An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer (original) (raw)
Abstract
Genome-wide association studies (GWAS) routinely identify risk variants in noncoding DNA, as exemplified by reports of multiple single nucleotide polymorphisms (SNPs) associated with prostate cancer in five independent regions in a gene desert on 8q24. Two of these regions also have been associated with breast and colorectal cancer. These findings implicate functional variation within long-range _cis_-regulatory elements in disease etiology. We used an in vivo bacterial artificial chromosome (BAC) enhancer-trapping strategy in mice to scan a half-megabase of the 8q24 gene desert encompassing the prostate cancer-associated regions for long-range _cis_-regulatory elements. These BAC assays identified both prostate and mammary gland enhancer activities within the region. We demonstrate that the 8q24 cancer-associated variant rs6983267 lies within an in vivo prostate enhancer whose expression mimics that of the nearby MYC proto-oncogene. Additionally, we show that the cancer risk allele increases prostate enhancer activity in vivo relative to the non-risk allele. This allele-specific enhancer activity is detectable during early prostate development and throughout prostate maturation, raising the possibility that this SNP could assert its influence on prostate cancer risk before tumorigenesis occurs. Our study represents an efficient strategy to build experimentally on GWAS findings with an in vivo method for rapidly scanning large regions of noncoding DNA for functional _cis_-regulatory sequences harboring variation implicated in complex diseases.
Genome-wide association studies (GWAS) routinely implicate variation within gene deserts and other types of noncoding DNA in the etiology of disease (Houlston et al. 2008; Silverberg et al. 2009; Yang et al. 2009; Liu et al. 2010). A recent meta-analysis of ∼1200 disease-associated single nucleotide polymorphisms (SNPs) found that in 40% of cases, known exonic sequences were absent from the associated linkage disequilibrium (LD) blocks (Visel et al. 2009). While the presence of nonannotated transcripts or noncoding RNAs may explain some of the noncoding disease associations, these observations also have been interpreted as evidence that many of the associated noncoding regions harbor variants that alter the activity of long-range _cis_-regulatory elements controlling gene expression. Enhancers are one such type of long-range element, functioning over up to megabase-long genomic distances to regulate the temporal and tissue-specific expression patterns of their target gene(s) (Nobrega et al. 2003). A large number of genes with tissue- and temporal-specific expression patterns are known to be controlled by an array of enhancers, with each individual _cis_-regulatory element driving a subset of its gene's entire expression profile (Carroll 2008). This modular nature of enhancer activity makes them ideal candidates for involvement in complex diseases, as functional variants in an individual _cis_-element would result in changes to gene expression only in specific organs/tissue types.
Despite the plethora of GWAS signals implicating noncoding regions in complex disease risk, strategies to experimentally follow up on such findings are lacking. This deficiency stems principally from the difficulty in identifying functional noncoding sequences that map remotely from their target genes. Programs such as ENCODE have been addressing this deficiency by developing and applying technologies to identify these elusive types of long-range regulatory elements (The ENCODE Project Consortium 2007). While these technologies have been invaluable in the identification of putative functional noncoding sequences, they rely heavily on cell culture and other in vitro and in silico methodology to identify and experimentally validate enhancers and other elements. Thus, although these techniques are ideal for functionally following up on noncoding GWAS results when the relevant cell type of interest is obvious and accessible, problems can arise if the putative element under investigation imparts its transcriptional regulatory effects in a cell type of unpredicted origin or one that is not amenable to routine culture. Necessary, but lagging, is the development of simpler in vivo strategies that can concurrently query the spatial and temporal properties of functional _cis_-regulatory sequences within large segments of noncoding DNA. Our goal in this study is to describe one such strategy for following up on GWAS results, and to test its ability to uncover noncoding risk variants in loci associated with complex diseases.
