Cajal body surveillance of U snRNA export complex assembly (original) (raw)
Passage of transcribed U snRNA precursors through Cajal bodies ensures that they are properly bound to the PHAX adaptor protein required for nuclear exit.
Abstract
Phosphorylated adaptor for RNA export (PHAX) is the key export mediator for spliceosomal U small nuclear RNA (snRNA) precursors in metazoa. PHAX is enriched in Cajal bodies (CBs), nuclear subdomains involved in the biogenesis of small ribonucleoproteins. However, CBs’ role in U snRNA export has not been demonstrated. In this study, we show that U snRNA precursors microinjected into Xenopus laevis oocyte nuclei temporarily concentrate in CBs but gradually decrease as RNA export proceeds. Inhibition of PHAX activity by the coinjection of a specific anti-PHAX antibody or a dominant-negative PHAX mutant inhibits U snRNA export and simultaneously enhances accumulation of U snRNA precursors in CBs, indicating that U snRNAs transit through CBs before export and that binding to PHAX is required for efficient exit of U snRNAs from CBs. Similar results were obtained with U snRNAs transcribed from microinjected genes. These results reveal a novel function for CBs, which ensure that U snRNA precursors are properly bound by PHAX.
Introduction
In the course of the maturation of spliceosomal U small nuclear RNP (snRNP) in metazoa, U small nuclear RNA (snRNA) precursors are initially exported from the nucleus after transcription (Mattaj, 1988; for review see Will and Luhrmann, 2001). This export is mediated by CRM1, a member of the importin-β family (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Stade et al., 1997). CRM1, known as the export receptor for proteins carrying a leucine-rich nuclear export signal (NES; Fischer et al., 1995; Wen et al., 1995), binds directly to NES but indirectly to U snRNA.
Two adaptor proteins are involved in the interaction between CRM1 and U snRNAs. One is the heterodimeric cap-binding complex (CBC), which binds specifically to the essential export signal of U snRNA, the m7G-cap structure (Ohno et al., 1990; Izaurralde et al., 1994, 1995; Kataoka et al., 1995). The other adaptor is a phosphorylated protein termed phosphorylated adaptor for RNA export (PHAX) that acts as a bridge between CRM1 and the CBC–RNA complex (Ohno et al., 2000). PHAX has a leucine-rich NES to which CRM1 binds cooperatively with RanGTP. The NES sequence in PHAX is functional only when PHAX is properly phosphorylated (Ohno et al., 2000). In this way, these five proteins and a U snRNA molecule assemble into the export complex in the nucleus, and this complex subsequently moves to the cytoplasm. Thus, the composition of the U snRNA export complex is relatively well characterized. However, very little information is available regarding when and where these factors associate with U snRNA precursors in the nucleus.
After being exported to the cytoplasm, U snRNAs associate with a group of Sm proteins with the help of molecular chaperons termed the survival of motor neurons complex (for review see Kolb et al., 2007; Chari et al., 2009). The m7G-cap structure of the U snRNAs is subsequently trimethylated, and the trimethylated cap is recognized by snurportin-1 (Huber et al., 1998). The RNA-bound Sm proteins and snurportin-1 constitute a bipartite nuclear import signal that directs the nuclear import of U snRNPs by importin-β. Before or during the nuclear import process, a few nucleotides at the 3′ terminus are trimmed (for review see Will and Luhrmann, 2001). Once imported into the nucleus, the U snRNPs associate with Cajal bodies (CBs), the site for RNA modifications such as 2’-_O_-ribose-methylation and pseudouridylation (Jady et al., 2003; for review see Cioce and Lamond, 2005). Specific RNP assembly is also thought to occur within CBs, including the assembly of the 17S U2 snRNP (Nesic et al., 2004) and of tri-snRNPs after each round of splicing (Schaffert et al., 2004; Stanek and Neugebauer, 2004; for review see Stanek and Neugebauer, 2006). All these events represent critical steps in the making of a fully functional snRNP. Finally, snRNPs are recruited to nuclear speckles, where the final stages of maturation like further packaging may occur (for review see Patel and Bellini, 2008).
Thus, the role of CBs in the maturation of U snRNPs, after their reimport into the nucleus, is relatively clear. However, the following several studies have suggested that CBs also have roles in the processes before the initial nuclear export of U snRNA precursors. First, U snRNA genes are often found adjacent to CBs (Frey and Matera, 1995; Smith et al., 1995). The U snRNA transcription factor proximal sequence element-binding transcription factor is enriched in CBs (Schul et al., 1998), and active transcription of U snRNA genes frequently occurs in association with CBs (Jacobs et al., 1999; Frey and Matera, 2001; Dundr et al., 2007). It was also shown that the nascent U snRNA transcripts themselves mediate the association between U snRNA genes and CBs (Frey et al., 1999; Frey and Matera, 2001). Second, U snRNA precursors before nuclear export can be found in CBs. U snRNA precursors before 3′ end trimming were detected in CBs of HeLa cells by RNA FISH (Smith and Lawrence, 2000). Also, it was serendipitously observed in Xenopus laevis oocytes that a fraction of microinjected U2 snRNA localized to CBs before export (Yu et al., 2001). Third, both PHAX and CRM1, critical U snRNA export factors, are enriched in CBs (Frey and Matera, 2001; Massenet et al., 2002; Boulon et al., 2004; Renvoise et al., 2009). Collectively, these studies suggest that transcribed U snRNA precursors may first transit through CBs before their passage through the nuclear pore complexes (NPCs; for review see Matera et al., 2007). However, such a scenario is yet to be supported by strong experimental evidence.
