The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals (original) (raw)
. Author manuscript; available in PMC: 2011 Jan 1.
Abstract
The conserved Hippo signaling pathway regulates organ size in both Drosophila and mammals. While a core kinase cascade leading from the protein kinase Hippo (Hpo) (Mst1 and Mst2 in mammals) to the transcription coactivator Yorkie (Yki) (YAP in mammals) has been established, upstream regulators of the Hippo kinase cascade are less well defined, especially in mammals. Using liver-specific conditional knockout mice, we demonstrate that the Merlin/NF2 tumor suppressor and the YAP oncoprotein function antagonistically to regulate liver development. While inactivation of Yap led to loss of hepatocytes and biliary epithelial cells, inactivation of Nf2 led to hepatocellular carcinoma and bile duct hamartoma. Strikingly, the _Nf2_-deficient phenotype was largely suppressed by heterozygous deletion of Yap, suggesting that YAP is a major effector of Merlin/NF2 in growth regulation. Our studies link Merlin/NF2 to mammalian Hippo signaling and implicate YAP activation as a mediator of pathologies relevant to Neurofibromatosis 2.
Introduction
The Hippo signaling pathway was initially discovered in Drosophila as a potent mechanism that restricts organ size by impinging on cell growth, proliferation and apoptosis. At the core of the Hippo signaling pathway is a kinase cascade comprised of four tumor suppressor proteins, including the Ste20-like kinase Hippo (Hpo) (Wu et al., 2003; Udan et al., 2003; Harvey et al., 2003; Pantalacci et al., 2003; Jia et al., 2003) and its regulatory protein Salvador (Sav) (Tapon et al., 2002; Kango-Singh et al., 2002), the NDR family kinase Warts (Wts) (Justice et al., 1995; Xu et al., 1995) and its regulatory protein Mats (Lai et al., 2005). The Hpo-Sav complex phosphorylates and activates the Wts-Mats complex (Wu et al., 2003; Wei et al., 2007), which in turn phosphorylates and inactivates the oncoprotein Yki (Huang et al., 2005) by excluding the latter from the nucleus (Dong et al., 2007; Oh and Irvine, 2008), where it normally functions as a coactivator for the TEAD/TEF family transcription factor Scalloped (Sd) (Wu et al., 2008; Zhang et al., 2008; Goulev et al., 2008). Inactivation of the Hippo pathway tumor suppressors or overexpression of the Yki oncoprotein results in tissue overgrowth, characterized by excessive cell proliferation and diminished apoptosis. The Hippo pathway components are highly conserved from Drosophila to mammals, and mammalian homologues of Hpo (Mst1/2), Sav (WW45), Wts (Lats1/2), and Yki (YAP) constitute an analogous kinase cascade (Dong et al., 2007; Zhao et al., 2007; Praskova et al., 2008). The importance of Hippo signaling in mammalian growth control is supported by reports that YAP is amplified in certain tumors and can transform immortalized mammary epithelial cells in vitro (Zender et al., 2006; Overholtzer et al., 2006) and that transgenic overexpression of YAP, or loss of Mst1/2, leads to massive hepatomegaly and rapid progression to hepatocellular carcinoma (HCC) (Dong et al., 2007; Camargo et al., 2007; Zhou et al., 2009; Lu et al., 2010; Song et al., 2010). However, whether YAP is normally required for liver development or homeostasis has not been established.
Compared to the core kinase cascade leading from Hpo to Yki phosphorylation, proteins acting upstream of the Hippo kinase cascade are less well defined. Earlier studies in Drosophila have implicated the apical membrane-associated FERM-domain proteins Merlin (Mer) and Expanded (Ex) as pathway components upstream of Hpo (Maitra et al., 2006; Hamaratoglu et al., 2006; Pellock et al., 2006; Tyler and Baker, 2007), although how Mer and Ex regulate Hpo activity was not established. Most recently, the WW and C2 domain-containing protein Kibra was identified as another apical membrane-associated tumor suppressor protein that functions together with Mer and Ex (Yu et al., 2010; Baumgartner et al., 2010; Genevet et al., 2010). Moreover, it was shown that these apical tumor suppressor proteins regulate the Hippo kinase cascade via direct binding to the Hpo-Sav complex (Yu et al., 2010). Recent studies further implicated the apical transmembrane protein Crumbs (Crb) as a cell surface protein that regulates Hippo signaling (Grzeschik et al., 2010; Ling et al., 2010; Robinson et al., 2010) by binding directly to Ex (Ling et al., 2010). Besides Kibra, Ex, Mer and Crb, the atypical cadherin Fat (Ft) (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006; Cho et al., 2006), the CK1 family kinase Disc overgrown (Dco) (Sopko et al., 2009; Feng and Irvine, 2009) and the myosin-like protein Dachs (Mao et al., 2006) have also been proposed as modulators of the Hippo pathway, although it is unresolved as to whether these proteins function upstream of Hpo in a linear pathway (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006), or act in parallel with the Hippo kinase cascade by influencing the proteins levels of Wts (Cho et al., 2006).
Among the proteins that have been implicated as upstream regulators of the Hippo signaling pathway, Mer is of particular clinical relevance since mutations of the mammalian gene encoding Mer, also known as Nf2, underlies Neurofibromatosis 2, an autosomal dominant disorder characterized by the development of benign schwannomas and other Schwann-cell-derived tumors associated with the nervous system (McClatchey and Giovannini, 2005; Okada et al., 2007). While the recent studies in Drosophila linking Mer to Hippo signaling suggests a potential mechanism by which Mer/NF2 may function as a tumor suppressor in mammals, it remains to be established whether Mer/NF2 regulates Hippo signaling in the context of normal mammalian physiology, and furthermore, to what extent the supposed Mer/NF2-Hippo connection can account for Mer/NF2's tumor suppressor function. The latter point is especially relevant given that besides the Hippo pathway, Mer/NF2 has been linked to a wide spectrum of effector pathways such as Ras, Rac, STAT or PI3K signaling, as well as contact inhibition mediated by cell-surface receptors or adherens junctions, and endocytosis/degradation of various membrane proteins (McClatchey and Giovannini, 2005; Okada et al., 2007). Most recently, it was suggested that Mer/NF2 suppresses tumorigenesis in mammalian cells by translocating the nucleus to inhibit a specific E3 ubiquitin ligase (Li et al., 2010). Notably, the contributions of these effector pathways to Mer/NF2-mediated tumorigenesis have not been defined under physiological conditions such as within intact mammalian tissues and organs.
