cells [56]. As illustrated in Figure 6, the pathways involved are good candidates for future clinical manipulation to favor either inhibition or activation in efforts to determine their effects on the pathogenesis of HIV/SIV infection.
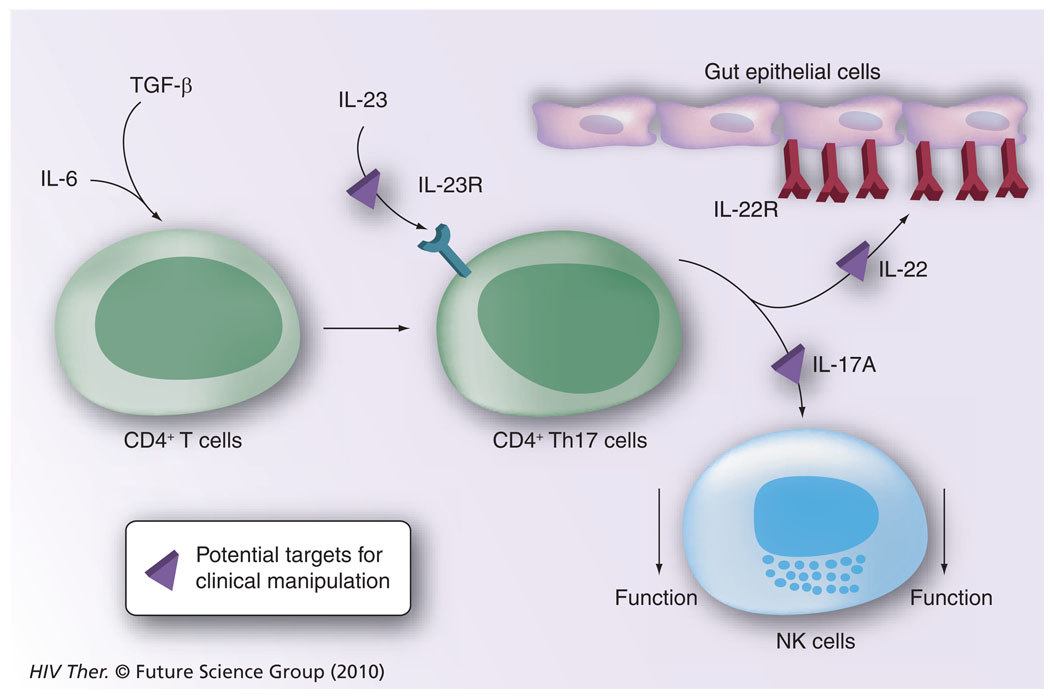
Figure 6. The potential role of CD4+-Th17 cells in downregulating natural killer cell function.
IL-6 and TGF-β signaling of CD4+ T cells leads to the generation of CD4+ Th17 cells, which, following stimulation with IL-23, synthesize IL-22 and IL-17A. While IL-22 binds to its receptor expressed by gastrointestinal epithelial cells leading to the synthesis of IL-10, IL-17A is known to downregulate NK-cell function. Pathways that are presumably amenable to clinical manipulation with the use of inhibitors and blocking agents are highlighted.
NK: Natural killer; R: Receptor.
Mechanisms of NK-cell signaling & cytolysis
Ever since the discovery that a subset of lymphoid cells possess the potential to kill target cells without prior priming, efforts into defining the following questions have been intensified: how do these NK cells seek out their target; what is the nature of the dialog at the cell surface that generates the killing mechanisms; and what are the biochemical entities that mediate such killing. One of the areas of focus has been the study of the immune synapse involved in NK-cell killing. The NK cell immunological synapse is considered to be a complex and dynamic structure with differences in the types of proteins that accumulate that lead to a cascade of activation signals as compared with inhibitory signals [59]. Thus, the activating receptors involved in NK cell-mediated killing are known to accumulate at specific regions of the cell membrane via an actin-dependent process where they appear to induce synergistic signaling that finally translates into synthesis and secretion of cytotoxic granules, which cause death of the target cell. It has been reasoned that the inhibitory molecules within NK cells, such as KIRs also accumulate at inhibitory synapses and block actin dynamics and prevent actin-dependent phosphorylation of NK-cell activation receptors. Recently, however, it has been shown that KIRs, in fact, do not prevent the accumulation and colocalization of the activating receptors, such as CD2 and 2B4, with the KIRs at this synapse, but they function by inhibiting the ability of the activating receptors to signal within the inhibitory synapses [60]. Studies of downstream signaling events have led to the finding that NK cells utilize both the linker for activation of T cells (LAT) and B cells (LAB), providing NK cells with greater plasticity of transducing signals than T or B cells. Thus, the NK cells have the potential to use both the ITAM-Syk-LAB/LAT and the ITAM/ ZAP70/LAT signaling cassettes with a greater dependency on LAT when IL-2 is utilized for activation due to the suppression of Syk signaling by IL-2 [61]. This dual signaling potential provides NK cells with the flexibility to respond in a variety of environmental conditions. In terms of the biochemical nature of molecules involved in mediating cytolysis, the granules secreted by NK cells have been shown to contain perforin, granzymes and granulysin [62], and of interest, the Wiskott-Aldrich syndrome protein (WASp) interacting protein (WIP) has been shown to be involved in the transport of the lytic granules and, thus, is essential to NK cell cytolytic function [59]. It is important to keep in mind that most of these studies have been conducted on NK cells from the blood and whether the molecular nature of the synapse, its signaling and the downstream events are the same for all NK-cell subsets remains to be determined.
NK-cell subsets in nonhuman primates
A variety of species of NHPs became the focus of intensive studies ever since the discovery that a select species of these monkeys, primarily from Africa, are naturally infected with the SIV, which shares approximately 70% homology with the etiological agent of human AIDS [63]. These NHPs naturally infected with SIV do not show any detectable signs of disease or immune deficiency, even though they carry plasma and cellular viral loads that result in disease and death in the SIV infected non-natural hosts. The fact that the virus that originated from and/or was isolated from these SIV-infected African monkeys when used to experimentally infect Asian species of NHPs, such as rhesus or pig-tailed macaques, led to a spectrum of disorders and disease remarkably similar to HIV-1-infected humans [64], has led to their use as the primary animal model for the study of AIDS. To date, the list of naturally infected species originating from sub-Saharan Africa has grown extensively to some 40 such species so far identified [65]. It was documented that SIV from sooty mangabeys was a predecessor of HIV-2, which developed following the inter-species transmission to humans [66]. It just so happens that the naturally SIV-infected sooty mangabeys are one of the NHP species that is bred in captivity by the Yerkes National Primate Center of Emory University, GA, USA, and the only current source of sooty mangabeys in the USA. Sooty mangabeys thus became a focus of intense studies by a number of groups, including ours, offering a unique opportunity to delineate the processes that underlie SIV-disease resistance in this species. Recent interest in the role of NK cells in the response to HIV infection in humans prompted us, and others, to begin to characterize the NK-cell phenotypes in NHPs. Two major studies attempted to rigorously phenotypically characterize NK-cell populations in rhesus macaques and sooty mangabeys and correlate these phenotypes with their respective functions. Webster et al. used multiparametric analysis of surface markers expressed by peripheral blood mononuclear cell and demonstrated that rhesus macaque NK cells can be accurately identified as CD3−CD8brightCD20−/dim-expressing cells [67]. Further analysis of NK-cell markers typical for NK subsets in humans showed that within this NK-cell population there are three distinct subpopulations of cells. These include CD16+CD56−, CD16−CD56− and CD16−CD56bright populations. The major CD16+ subpopulation showed clear NK cell cytotoxic activity, while the two relatively minor CD16−subpopulations expressed homing markers, similar to the corresponding populations found in human NK cells and are reasoned to be a source of cytokines upon activation. It is important to note that while the predominant NK-cell subset is the CD16+CD56− subset in the peripheral blood, it is the CD16−CD56− and the CD16−CD56− (double negative subset) that are predominantly present in the LNs [68]. The second study conducted later on in our laboratory utilized a slightly different gating strategy. Thus, first of all, the gates for the size of cells based on forward and side scatter were opened wider to allow for large granular lymphocytes, which are included within the NK-cell lineage. Subsequently, the cells expressing CD3, CD14 and CD20 were gated out and the remaining population gated on the expression of CD8α/α and NKG2A for the analysis of peripheral blood mononuclear cell from both rhesus macaques and sooty mangabeys. The NK cells were thus identified as CD3−CD8α/α+, NKG2A+, which could be further divided, similar to the aforementioned study, into three subpopulations. These included the CD16+CD56−, CD16−CD56−and CD16−/dimCD56bright subpopulations. Interestingly, there was a significant increase in the frequency of the CD16dimCD56bright subset of cells consistently observed in sooty mangabeys as compared with rhesus macaques [69]. Similar cell-surface markers have been used by others to identify NK-cell subpopulations in other species, such as cynomolgous macaques and chimpanzees [70,71]. In the case of chimpanzees, it is of interest to note that this species is characterized as a species whose NK-cell subsets cannot be distinguished on the basis of CD56 bright versus dim [71], which is also probably the case in all NHP species. Furthermore, in the case of the chimpanzees, the NK-cell subsets have been subdivided into functionally different subsets based on the expression of CD8. Thus, the CD8+ NK-cell subset appears to express significantly higher levels of NKp46 than the CD8− subset. In addition, upon activation, while the CD8+ NK subset expressed IFN-γ, TNF-α, CD107 and increased densities of KIRs, the CD8− subset only expressed HLA-DR with no change in the density of KIR expression [71]. While these findings do document differences in NK-cell subsets based on phenotypic markers in the various NHPs, these differences may in fact be merely due to differences in reagents being utilized and the strategies used for identifying the subsets by the various laboratories. Clearly, attempts need to be made by a single laboratory using the same set of reagents and gating strategies in efforts to address the issue of whether these differences are in fact real or due to technical differences.