A striking example of GWAS implicating noncoding variants in the etiology of complex diseases can be seen on chromosome 8q24, where numerous studies have reported associations between multiple types of cancer—including prostate, colorectal, breast, and urinary bladder—and variants concentrated within 620 kb of a 1.2-Mb gene desert (Amundadottir et al. 2006; Easton et al. 2007; Gudmundsson et al. 2007; Haiman et al. 2007; Tomlinson et al. 2007; Zanke et al. 2007; Ghoussaini et al. 2008; Kiemeney et al. 2008; Al Olama et al. 2009). Evidence for prostate cancer association within the region is particularly strong, with five distinct LD blocks spanning a 440-kb interval on 8q24 harboring risk variants (Fig. 1A, all shaded regions; Ghoussaini et al. 2008; Al Olama et al. 2009). One of these prostate cancer-associated variants, rs6983267, is independently associated with colorectal cancer (Fig. 1A, green; Tomlinson et al. 2007), and a second prostate cancer-associated LD block harbors a distinct SNP (rs13281615) that shows association with breast cancer (Fig. 1A, pink; Easton et al. 2007). Although no well-annotated genes lie within this interval, the independent associated variants (or linked functional elements within the associated regions) may all be regulating the expression patterns of a single gene involved in cancer tumorigenesis and/or progression in various tissue types. The proto-oncogene MYC lies immediately downstream of this gene desert, raising the possibility that the associated regions of risk may harbor long-range _cis_-regulatory elements involved in the tissue-specific transcriptional regulation of MYC expression; under this hypothesis, each distinct association interval might harbor a functional noncoding element involved in regulating MYC expression in the corresponding tissue type for each implicated cancer. A summary of the 8q24 gene desert and its numerous cancer loci is shown in Figure 1. Here, we have chosen to specifically focus on the multiple independent associations between this 8q24 gene desert and prostate cancer.
Figure 1.
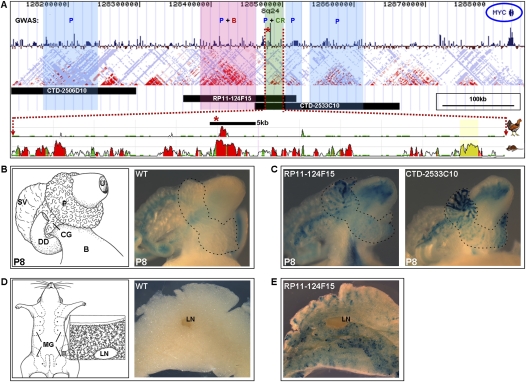
The 8q24 MYC gene desert harbors prostate and mammary gland transcriptional enhancers. (A) Five susceptibility loci within the 440-kb interval shown to be associated with prostate cancer (all shaded regions; blue denotes a prostate-only association), with one locus independently associated with breast cancer (pink) and a second associated with colorectal cancer (green). (B) Breast cancer–associated region, (CR) colorectal cancer–associated region, (P) prostate cancer–associated region. (Blue circle) MYC, (red asterisk) SNP rs6983267. (Below) The three human _lacZ_-tagged BACs encompassing the prostate cancer risk regions. (Red dotted lines) The LD block containing SNP rs6983267—associated with both prostate and colorectal cancers and contained within BACs RP11-124F15 and CTD-2533C10—is shown in detail. Sequence conservation is shown in chicken and mouse genomes (human genome used as reference). (B) The male genitourinary apparatus in P8 mice, shown as a cartoon (left) and in wild-type, nontransgenic mice (right). (Dashed line, right) Outline of the prostate. (B) Bladder, (CG) coagulating gland, (DD) ductus deferens, (P) prostate, (SV) seminal vesicle, (U) urethra. There is endogenous X-gal staining in the SV and DD. (C) Representative P8 prostates from transgenic mice containing BAC RP11-124F15 or CTD-2533C10 showing prostate and urogenital apparatus enhancer activity. (Dashed lines) Outlines of prostates. (D) The mammary gland in midgestational pregnant females, shown as a cartoon (left) and in wild-type, nontransgenic mice (right). The enlargement (left) illustrates a lymph node, ducts, and alveoli and in a mammary fat pad. (LN) Lymph node, (MG) mammary gland. (E) Representative mammary fat pad from a day 14.5 pregnant female harboring BAC RP11-124F15.