In this study, in vitro–transcribed U snRNA precursors were microinjected into the nuclei of Xenopus oocytes to examine whether they transit through CBs before their nuclear export. The signals from the injected U snRNAs temporarily accumulated in CBs soon after the microinjection and gradually decreased as the RNA export proceeded. Inhibition of PHAX activity by either an anti-PHAX antibody or a dominant-negative PHAX mutant led to inhibition of U snRNA export and simultaneously enhanced the accumulation of U snRNAs in CBs. In contrast, inhibition of CRM1 activity by excess NES sequences or inhibition of the function of NPCs by WGA resulted in the accumulation of U snRNAs in the nucleoplasm but not in CBs. Moreover, a combination of the inhibitors confirmed that U snRNAs transit through CBs before passing through NPCs and that proper binding of PHAX but not CRM1 to U snRNAs is required for the efficient exit of U snRNAs from CBs. From these results, it is suggested that one function of CBs is to check whether U snRNA precursors are properly bound by PHAX.
Results
Injected U snRNA precursors associate with CBs before nuclear export
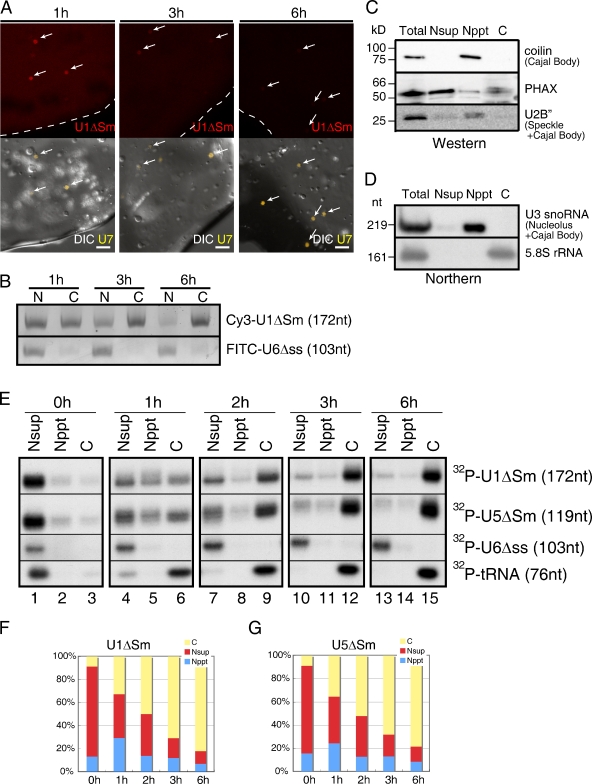
To examine whether U snRNA precursors transit through CBs before their nuclear export, an attempt was made to visualize the distribution of the precursors in Xenopus oocyte nuclei. Cy3-labeled U1ΔSm RNA, which has mutations in the Sm-binding site and therefore cannot be reimported into the nucleus, was microinjected into the nuclei of Xenopus oocytes. After incubation for certain periods, the nuclei were manually isolated in a mineral oil and observed under a fluorescence microscope as described previously (Patel et al., 2008). After 1 h of incubation, the signal of U1ΔSm RNA was highly enriched in specific dots with a relatively homogeneous nucleoplasmic signal (Fig. 1 A, top left).
Figure 1.
Transport of Cy3-labeled U1ΔSm RNA microinjected into the Xenopus oocyte nuclei. (A) In vitro–transcribed Cy3-labeled m7G-capped U1ΔSm RNA was microinjected into Xenopus oocyte nuclei that had been cytoplasmically preinjected with Cy5-labeled U7 RNA as a marker of Cajal Bodies 12 h earlier. The nuclei were manually isolated under mineral oil at 1 (left), 3 (middle), and 6 h (right) after injection. The nuclei were observed by fluorescent microscopy as described in Materials and methods. Red, Cy3-U1ΔSm; yellow, Cy5-U7. The arrows and the dotted lines represent Cajal bodies and the nuclear envelope, respectively. Bars, 20 µm. (B) A mixture of in vitro–transcribed Cy3-labeled U1ΔSm and FITC-labeled U6Δss RNAs was microinjected into the Xenopus oocyte nuclei. RNA was extracted from nuclear (N) and cytoplasmic (C) fractions 1, 3, and 6 h after microinjection and analyzed by 8% denaturating PAGE followed by Typhoon scan. (C) Xenopus oocytes were fractionated into Nsup, Nppt, and cyctoplasmic fractions as described in Materials and methods. Western blotting was performed with total oocyte (total), Nsup, Nppt, and cytoplasmic fractions using antibodies against coilin, PHAX, and U2B”. Each lane was loaded with materials from 2.5 oocytes. (D) Northern blotting was performed with RNA prepared from total, Nsup, Nppt, and cytoplasmic fractions using probes against U3 snoRNA and 5.8S rRNA. Each lane was loaded with RNA from 2.5 oocytes. nt, nucleotide. (E) A mixture of 32P-labeled m7G-capped U1ΔSm, m7G-capped U5ΔSm, U6Δss, and initiator methionyl tRNA was injected into the nucleus of Xenopus oocytes. RNA was extracted from Nsup, Nppt, and cytoplasmic fractions immediately at 0 (lanes 1–3) 1 (lanes 4–6), 2 (lanes 7–9), 3 (lanes 10–12), and 6 h (lanes 13–15) after microinjection and analyzed by 8% denaturating PAGE followed by autoradiography. (F and G) Quantification of the radioactivity of U1 and U5 bands in E, respectively. Yellow, C fraction; red, Nsup fraction; blue, Nppt fraction. Black lines indicate that intervening lanes have been spliced out.
To examine whether these specific dots represented CBs, Cy5-labeled U7 snRNAs, known to stably localize in CBs after microinjection (Wu et al., 1996), were preinjected as a marker. In fact, the two signals merged (Fig. 1 A, left), indicating that the injected U1ΔSm was concentrated in CBs at this time point. The U1ΔSm RNA signal was not enriched in other nuclear domains such as chromosomes, nucleoli, nuclear speckles (B snurposomes), or the nuclear envelope (Fig. 1 A and not depicted). At later time points (3 and 6 h after microinjection), the Cy3 signal of U1ΔSm RNA gradually decreased in both CBs and the nucleoplasm (Fig. 1 A, middle and right).