The fact that the GI tissues have been identified as the major target of both HIV and SIV infection during the acute infection period has prompted interest in the cells, including NK cells, that home to the GI tissues. This issue led our laboratory to study the expression of α4β7 gut-homing marker expressing CD4+ T cells and NK cells in the disease-resistant SIV-infected sooty mangabeys and the disease susceptible, SIV-infected rhesus macaques [69]. Flow cytometric analyses of peripheral blood mononuclear cell samples from a cohort of uninfected and SIV-infected sooty mangabeys and rhesus macaques revealed that neither NK cells as a whole or when isolated into subsets based on the differential expression of CD16 and CD56 expressed uniform relatively low levels of the α4β7 integrin. Thus, there was no subset of NK cells, unlike the central memory CD4+ T cells [69], that expressed high mean densities of the α4β7 integrin molecule. Table 2 summarizes our findings. As seen, while there were no major differences in the frequencies of total NK and the NK-cell subsets that expressed α4β7 within the species, there was a major difference (p < 0.0001) in the frequencies of α4β7 cells expressing NK cells in the sooty mangabey as compared with rhesus macaques. Of interest was the observation that whereas SIV infection did not lead to changes in the frequencies of α4β7 expressing cells in rhesus macaques, SIV-infected sooty mangabeys tended to show a lower frequency of total and NK-cell subsets that expressed α4β7. The significance of these data are not clear at present and studies are currently underway to determine whether such α4β7 expressing NK cells home to the gut or not in the two species. It is important to note that unlike CD4+ T cells, which upon incubation with retinoic acid show markedly increased mean frequencies and densities of α4β7 expression, such increases were not seen in NK cells after similar treatment [69]. These data suggest that since the GI tissues are known to release retinoic acid, which is one pathway that leads to upregulation of α4β7 on CD4+ T cells leading to their recruitment to the gut, this is probably not operational in the case of NK cells.
Table 2.
Frequency of α4β7+ major natural killer-cell subsets in uninfected and SIV-infected rhesus macaques and sooty mangabeys†.
| PBMCs from (n = 9each) | SIV status | CD3−CD8α/α+ NKG2a+ total | CD3−CD8α/α+ NKG2a+CD16+CD56− | CD3−CD8α/α+ NKG2a+CD16−CD56+ |
|---|---|---|---|---|
| RM | − | 55.1 ± 10.8 | 59.7 ± 9.8 | 35.7 ± 11.9 |
| RM | + | 55.6 ± 7.3 | 60.4 ± 7.2 | 39.1 ± 12.7 |
| SM | − | 30.4 ± 7.2 | 33.6 ± 8.4 | 23.1 ± 6.4 |
| SM | + | 23.6 ± 6.4 | 24.5 ± 4.7 | 16.8 ± 5.7 |
NK cells in humans: receptors & the diversity of KIRs
The majority of NK cells possess one critical function – non-HLA-restricted killing of infected or malignant cells. While at the beginning this function was thought to be regulated simply by the absence of self MHC on target cells (consistent with the missing self hypothesis), it turned out that it is regulated by a complex array of activating and inhibitory signals delivered by an ever growing family of NK-cell receptors summarized in Tables 3 & 4. The net effect is then dictated by the prevailing signaling mode – activating or inhibitory. This article will focus only on select receptor families; for a more complete description of known NK-cell receptors the reader is referred to several excellent reviews [72–74].
Table 3.
Select activating natural killer cell receptors, their ligands and function.
| Family | Receptor | Ligand | Function |
|---|---|---|---|
| NCR | NKp30 (CD377) | BAT3 | Surveillance of cell transformation |
| NKp46 (CVD335) | Viral HA | Surveillance of dividing cells | |
| CD16 | IgG | Antibody-dependent cell cytotoxicity | |
| KIR | KIR2DS1–2 | HLA-C | ? |
| KIR2ds3–6, KIR3DS | ? | ? | |
| KIR2DL4 (CD158d) | HLA-G (soluble) | Maternal–fetal interaction | |
| NKG2D (CD314) | ULBP, MICA, MICB | Surveillance of tumor cells, stress | |
| NKG2C (CD159c)/CD94) | HLA-E | ? | |
| 2B4 (CD244) | CD48 | Cell interactions | |
| Integrin | CD11a,b (α4β1, αMβ2) | ICAM, fibrinogen | Cell recruitment and activation |
| CD11c | ICAM, iC3b | Cell recruitment and activation | |
| CD49d,e (α4β1, α5β1) | VCAM, fibronectin | Cell recruitment and activation |
Table 4.
Select inhibitory natural killer-cell receptors, their ligands and function.
| Receptor | Ligand | Function |
|---|---|---|
| IL-T2/LIR-1 (CD85j) | HLA class I | Survey loss of HLA I |
| KIR3DL1 | HLA-B | Survey loss of HLA I |
| KIR3DL2 | HLA-A | Survey loss of HLA I |
| KIR2DL1, 2/3 (CD158a,b) | HLA-C | Survey loss of HLA I |
| NKG2A (CD159a/CD94) | HLA-E | Survey loss of HLA I |
| KLRG | Cadherins | Survey loss of tissue integrity |
| Siglec-7,9 (CD328,329) | Sialic acid | ? |
| LAIR-1(CD305) | Collagen | Control extracellular activation |
Among the NK-cell receptors, the CD16 molecule has received considerable attention. There are, in addition, three larger receptor families and a number of other signaling molecules, such as TRAIL, 2B4 (CD244), SIGLECs and others, expressed by NK cells. CD16, as mentioned previously, is a receptor for Fcγ and plays an important role in ADCC. The three major receptor families are the KIR, natural cytotoxicity receptors (NCRs) and C-lectin type receptors of NKG2 superfamily. The NCR represent a relatively well-defined family of nonpolymorphic activating receptors, such as NKp30, NKp44 and NKp46. The natural ligands for the NCR remain undefined and are potentially thought of as receptors for viral proteins such as hemagglutinins. The NKG2 family of molecules include NKG2A, B, C, E and F molecules, which are expressed primarily as dimers and have been shown to serve as both activating and inhibitory receptors based primarily on the nature of the molecule they dimerize with. Thus, NKG2A is expressed as a heterodimer with CD94 and induces an inhibitory signal upon ligation [75]. The major known ligands for the NKG2A/ CD94 heterodimer are the relatively less polymorphic nonclassical MHC class I molecules, such as HLA-E in humans [76]. The other known members of the NKG2 family possess activating function and are expressed as heterodimers with CD94, except for NKG2D. This receptor engages ligands such as the nonclassical MHC class I molecules MIC-A, MIC-B and ULBP, which are typically expressed on cells undergoing stress [77].
