Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP (original) (raw)
. Author manuscript; available in PMC: 2011 Aug 1.
Summary
We examined the role of clock genes in the diurnal regulation of plasma triglyceride-rich apolipoprotein B-lipoproteins and their biosynthetic chaperone, microsomal triglyceride transfer protein (MTP). Clock mt/mt mice showed sustained hypertriglyceridemia and high MTP expression. CLOCK knockdown activated MTP promoter, and reduced small heterodimer partner (SHP, NROB2). CLOCK up-regulated SHP by binding to its E-box. SHP suppressed MTP expression by binding to the HNF4α/LRH-1 at the MTP promoter. Cyclic expression of MTP after serum shock was abrogated by siCLOCK and siSHP. Plasma triglyceride and MTP showed reduced diurnal variations in Shp −/− mice. Whereas, peaks and nadirs in SHP expression were inversely correlated with those of MTP, these changes were reduced in Clockmt/mt mice. Expression of Shp abrogated hypertriglyceridemia in Clockmt/mt mice. Together, these studies describe a role of Clock/Shp in the diurnal regulation of MTP and plasma triglyceride and indicate that disruptions in circadian regulation might cause hyperlipidemia.
Introduction
Several biological, physiological, and behavioral activities show a characteristic recurrence with an interval of 24 h attuned to the light/dark cycle associated with sunrise and sunset. Light entrains central clocks present as 2 lateral suprachiasmatic nuclei in the hypothalamus via the retinohypothalamic tract (Dunlap, 1999; Hastings et al., 2003; Lowrey and Takahashi, 2004). The circadian clock arises from autoregulatory transcriptional, translational, and posttranslational feedback loops of few transcription factors known as clock genes, including circadian locomotor output cycles kaput (Clock), brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (Bmal1), neuronal PAS containing protein 2 (NPAS2), period genes (Per1, Per2, Per3), and cryptochrome genes (Cry1 and Cry2) (Dunlap, 1999; Reppert and Weaver, 2001; Lowrey and Takahashi, 2004).
Clock/Bmal1 or Npas2/Bmal1 heterodimers bind E-box sequences present in the promoter regions of Per1/2/3 and Cry1/2 genes and enhance their expression constituting a positive loop (Gekakis et al., 1998). When concentrations of Per and Cry proteins increase, they form heterodimers and suppress transcription of Bmal1 forming a negative feedback loop (Darlington et al., 1998). Besides this circuitry, circadian clock signals are transmitted to other clock-controlled genes via different transcription factors, leading to circadian variations in various biological and behavioral activities. For example, Clock/Bmal1 enhance expression of Rev-erbα and albumin D-element binding protein (DBP) that regulate bile acid and drug metabolisms (Green et al., 2008; Lowrey and Takahashi, 2004; Hussain and Pan, 2009). Rev-erbα regulates bile acid metabolism by regulating CYP7A1, via regulation of repressors, Shp or E4bp4, or by an indirect mechanism involving Insig2 (Duez et al., 2008; Le et al., 2009). Similarly, DBP is upregulated by the binding of Clock/Bmal1 to its E-box sequences (Ripperger et al., 2000). DBP and its homologs play an important role in drug detoxification (Gachon et al., 2006). Besides being a clock-controlled gene, Rev-erbα also downregulates Bmal1, and therefore, is a major component of the central clock genes (Preitner et al., 2002). In contrast to the suppression of Bmal1 by Rev-erbα, ROR transcription factors up-regulate Bmal1; therefore, Rev-erbα/ROR/Bmal1 also constitute an additional, regulatory loop that stabilizes the central circadian loops of Clock/Bmal1/NPAS2 and Crys/Pers (Ueda et al., 2002; Gachon et al., 2006; Liu et al., 2008). Aside from suprachiasmatic nuclei, clock and clock-controlled genes also are expressed in other tissues (Sakamoto et al., 1998; Yamamoto et al., 2004; Lowrey and Takahashi, 2004). Moreover, rhythmic expression of genes can be induced in cultured cells after brief exposure to high serum concentrations (Balsalobre et al., 1998; Balsalobre et al., 2000a; Balsalobre et al., 2000b). Thus, several cells are capable of expressing self-sustained circadian rhythms after appropriate entrainment. It is believed that the master clock synchronizes peripheral clocks involving neural connections and humoral chemicals.
The role of Clock in regulating circadian rhythms has been derived using Clock mutant mice (Clockmt/mt) that express a dominant-negative protein (Vitaterna et al., 1994; King et al., 1997). Clockmt/mt are arrhythmic exhibiting longer circadian periods and decreased amplitude of locomotors activity (Vitaterna et al., 1994). They entrain during normal light/dark cycles, but lose this ability in constant darkness (Vitaterna et al., 1994). These mice show several physiologic abnormalities, such as reduced fertility (Miller et al., 2004), obesity, and the metabolic syndrome (Turek et al., 2005). It is unclear how expression of a mutant Clock protein causes these metabolic abnormalities. Moreover, polymorphisms in CLOCK gene are associated with metabolic syndrome and obesity in humans (Garaulet and Madrid, 2009), but molecular mechanisms have not been elucidated. Our aim was to find out how CLOCK regulates plasma lipids to gain insights into hyperlipidemia that might contribute to metabolic syndrome and obesity.
Several studies have shown that plasma triglycerides show diurnal variations (Seaman et al., 1965; Schlierf and Dorow, 1973; Rudic et al., 2004; Pan and Hussain, 2007; Pan and Hussain, 2009). Plasma triglycerides show persistent diurnal rhythmicity in rats fasted for 96 hours (Escobar et al., 1998). The extent of the increases in triglycerides were, however, lower than those observed in the fed state. Plasma triglycerides are carried in apolipoprotein B-lipoproteins. We showed that plasma triglycerides exhibit diurnal variations due to changes in apoB-lipoproteins (Pan and Hussain, 2007). However, little is known about the mechanisms that control diurnal changes in plasma triglycerides and apoB-lipoproteins.
Biosynthesis of apoB-lipoproteins requires a dedicated chaperone, microsomal triglyceride transfer protein (MTP), which transfers lipids in vitro between membranes. Mutations in the MTTP gene are associated with an absence of plasma apoB-lipoproteins in abetalipoproteinemia. MTP expression is regulated at transcriptional level (Hussain et al., 2008). It is known that proximal 150 bp sequences in the MTTP promoter contain several regulatory sites (Hagan et al., 1994; Hussain et al., 2008). The binding of HNF4α to the HNF4 site is critical for MTTP expression, as _Hnf4α_−/− mice do not express MTP (Hayhurst et al., 2001). HNF1α synergistically activates HNF4α-mediated MTTP expression (Sheena et al., 2005). Binding of RXR/PPARs and FOXA2 to DR1 element enhances, whereas binding of NR2F2 to the same element reduces, MTTP expression (Kang et al., 2003). Binding of NR2F1 to DR1 has been suggested to suppress MTTP expression in undifferentiated intestinal cells (Dai et al., 2010). SREBP1c decreases expression by interacting with IRE/SRE elements (Sato et al., 1999). Furthermore, different activators and repressors modulate MTTP expression. For example, PGC1α and PGC1β modulate MTTP transcription by interacting with transcription factors that bind to DR1 element (Wolfrum and Stoffel, 2006). SHP reduces MTP expression by interacting with HNF4α and LRH-1 (Hirokane et al., 2004; Huang et al., 2007).
Several studies show a correlation between MTP expression and plasma lipids (Lin et al., 1994; Pan and Hussain, 2007). Changes in plasma lipids/lipoproteins and MTP expression are altered by restricted feeding (Pan and Hussain, 2007). Extended exposures to light and dark abolished rhythmicity in MTP and plasma lipids (Pan and Hussain, 2007). We observed that Clock is important for calorie restricted, food entrainment of intestinal nutrient absorption, and expression of different nutrient transporters (Pan and Hussain, 2009). These studies led to the hypothesis that visual cues and clock genes might play a role in the circadian regulation of MTP and plasma triglycerides. To test this hypothesis, we studied changes in plasma lipids and hepatic MTP in wildtype and Clockmt/mt mice fed ad libitum and subjected to food entrainment, and examined mechanisms by which Clock regulates hepatic MTP and plasma triglyceride.