Encoding a well-known transcription factor essential to the regulation of cell proliferation and growth, MYC is up-regulated at both the mRNA and protein levels in aggressive prostate cancers (DeMarzo et al. 2003). In addition, copy-number analyses in prostate cancer specimens have identified the 8q24 region surrounding MYC as the most common recurrent region of chromosomal gain (Lapointe et al. 2007). These findings show that prostate cancers employ multiple mechanisms for achieving MYC overexpression, through transcriptional up-regulation or through amplification of gene copy number. We hypothesized that variation within MYC's long-range _cis_-regulatory elements could disrupt the quantitative, temporal, or spatial expression patterns of MYC in the prostate, possibly underlying the GWAS signals identified in the 8q24 gene desert. In this study, we describe how an in vivo bacterial artificial chromosome (BAC) enhancer-trapping strategy efficiently scanned the 8q24 gene desert for _cis_-regulatory sequences, and report on the identification of both prostate and mammary gland enhancer activities within the assayed regions. We further refined the prostate enhancer interval, showing that it harbors the prostate cancer risk SNP rs6983267, and demonstrate that the two resultant allelic variants display functionally polymorphic prostate enhancer properties in vivo.
Results
Surveying the regulatory landscape of the 8q24 gene desert
To initially examine the 8q24 gene desert for regulatory elements, we surveyed the region using a broad-scale BAC scan approach (Spitz et al. 2003). This strategy allows for the rapid and effective examination of large genomic regions for _cis_-regulatory elements, and can be readily applied to any locus of interest. We identified three overlapping human BACs encompassing the prostate cancer risk regions (Fig. 1A), which together span 480 kb of noncoding DNA. Each BAC carried the prostate cancer-associated risk haplotype and was tagged through a Tn7 transposon-mediated random insertion of a beta-galactosidase (lacZ) gene driven by a beta-globin minimal promoter (Spitz et al. 2003). The transposon-mediated insertion was performed using simple, commercially available kits (see Methods) and occurs in vitro; the protocol yields rapid results and can be easily scaled up for the simultaneous tagging of numerous BACs.
The lacZ cassette integration converts the BACs into enhancer-trapping systems, whereby any long-range enhancer(s) contained within each ∼180-kb BAC can act upon the reporter gene to drive tissue- and temporal-specific beta-galactosidase expression. Any enhancers present within a given BAC are then simultaneously interrogated using a reporter assay system, allowing for the concurrent examination of large genomic regions for functional noncoding elements. The design of overlapping BACs aids in the efficiency of the system to narrow the critical region of interest, as expression profiles unique to only one BAC must be due to uniquely contained sequences; conversely, identical expression patterns present in overlapping BACs suggest that the functional element driving beta-galactosidase expression must be contained in the shared genomic region. Modified BACs were analyzed by PCR and pulsed-field gel electrophoresis to confirm the integration of the Tn7β-lacZ reporter cassette. To mitigate any possible effects of unknown insulator or silencer elements within the BAC sequence, we selected clones with at least two Tn7β-lacZ integration events. Each BAC was then injected into fertilized mouse oocytes to generate transgenic mice in accordance with IACUC regulatory standards. For each BAC, a minimum of two independent transgenic founders were obtained and studied; this is necessary to overcome potential position-dependent expression effects resulting from random integration of the transgene (BAC).
We assayed lacZ expression at multiple points in prostate organogenesis and maturation; postnatal days 0 and 8 (P0 and P8) during prostate development, and P21, when prostate maturation is virtually complete (Sugimura et al. 1986). At each developmental stage, prostates were dissected and stained for beta-galactosidase expression using X-gal (Fig. 1B,C; Kothary et al. 1989).
