Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress (original) (raw)
Abstract
Phenylalanine ammonia-lyase (PAL) catalyzes the first step of the phenylpropanoid pathway, which produces precursors to a variety of important secondary metabolites. Arabidopsis (Arabidopsis thaliana) contains four PAL genes (PAL1_–_PAL4), but there has been no genetic analysis to assess the biological functions of the entire gene family. Here, we report the generation and analysis of combined mutations for the four Arabidopsis PAL genes. Contrary to a previous report, we found that three independent pal1 pal2 double mutants were fertile and generated yellow seeds due to the lack of condensed tannin pigments in the seed coat. The pal1 pal2 double mutants were also deficient in anthocyanin pigments in various plant tissues, which accumulate in wild-type plants under stress conditions. Thus, PAL1 and PAL2 have a redundant role in flavonoid biosynthesis. Furthermore, the pal1 pal2 double mutants were more sensitive to ultraviolet-B light but more tolerant to drought than wild-type plants. We have also generated two independent pal1 pal2 pal3 pal4 quadruple knockout mutants, which are stunted and sterile. The quadruple knockout mutants still contained about 10% of the wild-type PAL activity, which might result from one or more leaky pal mutant genes or from other unknown PAL genes. The quadruple mutants also accumulated substantially reduced levels of salicylic acid and displayed increased susceptibility to a virulent strain of the bacterial pathogen Pseudomonas syringae. These results provide further evidence for both distinct and overlapping roles of the Arabidopsis PAL genes in plant growth, development, and responses to environmental stresses.
When the first land plants appeared about 500 million years ago from a pioneer green algal ancestor, they had to face harsh terrestrial environmental conditions, including desiccation, UV radiation, and attack of microbial pathogens (Kenrick and Crane, 1997). The emergence of the phenylpropanoid pathway is among a number of important adaptations that allow land plants to survive under these important stresses (Ferrer et al., 2008). Phenylpropanoid compounds are precursors to a wide range of phenolic compounds with many functions in plants. Lignin, which is synthesized from phenylpropanoid compounds, is a major structural component of secondarily thickened cell walls in the plant vascular system essential for stem rigidity and for conducting water, minerals, and photosynthetic products through the plant. Phenylpropanoids are also precursors to flavonoids, isoflavonoids, cumarins, and stilbenes. These compounds have important functions in plant defense against pathogens and other predators, as UV light protectants, and as regulatory molecules in signal transduction and communication with other organisms (Ferrer et al., 2008).
Phenylalanine ammonia-lyase (PAL; EC 4.3.1.5) catalyzes the deamination of Phe to give cinnamic acid, which is the first step in the phenylpropanoid pathway and an important regulation point between primary and secondary metabolism. PAL is encoded by a small gene family in plants with four members in Arabidopsis (Arabidopsis thaliana; PAL1_–_PAL4; Raes et al., 2003). A large number of studies have shown that PAL gene expression is responsive to a variety of environmental stimuli, including pathogen infection, wounding, nutrient depletion, UV irradiation, extreme temperatures, and other stress conditions (Lawton et al., 1983; Edwards et al., 1985; Liang et al., 1989a, 1989b; Dixon and Paiva, 1995). A number of studies have also employed molecular and genetic approaches to silence or disrupt the PAL genes for functional analysis of the gene family in plant growth, development, and responses to environmental stresses. Tobacco (Nicotiana tabacum) plants epigenetically suppressed in PAL expression exhibited unusual phenotypes such as localized fluorescent lesions, altered leaf shape and texture, reduced lignification in xylem, stunted growth, reduced pollen viability, and altered flower morphology and pigmentation (Elkind et al., 1990). These plants did not develop systemic acquired resistance in response to infection by Tobacco mosaic virus (Pallas et al., 1996). More recently, genetic analysis has been conducted on Arabidopsis PAL1 and PAL2 genes through phenotypic characterization of both single and double mutants (Rohde et al., 2004). While the pal1 and pal2 single mutants have no obvious visible phenotypes in growth and development, the pal1 pal2 double mutant has limited phenotypic alterations, including infertility, significant reduction in lignin accumulation, and alteration in secondary cell wall ultrastructure (Rohde et al., 2004). Molecular phenotyping revealed significant modifications in the transcriptome and metabolome of the pal1 pal2 mutant (Rohde et al., 2004).
The abolished systemic acquired resistance in the transgenic tobacco plants with cosuppressed PAL expression is associated with reduced accumulation of salicylic acid (SA) in both lower inoculated and upper uninoculated systemic leaves (Pallas et al., 1996). Furthermore, the PAL inhibitor 2-aminoindan-2-phosphonic acid reduced pathogen- or pathogen elicitor-induced SA accumulation in potato (Solanum tuberosum), cucumber (Cucumis sativus), and Arabidopsis (Meuwly et al., 1995; Mauch-Mani and Slusarenko, 1996; Coquoz et al., 1998). In Arabidopsis, treatment of the PAL inhibitor made the plants completely susceptible to the downy mildew oomycete Hyaloperonospora parasitica, which could be restored by SA (Mauch-Mani and Slusarenko, 1996). Based on these results, the authors have suggested that production of SA precursors is a major function of PAL in Arabidopsis downy mildew resistance (Mauch-Mani and Slusarenko, 1996). However, genetic and molecular analyses have indicated that the bulk of SA is synthesized from the isochorismate pathway. Arabidopsis contains two genes encoding isochorismate synthase (ICS). In ics1 mutants, total SA accumulation is only about 5% to 10% of wild-type levels after infection by the virulent biotroph Erysiphe or avirulent strains of Pseudomonas syringae (Wildermuth et al., 2001). Upon UV light exposure, the ics1 mutant accumulated roughly 10% and the ics1 ics2 double mutant accumulated about 4% of wild-type SA levels (Garcion et al., 2008). SA accumulation in Nicotiana benthamiana is also dependent on ICS (Catinot et al., 2008). If a vast majority of SA is synthesized from the ICS pathway, it is unclear how silencing or inhibition of PAL leads to a substantial reduction in SA accumulation and enhanced pathogen susceptibility.
Since only two of the four PAL genes in Arabidopsis have been genetically analyzed (Rohde et al., 2004), the biological functions of the remaining two PAL genes individually or in combination are still unknown. Here, we report the generation and analysis of two independent sets of single, double, triple, and quadruple T-DNA or transposon insertion mutants for the four Arabidopsis PAL genes. We found that pal1 pal2 double mutants were fertile and generated yellowish seeds due to the lack of condensed tannin pigments in the seed coat. The pal1 pal2 double mutants were also deficient in anthocyanin pigments in various plant tissues and were highly sensitive to UV-B light. Thus, PAL1 and PAL2 have a particularly important and redundant role in flavonoid biosynthesis. The two pal1 pal2 pal3 pal4 quadruple mutants, which contained almost no detectable transcripts for the four PAL genes but still had about 10% of the wild-type PAL activity, were stunted and sterile. The quadruple mutants had greatly reduced levels of lignin and accumulated reduced levels of SA after pathogen infection. These results provide new information about both the distinct and overlapping roles of the Arabidopsis PAL genes in plant growth, development, and responses to environmental stresses.