Results
Clock is important for diurnal regulation of plasma triglycerides and hepatic Mtp
We studied diurnal regulation of plasma lipids in C57BL/6J mice fed ad libitum and kept in 12 -h light-dark cycle (lights on 7 AM to 7 PM). Triglycerides in total plasma varied significantly within 24 hours owing to changes in apoB-lipoproteins (Fig S1A) consistent with rat studies (Pan and Hussain, 2007); they were high at night and low during the day. In contrast to triglycerides, plasma cholesterol did not change significantly (Fig. S1B). Hepatic MTP activity (Fig S1C), mRNA (Fig S1D), and protein (Fig S1E) showed diurnal variations consistent with earlier studies in rats (Pan and Hussain, 2007) and in mouse in testinal Mtp (Pan and Hussain, 2009). Therefore, Mtp expression in the liver and intestine as well as plasma triglycerides exhibit diurnal variations.
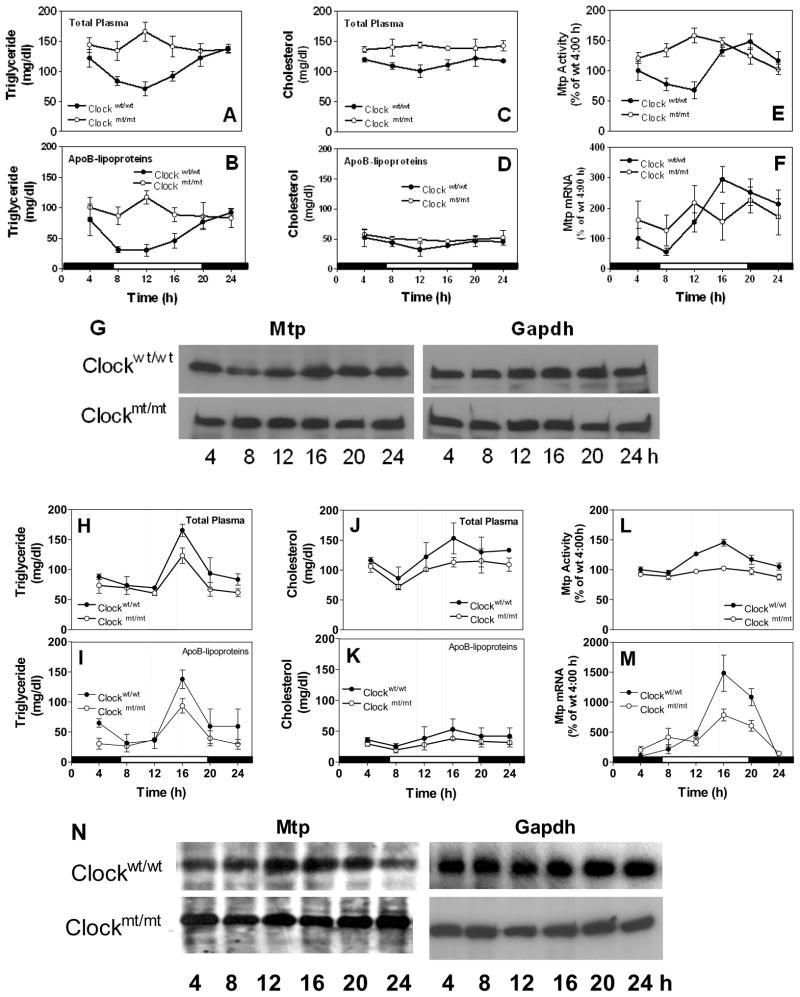
Clock is a critical transcription factor controlling circadian rhythms. Clock protein and mRNA were high just before the lights went on (Fig S2A–B). To evaluate the role of Clock in regulating plasma lipids and hepatic MTP, we used Clockmt/mt(mutant) and their wildtype (Clockwt/wt) siblings. Clock mt/mt mice expressed low levels of Clock that did not show circadian variations seen in wildtype mice (Fig S2C–D). Similarly, expression of several other clock genes, including Npas2, Bmal1, Rev-erbα, Dbp, Pers, was repressed in mutant mice (Fig S3). Plasma triglycerides in wildtype were 2-to 3 -fold higher at 24 h compared to 12 h (Fig 1A) owing to changes in apoB-lipoproteins (Fig 1B). In Clockmt/mt, plasma triglycerides (Fig 1B) and apoB100 (Fig S4A) did not change as in Clockwt/wt. These mice had higher plasma triglycerides at all times seen normally at night in wildtype mice due to the absence of nadirs in the day. In contrast to triglycerides, changes in plasma and apoB-lipoprotein cholesterol were less prominent (Fig 1C–D). Hepatic triglycerides and cholesterol were high in mutant than in wildtype mice (Fig S4B–C). Hepatic triglycerides showed diurnal changes in wildtype, but not in mutant mice (Fig S4B). Hepatic cholesterol did not show significant variations throughout the day in these mice (Fig S4C). In the Clockwt/wt liver, MTP activity (Fig 1E), mRNA (Fig 1F), and protein (Fig 1G) showed diurnal variations as in C57BL/6 wildtype mice shown in Fig S1C–E. Although experimental variations may occur due to different genetic background, the temporal expression pattern of Mtp was similar (Fig S1C-E vs Fig 1E–G). In Clockmt/mt, however, hepatic MTP did not show any significant variations within 24 h. Overall, these data indicate that Clock is important for daily regulation of plasma triglycerides and hepatic Mtp.
Figure 1. Changes in plasma lipids and hepatic MTP in Clockmt/mt and Clock wt/wt mice fed chow ad libitum or subjected to food restriction.
(A-G) Clockmt/mt mice and their wildtype siblings (Clockwt/wt) were kept in 12 h light-dark cycle and fed chow at ad libitum. Triglycerides and cholesterol were measured in total plasma (A, C) and apoB-lipoproteins (B, D). Liver samples were used to measured MTP activity (E), mRNA (F), and protein (G). Each time represents mean ±SD, n=6; western blot is representative of n=3
(H-N)Clock mt/mt mice and Clockwt/wt siblings were kept in 12 h light-dark cycle and fed chow from 11 AM to 3 PM for 10 days. Triglycerides and cholesterol were measured in total plasma (H, J) and apoB-lipoproteins (I, K). Liver samples were used to measured MTP activity (L), mRNA (M), and protein (N). Each time point represents mean ±SD, n=6; western blotting representative of n=3
Clock is required for optimal food entrainment of plasma triglycerides and hepatic Mtp
To understand mechanisms behind high plasma triglycerides in mutant mice, we studied food intake, hepatic lipoprotein assembly, and response to food entrainment. Clockmt/mt mice consumed more food (Fig S5A), and synthesized and secreted more triglyceride-containing lipoproteins (Fig S5B). To examine whether shifting mice from unrestricted feeding to daytime scheduled feeding alters daily changes in plasma lipids, mice were provided food between 11 AM and 3 PM for 10 days. In Clockwt/wt mice, plasma triglycerides (Fig 1H) and cholesterol (Fig 1J) were significantly higher at the time of food availability due to increases in apoB-lipoproteins (Fig 1I, K). The food entrainment response was significantly reduced in mutant mice (Fig 1H–K). Expression of hepatic MTP activity (Fig 1L), mRNA (Fig 1M), and protein (Fig 1N) was increased at the time of food availability, and these altered expressions were significantly dampened in mutant mice. All the clock genes did respond to food entrainment, and their mRNA levels either increased or decreased at the time of food availability in wildtype mice (Fig S6). Clock genes did not respond to, or responded poorly to, food entrainment in mutant mice. These studies indicate that food entrainment enhances plasma triglycerides and hepatic Mtp at the time of food availability and the optimum responses require normal Clock expression. Therefore, both food availability and Clock are important modifiers of MTP expression.