The third relatively large family of NK receptors is the KIRs. These receptors, unlike most other NK-cell receptors, are highly polymorphic, similar to HLA. Ligands for KIRs are the classical MHC class I molecules, such as HLA-A, B and C, and can perform both inhibitory and activating functions. Structurally, the KIR receptors consist of the extracellular portion, containing one or more Ig-like domains, the transmembrane part and the cytoplasmic tail, which performs the signaling function. The nomenclature of KIR molecules is based on the number of the extracellular Ig-like domains (i.e., one, two or three) and the length of the cytoplasmic tail (i.e., long or short). Thus, for example, the KIR1D, KIR2DL and KIR3DL receptors have one, two or three Ig-like domains, respectively, and a short or long cytoplasmic tail reflected as KIR1DS or KIR3DL, respectively. The cytoplasmic tail of the inhibitory KIRs contains immunoreceptor tyrosine-based inhibitory motifs (ITIMs), which upon signaling engage phosphatase (SHP-1), which then precludes the delivery of an activating signal from the neighboring activating receptors (provided in the form of activation by phosphorylation). The signaling by the activating KIRs is performed either as a result of the absence of the inhibitory signal (KIRs with the short tail), or by an unknown mechanism (KIR2DL4). The human KIR gene complex is located on chromosome 19q13.4. To date, and according to the current nomenclature, 15 different KIR gene loci are known and the KIR haplotypes are then defined by the absence or presence of these loci (reviwed in [78]). Adding to the diversity and complexity is the fact that each of these loci can be represented by different alleles, which may exhibit varying capacities to bind the ligand or transmit the signal, in addition to the duplication or deletion of individual genes. The KIR genes are inherited basically in two broad haplotypes – A and B. Both haplotypes contain four ‘framework’ gene loci – KIR3DL3, KIR3DL2, KIR2DL4 and KIR3DP1 (the P representing a pseudogene), which are almost always expressed except for a few rare cases of gene deletion. The A haplotype is characterized by the presence of up to four additional gene loci – KIR2DL1, KIR2DL3, KIR 2DS4 and KIR 3DL1. Most of the A haplotypes currently characterized contain all eight gene loci and the variability of this haplotype is mostly at the level of the allelic content for the individual loci. The B haplotype is defined by the presence of framework genes and one or more loci coding for the activating KIR molecules, such as KIR2DS1/2/3/5 and KIR3DS1, or the inhibitory KIR2DL5A/B and KIR2DL2 receptors. The variability of this haplotype is thus mostly achieved by the selection of loci, which are expressed creating many different combinations. The individual KIR receptors also appear to have variable degrees of affinity for binding to their cognate MHC class I ligands. Thus, the KIR2DL1, 2 and 3 gene products bind to HLA-C, but with varying affinity depending on the presence of asparagine or lysine at position 80 in the HLA-C molecule. KIR3DL1 exhibits affinity for the HLA-Bw4 epitope. Of interest is the finding that the KIR2DL4 is fairly conserved among species and has HLA-G as its ligand [79]. Interestingly, there seems to be a hierarchy of the signal strength and subsequent effector response to the cells lacking the self-MHC class I determinants between the different NK-receptor families, with the inhibitory KIR delivering the strongest signals [80]. It is also known that soluble forms of HLA-E and -G are present in the plasma at varying levels, with high levels being expressed during pregnancy in women [81,82]. The precise role such soluble HLA-E and -G molecules play in enhancing or blocking NK-cell function remains to be determined.
KIRs in NHPs
With increased interest in NHPs as models for a variety of human diseases, especially AIDS, came the need to characterize the immune system in more detail than previously documented. First reports focused on characterization of the NK activity in the peripheral blood of NHP [83]. Subsequently, the NK-cell receptors started to be characterized in more detail. This characterization included the studies of the NCR receptors NKp46 and NKp30 of Macaca fascicularis, which were shown to possess a relatively high degree of sequence homology and functional similarity to their human homologs [84]. LaBonte et al. characterized the CD94 and the NKG2 family members in rhesus macaques [85], and in subsequent reports presented the characteristics of the other members of NK cell and NCR families expressed by cells from M. mulatta and M. fascicularis [70,86]. The KIR receptors in NHP received more attention than the other NK-cell receptors, although they are not as well defined as in humans. The first study from Herschberger utilized expression cloning to characterize five families of KIR receptors in rhesus macaques – KIR3DL, KIR2DL4, KIR2DL5, KIR3DH and KIR1D [87]. Although the function of these receptors was not assessed, it is presumed that those, as homologs to the human counterparts, exhibit similar function. KIR3DH is a hybrid molecule consisting of the extracellular domains of the KIR3DL molecule and the KIR2DL4 tail containing a stop codon, which terminates the tail early, and the molecule is therefore presumed to have an activation function. KIR1D is a molecule rarely detected, which lacks the transmembrane and tail domains. The KIR receptor expression was subsequently characterized in other primate species, such as sebaeus monkeys [88], orangutans [89] and owl monkeys [90]. Several studies attempted to analyze the KIRs at the genomic level and defined several haplotypes in primates. The first haplotypes were described by Sambrook et al. in chimpanzees, rhesus macaques and orangutans [89,91], indicating rapid diversification of this particular locus in primates. The subsequent elegant study utilized relatively small and isolated population of Mauritian cynomolgous macaques and reported that there were in fact a total of at least eight loci in this species, containing loci for genes previously defined by Herschberger [92]. Further analysis from our laboratory performed on a cohort of 38 rhesus macaques has led to the identification of several alleles and variants for each of these loci [46,93]. Two more recent reports by Blokhius et al. [94] and Kruse et al. [95] used a combined approach consisting of both the cDNA analysis and genomic DNA analysis in the families of rhesus macaques present at the respective primate centers in the The Netherlands and Germany to identify potential haplotypes present in this species. The analysis of KIR genomic sequences, together with expressed polymorphic KIR sequences, led to the identification of 14 and 21 putative haplotypes, respectively. The haplotypes consisted of both inhibitory and activating KIR genes with 5–11 KIR genes per haplotype, with the finding that a given gene may be present on one halpotype but not on another. These studies also confirmed that some KIR genes, specifically the KIR3DL gene, exhibit great variability with regard to their duplication. In addition, these data underscore the high degree of diversity and complexity of KIR genes in rhesus macaques similar to that previously published for humans. These recent findings provide a foundation for the establishment of KIR genotyping and potentially for the identification of alleles for each of these genes with the hope to define the influence of the haplotypes, genes and alleles on the pathogenesis of disease, such as SIV.
Effect of lentivirus infection on NK cells & their potential role in virus control
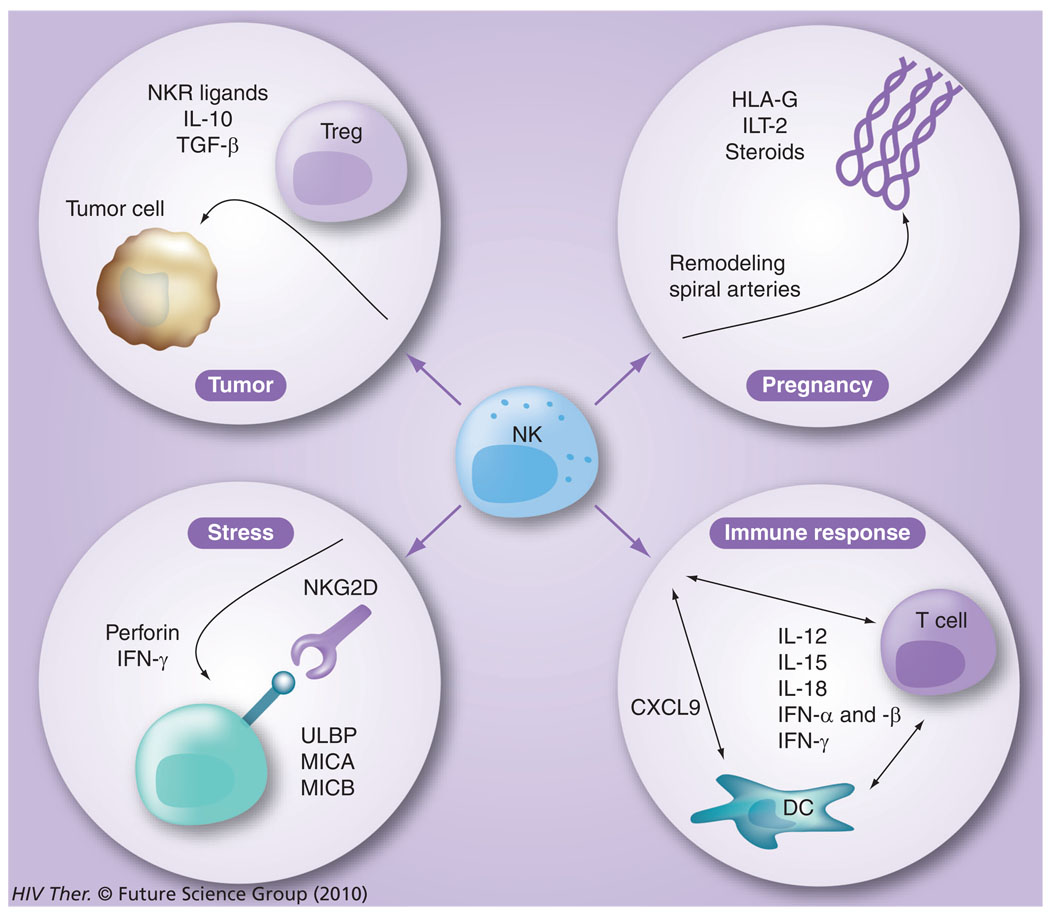
Besides the capacity of the NK-cell lineage to be involved in mediating immune responses against virally infected cells, they are also known to be involved in the suppression of cancer cell growth/immune surveillance, as well as their role in pregnancy and response to stress (Figure 7). Their role in influencing antiviral immunity is highlighted by the finding of recurrent herpetic infections of patients that have a genetically based disorder that leads to the absence of functional NK cells [96]. Therefore, not surprisingly, NK cells have recently been shown to play a significant role in HIV-1 infection and, conversely, HIV affects the function of NK cells. Proliferation and activation of NK cells are induced early after the infection and results in both killing of the infected cells and secretion of massive amounts of IFN-γ, TNF-α and chemokines, which subsequently drives the T-helper 1 antigen-specific T- and B-cell responses [97]. The importance of these early events associated with NK cells in HIV infection has been highlighted by the fact that the kinetics of viremia shows an eclipse phase (lasting from day 0 to day 7–9), a log phase (occurring from day 7–21) and the viral load set point phase (>21–35 days postinfection), a time period during which the adaptive immune responses have not had the time to develop [98]. Thus, the fact that viral load shows a sharp decrease following the log phase clearly indicates a prominent role for effector cells that comprise the innate immune system and most likely include NK cells. Hence, the role of the innate immune system, in particular the NK-cell lineage, has now taken center stage in terms of importance as a research target for the study of the pathogenesis of human HIV and NHP SIV infections.