RESULTS
Fertility and Pigmentation of the pal1 pal2 Double Mutants
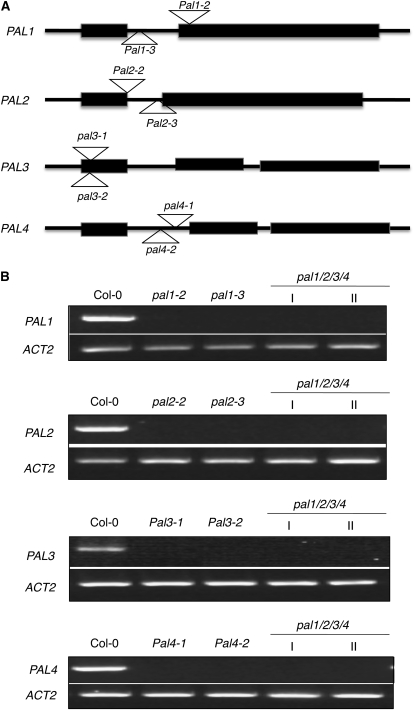
T-DNA insertion mutants pal1-1 and pal2-1 for PAL1 and PAL2, respectively, have been previously isolated and found to have no visible phenotypic alterations (Rohde et al., 2004). The pal1-1 pal2-1 double mutant was also generated through genetic cross and found to display no major morphological phenotypes either (Rohde et al., 2004). However, the double mutant was male sterile and could not set seeds (Rohde et al., 2004). Since these two reported pal mutants were in the Arabidopsis ecotype C24 (Rohde et al., 2004), we identified two new T-DNA insertion mutants each for PAL1 and PAL2 in the Arabidopsis ecotype Columbia (Col-0), so their genetic background is identical to that of later isolated pal3 and pal4 mutants (see below). The pal1-2 (Salk_022804) and pal1-3 (Salk_096474) mutants contain a T-DNA insertion in the second exon and first intron of PAL1, respectively (Fig. 1A). Both pal2-2 (Salk_092252) and pal2-3 (GABI_692H09) contain a T-DNA insertion in the first intron of PAL2 (Fig. 1A). Semiquantitative PCR analysis failed to detect PAL1 or PAL2 transcripts in the respective homozygous mutants (Fig. 1B). Real-time PCR showed that the mutants contained less than 1% of the wild-type levels of the PAL transcripts (Supplemental Table S1). Like pal1-1 and pal2-1 mutants, pal1-2, pal1-3, pal2-2, and pal2-3 single mutants had no visible morphological phenotypes (data not shown).
Figure 1.
Structures and mutants for the PAL genes. A, Exon and intron structures of the four PAL genes. The exons are indicated with rectangles and the introns with lines. The locations of T-DNA or transposon insertions in the pal mutants are indicated. B, Expression of PAL genes in the wild type and pal mutants. Semiquantitative RT-PCR was performed with total RNA isolated from 10-week-old inflorescence stems of the wild type and pal single and quadruple mutants. The experiment was repeated twice with similar results.
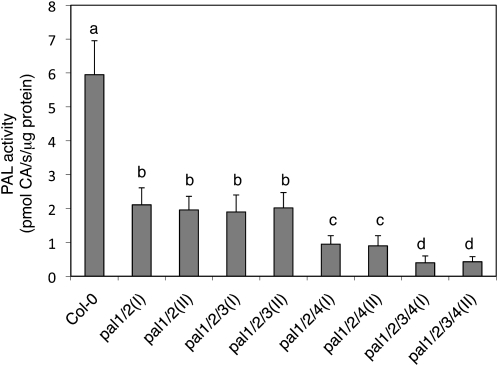
We also generated two independent pal1 pal2 double mutants through genetic crosses. The PAL activity in inflorescence stems of the double mutants was only about 25% to 30% of that of wild-type plants (Fig. 2), similar to that observed in the previously reported pal1-1 pal2-1 double mutant (Rohde et al., 2004). In addition, like the pal1-1 pal2-1 double mutant, the pal1-2 pal2-2 and pal1-3 pal2-3 double mutants displayed no major difference in morphology from the wild type or their respective single mutant plants. However, the double mutants grew significantly slower than wild-type plants, and this difference in growth rate also appeared to be affected by growth conditions. When grown in growth chambers at Purdue University, the pal1 pal2 double mutants were only slightly smaller than the wild type during the first 3 weeks after germination but could reach the same size as the wild type as they grew into mature stages. When grown in a growth room at Zhejiang University, the double mutant plants were significantly smaller than wild-type plants throughout the growth stages. Furthermore, while leaves and inflorescences of wild-type plants accumulated anthocyanin pigments at adult and flowering stages, particularly when underfertilized and at a relatively low temperature, no such pigments were observed in the pal1 pal2 double mutant plants (Fig. 3A).
Figure 2.
PAL activity in the wild type and pal mutants. Crude extracts were prepared from 10-week-old inflorescence stems of the wild type and indicated pal mutants, and PAL activity was determined with [14C]Phe and is expressed in pmol cinnamic acid (CA) s−1 mg−1 protein. Means and se were calculated from average PAL activities determined from three experiments with five plants per experiment for each genotype. According to Duncan's multiple range test (P = 0.05), means of PAL activities do not differ significantly if they are indicated with the same letter.
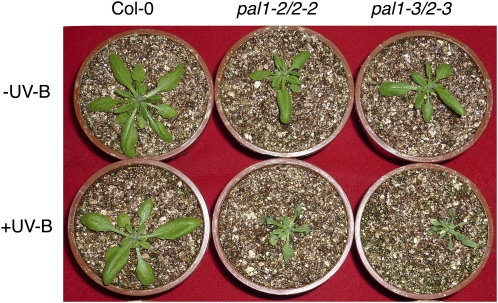
Figure 3.
Phenotypes of the pal1 pal2 double mutants. A, Lack of anthocyanin pigments in the pal1 pal2 double mutants. Wild-type and pal1 pal2 double mutant plants were grown in soil without fertilization at 19°C under a 12-h-light/12-h-dark photoperiod. The photograph was taken about 8 weeks after germination. B, Inflorescences and siliques of wild-type and pal1 pal2 double mutant plants. C, Seeds of wild-type and pal1 pal2 double mutant plants. D, Vanillin staining of immature seeds for detection of catechins and proanthocyanidins.
It has been previously reported that the pal1-1 pal2-1 double mutant is sterile (Rohde et al., 2004). Surprisingly, the pal1-2 pal2-2 and pal1-3 pal2-3 double mutants were fertile when grown in growth chambers at Purdue University and at Zhejiang University (Fig. 3B). The double mutants generated yellow seeds (Fig. 3C) due to the lack of tannin pigments in the seed coat. Tannin pigments are produced from polymerization and oxidation of uncolored proanthocyanidins, which can be detected in immature seeds by their dark red staining with vanillin (Debeaujon et al., 2000). Indeed, immature seeds from wild-type plants turned dark red, but the immature seeds from the pal1 pal2 double mutants showed very little staining when incubated with acidic vanillin (Fig. 3D). The yellow seeds from the double mutants germinated and grew into fertile plants, which again produced yellow seeds.