Clock suppresses MTP expression in hepatoma cells
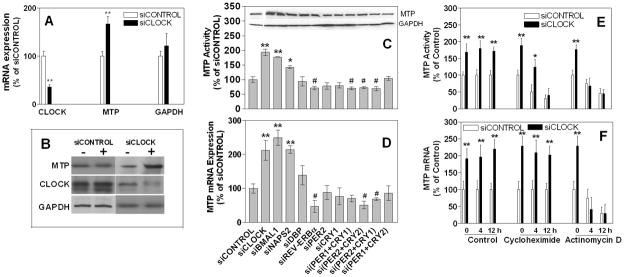
The above studies in Clockmt/mt could represent direct regulation of Mttp by Clock, or they might represent an accommodation to the long-term expression of the dominant-negative Clock protein. To examine the role of CLOCK in MTP regulation in human cell lines, we reduced its expression using RNAi in Huh-7 cells. siCLOCK significantly reduced CLOCK mRNA and protein, increased MTTP mRNA, protein and activity, and had no effect on GAPDH mRNA and protein (Fig 2A–B) compared with siCONTROL indicating that CLOCK is a negative regulator of MTTP expression.
Figure 2. Clock knockdown increases MTP expression.
**(A-B)**Huh -7 cells (n=3) were treated with siCLOCK or siCONTROL for 72 h. CLOCK, MTTP, and GAPDH mRNA (A) and protein (B) were determined by qRT-PCR and immunoblotting, respectively. Changes in mRNA with respect to siCONTROL treated cells are presented. **P< 0.05, compared to siCONTROL
**(C-D)**Huh -7 cells were treated with different siRNA for 72 h. Cells were used to measure triglyceride transfer activity (C), protein (inset), and mRNA (D) levels of MTP.
**(E-F)**Huh -7 cells (n=3) were treated with siCLOCK or siCONTROL for 72 h and treated with either cycloheximide [20 μM] or actinomycin D [2 μM] for 4 or 12 h and used to measure MTP activity (E) and mRNA (F). *P< 0.5, ** P< 0.05, compared to siCONTROL
To explore the role of other clock genes, these cells were exposed to siRNAs against different clock genes (Fig 2C–D). Not only CLOCK, but also BMAL1 and NPAS2 negatively regulated MTTP (Fig 2C–D). On the other hand, DBP, PER2, and CRY1 had no effect on MTP expression, while REV-ERBα, PER1+CRY1, and PER2+CRY2 positively regulated MTTP expression. These studies show that the positive loop of the circadian clock negatively regulates MTP expression.
Experiments were then performed to determine whether MTTP induction involves transcriptional and/or translational mechanisms. Cycloheximide, a protein translation inhibitor, and actinomycin D, an mRNA transcription inhibitor, decreased MTP activity with time (Fig 2E) in both siCONTROL-and siCLOCK -treated cells. MTTP mRNA remained elevated in cycloheximide supplemented siCLOCK-treated cells indicating that CLOCK may not require protein synthesis for the transcriptional regulation of MTTP (Fig 2F). As expected, actinomycin D reduced MTP mRNA with time in siCONTROL cells. In siCLOCK-treated cells, MTP mRNA levels were not elevated in the presence of actinomycin D, suggesting that CLOCK might control MTTP expression involving transcriptional mechanisms.
CLOCK regulates MTP using SHP
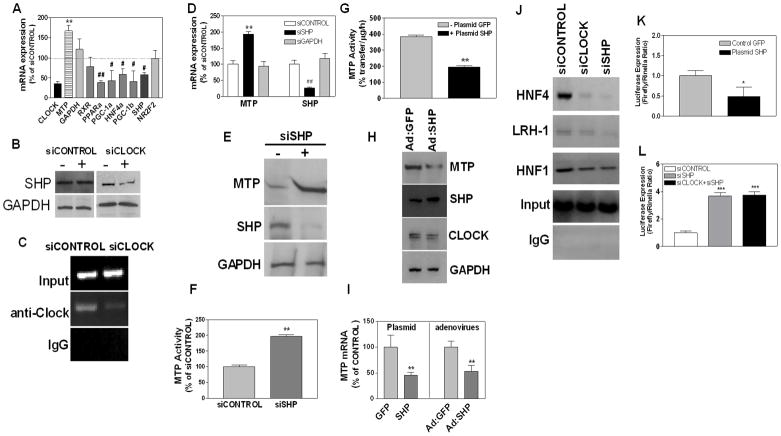
Attempts were made to determine how CLOCK suppresses MTTP transcription. Analysis of the MTTP promoter did not reveal any E-box sequences; therefore, we hypothesized that CLOCK might regulate expression of critical transcription factor(s) that control MTP expression (Hussain et al., 2008; Dai et al., 2010). Increased expression of activators or decreased expression of repressors in siCLOCK treated cells could explain higher MTP levels. siCLOCK increased MTP mRNA (Fig 3A), had no effect on RXRα(Fig 3A), but decreased PPARα, PGC-1α, HNF4α, and PGC-1β expression (Fig 3A). As these activators were not changed, or were reduced, we concluded that CLOCK might not regulate MTP by up-regulating activators. Therefore, we studied changes in MTTP repressors. siCLOCK reduced SHP mRNA (Fig 3A) and protein (Fig 3B), but had no effect on NR2F2 mRNA (Fig 3A), indicating that CLOCK might enhance MTTP expression by suppressing SHP. CLOCK has been shown to bind to the E-box sequences present in the SHP promoter and regulate its expression (Oiwa et al., 2007). We confirmed this by performing ChIP using anti-Clock antibodies (Fig 3C, siCONTROL). We detected significantly less CLOCK binding to SHP E-box after siCLOCK treatment compared with siCONTROL implying that CLOCK may directly up-regulate SHP. Therefore, SHP might be a clock-controlled gene responsible for Clock-mediated MTP regulation.
Figure 3. SHP suppresses MTP expression.
**(A-C)**Huh -7 cells were transfected with siCLOCK or siCONTROL. After 72 h, mRNA levels of activators and suppressors of MTTP expression were quantified by qRT-PCR (A). Data represent percentage of mRNA in siCLOCK-treated cells compared with siCONTROL. SHP and GAPDH protein were detected by immunoblotting (B). Cells were subjected to ChIP using anti-Clock antibodies and immunoprecipitates were used to amplify E-box sequences in the SHP promoter (C). Data are representative of n=3.# P< 0.5, ## P< 0.01, **P< 0.05, compared to siCONTROL
**(D)**Huh -7 cells were treated with or siGL2 (siCONTROL), siSHP or siGAPDH for 72 h and used to measure MTP and SHP mRNA.** P< 0.05, compared to siCONTROL
**(E-F)**Huh -7 cells were treated with siSHP for 72 h and used to measure MTP, SHP and GAPDH protein by immunoblotting (E) and MTP activity (F). **P< 0.05, compared to siCONTROL
(G) Huh-7 cells were transfected with plasmids expressing human SHP (+Plasmid SHP) or not (− Plasmid GFP). MTP activity was measured after 72 h.** P< 0.05
(H) Huh-7 cells were infected with adenoviruses expressing green fluorescent protein (Ad:GFP) or mouse SHP (Ad:SHP). After 48 h, cells were used to detect MTP, SHP, CLOCK and GAPDH proteins.
**(I)**Cells were transfected with indicated plasmids or infected with viruses. After 72 h, MTP mRNA were quantified by qRT-PCR.** P< 0.05
**(J)**Binding of SHP to different _cis_-elements in the MTTP promoter. Cells were treated with siCONTROL, siCLOCK, or siSHP. After 72 h, cells were subjected to ChIP using anti-SHP antibodies. Immunoprecipitates were used to amplify sequences corresponding to HNF4, LRH-1, and HNF1 _cis_-elements.