In this study, we use mouse genetics to dissect the physiological role of the Hippo signaling pathway, focusing on a single mammalian organ, the mouse liver. We found that loss of Yap in the liver led to not only defects in hepatocyte survival, as one would predict on the base of YAP's ability to induce hepatocyte proliferation and HCC formation, but also a profound defect in biliary epithelial cell development, suggesting that the Hippo-YAP pathway is normally required in these two major liver cell types. Interestingly, loss of Nf2 resulted in hyperplasia of both cell types evidenced by the formation of HCC and bile duct hamartoma, suggesting that NF2 and YAP function antagonistically to each other in the context of mammalian Hippo signaling. Strikingly, the _Nf2_-deficient liver phenotypes, as well as cataract formation caused by _Nf2_-deficiency in the lens epithelium, were largely suppressed by heterozygous deletion of the Yap gene, suggesting that YAP is a major effector of NF2 in growth regulation. Our studies link NF2 to mammalian Hippo signaling and implicate YAP as a potential drug target for the therapeutic intervention of Neurofibromatosis 2.
Results
Generation of mice lacking Yap or Nf2 in the liver
Complete knockout of Yap results in early embryonic lethality and precludes further analysis of its function at later stages of development (Morin-Kensicki et al., 2006). To investigate the role of YAP in peri- and postnatal liver development, we generated a floxed allele of Yap (Yapflox) by homologous recombination in embryonic stem (ES) cells (Figure S1A). In this allele, the first two exons of Yap were flanked by two LoxP sites. In addition, a neomycin resistance gene (Neo) flanked by two FRT sites was inserted before the second LoxP site for drug selection, and was removed by expressing Flp recombinase. Further expression of Cre recombinase would delete the targeted first two exons and generate a deletion allele (YapKO), disrupting the transcription of Yap (Figures S1A and S1B). Homozygous mice carrying the Yapflox allele and heterozygous mice carrying the YapKO allele were viable, fertile, and exhibited no overt abnormalities. However, homozygous mice carrying the YapKO allele were not recovered from heterozygous intercrosses (over 100 progeny genotyped), in agreement with the reported lethality of homozygous Yap mutant embryos (Morin-Kensicki et al., 2006). A floxed allele of Nf2 (Nf2flox2) has been described previously (Giovannini et al., 2000).
To analyze function of YAP and NF2 in mouse liver, Yapflox and Nf2flox2 were crossed to the Albumin-Cre (Alb-Cre) recombinase to obtain Alb-Cre;Yap flox/flox or Alb-Cre;Nf2 flox2/flox2 mice. Alb-Cre drives liver-specific Cre expression starting at E13.5 and achieves efficient deletion at perinatal stage (E18 to P1) in both hepatocytes and biliary cells (Xu et al., 2006; Geisler et al., 2008). Successful deletion of Yap and Nf2 in the liver was confirmed by PCR genotyping (Figure S1C) and western blotting (Figure 6A). Quantitative RT-PCR showed that the Yap mRNA expression level in the liver of embryonic day 18.5 (E18.5) Yap mutant embryos was ~30% of that in wildtype littermates. The expression level further decreased to 20% in 2-week-old mutant mice and 5% in 8-week-old mutant mice (Figure S1D).
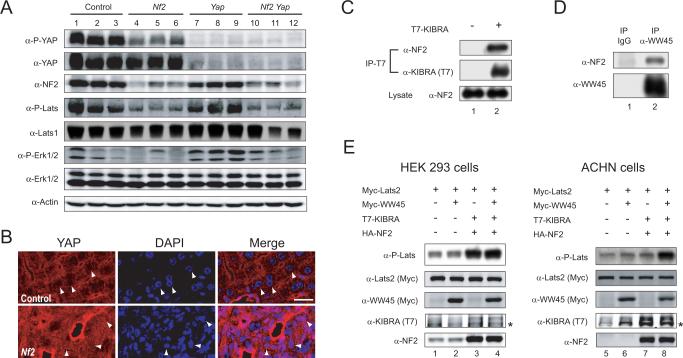
Figure 6. NF2 is required for Hippo signaling and physically associates with canonical pathway components.
(A) Western blot analysis of liver lysates from 2-month-old mice. Liver lysates from 3 mice of each indicated genotype were probed with antibodies against P-YAP, YAP, PLats, Lats1, P-Erk1/2, Erk1/2, NF2 and Actin. Note the decreased levels of YAP and Lats1/2 phosphorylation in Nf2 mutant livers.
(B) Confocal images of periportal areas from control and _Nf2_-deficient livers (at 2-month of age), stained for YAP (red) and DAPI (blue). Note that in wildtype hepatocytes (top), YAP staining shows a honeycomb-like pattern, with less YAP in the nucleus than in the cytoplasm (arrows). In contrast, _Nf2_-deficient cells (bottom) commonly show uniform distribution of YAP inside and outside of the nucleus. Representative cells are denoted by arrows. Scale bar = 20 μm.
(C) Physical association between NF2 and KIBRA. Cell lysate from HEK293 cells without (lane 1) or with (lane 2) T7-KIBRA expression was immunoprecipitated (IP) with α-T7 antibody and probed with antibody against endogenous NF2 and α-T7 antibody. Endogenous NF2 was detected in T7-KIBRA immunoprecipitates.
(D) Physical association between NF2 and WW45. α-WW45 antibody was used to immnoprecipitate endogenous WW45 from untransfected HEK293 cells and probed with α-NF2 or α-WW45 antibody. Endogenous NF2 was detected in WW45-immnoprecipitates, not in immunoprecipitates with control IgG.
(E) Lysates from HEK293 (left) or ACHN (right) cells expressing Myc-Lats2 with various combinations of Myc-WW45, T7-KIBRA and HA-NF2 were probed with the indicated antibodies. Note the stimulation of Lats2 phosphorylation by KIBRA-NF2 (lane 3) or KIBRA-NF2-WW45 (lane 4), but not WW45 (lane 2), in HEK293 cells. Also note the failure of KIBRA-NF2 (lane 7) to stimulate Lats2 phosphorylation, and the rescue of this defect by WW45 co-expression (lane 8). * denotes a non-specific band in α-KIBRA (T7) blots.
Impaired liver function in _Yap_-deficient livers
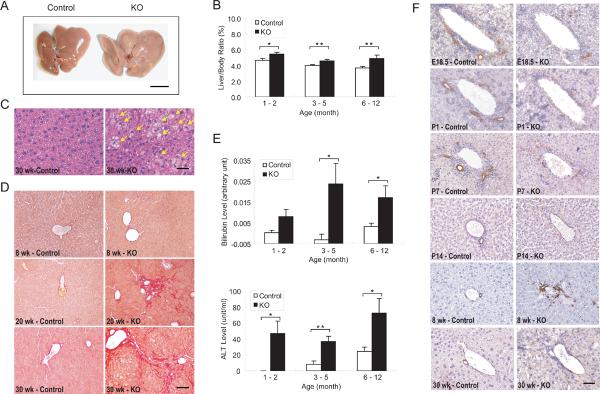
Alb-Cre;Yap flox/flox mice were born with the expected Mendelian ratio and no overt abnormalities. However, examination of the mutant mice revealed enlarged, pale livers (Figure 1A), and a moderate increase in liver/body weight ratio (Figure 1B). Histological analysis showed that the increased liver size resulted from macrovesicular steatosis (Figure 1C) and progressive fibrosis (Figure 1D). While no difference was seen between the mutant and control livers at 8 weeks of age, abnormal deposition of collagen was observed radiating from portal tracts in mutant livers at 20 weeks of age, indicating early periportal fibrosis. In mutant mice at 30 weeks of age, progressive fibrosis was evident as bridges of fibrosis that connected individual portal tracts and entrapped islands of hepatocytes, indicating a more advanced fibrosis with cirrhosis. Consistent with impaired liver function, elevated serum bilirubin levels and alanine aminotransferase (ALT) levels were detected in the mutant mice, which indicated an abnormal biliary system and hepatocellular injury, respectively (Figure 1E).