To confirm that the decrease in Cy3 signals was due not to RNA degradation but to U snRNA export, the stability and export kinetics of the injected U1ΔSmRNA were monitored in a parallel experiment. RNA was extracted from nuclear and cytoplasmic fractions at each time point after microinjection and analyzed by gel electrophoresis followed by visualization of the fluorescent RNA bands (Fig. 1 B). The stability and export kinetics of the injected Cy3-U1ΔSm RNA were similar to those of the 32P-labeled U1ΔSm RNA (unpublished data). The total U1ΔSm RNA signal was fairly constant, indicating that the injected U1ΔSm was highly stable in Xenopus oocytes. Moreover, the decrease in Cy3 signals in Fig. 1 A was well concomitant with the extent of RNA export. These results are consistent with the idea that U1ΔSm RNAs move to CBs before their nuclear export.
To obtain more quantitative information about the distribution of the injected U snRNA precursors, we fractionated the nuclei of Xenopus oocytes into insoluble (Nppt) and soluble (Nsup) fractions as described previously (Wu et al., 1996). The Nppt fraction contains nuclear organelles such as chromatins, CBs, nucleoli, and nuclear speckles, etc., whereas Nsup contains soluble materials of the nucleoplasm (Wu et al., 1996). Western blotting showed that coilin, a marker of CBs, and U2B”, a protein component of U2 snRNP found in both CBs and nuclear speckles, were mainly detected in the Nppt fraction (Fig. 1 C). Similarly, Northern blotting showed that U3 small nucleolar RNA (snoRNA), which is found mainly in nucleoli and partially in CBs, was also detected in the Nppt fraction, whereas 5.8S rRNA was mainly detected in the cytoplasmic fraction (Fig. 1 D). PHAX, the key U snRNA export mediator, was detected mainly in the Nsup fraction and partially in the Nppt and C fractions (Fig. 1 C), reminiscent of the fact that PHAX is found in CBs as well as the nucleoplasm in mammalian cell nuclei (Frey and Matera, 2001; Massenet et al., 2002; Boulon et al., 2004; Renvoise et al., 2009).
A mixture of 32P-labeled RNAs containing U1ΔSm, U5ΔSm, U6Δss, and initiator methionyl tRNA was microinjected into the nuclei of Xenopus oocytes, and the distribution of the RNAs was examined using the aforementioned fractionation method (Fig. 1 E). U6Δss RNA has been frequently used as a marker for the efficacy of nuclear microinjection because U6 RNA never leaves the nucleus (Vankan et al., 1990), and in addition, U6Δss RNA has a small deletion in the single-stranded region, therefore being defective in nuclear migration from the cytoplasm (Hamm and Mattaj, 1989). U6Δss RNA was consistently detected only in the Nsup fraction, whereas tRNA, which is rapidly exported, was mostly detected in the C fraction at all time points except 0 h (Fig. 1 E). However, both U snRNAs (U1ΔSm and U5ΔSm) were detected in Nppt and Nsup fractions maximally at 1 h (Fig. 1 E, lanes 4–6), and their signals gradually decreased as the RNA export proceeded (Fig. 1 E, lanes 7–15). Because the Nppt fraction contains CBs, and CBs are the only nuclear structures in which the injected U snRNAs are concentrated (Fig. 1 A), it is plausible that the signals of U snRNAs in the Nppt fraction mostly represent the signals in CBs. Note that the export of injected U snRNAs was completed within 6 h, and the injected U snRNAs were highly stable during the incubation. Given that ∼30% of the injected U snRNAs was found in the Nppt fraction at 1 h (Fig. 1, F and G), a significant fraction of the injected U snRNAs would transit through CBs before nuclear export.
Injected U snRNA precursors accumulate more strongly in CBs when PHAX activity is perturbed
The aforementioned results prompted us to examine the effect of various inhibitors of U snRNA export on the distribution of U snRNA in the nucleus. A mixture of 32P-labeled RNAs containing U1ΔSm, U5ΔSm, and U6Δss was microinjected into the nucleus of Xenopus oocytes in the presence or absence of various inhibitors of U snRNA export, and the distribution of the injected RNAs was examined (Fig. 2). Coinjection of WGA, which binds NPCs and therefore inhibits the passage of many nuclear transport cargoes through NPCs, strongly inhibited the export of both U snRNAs (Fig. 2 A, compare lanes 4–6 with lanes 7–9). Note that the injected U snRNAs mainly accumulated in the nucleoplasm (Nsup) in this case. Similarly, coinjection of an affinity-purified anti-PHAX antibody inhibited the export of U snRNAs (Fig. 2 A, lanes 13–15). However, in this case, a much higher percentage of U snRNAs accumulated in the Nppt fraction. In vitro gel mobility shift experiments revealed that this antibody inhibits the assembly of PHAX onto the U snRNA–CBC complex (Fig. 2 B). These results suggested that proper binding of PHAX is required for U snRNAs to efficiently exit from CBs into the nucleoplasm. When both WGA and the anti-PHAX antibody were coinjected, U snRNAs also accumulated in the Nppt fraction, which was nearly identical to the effect of the antibody alone (Fig. 2, A [lanes 16–21], C, and D [quantification]). These results confirmed that the PHAX antibody inhibited an earlier transport step than WGA. The accumulation of U snRNAs in CBs in the presence of WGA and the anti-PHAX antibody was confirmed by injecting Cy3-U1ΔSm and visualizing the Cy3 signals (Fig. 2 E).
Figure 2.