(K) pMTP-204 promoter construct expressing luciferase under the control of basal human MTTP promoter or pCMV-Renilla luciferase (control vector) were introduced in Huh -7 cells (n=3). In addition, cells also received a control (GFP) or SHP expressing plasmid. Luciferase activities were measured after 72 h. *P< 0.5
**(L)**pMTP -204 promoter construct or pCMV-Renilla luciferase (as a control vector) were transiently transfected in Huh-7 cells (n=3). In addition, these cells received siCONTROL, siSHP or siCLOCK +siSHP. Luciferase activity was measured after 48 h.*** P< 0.001, compared to siCONTROL
To evaluate the role of SHP in MTP regulation, Huh-7 cells were treated with siSHP. This treatment significantly reduced SHP mRNA (Fig 3D) and protein (Fig 3E), but increased MTP mRNA (Fig 3D), protein (Fig 3E), and activity (Fig 3F). siGAPDH had no effect on MTP and SHP mRNA (Fig 3D). To establish further a relation between SHP and MTP, we expressed SHP using adenoviruses or plasmids. Expression of SHP reduced MTP activity, mRNA, and protein (Fig 3G–I). These studies indicate that SHP represses MTP expression confirming earlier studies (Hirokane et al., 2004; Huang et al., 2007).
Attempts were then made to understand how SHP suppresses MTTP expression. SHP has been shown to reduce MTTP expression by binding to HNF-4α (Hirokane et al., 2004) and LRH-1 (Huang et al., 2007). Therefore, we studied the binding of SHP to HNF4α, LRH-1, and HNF-1α transcription factors associated with their putative binding sites on the MTTP promoter. Under normal conditions (siCONTROL), anti-SHP antibodies precipitated sequences occupied by HNF4α, LRH-1, and HNF1α (Fig 3J). The binding of SHP to the HNF4α binding site of the MTTP promoter was significantly reduced in siCLOCK-and siSHP -treated cells (Fig 3J). Compared to HNF4α, the binding of SHP to LRH-1 and HNF1 sites was decreased to a lesser extent in siCLOCK-treated cells. These studies indicate that knockdown of CLOCK and SHP significantly decreases the binding of SHP to the MTTP promoter. To substantiate the effect of SHP on MTTP promoter, we used constructs that express luciferase under the control of a basal MTTP promoter (Dai et al., 2010). Cells were cotransfected with pMTP-205-Luc (expresses firefly luciferase under the control of 205 bp MTTP promoter), Renilla luciferase (a transfection control), and plasmids expressing either GFP or SHP (Fig 3K). SHP expression reduced basal MTP promoter activity by 50%. Conversely, co-transfection of pMTP-204 with siSHP or siSHP/siCLOCK increased promoter activity by approximately 4-fold (Fig 3L). These studies indicate that SHP might suppress MTTP expression by binding to HNF4α and LRH-1. We posit that CLOCK increases SHP and enhanced SHP expression leads to decreased MTP expression.
Cyclic expression of MTP is regulated by CLOCK and SHP
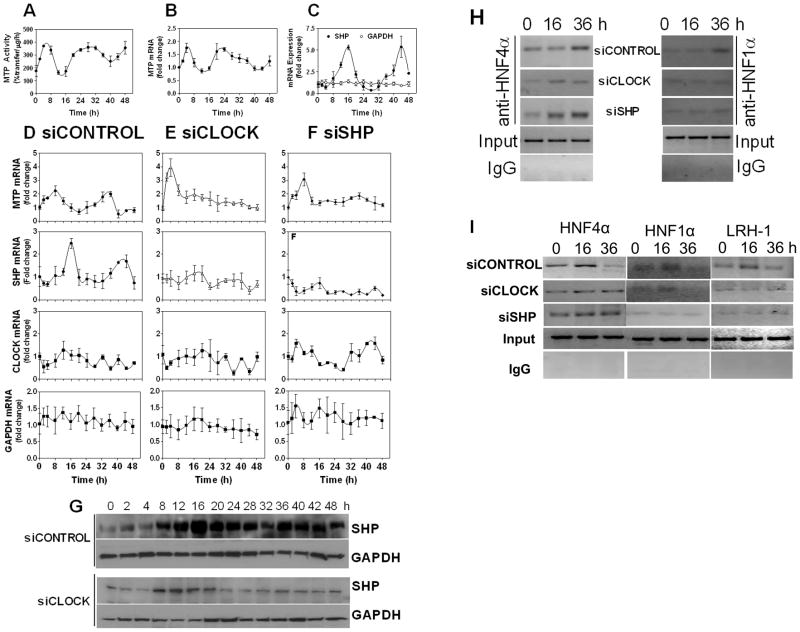
The above studies indicate that CLOCK regulates MTP expression by modulating SHP. However, these studies do not affirm whether these genes play a role in the rhythmic expression of MTP. It is known that brief exposure of cells to 50% horse serum induces circadian expression of various genes (Balsalobre et al., 1998). Therefore, Huh-7 (Fig 4), and HepG2 (Fig S7), cells were incubated in serum-free media for 18 h, exposed to 50% serum for 2 hours, washed, and incubated in serum-free media (Fig 4). MTP mRNA and activity were augmented immediately after serum supplementation. Subsequently, it showed rhythmic expression (Fig 4A–B). SHP mRNA (Fig 4C) also showed cyclic expression after 50% serum exposure; however, it was anti-phasic with MTP. First, there was no induction in SHP expression soon after serum shock. Second, peak expression of SHP occurred at times of low MTP expression. GAPDH mRNA did not change after serum shock and did not exhibit cyclic expression (Fig 4C). These studies show that serum shock induces cyclic expression of MTP and SHP. Their expression exhibits disparate peaks and nadirs.
Figure 4. CLOCK and SHP regulate cyclic expression of MTP in Huh-7 cells.
**(A-C)**Huh -7 cells were cultured in DMEM containing 10% serum. Confluent monolayers were incubated in serum free media for 18 h and for 2 h in media containing 50% fetal calf serum. Cells were washed and incubated in serum-free media. At indicated times, 3 wells were harvested to measure MTP activity (A), mRNA (B), and SHP/GAPDH mRNA (C) in triplicate.
(D-G)Huh -7 cells were transfected with siCONTROL (D), siCLOCK (E), or siSHP (F). After 48 h, cells were subjected to 2 h serum shock. At indicated times, mRNA levels of different genes were measured (D-F). For comparison, mRNA levels were normalized with 18S rRNA, and the values at 0 h were normalized to 1. Other values represent fold-change compared to 0 h. Cell lysates were also used to detect SHP and GAPDH protein (G).
(H) Huh-7 cells were treated with different siRNA and subjected to serum shock. Cells were harvested at 0, 16 and 36 h and subjected to ChIP assays using anti-HNF4α or anti-HNF1α antibodies. Immunoprecipitates were used to amplify HNF4 or HNF1 sites in the MTTP promoter. (I)Huh -7 cells were treated with siRNA and subjected to serum shock. Cells were harvested at 0, 16 and 36 h and used for ChIP assays using anti-SHP antibodies. The extracted DNAs from the immunoprecipitates were amplified using primers specific for human HNF4, HNF1 and LRH-1 sites in the MTTP promoter. Data are representative of n=3
Next, we explored the role of CLOCK and SHP in the cyclic regulation of MTTP mRNA by reducing their expression and then subjecting them to serum shock. Cells treated with siCONTROL showed high MTP expression soon after serum supplementation and a subsequent peak at 32-to 36 -h (Fig 4D). Expression patterns of MTP and SHP showed slightly different temporal expression than seen in Fig 4A, probably because these cells were subjected to transfections. CLOCK itself did not show significant rhythmic expression and is in concert with other studies (Balsalobre et al., 1998). Instead, its association with Bmal1 and transport to the nuclei show rhythmic changes (Tamaru et al., 2003; Gallego and Virshup, 2007). Again, GAPDH was resilient to serum shock, and its expression did not change with time. siCLOCK (Fig 4E) enhanced MTP expression (compare with Fig 4D) after serum shock and abrogated its subsequent rhythmic expression. siCLOCK reduced SHP as before (not shown) but abolished rhythmic changes in SHP mRNA (Fig 4E) and protein (Fig 4G), indicating that CLOCK is needed for the cyclic expression of SHP after serum shock. Enhanced expression of MTP after serum supplementation might be secondary to reduced SHP expression. siCLOCK reduced CLOCK (not shown), but the residual mRNA did not change significantly throughout the experiment compared to 0 time point. Similar to siCLOCK, siSHP also enhanced MTP expression soon after serum supplementation (Fig 4F). However, siSHP-treated cells failed to exhibit cyclic expression of MTTP. Cosinar analysis compared cyclic changes over 48 h and indicated a period of 23.7±0.55 and 25.2±1.07 h for Mtp and Shp mRNA, respectively (Fig 4D–F). Comparison of changes in two successive 24 h time intervals gave a P value of 0.003 for both Mtp and Shp indicating significant cyclic changes. Similar analyses in siShp and siClock treated cells gave a P value of 0.274 and 0.198 indicating loss of cyclic change in Mtp expression. These studies indicate that MTP exhibits a complex response to serum shock involving early up-regulation followed by rhythmic expression. The first immediate response is enhanced when CLOCK and SHP expressions are reduced, indicating that these proteins dampen serum response. However, these proteins are required for the subsequent cyclic expression of MTP.