Figure 1. Impaired liver function and bile duct development in _Yap_-deficient livers.
(A) Gross image of control and Yap mutant livers from 1-year-old littermates. Note the pale and enlarged mutant liver. Scale bar = 1 cm.
(B) Increased liver-to-body weight ratio in Yap mutant mice. Mutant and control littermates were analyzed at indicated ages (values are means ± SEM): 1–2 months, n = 6; 3–5 months, n = 20; 6–12 months, n = 14. *p < 0.05, **p < 0.01, t-test.
(C) Steatosis in Yap mutant livers, as revealed by H&E staining. Note the presence of fat accumulation in mutant hepatocytes (yellow arrows). Scale bar = 50 μm.
(D) Fibrosis in Yap mutant livers, as evaluated by Sirius Red staining, a specific method for collagen detection. Control and mutant livers were analyzed at the indicated ages. Scale bar = 100 μm.
(E) Increased serum bilirubin and alanine aminotransferase (ALT) levels in Yap mutant mice. Control and mutant livers were analyzed at the indicated ages (values are means ± SEM): 1–2 months, n = 6; 3–5 months, n = 14; 6–12 months, n = 9. *p < 0.05, **p < 0.01, t-test.
(F) Impaired bile duct development in Yap mutant livers, as revealed by cytokeratin (CK) staining at the indicated ages. While CK-positive BECs in normal livers progressed from ductal plates to tubular structures around the portal veins from E18.5 to P7, the CK-positive BECs in Yap mutant livers were scattered around the portal veins and did not form typical tubular structures, leading to an absence of bile ducts at P14. An overabundance of CK-positive cells was present in 8-week-old Yap mutant livers, but these cells appeared disorganized and not incorporated into the portal mesenchyme, and did not persist in older mice (30-week-old). Scale bar = 50 μm.
See Figure S1 for data supplemental to Figure 1.
YAP is required for bile duct development
To investigate the etiology of the elevated serum bilirubin levels, we examined the biliary system of the Yap mutant mice. We stained the _Yap_-deficient livers with cytokeratin (CK), which is normally expressed in the biliary epithelial cells (BECs or cholangiocytes) of the liver (Geisler et al., 2008). During the normal development of the intrahepatic bile ducts, hepatic progenitor cells adjacent to the portal veins undergo ductal commitment, forming a structure known as the ductal plate. Prior to birth, tubulogenesis occurs at discrete points along the ductal plates, giving rise to primitive ductal structures, while the remaining unincorporated BECs regress during the first 2 weeks of life (Lemaigre, 2003). We noted that at E18.5, while control mice displayed typical ductal plate morphology with CK-positive BECs forming tubular and non-tubular structures immediately adjacent to the portal vein, CK-positive BECs rarely formed such structures in _Yap_-deficient livers (Figure 1F). At this stage, loss of YAP did not affect the proliferation or apoptosis of BECs as revealed by Ki67 and TUNEL staining (data not shown). From postnatal day 1 (P1) to postnatal day 14 (P14), tubular structures in control livers progressed into mature bile ducts that integrate into the portal mesenchyme, while CK-positive BECs in _Yap_-deficient livers regressed without forming mature bile ducts in the portal mesenchyme (Figure 1F). During the postnatal 1–5 months period, while control mice maintained a single mature bile duct within most portal tracts, _Yap_-deficient livers contained an increased number of CK-positive BECs forming irregularly shaped and variably sized structures circumferentially distributed in the periportal region (Figure 1F). These structures most likely represents a compensatory response to the absence of functional bile ducts, as observed previously in Notch2-deficient livers (Geisler et al., 2008) and typically seen in human chronic biliary disease (Nakanuma and Ohta, 1986). However, these CK-positive cells did not persist, such that in mutant mice older than 30 weeks, there is a complete absence of any bile duct or bile duct-like structures leaving only fibrosis in the periportal region (Figure 1F). Taken together, these findings reveal a critical role for YAP in bile duct development.
YAP is required for hepatocyte survival
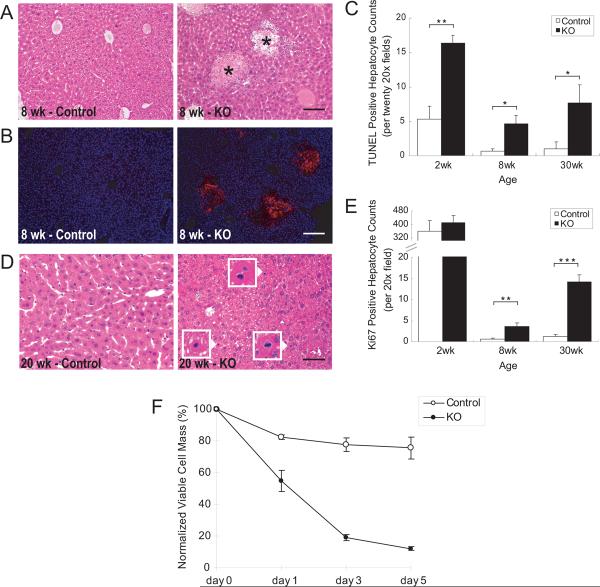
The increased serum ALT in Alb-Cre;Yap flox/flox mice suggests hepatocyte injury. Histological analysis of mutant livers revealed multiple areas of injured hepatocytes at the age of 1–2 months (Figure 2A). Hepatocytes in these injured areas were positive for cleaved Caspase-3 staining (Figure 2B), indicating that they were undergoing apoptosis. Although the visible apoptotic areas were mainly present in the mice at 1–2 months old, TUNEL staining showed increased numbers of apoptotic cells in Yap mutant livers at all ages examined (Figure 2C), suggesting that YAP is required for hepatocyte survival. Concomitant with increased hepatocyte death, _Yap_-deficient livers showed increased hepatocyte proliferation, as evidenced by increased mitotic figures (Figure 2D), Ki67 staining (Figure 2E), and BrdU labeling (data not shown). Thus, there appeared to be increased turnover of hepatocytes in _Yap_-deficient livers, with increased hepatocyte death accompanied with increased proliferation.
Figure 2. Increased hepatocyte turnover in _Yap_-deficient livers.
(A) H&E staining of control and Yap mutant livers, showing apoptotic areas of hepatocytes in the mutant liver (asterisks). Scale bar = 100 μm.