The difference in fertility between our double mutants and the previously reported pal1-1 pal2-1 double mutant could be due to differences in their genetic background (C24 versus Col-0) and T-DNA insertions. To distinguish among these possibilities, we requested and obtained pal1-1 pal2-1 double mutant seeds homozygous for pal1-1 but heterozygous for pal2-1. The double homozygous mutant plants were subsequently identified from their progeny by PCR using gene-specific primers flanking the insertion sites. Unlike Col-0 wild-type plants, C24 wild-type plants had little anthocyanin pigments in leaf veins of mature plants but accumulated visible pigments in the basal stem of inflorescence (Supplemental Fig. S1A). In the pal-1 pal2-1 double mutant, however no such anthocyanin pigments were observed (Supplemental Fig. S1A). Furthermore, the pal1-2 pal2-2 double mutants in the C24 background were also fertile when grown in growth chambers at Purdue University and generated yellow seeds just like the pal1-2 pal2-2 and pal1-3 pal2-3 double mutants in the Col-0 background (Supplemental Fig. S1, B and C). These results indicate that the fertility of the pal1 pal2 mutants is affected by the growth conditions.
We grew our Arabidopsis plants in both the United States and China at a light intensity of 120 _μ_E m−2 s−1, while the previously reported study of the pal1-1 and pal2-1 mutants used a lower light intensity (50 _μ_E m−2 s−1; Rohde et al., 2004). To determine whether the difference in light intensity is responsible for the fertility difference between the two studies, we first grew the Col-0 wild-type and pal1 pal2 double mutant plants at 120 _μ_E m−2 s−1 until they started to flower. The flowering plants were then divided into two groups, with one group kept at 120 _μ_E m−2 s−1 and the other group transferred to 50 _μ_E m−2 s−1. When grown at 120 _μ_E m−2 s−1, the pal1 pal2 double mutants produced approximately 11% of wild-type pollen grains (Supplemental Table S2) and were fertile (Supplemental Fig. S2). However, those pal1 pal2 double mutant plants that were transferred to 50 _μ_E m−2 s−1 produced few pollen grains (Supplemental Table S2) and were sterile (Supplemental Fig. S2).
Enhanced Sensitivity of pal1 pal2 Double Mutants to UV-B Light
UV-B radiation is an integral part of sunlight and can cause damage to the genome and photosynthetic machinery of plants. We compared the two pal1 pal2 double mutants with wild-type plants for sensitivity to short-term UV-B light treatment. As shown in Figure 4, exposure of wild-type plants to 1.24 _μ_mol m−2 s−1 UV-B light for 4 h did not cause apparent injury or foliar dehydration. By contrast, both pal1 pal2 double mutant plants exhibited extensive wilting due to foliar dehydration after such a short-term UV-B light exposure (Fig. 4). Thus, disruption of both PAL1 and PAL2 caused increased sensitivity to UV-B radiation.
Figure 4.
Enhanced sensitivity of the pal1 pal2 double mutants to short-term UV-B light treatment. Four-week-old wild-type and pal1 pal2 double mutant plants were irradiated with 1.24 _μ_mol m−2 s−1 UV-B light for 4 h. The photograph was taken 1 d after UV-B light treatment. The experiment was repeated twice with similar results.
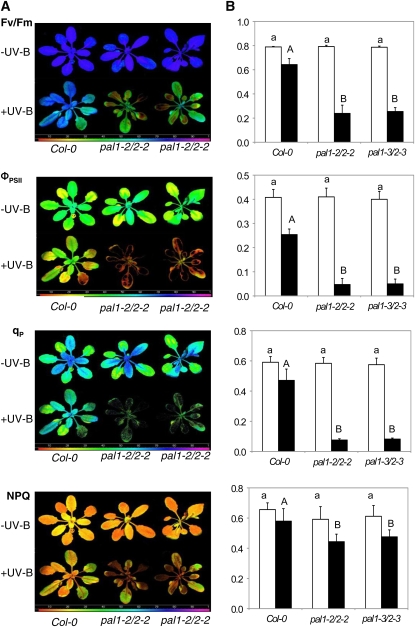
UV-B radiation has harmful effects on various biological processes in plants, including PSII. A number of studies have shown that UV-B light causes severe damage to PSII electron transport activity but no or little effects on PSI (Vass et al., 1999; Booij-James et al., 2000; Segui et al., 2000; Waring et al., 2006). To further determine increased sensitivity of the pal1 pal2 double mutants to UV-B light treatment, we compared wild-type and double mutant plants for the effects of UV-B on the efficiency of PSII photochemistry and photoprotection. As shown in Figure 5, without UV-B light treatment there was no significant difference between the wild type and double mutants in maximum quantum yield of PSII (_F_v/_F_m), quantum yield of PSII (ΦPSII), photochemical quenching (qP), and nonphotochemical quenching coefficient (NPQ). _F_v/_F_m, ΦPSII, and qP are indicators of PSII photochemistry, while NPQ is an indicator of photoprotection. After 4 h of UV-B light treatment, _F_v/_F_m, ΦPSII, qP, and NPQ of wild-type plants decreased by 22%, 38%, 18%, and 12%, respectively (Fig. 5). On the other hand, UV-B light treatment caused more than 70% reduction of _F_v/_F_m, almost 90% reduction in ΦPSII and qP, and more than 20% reduction in NPQ of the pal1 pal2 mutants (Fig. 5). Thus, the capacity of PSII photochemistry and photoprotection was much more compromised by UV-B light treatment in the double pal1 pal2 mutants than in wild-type plants.
Figure 5.
Enhanced sensitivity of the effects of UV-B light on the efficiency of PSII photochemistry and photoprotection between wild-type and double mutant plants. A, Images of _F_v/_F_m, ΦPSII, qP, and NPQ of plants before and after exposure to UV-B light at 1.24 _μ_mol m−2 s−1 for 4 h. Chlorophyll fluorescence images were taken immediately after UV-B light treatment. The false color code depicted at the bottom of the images ranged from 0 (black) to 1.0 (purple). B, Average values for the respective chlorophyll fluorescence images. Data are means of five replicates (±sd). According to Duncan's multiple range test (P = 0.05), means of the photosynthetic parameters do not differ significantly before UV-B light exposure if they are indicated with the same lowercase letter and do not differ significantly after UV-B light exposure if they are indicated with the same uppercase letter.
Enhanced Tolerance of pal1 pal2 Double Mutants to Drought
The availability of the pal1 pal2 double mutants allowed us to determine their phenotypes in tolerance to a variety of abiotic stress conditions. However, when assessed in a growth medium containing various concentrations of NaCl or polyethylene glycol, we found no significant difference between the wild type and double mutants in tolerance to salt or osmotic stress. For testing drought tolerance, we transferred 7-week-old Arabidopsis plants into a walk-in growth chamber with approximately 50% humidity. The pal1 pal2 double mutant plants were significantly small than wild-type plants during the first 3 weeks after germination, and we chose relatively old plants for the drought tolerance test to ensure that all tested plants had similar sizes and fresh weights. The plants were unwatered and observed for drought stress symptoms. As shown in Figure 6, wild-type plants showed extensive wilting about 2 weeks after watering was stopped. The two pal1 pal2 double mutants were still brightly green and exhibited only very minor wilting (Fig. 6). Thus, disruption of PAL1 and PAL2 increased plant tolerance to drought stress.