To gain biochemical insights into the rhythmic expression of MTP, we studied the temporal binding of 2 major activators, HNF4α and HNF1α, to the MTTP promoter. The binding of HNF4α and HNF1α to the MTTP promoter was low at 16 h and high at 36 h (Fig 4H). In siCLOCK-and siSHP -treated cells, binding of these transcription factors was similar at two different times. These data indicate that rhythmic expression of MTP might be related to the binding of different amounts of HNF4α/HNF1α to their respective sites on the MTTP promoter. We also evaluated the role of SHP in the rhythmic expression of MTP (Fig 4I) by performing ChIP using anti-Shp antibodies and amplifying sequences corresponding to different _cis_-elements. The binding of SHP to HNF4, HNF1 and LRH-1 sites in the MTTP promoter was high at 16 h and low at 36 h (Fig 4I, siCONTROL). These time-dependent changes were not evident in siCLOCK-and siSHP -treated cells. Based on these studies, we posit that rhythmic expression of MTP involves binding of different amounts of HNF4α and HNF1α to their respective sites and differential association of SHP at the HNF4/HNF1/LRH-1 sites in the MTTP promoter. At the time of high MTTP expression, its promoter has more HNF4α and HNF1α associated with it. At the same time, the amounts of SHP present with HNF4α and LRH-1 are low. At the time of low expression, the amounts of HNF4α and HNF1α associated with the MTTP promoter are low. But, at this time, there are higher amounts of SHP associated with the MTTP promoter.
Shp is important for the circadian regulation of plasma triglycerides and hepatic Mttp
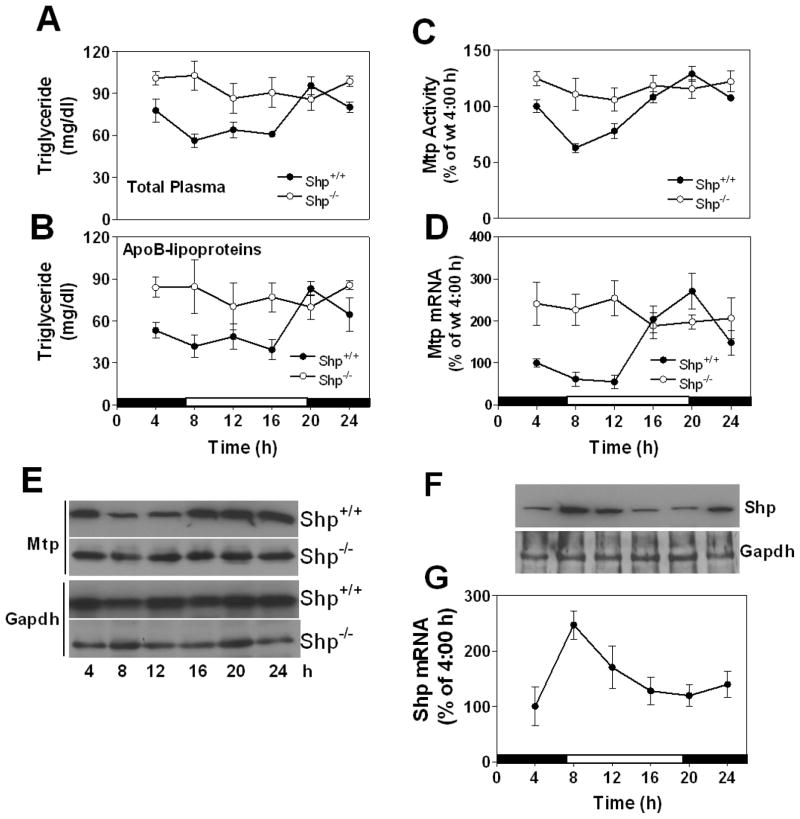
Our studies in Huh-7 cells indicate that SHP plays an important role in the cyclic expression of MTP. To determine whether Shp is essential for the circadian regulation of Mtp and plasma triglycerides in vivo, we studied their regulation in _Shp_−/− and Shp+/+ mice (Fig 5). As before, plasma triglycerides showed diurnal variations in Shp+/+ mice with higher levels at night (Fig 5A) owing to changes in apoB-lipoproteins (Fig 5B). In _Shp_−/− mice, plasma triglycerides did not show significant variations within 24 h and exhibited sustained hypertriglyceridemia (Fig 5A–B) similar to that seen in Clockmt/mt mice. Moreover, _Shp_−/− mice had high plasma apoB100 and apoB48 (Fig S8), absorbed more lipids (Fig S8B), and consumed more food (Fig S8C). Hepatic triglycerides and cholesterol levels were similar in wildtype and knockout mice (Fig S8D–E). In Shp+/+livers, MTP activity, protein, and mRNA (Fig 5C–E) showed diurnal variations with high levels seen at night. In _Shp_−/− liver, however, Mtp did not show significant variations during 24 h and they remained high at all times (Fig 5C–E). The expression profile of Mttp in _Shp_−/− was similar to that in Clockmt/mt. Therefore, consideration was given to the possibility that Shp might regulate Mttp involving clock genes. Many clock genes exhibited similar or reduced amplitudes in _Shp_−/− mice compared with Shp+/+mice, except for Rev -erbα and Dbp that showed a significant shift in their rhtymic expression (Fig S9). Certainly, their expression and diurnal variations were not as dampened as in Clockmt/mt mice (Fig S3). These studies indicate that Shp expression is important for the diurnal regulation of plasma triglyceride and hepatic Mttp, but not for clock genes.
Figure 5. Plasma triglyceride and hepatic Mtp do not change in Shp−/− mice.