(B) Cleaved Caspase-3 staining (red), showing apoptotic areas of hepatocytes in the mutant liver. Scale bar = 100 μm.
(C) Quantification of TUNEL-positive hepatocytes in control and Yap mutant livers. Values are means ± SEM (n = 3). *p < 0.05, **p < 0.01, t-test.
(D) Increased mitosis in Yap mutant livers, as evaluated by H&E staining. Arrowheads point to mitotic hepatocytes, which are shown in higher magnification in the insets. Scale bar = 50 μm.
(E) Quantification of Ki67-positive hepatocytes in control and Yap mutant livers. Values are means ± SEM (n = 4). **p < 0.01, ***p < 0.001, t-test.
(F) Decreased viability of Yap mutant hepatocytes. Hepatocytes isolated from control and Yap mutant livers were cultured in vitro. Viable cell numbers were measured at indicated time points and plotted as percentage of viable cells relative to day 0. Values are means ± SEM (n=3).
See Figure S2 for data supplemental to Figure 2.
The increased hepatocyte turnover that was observed in the Yap mutant mice could reflect either a requirement for YAP in hepatocytes or a secondary response to the biliary defects in the _Yap_-deficient livers. To distinguish these two possibilities, we isolated and cultured primary hepatocytes in vitro. Compared to wildtype hepatocytes, _Yap_-deficient hepatocytes showed significantly decreased viability (Figure 2F) due to increased apoptosis in vitro (Figure S2). Thus, inasmuch as overexpression of YAP in hepatocytes confers resistance to apoptosis (Dong et al., 2007; Camargo et al., 2007), loss of YAP results in poor survival of the hepatocytes.
Loss of NF2 leads to hepatocyte and biliary epithelium overproliferation
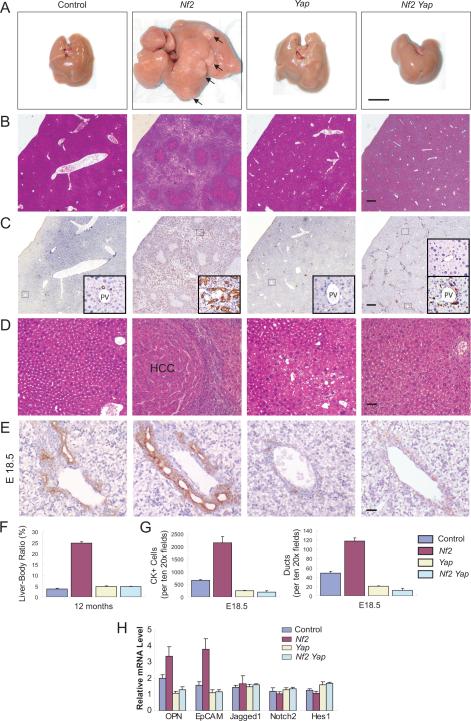
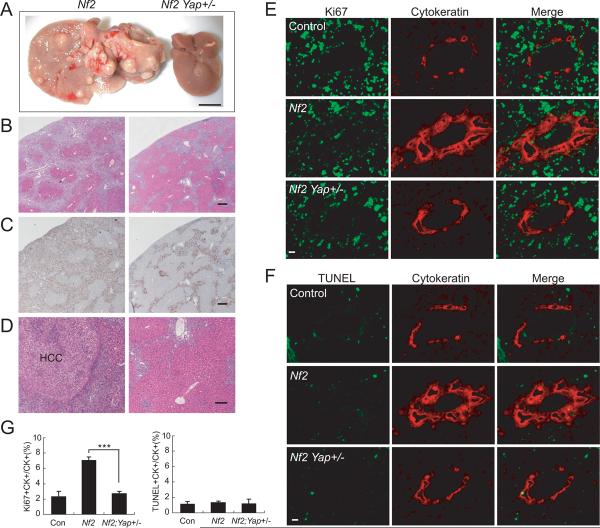
Heterozygous Nf2 knockout mice have been reported to develop a variety of tumors including hepatocellular carcinoma (9% incidence) (McClatchey et al., 1997), but the specific function of NF2 in liver growth has not been investigated in detail. To this end, we generated Alb-Cre;Nf2 flox2/flox2 mice. Strikingly, the Alb-Cre;Nf2 flox2/flox2 mice developed widespread bile duct hamartomas, which appeared microscopically as collections of irregularly shaped duct-like structures surrounded by stroma and which comprised of BECs without cellular atypia (Figures 3B and 3C). Cytokeratin staining confirmed that the proliferating cells were of biliary cell origin (Figure 3C). As is typical of bile duct hamartomas, they first appeared at the surface of the liver and were observed in all mutant livers at 1 month of age (Figure S4). These hamartomas continued to expand in adult life and extend from the hepatic capsule to the deep liver parenchyma, eventually dividing the liver into islands of hepatocytes (Figures 3B and 3C). In addition to bile duct hamartoma, hepatocellular carcinomas (HCC) developed in 100% of the mutant mice at 1 year of age (Figures 3A and 3D). The extensive proliferation of bile ducts and hepatocytes was so severe that the mutant livers represented ~25% of the body weight at 1 year of age (Figure 3F).
Figure 3. Loss of Yap suppresses Nf2 mutant phenotypes.
Unless otherwise indicated, livers from 1-year-old mice of the indicated genotypes were analyzed.
(A) Gross liver images. Note that the Nf2 mutant liver shows massive overgrowth and formation of hepatocellular carcinoma (HCC) (indicated by arrows and confirmed by H&E staining in D). Scale bar = 1 cm.
(B–C) H&E staining (B) and CK staining (C) showing the prevalence of bile duct hamartoma in Nf2 mutant livers and the absence of bile duct hamartoma in Nf2 Yap double mutant livers. Insets in C are higher magnifications of representative portal areas: normal bile duct in control liver; multiple bile ducts in Nf2 mutant liver; no bile duct in Yap mutant liver; some portal areas have no bile ducts, whereas others have CK-positive cells but do not form tubular structures in Nf2 Yap double mutant liver. Scale bars = 250 μm.
(D) H&E staining showing HCC in Nf2 mutant livers and the absence of HCC in Nf2 Yap double mutant livers. Scale bar = 60 μm.
(E) Comparison of bile ducts around portal veins in E18.5 livers by CK staining. Note the overabundance of CK-positive BECs and multiple focal dilations of ductal plates in Nf2 mutant livers, and the diminished BECs in Yap and Nf2 Yap double mutant livers. Scale bar = 30 μm.
(F) Quantification of liver-to-body weight ratio. Note the severe hepatomegaly of Nf2 mutant livers and the similar size of Yap and Nf2 Yap double mutant livers. Values represent means ± SEM (n = 3).
(G) Quantification of CK-positive BECs and ductule structures for E18.5 livers. Values represent means ± SEM (n = 3).