Figure 6.
Enhanced drought tolerance of the pal1 pal2 mutants. Seven-week-old Arabidopsis plants (eight plants for each genotype) were moved into a walk-in growth chamber with approximately 50% humidity. The photograph of representative plants was taken 2 weeks after withholding watering. The experiment was repeated twice with similar results.
Generation of pal1 pal2 pal3 pal4 Quadruple T-DNA Insertion Mutants
We also identified two T-DNA insertion mutants each for PAL3 and PAL4. Both pal3-1 (SM_3.39574) and pal3-2 (SM_3.19684) contain a transposon insertion in the first exon of PAL3 (Fig. 1A). Both pal4-1 (Salk_070702) and pal4-2 (Salk_022730) contain a T-DNA insertion in the first intron of PAL4 (Fig. 1A). Semiquantitative PCR analysis failed to detect PAL3 or PAL4 transcripts in the respective homozygous mutants, suggesting that they are null (Fig. 1B). Real-time PCR showed that the mutants contained 0.3% to 1.2% of the wild-type levels of the respective PAL transcripts (Supplemental Table S1). No visible morphological phenotypes were observed in these pal3 and pal4 single mutants (data not shown).
To analyze the biological functions of the entire PAL gene family, we generated triple and quadruple mutants for the four PAL genes through genetic crossing. The two pal1 pal2 pal3 triple mutants had levels of residual PAL activities similar to those of the pal1 pal2 double mutants (Fig. 2). Both triple mutants were also fertile when grown in growth chambers at Purdue University and generated yellow seeds (data not shown). On the other hand, the PAL activity in inflorescence stems of the pal1 pal2 pal4 triple mutants was only about 12% to 15% of wild-type levels, compared with about 25% of wild-type levels of PAL activity in the pal1 pal2 double and pal1 pal2 pal3 triple mutants (Fig. 2). In addition, the pal1 pal2 pal4 triple mutant plants were stunted and sterile.
We also generated two independent pal1 pal2 pal3 pal4 quadruple mutants. Reverse transcription (RT)-PCR analysis failed to detect transcripts for the four PAL genes in homozygous quadruple mutant plants (Fig. 1B). Real-time PCR showed that the mutants contained 0.4% to 1.1% of the wild-type levels of the respective PAL transcripts (Supplemental Table S1). However, in both quadruple mutants, there was still about 7% to 9% residual PAL activity in inflorescence stems (Fig. 2). Although stunted and sterile, both quadruple mutants were viable and exhibited no grossly altered morphological phenotypes (Fig. 7A).
Figure 7.
Phenotypes of the pal1 pal2 pal3 pal4 quadruple mutants. A, Six-week-old wild-type and pal1 pal2 pal3 pal4 quadruple mutant plants. B, Flowering plants of the wild type and pal1 pal2 pal3 pal4 quadruple mutants. C, Lignin staining with phloroglucinol/HCl in the basal part of 3-month-old inflorescence stems of wild-type and pal1 pal2 pal3 pal4 quadruple mutant plants. D, Lignin content of 3-month-old inflorescence stems of wild-type and pal1 pal2 pal3 pal4 quadruple mutant plants. Lignin content was determined as acetyl bromide-soluble lignin. Means and se were calculated from three average lignin contents determined from three experiments with five plants per experiment for each genotype. According to Duncan's multiple range test (P = 0.05), means of lignin contents do not differ significantly if they are indicated with the same letter.
Lignin Analysis of the pal1 pal2 pal3 pal4 Quadruple Mutants
The two quadruple mutant plants produced rigid inflorescence stems that were reduced in length (Fig. 7A). To analyze the defects of the quadruple mutants in lignin accumulation, we first used histochemical staining of hand sections of fresh basal rachis segments of 3-month-old plants using the phloroglucinol/HCl reagent. In the wild type, the walls of xylem cells exhibited an intensely bright red when stained with phloroglucinol/HCl (Fig. 7B). Both quadruple mutants showed a positive red color with distribution similar to that of the wild type (Fig. 7B). However, the color of the positively stained material was greatly reduced in the quadruple mutants when compared with that of the wild type (Fig. 7B).
To obtain a more quantitative analysis of the lignin content, we isolated cell wall and measured lignin contents using the acetyl bromide method of inflorescence stems of 10-week-old wild-type and pal1 pal2 pal3 pal4 quadruple mutant plants. As shown in Figure 7C, the two quadruple mutants had about 20% to 25% of wild-type levels of lignin contents. These results were consistent with those from the histochemical lignin staining: lignin was present but at greatly reduced levels in the quadruple mutants.
SA Analysis of the pal1 pal2 pal3 pal4 Quadruple Mutants
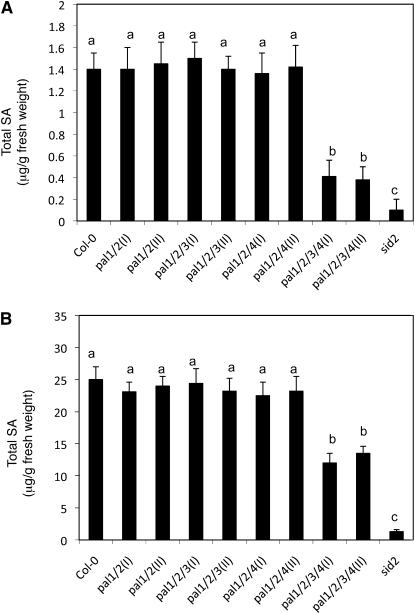
To determine the effect of PAL mutations on pathogen-induced SA accumulation, we compared the wild type and various pal double, triple, and quadruple mutants for total SA contents before and after infection of an avirulent strain of the bacterial pathogen P. syringae pv tomato DC3000 (Pst DC3000 avrRpt2). As shown in Figure 8A, before infection, there was no significant difference in total SA content between the wild type and the pal1 pal2 double or pal1 pal2 pla4 triple mutants. On the other hand, the basal SA levels in the two pal1 pal2 pal3 pal4 quadruple mutants were only about 25% of wild-type levels (Fig. 8B). At 24 h after infection by the avirulent pathogen, there was approximately 17-fold induction in the total SA levels in wild-type plants. Again, the total SA contents after the pathogen infection in the pal1 pal2 double or pal1 pal2 pla4 triple mutants were similar to those in the wild-type plants. In the two pal1 pal2 pal3 pal4 quadruple mutants, the total SA levels were about 50% of wild-type levels after the pathogen infection.
Figure 8.
Basal and pathogen-induced accumulation of total SA. A, Basal SA contents in wild-type (Col-0) and mutant plants. B, Pathogen-induced SA contents in wild-type (Col-0) and mutant plants. Plants were infiltrated with avirulent Pst DC3000 avrRpt2 (OD600 = 0.02 in 10 mm MgCl2). Inoculated leaves were harvested 24 h later for total SA determination. Means and se were calculated from average SA contents determined from three experiments with four to six plants per experiment for each genotype. According to Duncan's multiple range test (P = 0.05), means of SA contents do not differ significantly if they are indicated with the same letter.