(A-E) _Shp_−/− and control (Shp+/+) mice were kept in 12 h light-dark cycle and fed chow ad libitum. Triglycerides were measured in total (A) and apoB-lipoproteins (B). Liver samples were used to measured Mtp activity (C), mRNA (D), and protein (E). Each time point represents mean ±SD, n=4–6
(F-G) Livers were collected at indicated times from male C57BL/6J mice fed ad libitum and used to measure Shp and Gapdh protein (F), and Shp mRNA (G). They were plotted as line graphs and error bars, respectively. Mean ±SD, n=6
Circadian regulation of Shp in Clockwt/wt and Clockmt/mt siblings
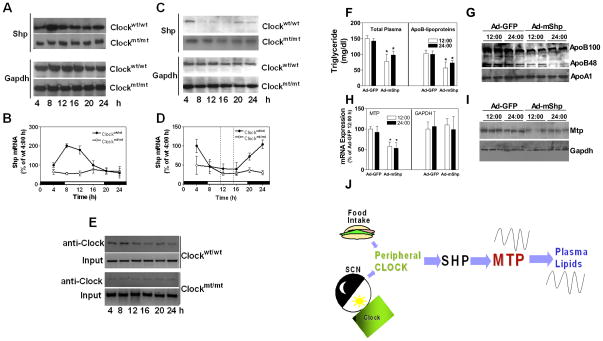
The studies described so far indicated that plasma triglycerides and hepatic Mtp profiles are similar in _Shp_−/− and Clockmt/mt mice. Furthermore, cell culture experiments indicate that CLOCK might regulate MTP via SHP. If SHP is the clock -controlled gene that suppresses MTTP, then it should show circadian rhythm out-of-sync with MTP. Tissue analyses showed that hepatic Shp expression in C57BL/6J was high in the day compared to the night (Fig 5F–G). Note that MTP expression is low at this time (Fig 1E–G, 5C–E). Shp mRNA and protein showed rhythmic expression in the livers of Clockwt/wt but not in Clockmt/mt siblings (Fig 6A–B). The expression of Shp in Clockmt/mt mice was significantly low compared to that seen in their wildtype siblings (Fig 6A–B). Food entrainment studies revealed that Shp levels were reduced at the time of food availability in Clockwt/wt, but not in Clockmt/mt mice (Fig 6C–D). These studies indicate that Shp shows daily variations with peak expressions occurring in the day when Mtp expression is low. Food entrainment reduces Shp while enhancing Mtp expression. Therefore, Mtp and Shp show anti-phasic expression profiles in wildtype mice fed ad libitum or subjected to food entrainment. However, similar to Mtp, Shp does not show daily variations in Clockmt/mt mice. In addition, we examined the association of Clock with the E-box of Shp in vivo by ChIP. The binding of Clock to Shp E-box sequences was high during 4 to 12 AM(Fig 6E) coincident with its high expression. In Clockmt/mt, the binding of Clock to Shp promoter was low and did not show rhythmic change within 24 h (Fig 6E). These data imply that the binding of Clock to Shp promoter is associated with increases in its expression.
Figure 6. Circadian regulation of Shp and effect of SHP expression in Clockwt/wt and Clockmt/mt siblings.
(A-B) Livers were collected at indicated times from male Clockmt/mt and Clockwt/wt siblings fed chow ad libitum to measure Shp/Gapdh protein (A) and Shp mRNA (B). Each time point for mRNA represents mean ±SD, n=6.
(C-D) Expression of Shp/Gapdh protein (C) and mRNA (D) in male Clockmt/mt and Clockwt/wt mice kept in 12 h light-dark cycle and fed chow from 11 AM to 3 PM for 10 days, n=6.
(E) Binding of Clock to the mouse Shp promoter in Clockmt/mt and Clockwt/wt was studied by ChIP. Livers collected at different times were immunoprecipitated using anti -CLOCK antibodies and used to amplify E-box sequences present in the Shp promoter. Representative electrophoresis image of the PCR products of clock binding (anti-Clock) and input is shown. This gel is a representative of n=3.
(F-I) Clockmt/mt mice were injected with Ad-GFP or Ad-mShp (1×1011 virus particles/mouse). Plasma and liver were collected after 72 h at 12:00 or 24:00 h. Triglycerides were measured in total plasma and apoB-lipoproteins (F). Plasma was used to detect apolipoproteins by Western blotting (G). Liver samples were used to measure Mtp mRNA and Gapdh mRNA (H), and Mtp protein (I). Each time point for mRNA represents mean ±SD, n=3. * P< 0.05, compared to Ad-GFP at 12 h.
**(J)**A schematic diagram suggesting that external stimuli such as food and light regulate cellular Clock. Clock regulates Shp. Shp then transmits this signal to Mtp. Changes in Mtp contribute to diurnal variations in plasma triglycerides.
Expression of SHP in Clockmt/mt mice abrogates hypertriglyceridemia
The above studies indicated that Shp expression is low in Clockmt/mt, and that it might contribute to high hepatic Mtp expression and hypertriglyceridemia. We then hypothesized that expression of SHP would decrease MTP expression and plasma triglycerides. Indeed, Shp expression significantly reduced triglycerides in plasma and apoB-lipoproteins (Fig 6F), plasma apoB100 and apoB48 (Fig 6G), and hepatic Mtp mRNA (Fig 6H) and protein (Fig 6I) without affecting GAPDH expression (Fig 6H–I). These studies show that Shp decreases Mtp expression and ameliorates hypertriglyceridemia.
Discussion
We previously reported that plasma triglycerides and MTP activity varies significantly within 24 h (Pan and Hussain, 2007). Here, we investigated the role of Clock in the diurnal and food entrained regulation of plasma lipids and hepatic MTP and explored molecular mechanisms underlying rhythmic expression of MTP using Clockmt/mt and _Shp_−/− mice fed ad libitum or subjected to food entrainment as well as human liver derived Huh-7 cells subjected to serum shock. These studies provide evidence that Clock plays an important role in the circadian and food entrained regulation of hepatic Mttp expression and plasma triglycerides. We further show that CLOCK regulates MTP involving SHP (Fig 6J). CLOCK temporally interacts with the E-box and increases SHP expression, whereas SHP reduces MTTP expression by differentially interacting with HNF4α and LRH-1. Decreased interaction of SHP with these transcription factors is associated with increased MTTP expression. Therefore, Shp is a clock-controlled gene that transmits information from Clock to Mttp (Fig 6J).
In Clockmt/mt mice, we found that smaller amounts of Clock bind to the Shp promoter and this binding does not fluctuate within a day. Moreover, this binding leads to low Shp expression and consequently high Mttp expression at all times and sustained hypertriglyceridemia similar to that seen at night in Clockwt/wt mice. The major effect of the Clock mutant protein is that reductions in the Mttp expression seen in Clockwt/wt mice at dawn are absent in Clockmt/mt mice.
CLOCK and other circadian genes in the regulation of MTP and plasma triglycerides
We combined Clockmt/mt mice and Clock knockdown approaches to explore circadian regulation of lipid metabolism. Clockmt/mt mice and Huh-7 cells exposed to siCLOCK express more MTTP indicating that Clockmt/mt mice have low Clock activity. This is in contrast to the observations that Clock mutant protein acts as a dominant negative in the SCN. However, our results are more consistent with the observations that Clock plays an important role in the circadian regulation of liver and lung rhythms (DeBruyne et al., 2007). In addition to CLOCK, our cell culture studies indicate that BMAL1 and NPAS2 play similar roles in the regulation of MTTP, that is, their knockdown enhances MTP expression, indicating that the positive loop of the circadian oscillators is important in reducing MTTP expression away from feeding times. In contrast, knockdown of the members of the negative circadian loop had no effect or reduce MTTP expression. It remains to be determined whether PERs and CRYs act directly on the MTTP promoter or reductions in Mttp expression after their knockdown is indirectly delivered via increased expression and activity of Clock/Bmal1. Thus, both forward and negative loops of the circadian clock might play a role in MTTP regulation and plasma triglyceride.
Diurnal regulation of SHP by CLOCK
SHP is a key transcriptional regulator of genes involved in diverse metabolic pathways and physiological functions (Chanda et al., 2008). Shp was one of the several transcription factors that showed diurnal variations during a global search of nuclear transcription factors (Yang et al., 2006). Oiwa and associates observed rhythmic expression of Shp in the mouse liver (Oiwa et al., 2007)and demonstrated that Clock/Bmal1, along with Lrh -1, synergistically activate Shp promoter in vitro, and this activation is suppressed by Shp itself. They also showed that Clock binds to E-box elements present in the Shp promoter. Here, we confirmed that Shp is rhythmically expressed in the liver (Fig 5). Moreover, we showed that Clock binds to the E-box in the Shp promoter and this binding is high in the day. The rhythmic binding of Clock to the Shp promoter was synchronous with mRNA and protein accumulation during daytime in the mouse liver. In Clockmt/mt mice, however, the binding of Clock to Shp promoter did not show cyclic change and Shp mRNA levels were relatively low and did not change (Fig 6). Similarly, levels of Per mRNA are low in Clockmt/mt(Fig S3). We propose that Shp is directly regulated by Clock similar to Per proteins.