Effect of anti-PHAX antibody on the distribution of injected U snRNA precursors. (A) A mixture of 32P-labeled m7G-capped U1ΔSm, m7G-capped U5ΔSm, and U6Δss was injected into the nucleus of Xenopus oocytes either alone (lanes 1–6) or together with 0.42 µg/oocyte WGA (lanes 7–9), 33 ng/oocyte control (ctr) IgG (lanes 10–12), or anti-PHAX antibody (lanes 13–15), WGA and control IgG (lanes 16–18), or WGA and anti-PHAX antibody (lanes 19–21). RNA was extracted and analyzed as in Fig.1 E at 0 and 2 h after injection. C, cytoplasmic fraction. (B) A 32P-labeled m7G-capped U1ΔSm was incubated either alone (lane 1) or together with 1 µM recombinant CBC (lane 2), 1 µM of each recombinant CBC and PHAX (lane 3), or CBC and PHAX and affinity-purified anti-PHAX antibody (lane 4) for 20 min at 25°C. The samples were fractionated by native 6% PAGE followed by autoradiography. Free RNA and major complexes are indicated on the right. Black lines indicate that intervening lanes have been spliced out. (C and D) The radioactivity of the bands of U1ΔSm and U5ΔSm RNAs, respectively, was quantified from three independent experiments as in A, and the ratio of the Nppt signal against the nuclear fraction total (Nppt + Nsup) signal was calculated. The means and standard deviations for WGA alone, WGA + control, IgG, and WGA + anti-PHAX antibody are shown. (E) The distribution of Cy3-labed m7G-capped U1ΔSm RNA was analyzed as in Fig.1 A at 2 h after microinjection in the presence of WGA + affinity-purified anti-PHAX antibody (right) or WGA + control (control) IgG (left). Bars, 20 µm.
Next, we examined the effects of coinjection of various PHAX mutant proteins (Fig. 3 A), some of which were already shown to act in a dominant-negative fashion on U snRNA export (Ohno et al., 2000; Segref et al., 2001; Kitao et al., 2008). Coinjection of the PHAX ΔCD mutant, having a small deletion in the RNA-binding domain (Fig. 3 A) and therefore unable to bind RNA (Segref et al., 2001), had no effect on the distribution of the U snRNAs (Fig. 3 B, lanes 7–9). Two other mutants, ΔNES and ΔST2, which had mutations in the NES and critical phosphorylation sites, respectively (Fig. 3 A), are both known to be defective in binding to CRM1 (Ohno et al., 2000; Kitao et al., 2008). Microinjection of the ΔNES or ΔST2 mutant protein inhibited U snRNA export as demonstrated previously (Ohno et al., 2000; Kitao et al., 2008), but the injected U snRNAs accumulated in the nucleoplasm (Nsup; Fig. 3 B, lanes 13–18). Another mutant protein, PHAX ΔN lacking the N-terminal 175 amino acid residues, can still bind to RNA but not stably to CBC (similar to the N4 mutant in Segref et al., 2001). This protein was not previously tested in microinjection experiments. Microinjection of PHAX ΔN also inhibited U snRNA export, but in this case, the injected U snRNAs strongly accumulated in the insoluble Nppt fraction (Fig. 3 B, lanes 10–12). Usage of both PHAX ΔN and WGA also resulted in enhanced accumulation of U snRNAs in the Nppt fraction (Fig. 3, B [lanes 22–24], C, and D). The longer the incubation, the more U snRNA accumulated in the Nppt fraction (Fig. 3 E, lanes 10–18). Note that the majority of the injected U snRNAs were found in the Nppt fraction at 3 h (Fig. 3 E, lane 17). Visualization of the U snRNAs confirmed that they in fact accumulated in CBs (Fig. 3 F). These results suggested that the ΔN mutant protein markedly slows down the exit of U snRNAs from CBs and confirmed that proper binding of PHAX is required for U snRNAs to exit CBs and that the transit through CBs is part of the major route of U snRNA export.
Figure 3.
Effect of PHAX mutant proteins on the distribution of injected U snRNA precursors. (A) Schematic representation of the domain structures of human PHAX protein and the features of the PHAX mutants used in this study. The numbers represent the amino acid numbers of the PHAX protein. WT, wild type. (B) The same mixture of 32P-labeled RNAs as in Fig. 2 A was injected into the nuclei of Xenopus oocytes either alone (lanes 1–6) or together with 25 ng/oocyte of recombinant PHAXΔCD (ΔCD; lanes 7–9), PHAXΔN (ΔN; lanes 10–12), PHAXΔNES (ΔNES; lanes 13–15), PHAXΔST2 (ΔST2; lanes 16–18), WGA and ΔCD (lanes 19–21), WGA and ΔN (lanes 22–24), WGA and ΔNES (lanes 25–27), or WGA and ΔST2 (lanes 28–30). RNA was analyzed as in Fig. 2 A. (C and D) The ratio of the Nppt signal against the nuclear fraction total signal of U1ΔSm and U5ΔSm, respectively, was calculated from three independent experiments as in A. The means and standard deviations for WGA + ΔCD, WGA + ΔN, WGA + ΔNES, and WGA + ΔST2 are shown. (E) The same RNA mixture was injected with either WGA + ΔCD (lanes 1–9), WGA + ΔN (lanes 10–18), or WGA + ΔNES (lanes 19–27). RNA was analyzed at 0, 1.5, and 3 h as in (A). (F) The distribution of Cy3-labeled m7G-capped U1ΔSm RNA was analyzed as in Fig. 1 A at 2 h after microinjection in the presence of WGA + ΔN (right) or WGA + ΔCD (left). C, cytoplasmic fraction. Bars, 20 µm.