These in vivo BAC transgenic reporter assays identified prostate enhancer activity contained within the 8q24 gene desert (Fig. 1C). While we did not observe beta-galactosidase prostate expression in BAC CTD-2506D10 transgenic mice (12 independent transgenics), animals harboring BACs CTD-2533C10 and RP11-124F15 displayed beta-galactosidase prostate expression at days P0 (data not shown), P8 (Fig. 1C), and P21 (data not shown). As illustrated in Figure 1C, the beta-galactosidase expression domain of both BAC RP11-124F15 and BAC CTD-2533C10 extends to other components of the urogenital system, including the coagulating glands, urethra, and the lining of the urinary bladder. While the seminal vesicles and ductus deferens also exhibit X-gal staining, we and others observed this expression pattern in both wild-type (Fig. 1B) and transgenic animals, reflecting the presence of endogenous beta-galactosidase in these structures (Wang et al. 2002; Krajnc-Franken et al. 2004). As 80% of the prostatic ducts are formed by day P15 in mice (Sugimura et al. 1986), our data indicate that the enhancer(s) contained within these two BACs are active both during and after prostate organogenesis and maturation.
Because some of the prostate cancer-associated regions also have been associated with breast and colorectal cancer (Fig. 1A), we chose to additionally assay the mammary glands, colon, and rectum of those animals transgenic for BACs containing the relevant regions (BAC RP11-124F15 for breast cancer, and both BACs RP11-124F15 and CTD-2533C10 for colorectal cancer). Mammary glands were examined at embryonic day 14.5 (E14.5), when the mammary buds have fully formed in female embryos, in 11-wk-old virgin females with mature branched glands, and in prelactating females 14 d after conception, when the mammary gland undergoes extensive hyperplasia and tissue remodeling (Hens and Wysolmerski 2005; Oakes et al. 2006; Sternlicht 2006).
We observed in vivo mammary gland enhancer activity in mice transgenic for BAC RP11-124F15 (Fig. 1E), which harbors associated intervals for not only prostate but also breast and colorectal cancer. Transgenic animals displayed beta-galactosidase expression in the epithelial compartment—ducts and alveoli (Hennighausen and Robinson 2005)—of the mammary glands of midgestational pregnant and 11-wk-old virgin females (Fig. 1E; data not shown). No enhancer activity was seen in E14.5 embryos. Of note, Jia et al. (2009) recently identified a noncoding element within this region capable of in vitro enhancer activity in breast cancer cell lines; this element should be viewed as a strong candidate for the mammary gland activity we see in vivo.
Characterizing the prostate enhancer
We next aimed to refine the location of the prostate enhancer(s) within the BACs driving prostate expression. Because of the highly similar reporter expression patterns obtained from BACs RP11-124F15 and CTD-2533C10, including prostate, coagulating gland, and urethral/bladder lining, we hypothesized that our BAC transgenic assays were identifying a single prostate enhancer within the 59-kb shared genomic segment of these two BACs. Interestingly, one of the most strongly associated prostate cancer risk SNPs, rs6983267, is contained within this 59-kb overlapping interval and disrupts an evolutionarily conserved sequence (Fig. 1A).
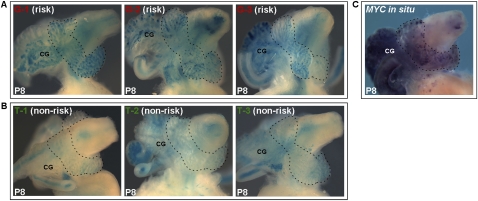
To directly test the rs6983267-containing evolutionarily conserved element for regulatory potential in vivo, we cloned a 5-kb DNA fragment containing each allele of this SNP in a lacZ reporter cassette using Invitrogen's Gateway cloning system (Kothary et al. 1989). Transgenic mice harboring either the risk or the non-risk variant of rs6983267 were generated and analyzed. We determined that the conserved sequence containing the prostate cancer GWAS SNP displayed allele-specific in vivo prostate enhancer properties (Fig. 2). Specifically, the risk allele, rs6983267-G, led to consistent, stronger beta-galactosidase expression in prostates and coagulating glands than the non-risk allele, rs6983267-T, in P0, P8, and P21 transgenic mice (Figs. 2A,B, 3B,C). The expression pattern driven by the rs6983267-G risk allele in three independent mouse transgenic lines closely resembled that observed in BACs RP11-124F15 and CTD-2533C10—both of which also harbor the risk allele. In contrast, the rs6983267-T non-risk allele led to weakened prostate and coagulating gland expression in three independent transgenic lines (Fig. 2B). For each allelic variant evaluated, those transgenic founders exhibiting enhancer activity showed highly concordant beta-galactosidase expression in the prostate, with a clear qualitative difference between the risk and non-risk variants.