Responses to Pathogen Infection
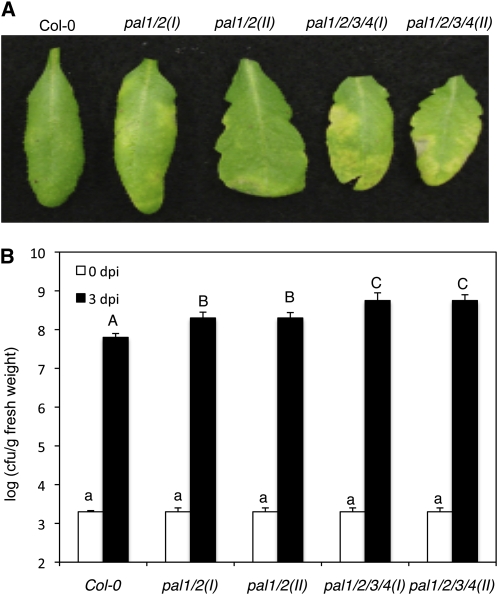
To determine whether mutations of the PAL genes affected plant disease resistance, we compared the wild type and the pal mutants for responses to microbial pathogens. First, we determined the pal mutants for responses to the necrotrophic fungal pathogen Botrytis cinerea but found no significant alteration with both the pal1 pal2 double and pal1 pal2 pal3 pal4 quadruple mutants when compared with wild-type plants (Supplemental Fig. S3). We also compared the pal double and quadruple mutants with wild-type plants for responses to the virulent strain of Pst DC3000. When inoculated at a relatively high dosage with the bacterial pathogen (optical density at 600 nm [OD600] = 0.001), we observed no significant difference between the wild type and the pal mutants in disease symptom development. However, at 3 d post inoculation with a low dosage of the bacterial pathogen (OD600 = 0.0002), the two pal1 pal2 double mutants had a slight but detectable enhancement in disease symptoms when compared with those in the wild type (Fig. 9A). The double mutant plants also displayed a significantly greater bacterial growth (4- to 5-fold) than wild-type plants (Fig. 9B). The two pal1 pal2 pal3 pal4 quadruple mutants exhibited even greater disease symptoms (Fig. 9A) and bacterial growth (10- to 12-fold; Fig. 9B) than wild-type plants.
Figure 9.
Responses of Arabidopsis pal mutants to P. syringae. A, Disease symptom development. Plants were infiltrated with a suspension of Pst DC3000 (OD600 = 0.0002 in 10 mm MgCl2). Photographs of representative inoculated leaves were taken at 3 d post inoculation (dpi). B, Altered bacterial growth. Pathogen inoculation was performed as in A. Samples were taken at 0 dpi (white bars) or 3 dpi (black bars) to determine the growth of the bacterial pathogen. Means and se were calculated from 10 plants for each treatment. According to Duncan's multiple range test (P = 0.05), means of colony-forming units (cfu) at 0 dpi do not differ significantly if they are indicated with the same lowercase letter and means of colony-forming units at 3 dpi do not differ significantly if they are indicated with the same uppercase letter.
DISCUSSION
PAL genes are among the most extensively studied plant genes, particularly with respect to their response in expression to a variety of environmental conditions. Attempts have also been made to address the biological functions of the PAL genes through silencing of PAL genes in tobacco and disruption of the PAL1 and PAL2 genes in Arabidopsis. To determine the biological functions of the entire PAL gene family, we have generated two independent sets of single, double, triple, and quadruple T-DNA/transposon insertion mutants for the four Arabidopsis PAL genes. Our analysis of these mutants has generated new and important information about the diverse roles of the gene family in plant growth, development, and responses to environmental conditions.
Roles of PAL1 and PAL2 in Flavonoid Synthesis and UV-B Light Protection
Previously, Rohde et al. (2004) isolated and characterized T-DNA insertion mutants pal1-1 and pal2-1 for Arabidopsis PAL1 and PAL2, respectively. These authors reported that the pal1-1 and pal2-1 single mutants lacked clear phenotypic alterations. The pal1-1 pal2-1 mutant had no gross morphological phenotypes either but became sterile. However, the transcriptome and metabolome of the pal mutants were significantly modified. Disruption of the two PAL genes altered the expression of genes involved in phenylpropanoid biosynthesis and carbohydrate and amino acid metabolism (Rohde et al., 2004). Phe was overaccumulated, but three major flavonol glucosides and lignin monomers were significantly reduced (Rohde et al., 2004). In this study, we isolated two additional T-DNA insertion mutants for PAL1 and PAL2 and provide new insights into the critical roles of the two PAL genes. Unlike previous reports with the pal1-1 pal2-1 double mutant, the pal1-2 pal2-2 and pal1-3 pal2-3 double mutants are fertile (Fig. 3B). The difference in the fertility among the pal1 pal2 double mutants is most likely caused by the difference in light intensity used in the two studies, because the pal1-1 pal2-1 double mutant was also fertile under our growth conditions (Supplemental Fig. S1B). Thus, disruption of PAL1 and PAL2 renders the fertility of the mutant plants particularly sensitive to environmental growth conditions.
The pal1 pal2 mutants had approximately 30% of wild-type PAL activity (Fig. 2; Rohde et al., 2004), and the pal1-1 pal2-2 double mutant contained about 30% of wild-type levels of lignin in the inflorescence stems (Rohde et al., 2004). Lignin plays a crucial role in conducting water in plant stems. The polysaccharide compounds of cell walls of water-conducting tracheids and vessel elements of xylem tissues in vascular plants are highly hydrophilic. The cross-linking of cell wall polysaccharides by lignin, which is more hydrophobic, reduces water absorption to the cell wall and makes it possible for the plant vascular tissues to conduct water efficiently (Boudet, 2007). The cross-linking also enhances both whole-plant structural support and resistance to cell collapse under the tension of water transport (Boyce et al., 2004). The reduced lignin contents in the double mutants might lead a plant's vascular tissue to transport water at a reduced efficiency and consequently to enhanced drought tolerance, as observed in the pal1 pal2 double mutants (Fig. 6). Further studies will be required to determine whether manipulation of lignin contents or composition in plant vascular tissue could provide a novel avenue for modifying the rates of plant transpiration and improving drought tolerance.
The most striking phenotype of the pal1 pal2 double mutants is the lack of flavonoid pigments. The seeds of three independent pal1 pal2 double mutants are yellow (Fig. 3C; Supplemental Fig. S1C), apparently due to the lack of condensed tannin pigments in the seed coat (Fig. 3D). Unlike the wild type, the pal1 pal2 mutant plants also accumulated little anthocyanin pigments when plants were grown in underfertilized and relatively low-temperature conditions (Fig. 3A; Supplemental Fig. S1A). Lack of condensed tannin and anthocyanin pigments in the pal1 pal2 double mutants indicate that PAL1 and PAL2 have an important and redundant role in flavonoid biosynthesis. This macroscopic phenotype of the mutants was consistent with the previous molecular phenotyping that the pal1-1 pal2-1 double mutant lacks three major flavonol glucosides (Rohde et al., 2004), which are the major UV light-absorbing compounds in Arabidopsis. Other UV light-protecting metabolites, such as sinapoyl Glc and sinapoyl malate, were not reduced in the leaves or inflorescence stem of the pal1-1 pal2-1 mutant plants (Rohde et al., 2004). Thus, lack of flavonoid synthesis in the pal1 pal2 double mutants is most likely responsible for its high sensitivity to UV-B light treatment. In addition, we have observed that the double mutant exhibited reduced tolerance to paraquat, which induces oxidative stress, and had substantially reduced seed longevity (Z. Chen, unpublished results).