(H) Quantitative real-time PCR analysis of selected genes from livers of control, Nf2, Yap and Nf2 Yap mice at the age of E18.5. OPN and EpCAM expression was increased in Nf2 livers, but decreased in Yap and Nf2 Yap livers. Jagged 1, Notch2 and Hes1 expression was not affected in any of the genetic background. Values are means ± SEM (n=3).
See Figure S3 for data supplemental to Figure 3.
Suppression of Nf2 mutant liver phenotypes by loss of YAP
The contrasting phenotypes of _Yap_- and _Nf2_-deficient livers suggest that NF2 and YAP act antagonistically with each other in a common pathway to regulate liver homeostasis. To further investigate this possibility, we generated Nf2 and Yap double mutant livers using Alb-Cre (Alb-Cre; Nf2 flox2/flox2; Yap flox/flox). In these mice, the overgrowth of Nf2 mutant livers was greatly suppressed by loss of Yap. First, Nf2Yap double mutant mice had liver sizes comparable to Yap mutant mice, while the Nf2 mutant livers were much larger (Figures 3A and 3F). Second, bile duct overproliferation of Nf2 mutant livers was suppressed in the Nf2Yap double mutant livers. At E18.5, while Nf2 mutant livers showed increased number of BECs and multiple tubular structures surrounding each portal vein, Nf2Yap double mutant livers showed no tubular structures and a decreased number of CK-positive cells similar to that observed in _Yap_-deficient livers (Figures 3E and 3G). In agreement with the histological analysis, Nf2 mutant livers showed increased mRNA levels of osteopontin (OPN) and EpCAM, two BEC-enriched markers (Zong et al., 2009), while Yap or Nf2Yap livers showed a similar decrease of these BEC markers (Figure 3H). At 1 year of age, while Nf2 mutant livers showed widespread bile duct hamartomas, hamartomas were not detected in Nf2Yap double mutant livers (Figures 3A–C). Third, while 100% of Nf2 mutant mice developed extensive HCC by 1 year of age, neither Yap nor Nf2Yap mutant mice developed HCC at appreciable frequencies (Figure 3D). Lastly, the suppression of Nf2 mutant phenotype by loss of Yap is specific, since _Yap_-deficiency did not suppress liver overgrowth and tumorigenesis induced by the expression of an oncogenic Kras mutant (G12D) (Tuveson et al., 2004) (Figures S3A–C). Thus, Yap is genetically epistatic to Nf2 in liver growth.
In the course of analyzing the genetic interactions between Nf2 and Yap, we noted that even heterozygous deletion of Yap, which normally has no phenotypic consequences in an otherwise wildtype genetic background, caused significant suppression of the Nf2 mutant phenotype. Thus, while Alb-Cre; Nf2 flox2/flox2 mice showed massive liver growth (with a liver/body ratio of over 30%) and tumor formation (bile duct hamartoma and HCC) at 16 months of age, livers from Alb-Cre; Nf2 flox2/flox2; Yap flox/+ littermates were only slightly larger than wildtype (with a liver/body ratio of 6.5% versus 5% in wildtype), had no apparent HCC, and displayed much reduced bile duct overproliferation (Figures 4A–D). The dominant suppression of _Nf2_-loss phenotype by Yap heterozygosity was also apparent at 1 month of age, when bile duct hamartomas appeared in 100% of Nf2 mutant livers but were absent in Nf2Yap+/− livers (Figure S4). To trace the developmental origin of this dominant suppression, we examined perinatal animals at P0. While Alb-Cre; Nf2 flox2/flox2 animals showed enhanced proliferation of the CK-positive BECs compared to the wildtype littermates, such overproliferation was greatly suppressed in Alb-Cre; Nf2 flox2/flox2; Yap flox/+ littermates (Figures 4E and 4G). In contrast, TUNEL staining
Figure 4. Heterozygosity of Yap suppresses Nf2 mutant liver phenotypes.
(A) Gross liver images from two littermates at 16 months of age. Left: Alb-Cre; _Nf2_flox2/flox2; Yap+/+. Right: Alb-Cre; _Nf2_flox2/flox2; _Yap_flox/+. Note the massive overgrowth and tumor formation in Nf2 mutant liver (left) and the normal appearance of Nf2, Yap+/− liver (right). Scale bar = 1 cm.
(B–C) H&E staining (B) and CK staining (C) showing the prevalence of bile duct hamartoma in Nf2 mutant livers (left) and the greatly diminished bile duct hyperplasia in Nf2 Yap+/− livers (right). Scale bars = 125 μm.
(D) H&E staining showing HCC in Nf2 mutant liver (left) and the absence of HCC in Nf2 Yap+/− liver (right). Scale bar = 60 μm.
(E) Control (top), Nf2 (middle) and Nf2 Yap+/− (bottom) livers from P0 animals were analyzed for Ki67 staining (green) and counterstained for the BEC marker cytokeratin (red). Note the abundance of Ki67-positive BECs in Nf2 livers, but not in control or Nf2 Yap+/− livers. Scale bar = 20 μm.
(F) Similar to (E) except that apoptosis was analyzed by TUNEL staining (green). Note the general absence of TUNEL-positive BECs in all genotypes.
(G) Quantification of Ki67- and TUNEL-positive BECs from (E–F). Values are means ± SEM (n = 5). ***, p < 0.001, t-test.
See Figure S4 for data supplemental to Figure 4.
revealed hardly any apoptosis in the BECs from wiltype control, Alb-Cre; Nf2flox2/flox2 or Alb-Cre; Nf2flox2/flox2; Yap flox/+ animals (Figures 4F and 4G). Thus, the suppression of _Nf2_-deficient phenotypes by Yap heterozygosity is not due to the removal of excess cells generated upon loss of Nf2, but rather indicative of a direct role for YAP in NF2-mediated signaling.
Yap heterozygosity suppresses cataracts induced by loss of Nf2
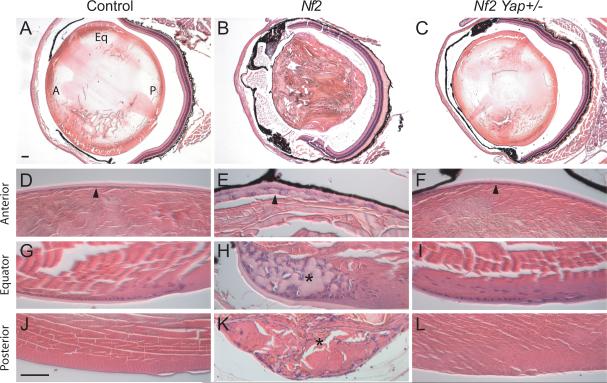
The striking suppression of Nf2 mutant liver phenotypes by heterozygosity of Yap prompted us to examine whether such dosage-sensitive interaction is a general feature of NF2-YAP signaling. For this purpose, we examined the functional relationship between NF2 and YAP in the lens, wherein nearly all neurofibromatosis 2 patients are known to exhibit ocular defects such as presenile cataracts, which serve as one of the diagnostic criteria for this disease (Bouzas et al., 1993). To model Neurofibromatosis 2-associated lens defects in mice, we conditionally deleted Nf2 using Nestin-Cre (Nes-Cre), which has been used for gene inactivation in the lens epithelium as early as E11.5 (Cang et al., 2006; Calera et al., 2006) (data not shown). Consistent with clinical findings, we found that loss of NF2 in the lens epithelium leads to the formation of different types of cataracts characterized by the disorganization of lens epithelium in the anterior region of the lens (compare Figures 5D and 5E), as well as accumulation of ectopic cells in the equator (compare Figures 5G and 5H) and posterior (compare Figures 5J and 5K) of the lens. Interestingly, all these defects were significantly suppressed by Yap heterozygosity (Figures 5F, 5I and 5L). Together with our findings in the livers, these results support a general epistatic relationship between Yap and Nf2 in mammalian tissue growth.