Binding of CRM1 is not required for U snRNAs to exit CBs
The results suggested that CBs check the binding of PHAX to U snRNAs. If PHAX is not bound properly, the RNA cannot exit CBs. However, CRM1, another U snRNA export factor, was also shown to be enriched in CBs (Boulon et al., 2004; Renvoise et al., 2009). To examine whether the binding of CRM1 is required for the exit of U snRNAs from CBs, we used BSA-NES, a conjugate of NES peptides coupled to BSA, which saturates CRM1 export (Fischer et al., 1995). Microinjection of BSA-NES but not a mutant version (BSAmut) inhibited U snRNA export, as described previously (Fornerod et al., 1997; Ohno et al., 2000), and U snRNAs accumulated in the nucleoplasm (Nsup) but not in Nppt (Fig. 4 A, compare lanes 22–24 with lanes 13–15). In contrast, coinjection of both BSA-NES and PHAXΔN resulted in enhanced RNA accumulation in Nppt, i.e., in CBs (Fig. 4, A [lanes 28–30], B, and C), indicating that BSA-NES blocked not the entry of U snRNAs into CBs but the step after the exit of U snRNAs from CBs, most likely the passage of RNA through NPCs. These results indicate that the binding of CRM1, unlike PHAX, is not required for U snRNA to exit CBs.
Figure 4.
Effect of CRM1 inhibition on the distribution of injected U snRNA precursors. (A) The same mixture of 32P-labeled RNAs as in Fig. 2 A was injected into the nuclei of Xenopus oocytes either alone (lanes 1–6) or together with ΔCD (lanes 7–9), ΔN (lanes 10–12), 0.3 µg/oocyte BSAmut (lanes 13–15), BSAmut + ΔCD (lanes 16–18), BSAmut + ΔN (lanes 19–21), 0.3 µg/oocyte BSA-NES (lanes 22–24), BSA-NES + ΔCD (lanes 25–27), or BSA-NES + ΔN (lanes 28–30). RNA was analyzed as in Fig. 2 A. (B and C) The ratio of the Nppt signal against the nuclear fraction total signal of U1ΔSm and U5ΔSm, respectively, was calculated from three independent experiments as in A. The means and standard deviations for BSA-NES alone, BSA-NES + ΔCD, and BSA-NES + ΔN are shown. C, cytoplasmic fraction.
Proper binding of PHAX is required for in vivo–transcribed U snRNAs to exit CBs
The aforementioned results were obtained from RNA microinjection experiments in which RNAs transcribed in vitro were microinjected into the nuclei of Xenopus oocytes and therefore in which RNA export was not dependent on transcription in vivo. This experimental system was advantageous for focusing on RNA transport by itself, but in some cases, might not reflect the true situation in vivo, where U snRNAs are transcribed from their genes. Therefore, we microinjected the plasmid containing the U1ΔSm and U2ΔSm genes into the nuclei of Xenopus oocytes and examined the distribution of U1ΔSm and U2ΔSm RNAs transcribed in vivo by Northern blotting. Coinjection of the anti-PHAX antibody or PHAX ΔN mutant protein, in the presence of WGA, led to enhanced RNA signals in the Nppt fraction as compared with the injection of the control IgG or other PHAX mutants (Fig. 5, A–C [quantification]). These findings indicated that proper binding of PHAX is required for U snRNAs transcribed in vivo to exit CBs efficiently and supported the results of the RNA microinjection experiments.
Figure 5.
Effect of the inhibitors on the distribution of in vivo–transcribed U snRNAs. A mixture of the plasmids harboring the U1ΔSm and U2ΔSm genes was microinjected into the nuclei of Xenopus oocytes either alone (lanes 1–3) or together with control (ctr) IgG (lanes 4–6), anti-PHAX antibody (lanes 7–9), ΔCD protein (lanes 10–12), ΔN protein (lanes 13–15), or ΔNES protein (lanes 16–18), and the nucleocytoplasmic distribution of the transcripts was analyzed by Northern blotting of the Nsup, Nppt, and cytoplasmic (C) RNA fractions after 1.5 h. Black lines indicate that intervening lanes have been spliced out. (B and C) The ratio of the Nppt signal against the nuclear fraction total signal of U1ΔSm and U2ΔSm, respectively, was calculated from three independent experiments as in A. The means and the standard deviations for WGA+ctr IgG, WGA+anti-PHAX antibody, WGA+ΔCD, WGA+ΔN, and WGA+ΔNES are shown.
Discussion
U snRNA transport from genes to cytoplasm
We have provided evidence that spliceosomal U snRNA precursors transit through CBs before their nuclear export. Part of the biological significance of this is confirmation that the precursors are properly bound by PHAX. The results of this and other experiments imply the following model as to the major route of U snRNA transport from genes to cytoplasm (Fig. 6).
Figure 6.
Model for the transport of U snRNA from genes to cytoplasm. See Results for details.
It was previously shown that actively transcribed U snRNA genes are frequently associated with CBs in mammalian cell nuclei and that the transcription by itself, i.e., the presence of the nascent U snRNA transcript, is required for this association (Frey and Matera, 1995, 2001; Frey et al., 1999). Thus, when a U snRNA gene starts transcription, it probably associates with a CB via a process mediated by the nascent U snRNA transcript that has not been released from the gene. Because the U snRNA–specific transcription factor proximal sequence element-binding transcription factor is abundant in CBs (Schul et al., 1998), this association should enhance the transcription itself. This seems to be part of the biological significance of this association.