Figure 2.
SNP rs6983267 mediates allelic-specific enhancer activity in mouse prostates. Three independent transgenic founders harboring reporter plasmids driven by either the G (risk) allele (A) or T (non-risk) allele (B) are shown at P8. (Dashed lines) Outlines of prostates; (CG) coagulating glands. The prostate cancer risk allele leads to consistently stronger beta-galactosidase expression in prostates and coagulating glands than the non-risk allele in vivo. (C) MYC in situ hybridization at P8 correlates with the reporter expression pattern driven by the rs6983267-containing enhancer.
Figure 3.
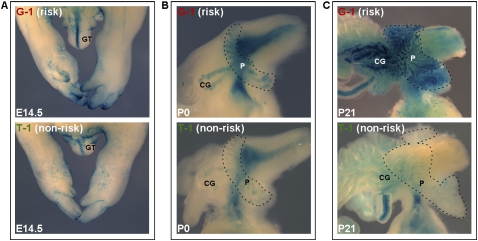
The rs6983267-containing enhancer demonstrates distinct temporal regulatory abilities. Representative G (risk, top) and T (non-risk, bottom) transgenics are shown at a series of developmental time points. (A) E14.5 transgenic embryos exhibit beta-galactosidase expression in the genital tubercle and limbs, with no apparent allele-specific enhancer activity. (GT) Genital tubercle. (B,C) Allele-specific regulatory ability is visible in neonatal P0 pups (B) and P21 adolescent mice (C), with in vivo prostate and coagulating gland beta-galactosidase expression qualitatively stronger in the risk allele (top) line than the non-risk variant (bottom). (CG) Coagulating gland, (P) prostate.
To test whether this spatial reporter expression pattern of the rs6983267-containing enhancer correlates with endogenous MYC expression in prostate and other components of the urogenital system, we performed whole mount in situ hybridizations using a full-length Myc probe in mouse prostates at P8 (Wilkinson and Nieto 1993). We observed Myc expression in the male genitourinary apparatus, including the prostate, in a pattern closely mimicking the reporter expression of the rs6983267-G enhancer and BACs CTD-2533C10 and RP11-124F15, both of which harbor the G risk allele as well (Fig. 2C).
This same prostate enhancer that we have characterized also has been shown to act as an allelic-specific long-range MYC enhancer in colorectal cancer cells (Jia et al. 2009; Pomerantz et al. 2009a; Tuupanen et al. 2009; Wright et al. 2010). Although we did not observe colorectal enhancer activity in our initial BAC screen of the region, we again assayed transgenic animals harboring either the risk or non-risk rs6983267-containing enhancer element for in vivo enhancer activity in the colorectal area at three developmental time points. We observed no beta-galactosidase expression in E14.5 intestines for either construct tested, and colorectal X-gal staining at P8 and P21 was indistinguishable between wild-type mice and transgenic animals harboring either enhancer variant (Supplemental material). Strong endogenous beta-galactosidase expression is observed in intestines of both wild-type and transgenic animals starting at E15.5, limiting our ability to identify in vivo colorectal enhancers in late embryogenesis and postnatally. These findings highlight the difficulty in assaying postnatal in vivo intestinal enhancers using lacZ reporter assays.
Investigations into the embryonic activity of the rs6983267-containing element demonstrated that while this enhancer has several spatial domains of expression, its allele-specific activity is restricted to the prostate and coagulating glands. Both the rs6983267-G and rs6983267-T enhancer elements drove expression in several spatial domains of E11.5 and E14.5 embryos, with no apparent allelic-specific enhancer activity (Fig. 3A). Transgenics harboring either haplotype variant showed similar X-gal staining in the limbs and tail at E11.5, consistent with previously reported patterns (data not shown; Tuupanen et al. 2009). We also observed enhancer activity in the developing urinary bladder, genital tubercle, and limbs in the E14.5 embryos. This pattern, which precedes prostate development, is also indistinguishable between the allelic variants of this enhancer (Fig. 3A).