Figure 5. Yap heterozygosity suppresses cataracts induced by loss of Nf2.
(A–C) H&E staining of lens from 2-month old littermates with the following genotypes: (A) _Nf2_flox2/flox2 (as control), (B) Nes-Cre; _Nf2_flox2/flox2, and (C) Nes-Cre; _Nf2_flox2/flox2; _Yap_flox/+. Note the general loss of structural integrity in (B), but not in (A) or (C). In (A), the anterior, equator and posterior regions of lens are marked by A, Eq and P, respectively.
(D–L) High magnification views of lens in (A–C) highlighting anterior, equator and posterior regions of the lens. Note the disorganization of anterior lens epithelium in Nes-Cre; _Nf2_flox2/flox2, and the normal monolayer cuboidal epitheliaium in Nes-Cre; _Nf2_flox2/flox2; _Yap_flox/+ lens (compare arrowheads in D–F). Also note the accumulation of ectopic cells and capsular material at the equator region of Nes-Cre; _Nf2_flox2/flox2 (asterisk, H), but not Nes-Cre; _Nf2_flox2/flox2; _Yap_flox/+ (I) lens. Furthermore, the posterior lens, which is normally accellular (J; appearing as hematoxylin-negative), shows abberant accumulation of cells in Nes-Cre; _Nf2_flox2/flox2 (asterisk, K), but not Nes-Cre; _Nf2_flox2/flox2; _Yap_flox/+ (L) lens.
Scale bars = 100 μm.
NF2 is required for Hippo signaling and physically associates with canonical pathway components
To further corroborate a functional relationship between NF2 and YAP, we examined YAP S112 (corresponding to S127 of human YAP) phosphorylation, a specific Hippo signaling readout, in _Nf2_-deficient livers. _Nf2_-deficient livers showed reduced YAP S112 phosphorylation (Figure 6A). Consistent with this finding, _Nf2_-deficient livers showed increased nuclear YAP and elevated YAP protein levels, which was seen most prominently in the periportal areas (Figure 6B). Furthermore, _Nf2_-deficient livers displayed reduced Lats1/2 activity, as revealed by a phospho-specific antibody against its hydrophobic motif (Yu et al., 2010) (Figure 6A). The reduction of YAP and Lats1/2 phosphorylation in _Nf2_-deficient livers was not merely a non-specific consequence of cell overproliferation, as their phosphorylation levels were not reduced in liver overgrowth driven by oncogenic KrasG12D (Figure S3D). Conversely, KrasG12D, but not loss of NF2, led to activation of MAP kinase (Erk1/2) (Figure 6A and Figure S3D). Taken together, these results demonstrate that endogenous NF2 is normally required for Hippo signaling in liver development and physiology.
A recent study in Drosophila showed that Mer/NF2 can directly associate with Kibra, a newly identified tumor suppressor of the Hippo pathway, and Sav, a component of the Hpo-Sav kinase complex (Yu et al., 2010). To test whether such protein-protein interactions are conserved in their mammalian counterparts, we conducted co-immunoprecipitation assays to examine physical interactions between these mammalian proteins. Consistent with the findings in Drosophila, we found that epitope-tagged human KIBRA protein immunoprecipitated endogenous NF2 (Figure 6C), and endogenous NF2 and WW45 (the mammalian Sav homologue) co-immunoprecipitated with each other (Figure 6D).
The NF2-KIBRA interactions described here is consistent with our previous findings that co-expression of mammalian NF2 and KIBRA resulted in synergistic activation of Lats1/2 hydrophobic motif phosphorylation in HEK293 cells (Yu et al., 2010; see also Figure 6E). To probe the functional significance of the observed NF2-WW45 interactions, we took advantage of the renal cancer cell line ACHN, which is known to have deleted WW45 (Tapon et al., 2002). While co-expression of NF2 and KIBRA stimulated Lats2 hydrophobic motif phosphorylation in HEK293 cells (Figure 6E), which are known to express WW45 (Dong et al., 2007), NF2-KIBRA expression failed to stimulate Lats2 phosphorylation in the WW45-deficient ACHN cells (Figure 6E). Importantly, re-introducing WW45 in ACHN cells rescued the ability of NF2-KIBRA to stimulate Lats2 phosphorylation, even though expression of WW45 by itself had no effect on Lats2 phosphorylation in either cell types (Figure 6E). Thus, at least in this cell culture context, WW45 is a critical mediator of NF2 input into Hippo signaling.
Discussion
YAP is required for liver homeostasis
First discovered in Drosophila, the Hippo signaling pathway has recently emerged as a conserved mechanism that regulates tissue homeostasis in metazoans. In Drosophila and mammals, the Hippo pathway regulates growth by phosphorylating and inactivating the conserved transcriptional coactivator Yki (Drosophila) or its mammalian counterpart YAP. Consistent with its pivotal position in Hippo signaling, it was shown in Drosophila that gain-of-function of Yki, as exemplified by Yki overexpression or inactivation of tumor suppressors upstream of Yki, results in tissue overgrowth, whereas loss-of-function of Yki results in poor cell survival and tissue atrophy (Huang et al., 2005). Interestingly, the evidence so far implicating Hippo signaling in mammalian tissue homeostasis are largely based on YAP gain-of-function contexts, wherein gene amplification (Zender et al., 2006; Overholtzer et al., 2006), transgenic overexpression of YAP (Dong et al., 2007; Camargo et al., 2007), or loss of Mst1/2 (Zhou et al., 2009; Lu et al., 2010; Song et al., 2010), all lead to tissue overgrowth or cell transformation. Whether YAP is normally required for mammalian tissue development or homeostasis has not been examined.
Using a conditional mouse knockout model, we investigated the function of YAP in mouse livers. We found that loss of YAP in the liver not only results in defects in hepatocyte survival, but also leads to a profound defect in bile duct development. While primitive ductal plates can form in the vicinity of portal veins in Alb-Cre; Yapflox/flox livers, they never undergo dilation and progression into tubular bile ducts, thus resulting in bile duct paucity by 2 weeks of age. Despite the compensatory proliferation of periportal BECs, no bile ducts within or surrounding the portal tracts can persist in _Yap_-deficient livers older than 30 weeks of age. Thus, YAP is required for normal bile duct development.