Figure 7. The various functions of natural killer cells.
NK cells are not only involved in elimination of virus-infected cells and influencing the generation of adaptive immune responses, they are also involved in tumor surveillance, pregnancy and stress responses.
DC: Dendritic cell; NK: Natural killer; NKR: NK receptor; Treg: T-regulatory cell.
Since the 1980s, it has been well documented that HIV-1 infection leads to significant decreases of NK-cell function in infected individuals and that the institution of ART leads to varying degrees of normalization of some of the affected NK-cell functions [99]. The fact that HIV-1-infected chimpanzees who rarely develop disease maintain the phenotype and function of NK cells suggests that perturbation of NK cells is a function of pathogenic but not apathogenic infection [71]. NK cells employ several mechanisms to control infection by viruses, including HIV (reviewed in [100,101]). First, they can eliminate the infected cells by direct cytotoxicity. This would presumably be mediated by a decrease and/or loss of MHC class I expression or induction of surface stress markers on the infected cells, which would both activate the NK-cell function. Interestingly, HIV proteins Vpr and Nef have been shown to induce the expression of stress markers MICA/B and downregulate the expression of MHC class I, respectively. However, at the same time, Nef also possesses the capacity to inhibit the expression of stress markers and, while it clearly downregulates HLA-A and B, it usually leaves the expression of HLA-C intact [102,103]. Clearly, these effects can lead to the escape of the infected cells from NK surveillance, but in addition, NK cells from patients with HIV viremia showed decreased cytotoxic function. NK cells can also kill the infected cells by ADCC and several studies have documented a decrease of ADCC associated with HIV infection [104–107]. In this regard, it is of interest to note that HIV-1 infection has been shown to induce the release of matrix metalloproteinases that cleave CD16 from the cell surface of NK cells, leading to their inability to execute ADCC function and contribute to the overall immune dysfunction in HIV-1-infected individuals [108]. Furthermore, NK cells, when activated, secrete various cytokines and chemokines, including CCL3, 4 and 5, which are ligands for the HIV-1 coreceptor CCR5, and can thus block virus entry by binding this coreceptor. The ability to secrete these chemokines was also shown to be decreased in HIV-infected individuals [109]. As previously mentioned, NK cells represent an important regulatory role affecting the development of antigen-specific responses by secreting the cytokines as well as by their cross-talk and regulation of DCs, which are important in antigen presentation. Therefore, any dysregulation of NK cells by HIV clearly affects these critical functions as well. Furthermore, NK cell-mediated response, unrestricted by previous exposure to the antigen, represents a major line of defense preventing the entry of the virus and acquiring the infection.
A number of studies have been conducted that are focused on changes in the NK-cell subsets secondary to HIV-1 infection. Thus, a progressive decrease in both the CD56dim and CD56bright subset with a concomitant increase in the CD16+CD56− subset was noted following HIV-1 infection, with a major effect primarily in the CD56bright subset [110], which is partially reversed by HAART. A more recent study attempted to characterize differences in the major NK-cell subsets based on clinical status (progressors versus long-term nonprogressors [LTNPs] and elite controllers) and viral loads utilizing CD16 and CD56 as markers and a large-scale analysis of 21 different phenotypic markers [111]. Results of these studies indicated:
- ▪
The CD3−CD56dim NK cells from elite controllers showed a profile distinct from those displayed by the same subset from LTNPs and progressors; - ▪
The CD56bright subset profile was similar in samples from elite controllers and LTNPs than those from progressors and controls; differences in viral loads influenced the profile of the CD16+CD56− subset; - ▪
The NK receptor profiles expressed by CD56dim and CD56bright subsets were distinct in the elite controllers and LTNPs as compared with the progressors and control subjects; - ▪
A key role for NKp44L in mediating cytolysis of autologous CD4+ T cells was noted in samples from progressors and LTNP as compared with elite controllers and controls.
These data appear to suggest that HIV infection induces distinct changes in the NK-cell subsets of HIV-1-infected individuals based on clinical outcome and viral loads. It would be important to conduct similar studies in SIV-infected rhesus macaques as a function of time postinfection, viral loads and disease outcome, with the objective to determine whether these changes occur during the acute infection period or are a consequence of chronic infection. Use of additional markers, such as the expression and density of CD57 on NK cells in this regard may also add to our knowledge since a recent study noted that HIV-1 infection is associated with a preferential loss in the frequency of the less differentiated CD56dimCD16+ NK-cell subset [112]. Additional effects of HIV infection on the NK-cell compartment include perturbations of not only NK-cell subsets but also the expression of activating/inhibitory receptors. The former effect is characterized by a decrease in CD16+CD56low cells and selective proliferation of the CD16+CD56− subset, which expresses higher density of inhibitory KIRs and is dysfunctional as measured by both cytotoxicity and cytokine secretion [113,114]. While the reported studies differ in their assessment of overall expression of the inhibitory KIR receptors in HIV-infected individuals (showing increase in some and no detectable change in others), there is clear evidence that the HIV infection leads to an overall decrease of activating NCRs, which include NKp30, NKp44 and NKp46 [115].
All the mechanisms presented previously show clear evidence that NK cell-mediated immune function is indeed important for the control of HIV and the fact that HIV-1-infected patients go on to develop chronic viremia suggests that the virus employs various strategies to evade NK-cell responses. Another line of evidence comes from the genetic-based studies, which document an association of certain combinations of KIR alleles and HLA class I haplotypes with altered course of HIV-1 infection. Thus, the HLA-B*57 expression in combination with certain KIR3DL1 allotypes has been shown to associate with a favorable course of HIV infection [116]. Results from a large epidemiological study performed by Martin et al. has shown that individuals who express a certain allele of the activating KIR3DS1 receptor in combination with HLA-Bw4081 (a variant of HLA-Bw4) exhibit significantly slower progression than those expressing only one of them [117]. Interestingly, another study has shown an association of the expression of certain alleles of the inhibitory KIR3DL1, with a favorable course of the HIV disease [17]. The mechanism of this effect is unknown, but potentially, the fact that different KIR3DL1 alleles show differential rates of surface expression could be involved.
In SIV infection, multiple effects on NK-cell function have also been observed. La Bonte et al. demonstrated decreased ability of NK cells from SIV-infected rhesus macaques to secrete IFN-γ, TNF-α and IL-2 [118]. However, the long-term nonprogressor animals retained this ability at a level similar to that noted in uninfected monkeys. This decrease in NK-cell function correlated with a decrease in detectable transcripts for NKG2C. The study conducted in our laboratory has demonstrated that the CD16+CD56- cytolytic NK-cell subset was preferentially depleted in the SIV-infected rhesus macaques, but, of interest, increased in the disease resistant SIV-infected sooty mangabeys [69]. These findings further underlined the importance of early NK-cell responses, showing early NK-cell activity in SIV-infected sooty mangabeys and those SIV-infected rhesus macaques, which eventually controlled viral load during the chronic infection. While the aforementioned data point towards the importance of NK cells in the immune response to SIV infection, surprisingly, Choi et al. have shown that the experimental depletion of NK cells during either chronic or primary infection by the in vivo administration of monoclonal antibodies to CD16 did not lead to overall changes in the levels of SIV viremia in chronically infected rhesus macaques and minimal effects during primary infection [119]. However, there are several caveats to these findings and further studies are needed in order to resolve the issues involved. First of all, it is well recognized that CD16 is not expressed by all NK cells. Second, the degree of depletion in the mucosal tissues was not studied and since SIV primarily targets the mucosal lymphoid tissues, it would have been important to document if the NK cells were indeed depleted at this site. Third, use was made of a murine xenogeneic antibody that is immunogenic in monkeys and makes it difficult to evaluate the role of the xenogeneic response in concert with the effect of CD16+ cell depletion. The recent findings by our laboratory of a strong association between select KIR loci alleles and levels of SIV viremia in addition to the analysis of gut-associated NK cells suggest a re-evaluation of the potential role of NK cells during acute SIV infection is in order.