The severe phenotypes of the pal1 pal2 double mutants in flavonoid pigment accumulation illustrate the differential roles of different PAL isoforms in different branches of the phenylpropanoid pathway. It has been previously shown that increased flavonoid synthesis in response to nitrogen deficiency and reduced temperature in Arabidopsis is associated with increased PAL activity and elevated levels of PAL1 and PAL2 transcripts (Olsen et al., 2008). Thus, PAL1 and PAL2 functional specialization in abiotic environment-triggered flavonoid synthesis may be attributed to their responsive expression, which would lead to increased PAL activity and elevated synthesis of trans-cinnamic acid. However, we were unable to rescue the phenotype of flavonoid pigment deficiency of the pal1 pal2 double mutants by repeatedly feeding trans-cinnamic acid to the mutant plants. Likewise, feeding of trans-cinnamic acid or _para_-coumaric acid could not restore the kaempferol glycoside peaks in the pal1-1 pal2-1 double mutant (Rohde et al., 2004). Thus, environment-triggered or tissue-specific flavonoid synthesis may require coordinated expression of not only PAL1 and PAL2 but also other genes involved in this specific branch of the phenylpropanoid pathway. Different isoforms of PAL may also differ in metabolite sensitivity, localization, and metabolite-channeling complexes, which could contribute to the differential roles of the protein family members in different branches of the phenylpropanoid pathway.
Roles of the PAL Gene Family in Plant Growth and Development
We have generated two independent sets of T-DNA/transposon insertion mutants for the four Arabidopsis PAL genes. Surprisingly, both quadruple mutants contained 7% to 9% of wild-type PAL activity (Fig. 2). The residual PAL activity in the quadruple mutants could be due to incomplete knockout of one or more of the four PAL genes in the mutants. Real-time PCR analysis detected very low levels of transcripts for the PAL genes (mostly less than 1%) in their respective homozygous single mutants and in the homozygous quadruple mutants (Fig. 1B). As a result, we were unable to identify any possible leaky mutants responsible for the residual PAL activity in the quadruple mutants. The very low levels of PAL transcripts in the mutants might be residual degradation products of PAL transcripts and, as such, may not even be functional mRNA. Thus, there is also a possibility that the residual PAL activity in the quadruple mutants is due to a novel plant PAL activity. The pal1 pal2 pal3 pal4 quadruple mutant plants will facilitate the characterization and purification of the novel PAL activity, if it is indeed present in plants.
Although we failed to generate mutants completely deficient in PAL activity, the phenotypes of the two independent quadruple mutants with severely reduced PAL activity provided a glimpse into the roles of PAL in plant growth, development, and responses to environmental conditions. The quadruple mutants were severely stunted, particularly at early seedling stages, even with approximately 10% residual PAL activity. It is quite likely, therefore, that disruption of all PAL genes that completely eliminates PAL activity may very well cause a lethal phenotype. In addition, we could not find any significant number of pollen grains from the pal1 pal2 pal4 triple mutants and the pal1 pal2 pal3 pal4 quadruple mutants, and these mutants are all sterile. Previously, it has been shown that antisense inhibition of flavonoid biosynthesis in petunia (Petunia hybrida) anthers causes male sterility (van der Meer et al., 1992). Both greatly reduced flavonoid biosynthesis in the pal1 pal2 pal4 triple mutants and the pal1 pal2 pal3 pal4 quadruple mutants could account for the sterile phenotypes of the mutants. These results support that the phenylpropanoid pathway is essential for plant growth and development.
Lignin has a negative effect on processing of plant biomass for biofuel production. Lignin contents in the pal1 pal2 double and pal1 pal2 pal3 pal4 quadruple mutants were reduced to only about 25% to 30% of wild-type levels in inflorescence stems (Rohde et al., 2004; Fig. 7), which would be beneficial for conversion to ethanol for biofuel crops. Despite the great reduction in lignin content, the mutants still produced rigid inflorescence stems (Figs. 3 and 7). Thus, a major reduction in lignin and other phenolic compounds in plant stem through manipulation of PAL genes did not appear to compromise severely the important mechanical property of the lignified tissues. These mutants, however, were compromised in growth, fertility, and tolerance to environmental stress. Many of these growth and developmental defects probably resulted from reduced phenolic compounds (e.g. flavonoids) derived from the phenylpropanoid pathway in nonlignified cells. Thus, tissue-specific manipulation of PAL genes in plant vascular tissues may improve plant biomass for biofuel such as ethanol production.
Phenylpropanoid Pathway and SA Biosynthesis
Several studies have shown that a high PAL activity is important for pathogen-induced SA formation in plants. In tobacco, the levels of free SA produced in both pathogen-inoculated and upper systemic leaves of _PAL_-silenced plants are roughly 4-fold lower than those in control plants (Pallas et al., 1996). Furthermore, the PAL inhibitor 2-aminoindan-2-phosphonic acid reduced pathogen- or pathogen elicitor-induced SA accumulation in potato, cucumber, and Arabidopsis (Meuwly et al., 1995; Mauch-Mani and Slusarenko, 1996; Coquoz et al., 1998). In this study, we observed that all tested pal single, double, and triple mutants accumulated normal basal and pathogen-induced SA levels (Fig. 8). On the other hand, the basal and pathogen-induced SA levels in the two pal1 pal2 pal3 pal4 quadruple mutants were only about 25% and 50% of wild-type levels, respectively (Fig. 8). Since the quadruple mutants still contained about 10% of wild-type PAL activity, one can expect further reduction of SA accumulation if the PAL activity can be further reduced or completely abolished. Thus, our genetic analysis supports that the PAL activity is important for both basal and pathogen-induced SA accumulation. However, since SA accumulation is affected only when the PAL activity is severely reduced in Arabidopsis (Fig. 8), it appears that the reaction catalyzed by PAL is not a rate-limiting step for the accumulation of SA.