We note that the paucity of intrahepatic bile ducts in _Yap_-deficient livers resembles that observed in Alagille syndrome, a human disease caused by defective Notch signaling, as well as various mouse models with defective Notch signaling in the liver (Geisler et al., 2008). However, defective Notch signaling is unlikely to account for the Yap mutant phenotype, since we observed no changes in the expression levels of various Notch pathway components (Figure 3H). Instead, the reciprocal bile duct defects in _Yap_- and _Nf2_-deficient livers implicate Hippo signaling as a pathway that regulates bile duct development independent of Notch.
A functional link between Merlin/NF2 and Hippo signaling in mammals
Since its initial discovery as a human disease gene underlying Neurofibromatosis 2, the Merlin/NF2 tumor suppressor has been the subject of intense investigation. While studies in mammalian cell cultures have linked a number of effector pathways to NF2 (McClatchey and Giovannini, 2005; Okada et al., 2007; Li et al., 2010), the relative contribution of these effector pathways to NF2 function in intact tissues has not been defined. In this study, we provide multiple lines of evidence implicating a functional link between NF2 and the Hippo signaling pathway. First, inactivation of NF2 results in hyperplasia of hepatocytes and BECs accompanied by the formation of HCC and bile duct hamartoma, whereas inactivation of YAP results in the loss of both cell types, suggesting that NF2 and YAP act antagonistically to regulate tissue homeostasis in vivo. Second, loss of NF2 in vivo results in a decrease of YAP and Lats1/2 phosphorylation, as one would predict if NF2 functions as an upstream regulator of Hippo signaling in mammals. Third, in mammalian cell culture, NF2 physically associates with KIBRA and WW45, two known components of the Hippo pathway, and can stimulate Hippo signaling in a WW45-dependent manner. Last and foremost, the loss-of-Nf2 phenotype, both in the liver and in the lens, is largely suppressed by heterozygous deletion of Yap. This observation strongly implicates YAP as a major effector downstream of the NF2 tumor suppressor in mammalian growth control. To our knowledge, genetic interactions of such nature have not been reported for any other purported effector pathways downstream of NF2. That Yap heterozygosity, which is inconsequential in an _Nf2_+ genetic background, protects _Nf2_-deficient cells from tumor development further suggests that YAP may represent a potential drug target for the development of therapeutics against Neurofibromatosis 2, for which no effective treatments are available at present (Evans et al., 2009).
Recent studies in Drosophila showed that Mer/NF2 functions together with two other apical membrane-localized proteins Ex and Kibra to regulate the Hpo-Sav kinase complex (Yu et al., 2010). Our current study, together with the functional conservation of Mer/NF2 activity between Drosophila and mammals (LaJeunesse et al., 1998), suggests that this aspect of Hippo signaling is likely conserved in mammals. How these apical proteins are regulated in either Drosophila or mammals remains an important and unanswered question that will likely be the focus of future investigation.
Experimental Procedures
Generation of Yap conditional knock-out (CKO) mice
A targeting vector containing the first two exons of the Yap gene was generated by recombineering as described previously (Liu et al., 2003). Transformed ES colonies were screened by long-template PCR with the following primer sets: P5F (5'-TTCCTGGAACTTAAGGTTCTGCCTCCGAGCTTGCTTGCCTTTTAG-3') and P5R (5'-TTAAGGGTTATTGAATATGATCGGAATTGGGCTGCAGGAA-3') to generate a 6.3-kb band for positive clones; P3F (5'-GCTCTATGGCTTCTGAGGCGGAAAGAACCAGCTGGGGCTCGAC-3') and P3R (5'-TACTGGATCACATCTGGCACATCAAATTCCGACGCCTCCGTGGTA-3') to generate a 3-kb band for positive clones.
Successfully targeted ES clones (confirmed by both 5'PCR and 3'PCR) were microinjected into C57BL/6 blastocysts. Germline transmission from generated chimeric offspring was confirmed by long-template PCR (Figure S1B). Mice carrying the targeted allele were bred to Flp recombinase transgenic mice (kindly provided by Dr. Jeremy Nathans, Johns Hopkins University School of Medicine) to remove the FRT-flanked Neo cassette and to generate the Yapflox mice.
Genomic DNAs extracted from tail biopsies were genotyped with a PCR primer set (P1: 5'-CCATTTGTCCTCATCTCTTACTAAC-3', P2: 5'-GATTGGGCACTGTCAATTAATGGGCTT-3', P3: 5'-CAGTCTGTAACAACCAGTCAGGGATAC-3') that generated a 498-bp band from the wild-type allele, a 597-bp band from the floxed allele, and a 697-bp band from the knockout allele (Figure S1C).
To achieve liver-specific gene deletion, _Yap_flox/flox mice were bred to Albumin-Cre transgenic mice (003574) from the Jackson Laboratory (Bar Harbor, USA). To avoid potential variation related to gender, all experiments were performed in male mice with paternal inheritance of Alb-Cre.
Hepatocyte isolation and culture
All animal experiments were carried out in accordance with NIH guidelines for the care and use of experimental animals. Hepatocytes were isolated by two-step collagenase perfusion of 10- to 12-week old Yap mutant mice and the littermate controls (Kreamer et al., 1986). Cells were plated at a density of 1×104 cells per well of a 96-well plate and grown in D-MEM/F-12 medium (Invitrogen) supplemented with 1× ITS (Insulin-transferrin-sodium selenite, Sigma), 30ug/ml proline, 1mg/ml galactose, 7.5% NaHCO3, and 1× penicillin/streptomycin at 37°C with 5% CO2 as described(Kreamer et al., 1986). The number of live cells was determined by the CellTiter 96® Non-Radioactive Cell Proliferation Assay (Promega; G4000). The experiments were performed in triplicate and repeated independently in three pairs of animals.
Quantitative real-time PCR
Total liver cellular RNA was extracted using the TRIzol reagent (Invitrogen). RNA was reverse-transcribed with random primers using the Affinitycript multiple temperature cDNA synthesis kit (Stratagene). Real-time quantitative PCR (Q-PCR) was performed using the Quantitect SYBR Green PCR kit (Qiagen) on a StepOnePlus PCR System (Applied Biosystems). Q-PCR was done in triplicate, using histone H2A.Z as a housekeeping control. Relative differences in the expression of the candidate genes in control and mutant livers were determined using the 2 -ΔΔCt method. The mouse primer sequences used are available on request.