Therapeutic reconstitution of NK cells
The general finding of a deficiency in NK-cell function in both HIV-infected humans and SIV-infected non-natural hosts of SIV has prompted the need for identifying strategies aimed at reconstituting this cell lineage, which are based to a large extent on data derived from strategies that have already been utilized in patients with malignancies. Lessons learnt from these studies in cancer patients have therefore provided a note of caution, which is outlined below. Chief amongst these strategies has been the use of the cytokines that share the common γ chain, which includes the cytokines IL-2, IL-15 and IL-21 and others have included the use of IL-12 and IL-18. Of these cytokines, IL-2 has been the major cytokine used clinically. The results from the use of these cytokines have been mixed. The reasons for such mixed results have been attributed to the finding that: these cytokines have pleiotropic effects, which lead to different effects on different patients; the finding that cytokines such as IL-2 preferentially activate and expand Tregs in addition to NK cells [120]; most of these cytokines have associated toxicity chief among these being the ‘vascular leakage syndrome’ with IL-2 and toxicities with the use of other cytokines and immunostimulating agents [121,122]; and the dosing regimens that have maximal therapeutic effects have been difficult to define. Thus, while cytokines such as IL-2, IL-15 and IL-21 show great promise when used in vitro for the expansion of NK cells or in vivo in the SCID-Hu murine model [39,123,124], they have to be viewed as potentially interesting but will require modifications to reduce their inherent toxicity prior to their use in HIV-infected patients. The other strategy worth consideration is to expand the NK cells from the SIV-infected NHPs or HIV-1-infected humans in vitro and then use these for autotransfusions. In the case of NHPs, one could utilize cryopreserved cells obtained prior to experimental SIV infection to ensure the ‘normal’ nature of the expanded cells being used for autotransfusion. In addition, with the knowledge of subsets of NK cells it may be worthwhile to expand specific subsets and determine their therapeutic efficacy. In the case of HIV/SIV infection where the GI tract is the major target of infection during acute infection, it may be worthwhile to explore the potential use of _in vitro_-expanded gut resident NK-22 cells as an autotransfusion therapeutic tool. Conversely, the potential role of the NK cells and its subsets in conferring protection from disease in the natural hosts of SIV, such as sooty mangabeys and African Green monkeys, may be warranted.
Conclusion
The important role of NK cells in influencing the quality and quantity of both host innate and adaptive immune responses is beginning to be better understood [125]. This view has led to a new appreciation for a major role for the innate immune system on influencing the pathogenesis of human HIV-1 infection and SIV infection in the NHP models of HIV-1 infection. The fact that the GI tract becomes the primary target of HIV and SIV infection during the acute infection period, regardless of the route of infection, clearly dictates a need for a more thorough evaluation of how the cells of the innate immune system mediate their effect within the GI system. The NK cells are a major component of the innate immune system and the subject of this article. While considerable knowledge has been acquired on the heterogeneity of this cell lineage, the maturation and differentiation of NK cells, their effector mechanisms and the mechanisms by which their function is regulated, one of the major areas of focus has been the role of select cell-surface receptors termed KIRs. While considerable knowledge of KIR polymorphisms has been acquired in humans and their potential role in HIV-1 infection, unfortunately, there is limited data on the genes that code for KIRs in rhesus macaques and how these polymorphisms influence the early events of SIV infection. The new discovery of a NK-cell lineage, which is unique to the GI tract and synthesizes IL-22 (NK-22) following stimulation by IL-23, has added a new dimension to our understanding of the potential role of the NK cells in HIV/ SIV infection. At present, it is not clear whether peripheral NK cells traffic to the GI tissues and are part of the mechanisms of pathogenesis within the GI tract, although a significant frequency of NK cells do express the gut-homing α4β7 integrin. It appears that the gut resident NK-cell lineage is the likely candidate that plays an important role in maintaining homeostasis within the GI tract and either its function or the function of gut-homing NK cells influence the early events involving HIV/SIV infection. It is also important to keep in mind that other cell lineages, such as the plasmacytoid DCs are probably involved in either directing NK-cell function and/or modulating their function. Thus, a lot needs to be learnt of the role of these innate immune cell lineages within the GI tract, which can only best be learned in the SIV-infected NHP model. In this regard, it would be of great importance to understand the role of these innate immune cell lineages in rhesus macaques infected with predominantly nondisease-inducing attenuated viruses, such as SIVΔnef and the deglycosylated recombinant SIVΔ5G viruses. In addition, similar studies of these innate immune cell lineages in disease-resistant natural hosts of SIV during the acute infection period is likely to contribute greatly to our knowledge of how these cell lineages potentially contribute to disease resistance.
Future perspective
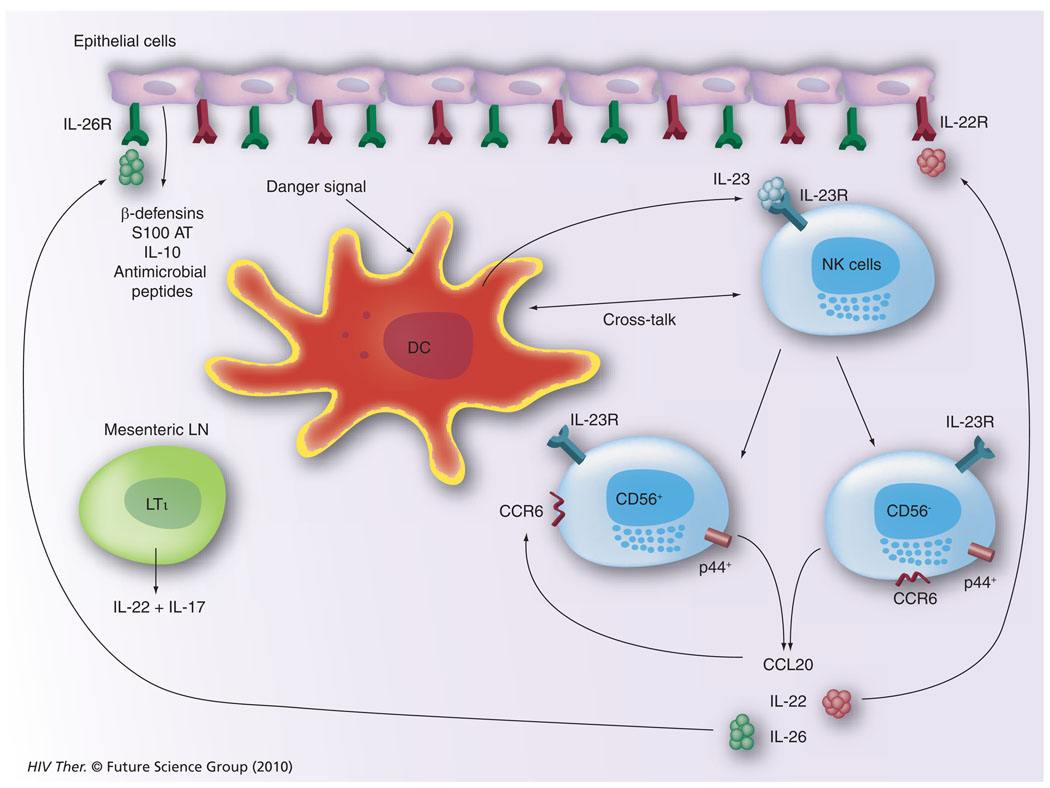
Our current understanding of the potential mechanisms by which cells of the innate immune system, such as the NK cell, function within the GI tissues is illustrated in Figure 8. It appears that GI tissue resident DCs or similar cell lineages initiate the cascade of immune reactivity by sensing a pathogenic organism, such as HIV/SIV. This leads to the synthesis of IL-23 (a cytokine belonging to the IL-12 family), which induces the activation of IL-23R–expressing NK cells. These NK cells can give rise to p44+, CD56+, CCR6+ or p44+, CD56− CCR6+ NK cells, which when activated synthesize CCL20, IL-22 and IL-26. The CCR6 is the cognate ligand for CCL20, which thus serves to self perpetuate its synthesis. Receptors for the cytokines IL-22 and IL-26 are uniquely expressed by gut epithelial cells and the ligation of these receptors leads to the synthesis of a large array of antimicrobial agents, including β defensins, S100 AT, IL-10 and a series of other antimicrobial peptides. Such a cycle of innate immune activation and epithelial cell synthesis of antimicrobial agents, including IL-10, keeps the invading organisms in check and maintains homeostasis in the GI tissues. There is also a rare frequency of lymphoid tissue inducer cells particularly resident within mesenteric LNs that are distinguished from NK cells by their ability to synthesize IL-17. Clearly, during acute HIV/SIV infection, this homeostatic balance is perturbed giving rise to GI pathology that can be reduced but clearly not completely reversible. Thus, the question that needs to be addressed is what are the changes that are induced by HIV/SIV infection in this cycle of dialog within the GI tissues and how can we best reverse this pathogenic event and return the GI tract back to function in a homeostatic manner?
Figure 8. Our current view of the interplay between natural killer cells, dendritic cells and the cytokine/chemokine network within the gastrointestinal tissues that leads to the generation of a variety of antimicrobial agents synthesized by the gastrointestinal epithelial cells.
DC: Dendritic cell; LN: Lymph node; R: Receptor.