Some factor in CBs should mediate the association with the gene through interaction with the nascent U snRNA transcript. PHAX is a good candidate for such a factor. The cap structure of the nascent U snRNA transcript should first become associated with CBC cotranscriptionally, and the complex should then recruit PHAX, forming a trimeric complex of U snRNA, CBC, and PHAX, termed the precomplex of U snRNA export (Ohno et al., 2000). Because PHAX is enriched in CBs, this should result in the association of the U snRNA genes with CBs. This association may be assisted and/or stabilized by the nuclear actin system (Dundr et al., 2007). Thus, when the U snRNA transcript is released from the gene into CBs, it is already in the form of the precomplex of U snRNA export. If this scenario is true, the binding of CBC to U snRNAs would be required for U snRNAs to accumulate in CBs. Moreover, the binding of PHAX to U snRNAs should take place simultaneously with the association of U snRNAs with CBs, meaning that CBs are the major site for association of PHAX with U snRNAs. Our findings that the injected U snRNAs first accumulated in CBs may reflect part of this process. It is also likely that U snRNA precursors are sometimes transcribed away from CBs. Such transcripts may actually go first to CBs. It should be pointed out that there is no evidence that U snRNA genes are associated with CBs in amphibian oocytes (Abbott et al., 1999; for review see Cioce and Lamond, 2005). Therefore, it is likely that U snRNA precursors themselves move to CBs in Xenopus oocytes.
We do not have direct evidence that the binding of CBC is required for the entry of U snRNAs into CBs, mainly because of unavailability of strong inhibitory antibodies against CBC. However, we do have supporting evidence. A-capped U snRNA precursors, which do not bind CBC (Ohno et al., 1990), did not accumulate in CBs after microinjection, unlike m7G-capped counterparts (Fig. S1), suggesting that the m7G-cap structure and probably also the binding of CBC to U snRNAs are important for the entry of U snRNAs into CBs. Moreover, our findings do not apparently support that the binding of PHAX is required for U snRNAs to enter CBs. The result that U snRNAs accumulated in CBs in the presence of the anti-PHAX antibody can be interpreted to mean that the antibody inhibited the exit from, but not the entry into, CBs. However, it is also possible that the antibody actually inhibits both the entry and exit to a certain extent but that its inhibitory effect on the exit is stronger for some reason. If so, U snRNAs should accumulate in CBs in the presence of the antibody. This is a mere assumption, but is consistent with the effect of the antibody being weaker than that of the PHAX ΔN mutant.
After their entry into CBs, U snRNAs can be released from the CBs into the nucleoplasm, provided that the precomplex is properly formed and maintained. However, if the binding of PHAX is affected for some reason, by the actions of a specific antibody for example, the U snRNAs cannot leave the CBs. Thus, we provide the first evidence that CBs function in the surveillance of the U snRNA preexport complex assembly. Regarding the actions of PHAX ΔN, this mutant can bind to RNA but not stably to CBC (Segref et al., 2001), and therefore, the precomplex containing this mutant is likely to be abnormal. This abnormal precomplex may be functional for U snRNA entry into CBs but not for their exit. The requirements of the quality of the precomplex may be more strict for the exit than entry. This notion is also consistent with the assumption that the effect of the PHAX antibody is stronger for the exit. The molecular mechanism to retain U snRNAs without properly bound PHAX is totally unknown at the moment.
Another U snRNA export factor, CRM1, is also concentrated in CBs, but we could not provide evidence to show that its binding to the precomplex, i.e., the formation of the full export complex, is required for the exit of U snRNAs from CBs. In relation to this, there is no evidence that RanGTP, the last known component of the export complex, is concentrated in CBs. Therefore, the binding of CRM1–RanGTP, i.e., the formation of the full export complex, may occur not in CBs but in the nucleoplasm. In any case, once the export complex is formed, it should be competent for passage through NPCs.
Comparison with intranuclear transport of snoRNAs
There are a group of small RNAs termed snoRNAs that finally accumulate in the nucleoli (for review see Matera et al., 2007). It is thought that a class of snoRNAs, including U3, also transit through CBs before going to the nucleolus after transcription, although snoRNAs are believed not to leave the nucleus, unlike U snRNAs (for review see Matera et al., 2007). Because some of the actively transcribed snoRNA genes are also associated with CBs (Gao et al., 1997; Jacobs et al., 1999), intranuclear transport of such snoRNAs and U snRNAs may overlap at least partially. Is it then possible to find a unified mechanism for these two intranuclear transport systems? It was previously shown with mammalian cells that PHAX is required for the entry of microinjected U3 snoRNAs into CBs (Boulon et al., 2004). Considering the possibility that the binding of PHAX may also be required for U snRNAs to enter CBs in our system, our results do not necessarily contradict those of Boulon et al. (2004). Requirement of PHAX for the exit of U3 snoRNA from CBs is still open to question because the antibody inhibited the entry itself in their system (Boulon et al., 2004). In any case, because the majority of the transcription of both small RNAs takes place adjacent to CBs, the binding of these RNAs to PHAX may take place simultaneously with their association with CBs.
However, the requirement of CRM1 for the exit from CBs seems clearly different between the two systems. The binding of CRM1 is not required for U snRNAs to exit CBs in our system (Fig. 4). In contrast, Boulon et al. (2004) showed that CRM1 is important for U3 snoRNAs to exit CBs in mammalian cells. This is a puzzling result because CRM1 is an export factor, and U3 snoRNAs are thought not to leave the nucleus. However, it should also be pointed out that some snoRNAs might actually leave the nucleus temporarily like U snRNAs (Watkins et al., 2007). If this is the case, the maturation pathways for U snRNAs and snoRNAs would be even more similar. More experiments are required to establish a plausible unified model.
CBs, dynamic structures for accelerating maturation of small RNPs
CBs are dynamic nuclear subdomains involved in the biogenesis of several classes of small RNPs. In CBs, extensive internal modifications of the U snRNAs by 2’-_O_-methylation and pseudouridylation, as well as specific assemblies of some U snRNPs, take place (for review see Stanek and Neugebauer, 2006). In this study, using the Xenopus oocyte system, we found another function of CBs, surveillance of the export U snRNP assembly. Our results also suggested that CBs are the major site for PHAX to associate with U snRNAs. Whether this is a general rule applicable to other cell types is still open to question. Xenopus CBs are known to be morphologically distinct from mammalian counterparts and could be somewhat special in certain aspects (for review see Cioce and Lamond, 2005).