In a number of plants, including tobacco, rice (Oryza sativa), and potato, biochemical studies using isotope feeding have suggested that plants can synthesize SA from benzoate, which is synthesized from chain-shortening reactions of cinnamate produced by PAL (Klambt, 1962; Yalpani et al., 1993; Leon et al., 1995; Silverman et al., 1995). In Arabidopsis, the ics1 ics2 double mutant still accumulated about 4% of wild-type SA levels upon UV light exposure, and the phenylpropanoid pathway might be responsible for the synthesis of the residual SA in the double mutant. Since inhibition, silencing, or disruption of PAL activity or genes all have a major impact on SA accumulation, there must be other ways in which the phenylpropanoid pathway participates in SA biosynthesis and accumulation. For example, the PAL-dependent products may be involved in the generation of a precursor for SA biosynthesis. In bacteria, two enzymes catalyze the synthesis of SA from chorismate (Serino et al., 1995). ICS catalyzes the synthesis of isochorismate from chorismate, and isochorismate pyruvate lyase catalyzes the conversion of SA from isochorismate. Although plant ICS have been identified and analyzed, no plant isochorismate pyruvate lyase has been reported; therefore, how isochorismate is converted into SA in plants is still unknown. It is possible that isochorismate from the ICS pathway might not be directly converted into SA, as in bacteria, but instead might be conjugated with an intermediate from the phenylpropanoid pathway to produce an unknown SA precursor, analogous to the way in which intermediates of two different pathways are involved in lignin biosynthesis in the form of the phenylpropanoid intermediate _p_-coumaroylshikimate (Hoffmann et al., 2003). Indeed, we have recently shown that an Arabidopsis gene (Enhanced Pseudomonas Susceptibility1) encoding a BAHD acyltransferase is important for pathogen-induced SA accumulation (Zheng et al., 2009). The critical role of an acyltransferase in pathogen-induced SA accumulation is consistent with a possible integrated grid through formation of an ester conjugate from intermediates synthesized from two distinct pathways (Chen et al., 2009). Alternatively, the phenylpropanoid pathway may be responsible for the production of a regulatory molecule for SA biosynthesis or accumulation.
MATERIALS AND METHODS
Plant Growth Conditions
The Arabidopsis (Arabidopsis thaliana) wild type and pal mutants were grown in growth chambers at Purdue University or in a growth room at Zhejiang University at 22°C and 120 _μ_E m−2 s−1 light on a 12-h-light and 12-h-dark photoperiod.
Isolation of pal Mutants
Homozygous pal1, pal2, pal3, and pal4 mutant plants were identified by PCR using pairs of primers corresponding to sequences flanking the T-DNA insertions (Supplemental Table S3). The pal double, triple, and quadruple mutants were generated by genetic crossing. Homozygous pal1 pal2 double mutant plants were identified from the F2 progeny by PCR genotyping using pairs of primers flanking the insertion sites. The pal1 pal2 double mutants were then crossed with pal3 and pal4 to generate pal1 pal2 pal3 and pal1 pal2 pal4 triple mutants, respectively. The pal1 pal2 pal3 triple mutants were crossed with pal4 single mutants to generate pal1 pal2 pal3 pal4 quadruple mutants. Since the pal1 pal2 pal4 triple mutants and the pal1 pal2 pal3 pal4 quadruple mutants are sterile, they were identified in the progeny of plants homozygous for the pal1 and pal4 mutations (for triple mutants) or homozygous for pal1, pal3, and pal4 mutations (for quadruple mutants) but heterozygous for the pal2 mutations. Heterozygous pal2 mutants were identified by PCR using a combination of a T-DNA-specific primer (5′-TTGATTTGGGTGATGGTTCA-3′ for pal2-2 and 5′-CCCATTTGGACGTGAATGTAGACAC-3′ for pal2-3) and a _PAL2_-specific primer (5′-CAATGGATCAAATCGAAGCA-3′ for pal2-2 and 5′-TATTCCGGCGTTCAAAAATC-3′ for pal2-3).
RNA Isolation and RT-PCR
Total RNA was extracted with the RNeasy Plant Mini kit (Qiagen). RNA preparations were treated with DNase using the DNA-free kit from Ambion to remove contaminating DNA, and the treated RNA (1–5 _μ_g) was subjected to RT using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). Real-time PCR was conducted using SYBR PCR Master Mix (Applied Biosystems) and run on the ABI Prism 7000 system as described previously (Kim et al., 2008). The primers for the four PAL genes and ACT2 (At3g18780) for semiquantitative PCR were as described previously (Raes et al., 2003; Rohde et al., 2004). The primers for the four PAL genes for real-time PCR are listed in Supplemental Table S3.
Assay of PAL Activity
PAL activity was determined as described previously (Rohde et al., 2004). Briefly, leaf and stem tissue samples were homogenized in 0.1 m sodium borate, pH 8.8, and centrifuged for 10 min at 4°C at 14,000 rpm. The supernatant was saved and determined for protein content using the Bradford procedure. The reaction mixture consisted of 25 _μ_L of protein supernatant (10–50 _μ_g of protein), 75 _μ_L of 0.1 m sodium borate, pH 8.8, 3 _μ_L of [14C]l-Phe (0.01 _μ_Ci), and 5 _μ_L of 2 mm l-Phe. The reaction mixture was incubated at 37°C for 3 h. The reaction was stopped by adding 10 _μ_L of 10 n H2SO4. After adding 400 _μ_L of water, the mixture was extracted by adding 750 _μ_L of ether:cyclohexane mixture (1:1) and vortexing for 30 s. The organic phase was measured by a scintillation counter for the [14C]trans-cinnamic acid.
Vanillin Assays
Immature seeds (approximately 7–10 d after pollination) were incubated in a solution of 1% (w/v) vanillin and 6 n HCl at room temperature for 10 to 20 min as described previously (Debeaujon et al., 2000).
Pollen Collection and Counting
Anthers were removed from a fully open flower, and pollen grains were released into 50 _μ_L of 0.01% Silwet L-77 by gently squashing and agitating with a forceps. Pollen grains were counted with a hemocytometer using a microscope. Pollen grains were counted from 12 flowers for each genotype to increase the accuracy of the data.
Analysis of Tolerance to UV-B Light Treatment
Four-week-old plants of Col-0 and pal1 pal2 double mutants were irradiated with 1.24 _μ_mol m−2 s−1 UV-B light from four UV-B lamps (model BLE-1T158; Spectronics) for 4 h and then analyzed by chlorophyll fluorescence. Chlorophyll fluorescence was determined with an imaging pulse amplitude-modulated fluorometer (IMAG-MAXI; Heinz Walz). For measurement of _F_v/_F_m, plants were dark adapted for 30 min. Minimal fluorescence (_F_o) was measured during the weak measuring pulses, and maximal fluorescence (_F_m) was measured by a 0.8-s pulse of light at about 4,000 _μ_mol m−2 s−1. An actinic light source was then applied to obtain steady-state fluorescence yield (_F_s), after which a second saturation pulse was applied for 0.7 s to obtain light-adapted maximum fluorescence (_F_m′). _F_v/_F_m, ΦPSII, qp, and NPQ were calculated as _F_m − _F_o/_F_m, (_F_m′ − _F_s)/_F_m′, (_F_m′ − _F_s)/(_F_m′− _F_o), and (_F_m/_F_m′) − 1, respectively (Genty et al., 1989; Bilger and Bjorkman, 1990). In all cases, the whole plant was used as the area of interest.
Analysis of Drought Tolerance
For testing drought tolerance, 7-week-old Arabidopsis plants (eight plants for each genotype) were transferred into a walk-in growth chamber with approximately 50% humidity. The plants were unwatered and observed for drought stress symptom development.