Taken together, our data posit that the rs6983267-containing enhancer is part of MYC's regulatory landscape, and that the variant within this enhancer may increase the risk of prostate cancer through its role in allelic-specific control of MYC expression in the prostate.
Discussion
The BAC enhancer-trapping strategy that we employed allowed us to rapidly interrogate the 440 kb of 8q24 prostate cancer-associated noncoding DNA for _cis_-regulatory elements. We effectively screened a half-megabase genomic interval in vivo using only three constructs, identifying the existence of mammary gland and prostate enhancers in the interval associated with each respective cancer type. We believe that this methodology provides a significant advance to current genomic techniques for following up on GWAS results in noncoding regions, as it can be easily adapted to examine loci in vivo on a megabase scale. As demonstrated by our results, this strategy can be used to concurrently identify spatially and temporally unique enhancers within a large sequence, and can be useful in refining the critical regions for enhancer mapping, while still permitting the use of a whole-systems, in vivo animal model.
These relatively straightforward BAC transgenic reporter assays also provide a way to more closely approximate the genomic context of relevant enhancers. By testing ∼200 kb of sequence simultaneously, enhancers are assayed in a context much closer to their true genomic environment, one where they are subjected to (largely unknown) modifications by neighboring repressors, insulators, chromatin changes, and/or various other interactions with nearby cis sequences. In traditional plasmid-based reporter assays, this important genomic context is lost. We conducted our clone selection strategy so as to minimize the potential negative effects of such insulators or repressors; tagged BACs containing at least two copies of the Tn7β-lacZ reporter cassette—integrated near each end of the BAC sequence—were selected for experimental use. We hypothesized that this would diminish false-negative results caused by repressive elements in a single-copy integration clone. When compared with BACs tagged with just a single Tn7β-lacZ cassette, we observed more reproducible results in mice transgenic for BACs harboring two Tn7β-lacZ integrations (M.A.N, unpubl.).
Because we observed the same urogenital system spatial pattern of expression in both of the overlapping BACs tested, we deduced that the enhancer was within the small interval shared between those BACs. However, it is possible that other prostate enhancers also exist within the BACs we tested. To formally exclude this possibility, other approaches could have been used, including the analysis of additional enhancer-trapping BACs with complementary overlapping patterns. Alternatively, BAC recombineering could have been employed to specifically delete our known enhancer from the BACs assayed. Both approaches are logical follow-ups to the in vivo BAC transgenic reporter assays, and would maintain the analytical strengths of assaying enhancers in their genomic environments.
Recent studies have reported on the colorectal and prostate enhancer activities of the rs6983267-containing sequence we describe here (Jia et al. 2009; Pomerantz et al. 2009a; Tuupanen et al. 2009; Sotelo et al. 2010; Wright et al. 2010). Using a combination of genome-wide in vitro assays, this sequence has been highlighted as possessing attributes of an enhancer, including specific chromatin modifications and binding of transcription factors. Several groups have demonstrated that in colorectal cancer cell lines, TCF7l2 (TCF4) binds preferentially to the risk allele (rs6983267-G) of this enhancer (Pomerantz et al. 2009a; Tuupanen et al. 2009; Wright et al. 2010). Reports regarding the enhancer properties of this sequence in prostate cancer cell lines have been mixed, however. When tested in LNCaP and PC3 prostate cancer cell lines, this sequence displayed enhancer properties only in the former, possibly due to the PC-3 line's lack of androgen receptor expression (Jia et al. 2009). In a second study, this rs6983267-containing enhancer was unable to drive luciferase expression above promoter-only levels in LNCaP or PC-3 cells, unless cells were cotransfected with Tcf4 and beta-catenin expression vectors (Sotelo et al. 2010). Under those conditions, the rs6983267-containing element demonstrated allelic-specific enhancer activity in LNCaP cells, but with the non-risk rs6983267-T variant driving stronger expression than the risk rs6983267-G allele.