It should also be pointed out that CBs seem not to be essential structures for all types of cells. For instance, CBs are found in oocytes, liver cells, and cultured cells like HeLa cells but not in certain cells in adult tissues, including smooth and cardiac muscle cells, skin cells, and spleen parenchymal cells (Young et al., 2000). Thus, CBs appear to be specialized organelles that accelerate the maturation of small RNPs in rapidly dividing cells and cells in preparation for dividing rapidly like oocytes. If some processes are accelerated to too great an extent, the quality of the products tends to decrease. Therefore, CBs may have acquired the function to check the quality of the RNP products simultaneously with the function to accelerate their maturation.
Materials and methods
DNA constructs and recombinant proteins
The Xenopus U7 gene was PCR amplified from Xenopus genomic DNA, and the amplified fragment was cloned in the HindIII–XhoI sites of pcDNA3. Recombinant CBC and various recombinant PHAX proteins were expressed and purified as described previously (Kitao et al., 2008).
Microinjection into Xenopus oocytes
32P-labeled RNAs for microinjection were prepared as described previously (Ohno et al., 2000). Fluorescent labeling of in vitro–transcribed RNAs was performed using LabelIT reagent (Mirus). Microinjection of RNA/DNA into Xenopus oocytes was performed as described previously (Jarmolowski et al., 1994; Fuke and Ohno, 2008). Detection of fluorescently labeled RNA bands was performed by scanning the gel with Typhoon (GE Healthcare). BSA-NES and BSAmut were prepared as described previously (Masuyama et al., 2004). WGA was purchased from EMD.
Microscopic observation of Xenopus oocyte nuclei
Nuclei of Xenopus oocytes were prepared for microscopy essentially as described previously (Patel et al., 2008). In short, the nuclei were manually isolated under mineral oil, and the sample was spotted onto a glass slide with a shallow depression of 20 µm together with 2 µl of mineral oil and mounted with a coverslip. All microscopic observations were performed at room temperature with an inverted microscope (Axio Observer Z1; Carl Zeiss, Inc.) with a Plan Neofluar 40× NA 0.75 objective in the ApoTome mode for high optical resolution. Images were captured using a camera (AxioCam MRm; Carl Zeiss, Inc.) driven by AxioVision software (version 4.7; Carl Zeiss, Inc.). Figures were processed using Photoshop (CS version 8.0; Adobe). Relative fluorescence intensity was measured using AxioVision.
Subfractionation of Xenopus oocyte nuclei
The subcellular fractionation of Xenopus oocyte nuclei was performed essentially as described previously (Wu et al., 1996). In short, the nucleus and the cytoplasm were manually separated in the isolation medium (Gall et al., 1991). The nuclei were collected in an Eppendorf tube, and isolation medium was added to 150 µl. The sample was spun down in a microfuge at 15,000 rpm for 15 min at 4°C, and the soluble fraction was recovered. The insoluble fraction was washed once with 150 µl of isolation medium, and the soluble fraction was combined with the previous soluble fraction. RNA was isolated from Nsup, Nppt, and the cytoplasmic fraction by phenol extraction and ethanol precipitation. RNA was analyzed by 8% polyacrylamide/7 M urea gel electrophoresis. Analysis and quantitation of 32P-labeled RNA bands were performed with BSA-2500 (Fujifilm) and Image Gauge (version 3.45; Fujifilm). Alternatively, RNA bands were visualized by Northern blotting with specific probes.
Band shift assay
The band shift assay was performed as described previously (Ohno et al., 2000). In short, 32P-labeled m7G-capped U1ΔSm was incubated with recombinant proteins in band shift buffer (40 mM Hepes KOH, pH 7.3, 110 mM KOAc, 6 mM Mg(OAc)2, 250 mM sucrose, and 0.8 mg/ml Escherichia coli tRNA) for 20 min at 25°C. The samples were fractionated by native 6% PAGE followed by autoradiography. Anti-PHAX antibody was affinity purified as described previously (Kitao et al., 2008).
Western and Northern blot analyses
Western blotting was performed as described previously (Kitao et al., 2008) with a monoclonal anti-coilin antibody (Invitrogen), a monoclonal anti-U2B” antibody (Progen), and an affinity-purified anti-PHAX antibody (Kitao et al., 2008). Northern blotting was performed as described previously (Fuke and Ohno, 2008) with 5′-CTTTCGAGCACATTTCACCAGG-3′ and 5′-CCAACTCCCGGATCCCCGGAGCTTGCCATTTAATA-3′ as probes for U3 snoRNA and U2ΔSm, respectively. The probes for 5.8S and U1ΔSm RNAs were as described previously (Fuke and Ohno, 2008).
Online supplemental material
Fig. S1 shows the effect of the m7G-cap structure on the distribution of injected U snRNA precursors. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004109/DC1.
Acknowledgments
We thank Drs. Akila Mayeda, Makoto Kitabatake, Ichiro Taniguchi, Saori Kitao, and Mr. Kotaro Fujii for suggestions and criticisms of the manuscript.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan. T. Suzuki was a Japan Society for the Promotion of Science Research Fellow.