There appear to be several avenues of approach that are likely to provide unique insights and be of therapeutic value. First of all, the important point to keep in mind is that clearly such irreversible pathology does not occur in natural hosts of SIV that appear to live to a large extent very normal lives despite plasma viral loads that exceed those that result in AIDS and death of the non-natural hosts of SIV. Thus, some major distinguishing event must occur within the GI tract of the natural as compared with the non-natural hosts of SIV. This reversible GI pathology also appears to likely occur in adult non-natural hosts, such as rhesus macaques, but not infant rhesus macaques infected with SIVΔnef and presumably in other models of attenuated recombinant SIV, such as the deglycosylated SIVΔ5G. Thus, a lot can be learnt from the differences of how the natural hosts versus non-natural hosts of SIV, and the recipients of SIVΔnef and SIVΔ5G as compared with wild-type and infant rhesus macaques, handle SIV infection within the GI tissues. It is reasoned that inhibiting trafficking of CD4+ T cells to the GI tissues is one major avenue of therapeutic approach that should limit the seeding of new targets of lentiviral infection and replication. This scenario assumes that the natural continuous trafficking of CD4+ T cells to the GI tissues following HIV/SIV infection not only contributes to gut pathology but also adds fire to the flame and therefore needs blocking. It is also hypothesized that perhaps plasmacytoid DCs that become mobilized and activated during acute HIV/SIV infection may home to the gut as would the α4β7 expressing NK cells, which probably results in the synthesis of α-IFN, which causes local immune activation and while reasonable for a short time period may in fact lead to overt pathology and contribute to GI tissue dysfunction. Thus, the trafficking of these plasmacytoid DCs and α4β7 expressing NK cells needs to blocked. If the GI tissues are spared from overt pathology, there would be an increased probability for the generation of effective HIV/SIV neutralizing antibodies and HIV/SIV-specific cell-mediated immune responses, especially within the mucosal tissues following vaccination (preventive vaccination). It is hypothesized that the administration of HAART-based therapy to HIV/SIV infected individuals following blocking of gut homing cell therapy may lead to the generation of more effective HIV/SIV-specific immune responses (therapeutic vaccination) and also limit the generation of ‘viral reservoirs’. The potential role of TH17 cells needs to be explored within this context. Thus, while TH17 cells have been generally viewed as being proinflammatory and potentially promoting cell activation and thereby increasing viral replication, there is also the potential role of TH17 cells in potentially maintaining gut tissue integrity and promotion of the synthesis of nonspecific antimicrobial agents. These findings seem to suggest a delicate balance between gut tissue integrity as compared with promoting increased viral replication. Strategies aimed at maintaining gut tissue integrity while limiting TH17 proinflammatory immune responses need to be explored.
Another approach could be to determine if the generation of immune tolerance to HIV/SIV can be successfully achieved. Thus, it seems that the natural hosts of SIV seem to live quite a normal life despite high viral loads. It seems that the natural hosts have developed ‘immunological accommodation’ against SIV. It would therefore seem logical that approaches to achieve similar ‘immunological accommodation’ against SIV in otherwise disease-susceptible Asian rhesus macaques need to be studied and exploited. This approach would clearly require a firm understanding of the mechanisms of ‘immunological accommodation’.
Executive summary.
- ▪
Innate immunity probably plays an important role during acute infection in both HIV/SIV infection. - ▪
Natural killer (NK) cells represent one of the key effectors of the innate immunity and the data suggest a correlation between NK function and AIDS progression. - ▪
NK-cell population consists of specialized subsets of cells, which can perform diverse effector functions (i.e., target cell lysis, cytokine secretion, play a role in embryo implantation and others). - ▪
Contrary to previous beliefs, NK cells exhibit ‘memory’ and require education. - ▪
NK cells are regulated by complex array of activating and inhibitory signals generated by several families of receptors. - ▪
Killer Ig-like receptors (KIRs) represent a diverse set of receptors and can deliver both inhibitory and activating signals. - ▪
KIRs engage MHC I class molecules and deliver mostly negative signals; in the absence of MHC I (‘missing self’), caused, for example, by virus-induced MHC I downregulation, negative signals are missing and NK cells target the cells for lysis. - ▪
Loss of NK-cell function inversely correlates with AIDS disease progression in both humans and the nonhuman primate models. - ▪
Certain genotypic characteristics of KIR and MHC I are significantly associated with an improved prognosis of lentivirus infection. - ▪
A potential important role for gut-resident IL-22 synthesizing NK cells is visualized in the pathogenesis of HIV/SIV infection during the acute infection period. - ▪
There is an association between plasmacytoid dendritic cells and NK cells within the mesenteric lymph nodes and probably within the gut tissues, and the relative levels of this interaction most likely dictate the course of HIV/SIV disease. Thus, an effective transient response leads to return to homeostasis but prolonged responses probably contribute to irreversible pathogenesis.
Clearly, the classical approach to generate an effective vaccine against HIV-1 has failed. It seems reasonable that in the next decade methods to reduce GI pathology will have been achieved and, if combined with effective HAART under the cover of such gut pathology inhibiting therapy, may lead to a therapeutic option in terms of letting the infected individual generate an effective immune response and thus a therapeutic vaccine. In addition, a similar approach of treating vaccine recipients with agents that would prevent gut homing coupled with a better gut tissue immune-adjuvanted vaccine may be developed (i.e., a preventive vaccine). It is also clearly possible that genes that affect the course of HIV and SIV infection and disease will have been identified, which could serve as a basis for exploitation for better vaccine formulations.
Acknowledgments
This work was supported by NIH NIAID RO1 AI078773 and by P304/10/1161 from the Grant Agency of the Czech Republic.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Christensen JE, Thomsen AR. Co-ordinating innate and adaptive immunity to viral infection: mobility is the key. APMIS. 2009;117:338–355. doi: 10.1111/j.1600-0463.2009.02451.x. [DOI] [PubMed] [Google Scholar]
- 2.Gregoire C, Chasson L, Luci C, et al. The trafficking of natural killer cells. Immunol. Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum KS, Pabst R. Lymphocyte numbers and subsets in the human blood. Do they mirror the situation in all organs? Immunol. Lett. 2007;108:45–51. doi: 10.1016/j.imlet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Kiessling R, Klein E, Pross H, et al. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 5.Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur. J. Immunol. 1975;5:112–117. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 6.Janeway CA. Natural killer cells: a primitive immune system. Nature. 1989;341:108. doi: 10.1038/341108a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 8.Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Hayakawa Y, Screpanti V, Yagita H, et al. NK cell TRAIL eliminates immature dendritic cells in vivo and limits dendritic cell vaccination efficacy. J. Immunol. 2004;172:123–129. doi: 10.4049/jimmunol.172.1.123. [DOI] [PubMed] [Google Scholar]
- 10.Piccioli D, Sbrana S, Melandri E, et al. Contact-dependent stimulation and inhibition of dendritic cells by natural killer cells. J. Exp. Med. 2002;195:335–341. doi: 10.1084/jem.20010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda K, Dennert G. The development of autoimmunity in C57BL/6 lpr mice correlates with the disappearance of natural killer type 1-positive cells: evidence for their suppressive action on bone marrow stem cell proliferation, B cell immunoglobulin secretion, and autoimmune symptoms. J. Exp. Med. 1993;177:155–164. doi: 10.1084/jem.177.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L, Ikizawa K, Hu D, et al. Regulation of activated CD4+ T cells by NK cells via the Qa-1-NKG2A inhibitory pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Leary JG, Goodarzi M, Drayton DL, et al. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 14.Sun JC, Lanier LL. Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur. J. Immunol. 2009;39:2059–2064. doi: 10.1002/eji.200939435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alter G, Malenfant JM, Delabre RM, et al. Increased natural killer cell activity in viremic HIV-1 infection. J. Immunol. 2004;173:5305–5311. doi: 10.4049/jimmunol.173.8.5305. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor GM, Holmes A, Mulcahy F, et al. Natural killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin. Immunol. 2007;124:277–283. doi: 10.1016/j.clim.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritz J, Schmidt RE, Michon J, et al. Characterization of functional surface structures on human natural killer cells. Adv. Immunol. 1988;42:181–211. doi: 10.1016/s0065-2776(08)60845-7. [DOI] [PubMed] [Google Scholar]