Lignin Analysis
Cell wall isolation was performed as described previously (Rohde et al., 2004). Briefly, the inflorescence stems of 3-month-old plants of Col-0 wild type and pal1 pal2 pal3 pal4 mutants were harvested individually. Stem pieces (1.5 g) were immediately frozen and ground in liquid nitrogen. After addition of 30 mL of 50 mm NaCl, the mixture was kept at 4°C overnight and then centrifuged for 10 min at 3,500 rpm. The pellet was extracted with 40 mL of 80% ethanol and sonicated for 20 min. This extraction was repeated twice. The same centrifugation, extraction, and sonication steps were performed with acetone, chloroform:methanol (1:1), and acetone. For measuring lignin content, the acetyl bromide-soluble lignin method was performed as described previously (Fukushima and Hatfield, 2001). The acetyl bromide-soluble lignin was calculated using an extinction coefficient of 17.2 (Rohde et al., 2004).
SA Analysis
Pathogen inoculations were performed by infiltration of leaves of at least six plants (6–7 weeks old) for each treatment with the avirulent Pseudomonas syringae pv tomato DC3000 (avrRpt2) suspension in 10 mm MgCl2 as described previously (Xu et al., 2006). Total SA content was determined with a biosensor strain Acinetobacter species, ADPWH_lux, as described previously (Defraia et al., 2008). SA concentrations in the leaf samples were calculated based on the SA standard curve, which was constructed using the sid2 mutant leaf extract.
Pathogen Inoculation
Pathogen inoculations were performed by infiltration of leaves of at least six plants for each treatment with a suspension of the virulent Pst DC3000 strain in 10 mm MgCl2. For determining the bacterial growth, inoculated leaves were harvested at indicated days post inoculation and homogenized in 10 mm MgCl2. Diluted leaf extracts were plated on King's B medium supplemented with rifampicin (100 _μ_g mL−1) and kanamycin (25 _μ_g mL−1) and incubated at 25°C for 2 d before counting the colony-forming units as described previously (Xu et al., 2006). Inoculation of Botrytis cinerea was performed as described previously (Zheng et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. Phenotypes of the pal1-1 pal2-1 double mutant.
- Supplemental Figure S2. Inflorescences and siliques of wild-type and pal1 pal2 double mutant plants grown under different light conditions.
- Supplemental Figure S3. Responses to B. cinerea.
- Supplemental Table S1. pal mutants analyzed in the study.
- Supplemental Table S2. Pollen numbers of the wild type and pal1 pal2 mutants.
- Supplemental Table S3. Primers used for mutant identification and real-time PCR.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank the Arabidopsis Biological Resource Center at The Ohio State University and the Nottingham Arabidopsis Stock Centre for the T-DNA and transposon insertion mutants. We thank Dr. Wout Boerjan (Ghent University) for the pal1-1 and pal2-1 mutants. We are grateful to Hui Wang of the Natural Environment Research Council/Centre for Ecology and Hydrology (Oxford University) for the SA biosensor strain and Dr. Zhonglin Mou (University of Florida) for the assay protocol.
References
- Bilger W, Bjorkman O. (1990) Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis . Photosynth Res 25: 173–185 [DOI] [PubMed] [Google Scholar]
- Booij-James IS, Dube SK, Jansen MA, Edelman M, Mattoo AK. (2000) Ultraviolet-B radiation impacts light-mediated turnover of the photosystem II reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiol 124: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet AM. (2007) Evolution and current status of research in phenolic compounds. Phytochemistry 68: 2722–2735 [DOI] [PubMed] [Google Scholar]
- Boyce CK, Zwieniecki MA, Cody GD, Jacobsen C, Wirick S, Knoll AH, Holbrook NM. (2004) Evolution of xylem lignification and hydrogel transport regulation. Proc Natl Acad Sci USA 101: 17555–17558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catinot J, Buchala A, Abou-Mansour E, Metraux JP. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett 582: 473–478 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquoz JL, Buchala A, Metraux JP. (1998) The biosynthesis of salicylic acid in potato plants. Plant Physiol 117: 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defraia CT, Schmelz EA, Mou Z. (2008) A rapid biosensor-based method for quantification of free and glucose-conjugated salicylic acid. Plant Methods 4: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Cramer CL, Bolwell GP, Dixon RA, Schuch W, Lamb CJ. (1985) Rapid transient induction of phenylalanine ammonia-lyase mRNA in elicitor-treated bean cells. Proc Natl Acad Sci USA 82: 6731–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ. (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87: 9057–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer JL, Austin MB, Stewart C, Jr, Noel JP. (2008) Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem 46: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD. (2001) Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J Agric Food Chem 49: 3133–3139 [DOI] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodiere E, Catinot J, Buchala A, Doermann P, Metraux JP. (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briatais JM, Baker NR. (1989) The relationships between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt HD. (1962) Conversion in plants of benzoic acid to salicylic acid and its _β_-D-glucoside. Nature 196: 491 [Google Scholar]
- Lawton MA, Dixon RA, Hahlbrock K, Lamb C. (1983) Rapid induction of the synthesis of phenylalanine ammonia-lyase and of chalcone synthase in elicitor-treated plant cells. Eur J Biochem 129: 593–601 [DOI] [PubMed] [Google Scholar]
- Leon J, Shulaev V, Yalpani N, Lawton MA, Raskin I. (1995) Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc Natl Acad Sci USA 92: 10413–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XW, Dron M, Cramer CL, Dixon RA, Lamb CJ. (1989a) Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J Biol Chem 264: 14486–14492 [PubMed] [Google Scholar]
- Liang XW, Dron M, Schmid J, Dixon RA, Lamb CJ. (1989b) Developmental and environmental regulation of a phenylalanine ammonia-lyase-beta-glucuronidase gene fusion in transgenic tobacco plants. Proc Natl Acad Sci USA 86: 9284–9288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ. (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwly P, Molders W, Buchala A, Metraux JP. (1995) Local and systemic biosynthesis of salicylic acid in infected cucumber plants. Plant Physiol 109: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C. (2008) Differential expression of four Arabidopsis PAL genes: PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Pallas JA, Paiva NL, Lamb C, Dixon RA. (1996) Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J 10: 281–293 [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui JA, Maire V, Gabashvili IS, Fragata M. (2000) Oxygen evolution loss and structural transitions in photosystem II induced by low intensity UV-B radiation of 280 nm wavelength. J Photochem Photobiol B 56: 39–47 [DOI] [PubMed] [Google Scholar]
- Serino L, Reimmann C, Baur H, Beyeler M, Visca P, Haas D. (1995) Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet 249: 217–228 [DOI] [PubMed] [Google Scholar]
- Silverman P, Seskar M, Kanter D, Schweizer P, Metraux JP, Raskin I. (1995) Salicylic acid in rice: biosynthesis, conjugation and possible role. Plant Physiol 108: 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JNM, Stuitje AR. (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass I, Kirilovsky D, Etienne AL. (1999) UV-B radiation-induced donor- and acceptor-side modifications of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 38: 12786–12794 [DOI] [PubMed] [Google Scholar]
- Waring J, Underwood GJ, Baker NR. (2006) Impact of elevated UV-B radiation on photosynthetic electron transport, primary productivity and carbon allocation in estuarine epipelic diatoms. Plant Cell Environ 29: 521–534 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Leon J, Lawton MA, Raskin I. (1993) Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol 103: 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qualley A, Fan B, Dudareva N, Chen Z. (2009) An important role of a BAHD acyl transferase-like protein in plant innate immunity. Plant J 57: 1040–1053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]