Footnotes
Abbreviations used in this paper:
CB
Cajal body
CBC
cap-binding complex
NES
leucine-rich nuclear export signal
NPC
nuclear pore complex
PHAX
phosphorylated adaptor for RNA export
snoRNA
small nucleolar RNA
snRNA
small nuclear RNA
snRNP
small nuclear RNP
References
- Abbott J., Marzluff W.F., Gall J.G. 1999. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol. Biol. Cell. 10:487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonné R., Bertrand E. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 16:777–787 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Chari A., Paknia E., Fischer U. 2009. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 21:387–393 10.1016/j.ceb.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Cioce M., Lamond A.I. 2005. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21:105–131 10.1146/annurev.cellbio.20.010403.103738 [DOI] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., Matera A.G. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179:1095–1103 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Huber J., Boelens W.C., Mattaj I.W., Lührmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 82:475–483 10.1016/0092-8674(95)90436-0 [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Frey M.R., Matera A.G. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., Matera A.G. 2001. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J. Cell Biol. 154:499–509 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., Bailey A.D., Weiner A.M., Matera A.G. 1999. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9:126–135 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- Fuke H., Ohno M. 2008. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 36:1037–1049 10.1093/nar/gkm1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 390:308–311 10.1038/36894 [DOI] [PubMed] [Google Scholar]
- Gall J.G., Murphy C., Callan H.G., Wu Z.A. 1991. Lampbrush chromosomes. Methods Cell Biol. 36:149–166 10.1016/S0091-679X(08)60276-9 [DOI] [PubMed] [Google Scholar]
- Gao L., Frey M.R., Matera A.G. 1997. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 25:4740–4747 10.1093/nar/25.23.4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Mattaj I.W. 1989. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 8:4179–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J., Cronshagen U., Kadokura M., Marshallsay C., Wada T., Sekine M., Lührmann R. 1998. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17:4114–4126 10.1093/emboj/17.14.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 78:657–668 10.1016/0092-8674(94)90530-4 [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I.W. 1995. A cap-binding protein complex mediating U snRNA export. Nature. 376:709–712 10.1038/376709a0 [DOI] [PubMed] [Google Scholar]
- Jacobs E.Y., Frey M.R., Wu W., Ingledue T.C., Gebuhr T.C., Gao L., Marzluff W.F., Matera A.G. 1999. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol. Biol. Cell. 10:1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E., Darzacq X., Tucker K.E., Matera A.G., Bertrand E., Kiss T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22:1878–1888 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W.C., Izaurralde E., Mattaj I.W. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627–635 10.1083/jcb.124.5.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Ohno M., Moda I., Shimura Y. 1995. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 23:3638–3641 10.1093/nar/23.18.3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S., Segref A., Kast J., Wilm M., Mattaj I.W., Ohno M. 2008. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol. Cell. Biol. 28:487–497 10.1128/MCB.01189-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S.J., Battle D.J., Dreyfuss G. 2007. Molecular functions of the SMN complex. J. Child Neurol. 22:990–994 10.1177/0883073807305666 [DOI] [PubMed] [Google Scholar]
- Massenet S., Pellizzoni L., Paushkin S., Mattaj I.W., Dreyfuss G. 2002. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 22:6533–6541 10.1128/MCB.22.18.6533-6541.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K., Taniguchi I., Kataoka N., Ohno M. 2004. RNA length defines RNA export pathway. Genes Dev. 18:2074–2085 10.1101/gad.1216204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., Terns M.P. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. 1988. U snRNP assembly and transport. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Birnstiel M.L., editor Springer-Verlag New York Inc., New York/Berlin: 100–114 [Google Scholar]
- Nesic D., Tanackovic G., Krämer A. 2004. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 117:4423–4433 10.1242/jcs.01308 [DOI] [PubMed] [Google Scholar]
- Ohno M., Kataoka N., Shimura Y. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18:6989–6995 10.1093/nar/18.23.6989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 101:187–198 10.1016/S0092-8674(00)80829-6 [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bachelerie F., Dargemont C. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 278:141–144 10.1126/science.278.5335.141 [DOI] [PubMed] [Google Scholar]
- Patel S.B., Bellini M. 2008. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 36:6482–6493 10.1093/nar/gkn658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Novikova N., Beenders B., Austin C., Bellini M. 2008. Live images of RNA polymerase II transcription units. Chromosome Res. 16:223–232 10.1007/s10577-007-1189-z [DOI] [PubMed] [Google Scholar]
- Renvoisé B., Colasse S., Burlet P., Viollet L., Meier U.T., Lefebvre S. 2009. The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy. Hum. Mol. Genet. 18:1181–1189 10.1093/hmg/ddp009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N., Hossbach M., Heintzmann R., Achsel T., Lührmann R. 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23:3000–3009 10.1038/sj.emboj.7600296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W., van Driel R., de Jong L. 1998. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol. Biol. Cell. 9:1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Mattaj I.W., Ohno M. 2001. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA. 7:351–360 10.1017/S1355838201002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.P., Lawrence J.B. 2000. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol. Biol. Cell. 11:2987–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.P., Carter K.C., Johnson C.V., Lawrence J.B. 1995. U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell. Biochem. 59:473–485 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C.S., Guthrie C., Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050 10.1016/S0092-8674(00)80370-0 [DOI] [PubMed] [Google Scholar]
- Stanĕk D., Neugebauer K.M. 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 166:1015–1025 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D., Neugebauer K.M. 2006. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 115:343–354 10.1007/s00412-006-0056-6 [DOI] [PubMed] [Google Scholar]
- Vankan P., McGuigan C., Mattaj I.W. 1990. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 9:3397–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Lemm I., Lührmann R. 2007. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol. Cell. Biol. 27:7018–7027 10.1128/MCB.00516-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell. 82:463–473 10.1016/0092-8674(95)90435-2 [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290–301 10.1016/S0955-0674(00)00211-8 [DOI] [PubMed] [Google Scholar]
- Wu C.H., Murphy C., Gall J.G. 1996. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 2:811–823 [PMC free article] [PubMed] [Google Scholar]
- Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. 2000. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 256:365–374 10.1006/excr.2000.4858 [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Shu M.D., Narayanan A., Terns R.M., Terns M.P., Steitz J.A. 2001. Internal modification of U2 small nuclear (snRNA) occurs in nucleoli of Xenopus oocytes. J. Cell Biol. 152:1279–1288 10.1083/jcb.152.6.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]