SHP expression did not change soon after serum supplementation in Huh-7 cells (Fig 4) indicating that it is not a serum-response gene. Later on, SHP displayed cyclic expression. The rhythmic expression of SHP was absent in siCLOCK-treated cells pointing to a critical role of CLOCK. In siCLOCK-treated cells, low levels of CLOCK might not be sufficient to support cyclic induction of SHP transcription. This interpretation is consistent with earlier conclusion that diurnal regulation of SHP is positively related with increased association of CLOCK with the SHP promoter. Although these studies indicate that changes in the association of CLOCK with SHP promoter regulate SHP expression, we did not observe comparable changes in CLOCK transcription consistent with several studies (Mehra et al., 2009; Gallego and Virshup, 2007). Therefore, increased association of CLOCK with the SHP promoter is probably not a direct consequence of increased amounts of CLOCK. Instead, posttranslational modifications and its association with BMAL1 might determine enhanced association and subsequent dissociation of CLOCK from the SHP promoter (Mehra et al., 2009; Gallego and Virshup, 2007).
Shp is negatively regulated by Rev-erbα (Duez et al., 2008). Hence, we hypothesized that Rev-erbα would up-regulate MTP expression. This is supported by several observations: siRev-erbα decreases Mtp expression in Huh-7 cells (Fig 2C–D); Clockmt/mt mice that have high MTP levels express low levels of Rev-erbα (Fig 1E–G & S3); both Mtp and Rev-erbα do not show temporal changes in Clockmt/mt mice (Fig 1E-G & S3). Next, we theorized that if Rev-erbα is directly involved in negative regulation of MTP, then their temporal expression would be anti-phasic, as we had seen between Mtp and Shp (Fig 6J). Instead, we found that both Rev -erbα and Mtp show peak expression at similar times. Therefore, it is likely that Rev-erbα indirectly regulates MTP via Shp. We theorize that both the positive and negative loops of the circadian clock are involved in the diurnal regulation of MTP (Fig S10). Clock/Bmal1/Npas2 enhance Shp expression and reduce MTP expression in the daytime. High levels of Rev-erbα expression at night might suppress Shp expression and enhance Mtp expression at night.
Mechanisms controlling cyclic expression of MTP in liver cells
Many studies have identified several transcription factors that regulate MTP expression (Hussain et al., 2008). Here, we recognized mechanisms that could contribute to rhythmic expression of MTP. First, our studies suggest that cyclic expression of MTTP involves differential association of HNF4α and HNF1α to its promoter. The bindings of these transcription factors to the promoter were high at the time of maximum MTTP expression. These transcription factors are known to synergistically activate MTTP expression (Sheena et al., 2005).
Second, binding of SHP to the MTTP promoter occurs in rhythmic fashion. At the time of high MTTP expression, lower amounts of SHP were found associated with HNF4α and LRH-1. In contrast, higher amounts of SHP were with MTTP promoter coincident with optimal SHP expression. Therefore, MTTP expression might be directly related to SHP concentrations. This was confirmed by over-expressing SHP using plasmids and adenoviruses as well as by reducing its expression using siRNA. As discussed above, Shp mRNA levels are high at the time of higher E-box occupancy by the Clock in mice. Therefore, increased binding of Clock might lead to enhanced Shp expression. High Shp levels facilitate its association with MTP promoter causing repression.
Third, the rhythmic binding of HNF4α and HNF1α to the MTTP promoter was not seen in siCLOCK-treated cells (Fig 4H) indicating that CLOCK might also play a role in the association of HNF4α with the MTTP promoter. How CLOCK modulates the cyclic bindings of HNF4α and HNF1α to the MTTP promoter remains unknown. Under normal conditions HNF4α expression shows no cyclic change [not shown and (Oiwa et al., 2007)]. Therefore, increased association of HNF4α with the MTTP promoter might involve posttranslational mechanisms such as differential binding of its ligands.
Regulation of plasma triglycerides
Metabolic syndrome and obesity are major metabolic disorders characterized by high plasma lipid concentrations. Plasma lipids are tightly controlled by mechanisms regulating their production and clearance. These processes are affected by several factors such as availability of food and hormonal fluctuations. Here, we show that light -entrained mechanisms involving clock genes also play a role in regulating plasma triglycerides. We demonstrate that Clock regulates circadian expression of Mtp by controlling Shp in mice. Changes in Mtp are correlated with changes in plasma triglycerides indicating that circadian changes in Mtp could contribute to changes in plasma apoB-lipoproteins and triglycerides. It is known that hepatic Mtp deficiency significantly lowers plasma triglycerides (Bjorkegren et al., 2002)and plasma triglycerides are very low in abetalipoproteinemia (Berriot-Varoqueaux et al., 2000). In Clockmt/mt mice, the amounts of plasma triglycerides were similar to those seen in Clockwt/wt siblings at night suggesting that nadirs seen in wildtype mice at dawn are absent in these mice. Most likely, the association of Clock with the Shp promoter, leading to increased synthesis of Shp, followed by reduced MTP expression, plays a role in the nadirs seen in plasma triglycerides.
Triglyceride transfer activity of MTP has been exploited to identify several potent antagonists, which inhibit triglyceride transfer activity in vitro, suppress lipoprotein assembly and secretion in vivo, and decrease plasma lipids in humans and animals (Hussain et al., 2008). These drugs show several adverse effects. Dosing MTP inhibitors away from food intake diminishes intestinal distress. Based on the daily changes reported here for MTP, it might be interesting to evaluate the time of dosing on the pharmacokinetics of drug availability and efficacy as well as in avoiding adverse effects.
In short, this study establishes a molecular link between circadian physiology and plasma lipid metabolism. These studies suggest that CLOCK regulates MTP expression via SHP. However, it is possible that other unidentified mechanisms, such as other transcription factors or modifying proteins, might be involved. Nonetheless, these studies point to the importance of both CLOCK and SHP in the circadian regulation of MTP and plasma triglycerides. Aberrations in the clock genes, as in Clockmt/mt, lead to sustained hypertriglyceridemia because changes associated with dawn, such as up-regulation of Shp, downregulation of Mttp and reductions in plasma triglycerides, are absent.
Experimental Procedures
Animals
Male 8–12 week old C57BL/6J mice from the Jackson Laboratory were maintained under a 12-h light/dark (LD) cycle (light on 7 AM to 7 PM) with free access to food and water. Heterozygous clock mutant breeding pairs (C57BL/6J-Clockm1Jt/J) from the Jackson Laboratory (Stock No. 002923) were bred at Downstate Medical Center. Shp+/+and _Shp_−/− mice on a C57BL/6J background were maintained at the University of Utah. Male Clockmt/mt mice and their Clockwt/wt siblings, as well as Shp+/+and _Shp_−/− mice were housed under a 12 h light/dark cycle. Dissected tissues were quickly frozen and stored at −80°C. The Animal Care Committees at SUNY Downstate Medical Center and the University of Utah approved animal protocols.
Plasma lipid measurements
Total triglycerides and cholesterol were measured using kits (ThermoTrace Ltd). Lipids in high density lipoproteins were measured after precipitating apoB lipoproteins. ApoB lipoprotein lipids were determined by subtracting HDL lipids from total lipid levels (Pan et al., 2007; Pan and Hussain, 2007).
Measuring MTP activity
Livers (100 μg) from different mice were homogenized in 1 ml of buffer K (1.0 mM EGTA, 1 mM Tris-HCl, and mM MgCl2) and centrifuged at 50,000 rpm for 1 h. The supernatant was used to measure MTP activity using a kit (Chylos Inc) as described (Athar et al., 2004; Rava et al., 2005).
RNA extraction and real-time PCR analysis
Total RNA was isolated from cells and tissues using Trizol reagent. Isolated RNA was reverse-transcribed, and their levels were quantified by SYBR Green using an ABI Prism 7000 (Applied Biosystems). The primers used to measure different transcripts have been described (Pan et al., 2007; Pan and Hussain, 2007) and in Table S1.
Small RNA interference
siRNA directed against different clock genes and nonspecific control siRNA were obtained from Santa Cruz. They were introduced into Huh-7 cells plated in 12-well plates using siRNA transfection reagent (SC-29528, Santa Cruz). After 72 h, cells were harvested for RNA or protein analysis.