- 19.Lanier LL, Testi R, Bindl J, et al. Identity of Leu-19 (CD56) leukocyte differentiation antigen and neural cell adhesion molecule. J. Exp. Med. 1989;169:2233–2238. doi: 10.1084/jem.169.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 21.Vossen MT, Matmati M, Hertoghs KM, et al. CD27 defines phenotypically and functionally different human NK cell subsets. J. Immunol. 2008;180:3739–3745. doi: 10.4049/jimmunol.180.6.3739. [DOI] [PubMed] [Google Scholar]
- 22.Fehniger TA, Cooper MA, Nuovo GJ, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 23.Ferlazzo G, Munz C. NK cell compartments and their activation by dendritic cells. J. Immunol. 2004;172:1333–1339. doi: 10.4049/jimmunol.172.3.1333. [DOI] [PubMed] [Google Scholar]
- 24.Romagnani C, Juelke K, Falco M, et al. CD56brightCD16 killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J. Immunol. 2007;178:4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 25.Ferlazzo G, Thomas D, Lin SL, et al. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 26.Bradley TP, Bonavida B. Mechanism of cell-mediated cytotoxicity at the single cell level. I V. Natural killing and antibody-dependent cellular cytotoxicity can be mediated by the same human effector cell as determined by the two-target conjugate assay. J. Immunol. 1982;129:2260–2265. [PubMed] [Google Scholar]
- 27.Maghazachi AA. Role of chemokines in the biology of natural killer cells. Curr. Top. Microbiol. Immunol. 2010 doi: 10.1007/82_2010_20. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 28.Yagel S. The developmental role of natural killer cells at the fetal-maternal interface. Am. J. Obstet. Gynecol. 2009;201:344–350. doi: 10.1016/j.ajog.2009.02.030. [DOI] [PubMed] [Google Scholar]
- 29.Kwak-Kim J, Park JC, Ahn HK, et al. Immunological modes of pregnancy loss. Am. J. Reprod. Immunol. 2010 doi: 10.1111/j.1600-0897.2010.00847.x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 30.Redman CW, Sargent IL. Immunology of pre-eclampsia. Am. J. Reprod. Immunol. 2010 doi: 10.1111/j.1600-0897.2010.00831.x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 31.Hanna J, Goldman-Wohl D, Hamani Y, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 32.Cella M, Fuchs A, Vermi W, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanos SL, Bui VL, Mortha A, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luci C, Reynders A, Ivanov II, et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 35.Satoh-Takayama N, Dumoutier L, Lesjean-Pottier S, et al. The natural cytotoxicity receptor NKp46 is dispensable for IL-22-mediated innate intestinal immune defense against. Citrobacter rodentium. J. Immunol. 2009;183:6579–6587. doi: 10.4049/jimmunol.0901935. [DOI] [PubMed] [Google Scholar]
- 36.Andrews DM, Smyth MJ. A potential role for RAG-1 in NK cell development revealed by ana lysis of NK cells during ontogeny. Immunol. Cell. Biol. 2010;88:107–116. doi: 10.1038/icb.2009.94. [DOI] [PubMed] [Google Scholar]
- 37.Haraguchi K, Suzuki T, Koyama N, et al. Notch activation induces the generation of functional NK cells from human cord blood CD34-positive cells devoid of IL-15. J. Immunol. 2009;182:6168–6178. doi: 10.4049/jimmunol.0803036. [DOI] [PubMed] [Google Scholar]
- 38.Kamizono S, Duncan GS, Seidel MG, et al. Nfl3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huntington ND, Legrand N, Alves NL, et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009;206:25–34. doi: 10.1084/jem.20082013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satoh-Takayama N, Lesjean-Pottier S, Vieira P, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 2010;207:273–280. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun S, Lee SH, Kang YH, et al. YC-1 enhances natural killer cell differentiation from hematopoietic stem cells. Int. Immunopharmacol. 2010;10(4):481–486. doi: 10.1016/j.intimp.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chinen H, Matsuoka K, Sato T, et al. Lamina propria c-kit+ immune precursors reside in human adult intestine and differentiate into natural killer cells. Gastroenterology. 2007;133:559–573. doi: 10.1053/j.gastro.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Lynch L, O’Donoghue D, Dean J, et al. Detection and characterization of hemopoietic stem cells in the adult human small intestine. J. Immunol. 2006;176:5199–5204. doi: 10.4049/jimmunol.176.9.5199. [DOI] [PubMed] [Google Scholar]
- 45.Cupedo T, Crellin NK, Papazian N, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 46.Bostik P, Kobkitjaroen J, Tang W, et al. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J. Immunol. 2009;182:3638–3649. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y, Sumitran S, Holgersson J. Direct binding of purified HLA class I antigens by soluble NKG2/CD94 C-type lectins from natural killer cells. Scand. J. Immunol. 1999;49:459–465. doi: 10.1046/j.1365-3083.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- 48.Pende D, Parolini S, Pessino A, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivori S, Vitale M, Morelli L, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J. Exp. Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitale M, Bottino C, Sivori S, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avril T, North SJ, Haslam SM, et al. Probing the cis interactions of the inhibitory receptor Siglec-7 with α2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by ana lysis of α2,8-sialyltransferase gene expression. J. Leukoc. Biol. 2006;80:787–796. doi: 10.1189/jlb.1005559. [DOI] [PubMed] [Google Scholar]
- 52.Betser-Cohen G, Mizrahi S, Elboim M, et al. The association of MHC class I proteins with the 2B4 receptor inhibits self-killing of human NK cells. J. Immunol. 2010;184:2761–2768. doi: 10.4049/jimmunol.0901572. [DOI] [PubMed] [Google Scholar]
- 53.Vales-Gomez M, Reyburn HT, Erskine RA, et al. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/ NKG2-A and the activating receptor CD94/ NKG2-C to HLA-E. EMBO J. 1999;18:4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryceson YT, Ljunggren HG, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghiringhelli F, Menard C, Terme M, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-β-dependent manner. J. Exp. Med. 2005;202:1075–1085. doi: 10.1084/jem.20051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawakami Y, Tomimori Y, Yumoto K, et al. Inhibition of NK cell activity by IL-17 allows vaccinia virus to induce severe skin lesions in a mouse model of eczema vaccinatum. J. Exp. Med. 2009;206:1219–1225. doi: 10.1084/jem.20082835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robson NC, Wei H, McAlpine T, et al. Activin-A attenuates several human natural killer cell functions. Blood. 2009;113:3218–3225. doi: 10.1182/blood-2008-07-166926. [DOI] [PubMed] [Google Scholar]
- 58.Tarazona R, Delgado E, Guarnizo MC, et al. Human prostasomes express CD48 and interfere with NK cell function. Immunobiology. 2010 doi: 10.1016/j.imbio.2010.03.002. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 59.Krzewski K, Chen X, Strominger JL. WIP is essential for lytic granule polarization and NK cell cytotoxicity. Proc. Natl Acad. Sci. USA. 2008;105:2568–2573. doi: 10.1073/pnas.0711593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schleinitz N, March ME, Long EO. Recruitment of activation receptors at inhibitory NK cell immune synapses. PLoS ONE. 2008;3:e3278. doi: 10.1371/journal.pone.0003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittaker GC, Burshtyn DN, Orr SJ, et al. Analysis of the linker for activation of T cells and the linker for activation of B cells in natural killer cells reveals a novel signaling cassette, dual usage in ITAM signaling, and influence on development of the Ly49 repertoire. Blood. 2008;112:2869–2877. doi: 10.1182/blood-2007-11-121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Q. NK cell assays in immunotoxicity testing. Methods Mol. Biol. 2010;598:207–219. doi: 10.1007/978-1-60761-401-2_15. [DOI] [PubMed] [Google Scholar]
- 63.Fultz PN, McClure HM, Anderson DC, et al. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys) Proc. Natl Acad. Sci. USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniel MD, Letvin NL, King NW, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 65.VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn BH, Shaw GM, De Cock KM, et al. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 67.Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology. 2005;115:206–214. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reeves RK, Gillis J, Wong FE, et al. CD16 natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood I. 2010;15(22):4439–4446. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira LE, Johnson RP, Ansari AA. Sooty mangabeys and rhesus macaques exhibit significant divergent natural killer cell responses during both acute and chronic phases of SIV infection. Cell. Immunol. 2008;254:10–19. doi: 10.1016/j.cellimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 70.Biassoni R, Fogli M, Cantoni C, et al. Molecular and functional characterization of NKG2D, NKp80, and NKG2C triggering NKcell receptors in rhesus and cynomolgus macaques: monitoring of NK cell function during simian HIV infection. J. Immunol. 2005;174:5695–5705. doi: 10.4049/jimmunol.174.9.5695. [DOI] [PubMed] [Google Scholar]
- 71.Rutjens E, Mazza S, Biassoni R, et al. CD8+ NK cells are predominant in chimpanzees, characterized by high NCR expression and cytokine production, and preserved in chronic HIV-1 infection. Eur. J. Immunol. 2010;40:1440–1450. doi: 10.1002/eji.200940062. [DOI] [PubMed] [Google Scholar]