Histological analysis and immunostaining
Paraffin-embedded liver tissue was cut into 5-μm sections, which were stained with hematoxylin-eosin for histologic examination or with Sirius Red to visualize fibrosis. For analysis of adult lens, mouse skulls were fixed and decalcified in Formical-4, and processed for paraffin embedding and sections. Immunohistochemical and immunofluorescent staining were performed according to the protocols provided by the manufacturers of the respective antibodies. The following primary antibodies were used: Ki-67 (DAKO; M7249, 1/25), wide spectrum screening cytokeratin (pan-CK) (DAKO; Z0622, 1/500), Yap (Sigma; WH0010413M1, 1/100), NF2 (Sigma; HPA003097, 1/150), and cleaved Caspase-3 (Asp175) (Cell Signaling; #9661, 1/100). The secondary antibodies used were: Envision™ anti-rabbit (DAKO; K4002), and rabbit anti-goat IgG HRP (Santa Cruz Biotechnology; SC-2768, 1/100). The DAB+ (DAKO; K3467) visualization system was used for immunohistochemical staining, and Cy3- or Alexa488-conjugated rabbit secondary antibody was used for immunofluorescent staining. TUNEL staining was performed with the DeadEnd™ colorimetric TUNEL system (Promega; G7130) or In Situ Cell Death Detection Kit (Roche) according to the manufacturers' instructions.
BEC and bile duct quantification
To quantify the pan-CK+ cells and bile ducts in the E18.5 embryo, we evaluated 10 randomly chosen 20× fields/section for each mouse. The numbers of pan-CK+ cells and bile ducts were quantified by adding the respective numbers for each 20× field.
Protein lysate and western blot analysis
Cells or tissues were lysed in RIPA buffer (150mM NaCl, 50mM Tris-HCl, pH7.4, 1% NP-40, 0.5% sodium-deoxycholate, 0.1% SDS, and 1 mM PMSF) with protease inhibitors (Roche). The proteins were separated on SDS-polyacrylamide gels and transferred onto PVDF membranes (Millipore). The blots were probed with antibodies against YAP (Cell Signaling; #4912), phospho-YAP (Ser127) (Cell Signaling; #4911), Lats1 (Cell Signaling; #3477), phospho-Lats1/2 (Yu et al., 2010), Erk1/2 (Cell Signaling; #4696), Phospho-Erk1/2 (Thr202/Tyr204) (Cell Signaling; #4370), NF2 (Sigma; HPA003097), and normalized by Actin (Millipore; MAB1501). Signals were detected and quantified by LI-COR Infrared Imaging system.
Mammalian cell culture and Immunoprecipitation
HEK293 cells and ACHN cells were maintained in DMEM medium supplemented with 10% fetal bovine serum and antibiotics (Invitrogen). The T7-KIBRA, Myc-Lats2, Myc-WW45 and HA-NF2 expression plasmids were described previously (Yu et al., 2010). Immunoprecipitation was carried out using RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH8.0) supplemented with 1 mM PMSF and protease inhibitors (Roche). Antibodies for western blot include: phospho-Lats (Yu et al., 2010), Myc (Santa Cruz, Sc40 HRP), T7 (Novagen, #69522), NF2 (Sigma; HPA003097), WW45 (Novus, H00060485-M02).
Serum ALT and direct bilirubin measurements
Serum levels of alanine aminotransferase (ALT) and direct bilirubin were measured using commercially available kits (product nos. 68-D and 020-A, respectively) from Biotron Diagnostic Inc. (Hemet, CA) according to the manufacturer's protocol.
Supplementary Material
01
Acknowledgments
We thank Dr. Jeremy Nathans for providing CMV-Cre and Flp transgenic mice, Dr. Kuan-Ting Kuo for discussion of the Yap mutant phenotype, and Kathleen McGowan for help with mouse genotyping. This study was support in part by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01 DK081417 01 to RAA. D.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of hippo in Drosophila. Dev. Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bouzas EA, Freidlin V, Parry DM, Eldridge R, Kaiser-Kupfer MI. Lens opacities in neurofibromatosis 2: further significant correlations. Br. J. Ophthalmol. 1993;77:354–357. doi: 10.1136/bjo.77.6.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calera MR, Topley HL, Liao Y, Duling BR, Paul DL, Goodenough DA. Connexin43 is required for production of the aqueous humor in the murine eye. J. Cell Sci. 2006;119:4510–4519. doi: 10.1242/jcs.03202. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cang Y, Zhang J, Nicholas SA, Bastien J, Li B, Zhou P, Goff SP. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell. 2006;127:929–940. doi: 10.1016/j.cell.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Kalamarides M, Hunter-Schaedle K, Blakeley J, Allen J, Babovic-Vuskanovic D, Belzberg A, Bollag G, Chen R, DiTomaso E, Golfinos J, Harris G, Jacob A, Kalpana G, Karajannis M, Korf B, Kurzrock R, Law M, McClatchey A, Packer R, Roehm P, Rubenstein A, Slattery W, III, Tonsgard JH, Welling DB, Widemann B, Yohay K, Giovannini M. Consensus recommendations to accelerate clinical trials for neurofibromatosis type 2. Clin. Cancer Res. 2009;15:5032–5039. doi: 10.1158/1078-0432.CCR-08-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11989–11994. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM, Siveke JT. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–616. doi: 10.1002/hep.22381. [DOI] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini M, Robanus-Maandag E, van d., V, Niwa-Kawakita M, Abramowski V, Goutebroze L, Woodruff JM, Berns A, Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kreamer BL, Staecker JL, Sawada N, Sattler GL, Hsia MT, Pitot HC. Use of a low-speed, iso-density percoll centrifugation method to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell Dev. Biol. 1986;22:201–211. doi: 10.1007/BF02623304. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, McCartney BM, Fehon RG. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J. Cell Biol. 1998;141:1589–1599. doi: 10.1083/jcb.141.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaigre FP. Development of the biliary tract. Mech. Dev. 2003;120:81–87. doi: 10.1016/s0925-4773(02)00334-9. [DOI] [PubMed] [Google Scholar]
- Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, Erdjument-Bromage H, Zhou P, Tempst P, Giancotti FG. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. U. S. A. 2010 doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Saotome I, Ramesh V, Gusella JF, Jacks T. The Nf2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanuma Y, Ohta G. Immunohistochemical study on bile ductular proliferation in various hepatobiliary diseases. Liver. 1986;6:205–211. doi: 10.1111/j.1600-0676.1986.tb01067.x. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, You L, Giancotti FG. Shedding light on Merlin's wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 2006 doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr. Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang CY, Gao B, Jiang J, Yang Y. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Silva E, Clayton L, Gardano L, Barrios-Rodiles M, Wrana J, Varelas X, Arbouzova NI, Shaw S, Saburi S, Matakatsu H, Blair S, McNeill H. Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr. Biol. 2009;19:1112–1117. doi: 10.1016/j.cub.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Bronson RT, Orkin SH, DePinho RA, Jacks T. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Xu X, Kobayashi S, Qiao W, Li C, Xiao C, Radaeva S, Stiles B, Wang RH, Ohara N, Yoshino T, LeRoith D, Torbenson MS, Gores GJ, Wu H, Gao B, Deng CX. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J. Clin. Invest. 2006;116:1843–1852. doi: 10.1172/JCI27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates hippo signaling in conjunction with Merlin and expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01