Serum shock
HepG2 and Huh-7 cells were grown to confluence in high glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were starved in the same medium with no FBS for 18 h, 50% horse serum was added for 2 h, and then the medium was changed back to starvation medium (Balsalobre et al., 1998). Cells were harvested at 4-h intervals for analyses.
Chromatin immunoprecipitation (ChIP) Assay
was performed according to the instructions (USB CHIP assay kit #78460) using goat polyclonal antibodies against CLOCK or SHP from Santa Cruz. DNA samples recovered after immunoprecipitation were subjected to PCR using primers described (Huang et al., 2007; Dai et al., 2010) and in Table S1. As negative controls, ChIP was performed in the absence of antibody or in the presence of rabbit IgG. These experiments were repeated 3 to 4 times with similar results. Data from a representative experiment are provided.
Plasmid expression and luciferase assay
The expression of SHP using plasmids and adenoviruses has been described (Huang et al., 2007). Plasmid pMTP204 was generated by cloning a 204 kb MTTP promoter in pBGL2 basic vector (Dai et al., 2010). Huh-7 cells transfected with either the pMTP204 construct or empty vector along with a reporter Renilla luciferase construct were assayed for luciferase activity, using the dual luciferase reporter assay system, according to the manufacturer’s instructions.
Statistical analyses
Values are expressed as mean ± S.D. For statistical analysis, one-way ANOVA was applied followed by Dunnett’s 2-tailed test, or a _t_-test. Cosinar analysis was performed using a program downloaded from http://www.circadian.org/softwar.html.
Highlights.
- Plasma triglycerides and tissue MTP activity show circadian changes
- These circadian changes are absent in Clock mutant mice
- Clock mutant mice have high plasma triglycerides at all times
- Knockdown of Clock decreases SHP, increases MTP, and abolishes cyclic expression of MTP in hepatocyte-derived cells
- Clock up-regulates SHP by binding to its E-box
- SHP down-regulates MTP expression by interacting with other nuclear receptors that activate MTP expression
- Over-expression of SHP avoids hypertriglyceridemia in Clock mutant mice
- Clock regulates MTP and plasma triglycerides via SHP
Supplementary Material
01
Acknowledgments
These studies were supported in part by NIH grants DK-81879 (MMH) and DK-080440 (LW) as well as Scientist Development Grant from the AHA (XP).
Abbreviations used
MTP
human microsomal triglyceride transfer protein
Mtp
mouse ortholog of MTP
MTTP
human MTP gene
Mttp
mouse Mtp gene
CLOCK
human circadian locomotor output cycles kaput gene
Clock
mouse Clock
Per
period
ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Athar H, Iqbal J, Jiang XC, Hussain MM. A simple, rapid, and sensitive fluorescence assay for microsomal triglyceride transfer protein. J Lipid Res. 2004;45:764–772. doi: 10.1194/jlr.D300026-JLR200. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000a;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000b;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Berriot-Varoqueaux N, Aggerbeck LP, Samson-Bouma M, Wetterau JR. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu Rev Nutr. 2000;20:663–697. doi: 10.1146/annurev.nutr.20.1.663. [DOI] [PubMed] [Google Scholar]
- Bjorkegren J, Beigneux A, Bergo MO, Maher JJ, Young SG. Blocking the secretion of hepatic very low density lipoproteins renders the liver more susceptible to toxin-induced injury. J Biol Chem. 2002;277:5476–5483. doi: 10.1074/jbc.M108514200. [DOI] [PubMed] [Google Scholar]
- Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor Small Heterodimer Partner. Endocr J. 2008;55:253–268. doi: 10.1507/endocrj.k07e-103. [DOI] [PubMed] [Google Scholar]
- Dai K, Khatun I, Hussain MM. NR2F1 and IRE1β suppress MTP expression and lipoprotein assembly in undifferentiated intestinal epithelial cells. Arterioscler Thromb Vasc Biol. 2010;30:568–574. doi: 10.1161/ATVBAHA.109.198135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- Duez H, van dV, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Bauge E, Havinga R, Bloks VW, Wolters H, van der Sluijs FH, Vennstrom B, Kuipers F, Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Escobar C, az-Munoz M, Encinas F, guilar-Roblero R. Persistence of metabolic rhythmicity during fasting and its entrainment by restricted feeding schedules in rats. Am J Physiol. 1998;274:R1309–R1316. doi: 10.1152/ajpregu.1998.274.5.R1309. [DOI] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Garaulet M, Madrid JA. Chronobiology, genetics and metabolic syndrome. Curr Opin Lipidol. 2009;20:127–134. doi: 10.1097/MOL.0b013e3283292399. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan DL, Kienzle B, Jamil H, Hariharan N. Transcriptional regulation of human and hamster microsomal triglyceride transfer protein genes - Cell type-specific expression and response to metabolic regulators. J Biol Chem. 1994;269:28737–28744. [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokane H, Nakahara M, Tachibana S, Shimizu M, Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J Biol Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- Hussain MM, Pan X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol Metab. 2009;20:177–185. doi: 10.1016/j.tem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MM, Rava P, Pan X, Dai K, Dougan SK, Iqbal J, Lazare F, Khatun I. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol. 2008;19:277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- Kang S, Spann NJ, Hui TY, Davis RA. ARP-1/COUP-TF II determines hepatoma phenotype by acting as both a transcriptional repressor of microsomal triglyceride transfer protein and an inducer of CYP7A1. J Biol Chem. 2003;278:30478–30486. doi: 10.1074/jbc.M304201200. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MG, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MCM, Arbeeny C, Bergquist K, Kienzle B, Gordon DA, Wetterau JR. Cloning and regulation of hamster microsomal triglyceride transfer protein -The regulation is independent from that of other hepatic and intestinal proteins which participate in the transport of fatty acids and triglycerides. J Biol Chem. 1994;269:29138–29145. [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun. 2007;353:895–901. doi: 10.1016/j.bbrc.2006.12.131. [DOI] [PubMed] [Google Scholar]
- Pan X, Hussain FN, Iqbal J, Feuerman MH, Hussain MM. Inhibiting proteasomal degradation of microsomal triglyceride transfer protein prevents CCl4 induced steatosis. J Biol Chem. 2007;282:17078–17089. doi: 10.1074/jbc.M701742200. [DOI] [PubMed] [Google Scholar]
- Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem. 2007;282:24707–24719. doi: 10.1074/jbc.M701305200. [DOI] [PubMed] [Google Scholar]
- Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Rava P, Athar H, Johnson C, Hussain MM. Transfer of cholesteryl esters and phospholipids as well as net deposition by microsomal triglyceride transfer protein. J Lipid Res. 2005;46:1779–1785. doi: 10.1194/jlr.D400043-JLR200. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, FitzGerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- Sato R, Miyamoto W, Inoue J, Terada T, Imanaka T, Maeda M. Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J Biol Chem. 1999;274:24714–24720. doi: 10.1074/jbc.274.35.24714. [DOI] [PubMed] [Google Scholar]
- Schlierf G, Dorow E. Diurnal patterns of triglycerides, free fatty acids, blood sugar, and insulin during carbohydrate-induction in man and their modification by nocturnal suppression of lipolysis. J Clin Invest. 1973;52:732–740. doi: 10.1172/JCI107235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman GV, Engel R, Swank RL, Hissen W. Circadian periodicity in some physicochemical parameters of circulating blood. Nature. 1965;207:833–835. doi: 10.1038/207833a0. [DOI] [PubMed] [Google Scholar]
- Sheena V, Hertz R, Nousbeck J, Berman I, Magenheim J, Bar-Tana J. Transcriptional regulation of human microsomal triglyceride transfer protein by hepatocyte nuclear factor-4alpha. J Lipid Res. 2005;46:328–341. doi: 10.1194/jlr.M400371-JLR200. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Isojima Y, van der Horst GT, Takei K, Nagai K, Takamatsu K. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells. 2003;8:973–983. doi: 10.1046/j.1365-2443.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Stoffel M. Coactivation of Foxa2 through Pgc-1beta promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metab. 2006;3:99–110. doi: 10.1016/j.cmet.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01