- 72.Vivier E, Tomasello E, Baratin M, et al. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 73.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bryceson YT, March ME, Ljunggren HG, et al. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 2006;214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brooks AG, Posch PE, Scorzelli CJ, et al. NKG2A complexed with CD94 defines a novel inhibitory natural killer cell receptor. J. Exp. Med. 1997;185:795–800. doi: 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Braud VM, Allan DS, O’Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 77.Steinle A, Li P, Morris DL, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 78.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu J, Heller G, Chewning J, et al. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 81.Tripathi P, Naik S, Agrawal S. HLA-E and immunobiology of pregnancy. Tissue Antigens. 2006;67:207–213. doi: 10.1111/j.1399-0039.2005.00550.x. [DOI] [PubMed] [Google Scholar]
- 82.Rebmann V, Pfeiffer K, Passler M, et al. Detection of soluble HLA-G molecules in plasma and amniotic fluid. Tissue Antigens. 1999;53:14–22. doi: 10.1034/j.1399-0039.1999.530102.x. [DOI] [PubMed] [Google Scholar]
- 83.Vowels BR, Gershwin ME, Gardner MB, et al. Natural killer cell activity of rhesus macaques against retrovirus-pulsed CD4+ target cells. AIDS Res. Hum. Retroviruses. 1990;6:905–918. doi: 10.1089/aid.1990.6.905. [DOI] [PubMed] [Google Scholar]
- 84.De Maria A, Biassoni R, Fogli M, et al. Identification, molecular cloning and functional characterization of NKp46 and NKp30 natural cytotoxicity receptors in Macaca fascicularis NK cells. Eur. J. Immunol. 2001;31:3546–3556. doi: 10.1002/1521-4141(200112)31:12<3546::aid-immu3546>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 85.LaBonte ML, Hershberger KL, Korber B, et al. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol. Rev. 2001;183:25–40. doi: 10.1034/j.1600-065x.2001.1830103.x. [DOI] [PubMed] [Google Scholar]
- 86.Mavilio D, Benjamin J, Kim D, et al. Identification of NKG2A and NKp80 as specific natural killer cell markers in rhesus and pigtailed monkeys. Blood. 2005;106:1718–1725. doi: 10.1182/blood-2004-12-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hershberger KL, Shyam R, Miura A, et al. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J. Immunol. 2001;166:4380–4390. doi: 10.4049/jimmunol.166.7.4380. [DOI] [PubMed] [Google Scholar]
- 88.Hershberger KL, Kurian J, Korber BT, et al. Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur. J. Immunol. 2005;35:922–935. doi: 10.1002/eji.200425408. [DOI] [PubMed] [Google Scholar]
- 89.Guethlein LA, Flodin LR, Adams EJ, et al. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J. Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 90.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 91.Sambrook JG, Bashirova A, Palmer S, et al. Single haplotype ana lysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bimber BN, Moreland AJ, Wiseman RW, et al. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J. Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chaichompoo P, Bostik P, Stephenson S, et al. Multiple KIR gene polymorphisms are associated with plasma viral loads in SIV infected rhesus macaques. Cell. Immuol. 2010;263(2):176–187. doi: 10.1016/j.cellimm.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blokhuis JH, van der Wiel MK, Doxiadis GG, et al. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. 2010;62:295–306. doi: 10.1007/s00251-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics. 2010;62:281–293. doi: 10.1007/s00251-010-0433-4. [DOI] [PubMed] [Google Scholar]
- 96.Orange JS. Human natural killer cell deficiencies. Curr. Opin. Allergy Clin. Immunol. 2006;6:399–409. doi: 10.1097/ACI.0b013e3280106b65. [DOI] [PubMed] [Google Scholar]
- 97.Mocikat R, Braumuller H, Gumy A, et al. Natural killer cells activated by MHC class Ilow targets prime dendritic cells to induce protective CD8 T cell responses. Immunity. 2003;19:561–569. doi: 10.1016/s1074-7613(03)00264-4. [DOI] [PubMed] [Google Scholar]
- 98.Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J. Intern. Med. 2009;265:29–42. doi: 10.1111/j.1365-2796.2008.02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weber K, Meyer D, Grosse V, et al. Reconstitution of NK cell activity in HIV-1 infected individuals receiving antiretroviral therapy. Immunobiology. 2000;202:172–178. doi: 10.1016/S0171-2985(00)80063-7. [DOI] [PubMed] [Google Scholar]
- 100.Iannello A, Debbeche O, Samarani S, et al. Antiviral NK cell responses in HIV infection: II. viral strategies for evasion and lessons for immunotherapy and vaccination. J. Leukoc. Biol. 2008;84:27–49. doi: 10.1189/jlb.0907649. [DOI] [PubMed] [Google Scholar]
- 101.Iannello A, Debbeche O, Samarani S, et al. Antiviral NK cell responses in HIV infection: I. NK cell receptor genes as determinants of HIV resistance and progression to AIDS. J. Leukoc. Biol. 2008;84:1–26. doi: 10.1189/jlb.0907650. [DOI] [PubMed] [Google Scholar]
- 102.Bonaparte MI, Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104:2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 103.Cohen GB, Gandhi RT, Davis DM, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 104.Brenner BG, Dascal A, Margolese RG, et al. Natural killer cell function in patients with acquired immunodeficiency syndrome and related diseases. J. Leukoc. Biol. 1989;46:75–83. doi: 10.1002/jlb.46.1.75. [DOI] [PubMed] [Google Scholar]
- 105.Ahmad A, Menezes J. Antibody-dependent cellular cytotoxicity in HIV infections. FASEB J. 1996;10:258–266. doi: 10.1096/fasebj.10.2.8641559. [DOI] [PubMed] [Google Scholar]
- 106.Ullum H, Gotzsche PC, Victor J, et al. Defective natural immunity: an early manifestation of human immunodeficiency virus infection. J. Exp. Med. 1995;182:789–799. doi: 10.1084/jem.182.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sirianni MC, Mezzaroma I, Aiuti F, et al. Analysis of the cytolytic activity mediated by natural killer cells from acquired immunodeficiency syndrome patients in response to phytohemagglutinin or anti-CD16 monoclonal antibody. Eur. J. Immunol. 1994;24:1874–1878. doi: 10.1002/eji.1830240824. [DOI] [PubMed] [Google Scholar]
- 108.Liu Q, Sun Y, Rihn S, et al. Matrix metalloprotease inhibitors restore impaired NK cell-mediated antibody-dependent cellular cytotoxicity in human immunodeficiency virus type 1 infection. J. Virol. 2009;83:8705–8712. doi: 10.1128/JVI.02666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kottilil S, Chun TW, Moir S, et al. Innate immunity in human immunodeficiency virus infection: effect of viremia on natural killer cell function. J. Infect. Dis. 2003;187:1038–1045. doi: 10.1086/368222. [DOI] [PubMed] [Google Scholar]
- 110.Mantegani P, Tambussi G, Galli L, et al. Perturbation of the natural killer cell compartment during primary human immunodeficiency virus 1 infection primarily involving the CD56bright subset. Immunology. 2009;129(2):220–233. doi: 10.1111/j.1365-2567.2009.03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vieillard V, Fausther-Bovendo H, Samri A, et al. Specific phenotypic and functional features of natural killer cells from HIV-infected long-term nonprogressors and HIV controllers. J. Acquir. Immune Defic. Syndr. 2010;53:564–573. doi: 10.1097/QAI.0b013e3181d0c5b4. [DOI] [PubMed] [Google Scholar]
- 112.Hong HS, Eberhard JM, Keudel P, et al. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J. Virol. 2010;84:1183–1188. doi: 10.1128/JVI.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mavilio D, Benjamin J, Daucher M, et al. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl Acad. Sci. USA. 2003;100:15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alter G, Teigen N, Davis BT, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106:3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 115.De Maria A, Fogli M, Costa P, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur. J. Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 116.Boulet S, Kleyman M, Kim JY, et al. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS. 2008;22:1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 117.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 118.LaBonte ML, McKay PF, Letvin NL. Evidence of NK cell dysfunction in SIV-infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur. J. Immunol. 2006;36:2424–2433. doi: 10.1002/eji.200635901. [DOI] [PubMed] [Google Scholar]
- 119.Choi EI, Reimann KA, Letvin NL. In vivo natural killer cell depletion during primary simian immunodeficiency virus infection in rhesus monkeys. J. Virol. 2008;82:6758–6761. doi: 10.1128/JVI.02277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Velilla PA, Shata MT, Lages CS, et al. Effect of low-dose IL-2 immunotherapy on frequency and phenotype of regulatory T cells and NK cells in HIV/HCV-coinfected patients. AIDS Res. Hum. Retroviruses. 2008;24:52–61. doi: 10.1089/aid.2007.0180. [DOI] [PubMed] [Google Scholar]
- 121.Locker GJ, Kapiotis S, Veitl M, et al. Activation of endothelium by immunotherapy with interleukin-2 in patients with malignant disorders. Br. J. Haematol. 1999;105:912–919. doi: 10.1046/j.1365-2141.1999.01453.x. [DOI] [PubMed] [Google Scholar]
- 122.Ponce R. Adverse consequences of immunostimulation. J. Immunotoxicol. 2008;5:33–41. doi: 10.1080/15476910801897920. [DOI] [PubMed] [Google Scholar]
- 123.Ramana Rao PV, Rajasekaran S, Raja A. Augumentation of natural killer activity with exogenous interleukins in patients with HIV and pulmonary tuberculosis coinfection. AIDS Res. Hum. Retroviruses. 2008;24:1435–1443. doi: 10.1089/aid.2008.0126. [DOI] [PubMed] [Google Scholar]
- 124.Strbo N, de Armas L, Liu H, et al. IL-21 augments natural killer effector functions in chronically HIV-infected individuals. AIDS. 2008;22:1551–1560. doi: 10.1097/QAD.0b013e3283089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hall LJ, Clare S, Dougan G. NK cells influence both innate and adaptive immune responses after mucosal immunization with antigen and mucosal adjuvant. J. Immunol. 2010;184:4327–4337. doi: 10.4049/jimmunol.0903357. [DOI] [PMC free article] [PubMed] [Google Scholar]