Our in vivo results—showing the cancer risk allele demonstrating stronger enhancer potential than the non-risk allele—corroborate those reported in colorectal cancer cell lines (Pomerantz et al. 2009a; Tuupanen et al. 2009; Wright et al. 2010), and are concordant with MYC's known role as a proto-oncogene. Our whole-animal experimental strategy obviated the experimental variation added by cell lines to clearly show that this element is a functional prostate enhancer in vivo, while also adding the ability to investigate enhancer activity throughout organogenesis. We believe that this broad spatial and temporal characterization of regulatory potential is ideally afforded by in vivo experimentation, and propose this as the standard in the follow-up to GWAS risk variants implicated in human disease.
The rs6983267-containing element physically interacts with MYC's promoter in both colorectal cancer and prostate cancer cell lines, providing evidence that this enhancer is involved in regulating MYC expression in these two tissue types (Pomerantz et al. 2009a; Sotelo et al. 2010; Wright et al. 2010). Despite these compelling findings and the fact that altered MYC expression has been implicated repeatedly in the pathogenesis of prostate cancers (Williams et al. 2005), no association has been seen between rs6983267 genotype and MYC mRNA levels in normal prostate cells or prostate tumors (Pomerantz et al. 2009b). This lack of genotype–phenotype correlation implies that steady-state MYC mRNA levels in adult prostate tissue may not be the correct biological entity underlying risk. Our findings demonstrate that the rs6983267-containing enhancer exhibits differential in vivo activity throughout prostate organogenesis, and raise the possibility that this variant asserts its influence on prostate cancer risk long before tumorigenesis occurs. With widely varying risk allele frequencies in different populations—from 49% in American Caucasians to 81% in African Americans (HapMap, merged Phase 1, 2, and 3 frequencies)—this SNP may also have an effect on the population prevalence of both prostate cancer and colorectal cancer (Jemal et al. 2009).
We have described how a noncoding SNP strongly associated with disease can in fact alter the in vivo activity of its encompassing _cis_-regulatory element, suggesting a possible impact on cancer risk before tumorigenesis actually occurs. Although further studies are warranted, our in vivo temporal data hint at an underlying molecular explanation for this nongenic SNP's contribution to prostate cancer risk. These findings emphasize the notion that thorough investigations into the regulatory impact of polymorphisms are an indispensable component to the functional follow-up of GWAS scans, and stress the importance of conducting these experiments using in vivo systems.
Methods
Transposon-mediated BAC modification
BACs CTD-2506D10, RP11-124F15, and CTD-2533C10 were modified by in vitro random transposition of Tn7β-lacZ (Spitz et al. 2003). BAC DNA was extracted by using the Nucleobond AX Kit (Macherey-Nagel). Twenty nanograms of Tn7β-lacZ vector was mixed with 20–40 ng of BAC DNA, GPS buffer, and TnsABC transposase (New England BioLabs), followed by incubation for 10 min at 37°C. Start solution was added and the reaction was extended for 1 h. After heat inactivation for 10 min at 75°C and a 1-h dialysis, electrocompetent DH10B cells were transformed with 2 μL of the transposition reaction. Cells were plated on LB agar containing 20 μg/mL kanamycin and 20 μg/mL chloramphenicol. Positive colonies were first identified by polymerase chain reaction (PCR) using beta-globin and lacZ primers (Tn7β-lacZ beta-globin F: AGCATCTATTGCTTACATTTGC; Tn7β-lacZ lacZ R: ATAGGTTACGTTGGTGTAGATGG). Modified BAC clones were then digested with NotI and separated by pulsed-field gel electrophoresis overnight on a 1% agarose gel to determine the number of copies and the position(s) of the integrated Tn7β-lacZ cassette. Clones with two copies of the cassette were chosen for further analysis to minimize the possible influence of silencer or insulator elements with the BACs.
lacZ plasmid generation
The 5 kb of sequence surrounding the rs6983267-containing conserved element was PCR amplified from human genomic DNA heterozygous for the rs6983267 SNP (rs6983267 F: TCTTGACCTGATTGCTGAAAAAT; rs6983267 R: TCTGGGGGTGAGTTAAATGATAA). The fragment was then purified using the QIAquick PCR Purification Kit (Qiagen) and cloned into the pDONR 221 Gateway entry vector (Invitrogen). Colonies were analyzed by restriction enzyme analysis for successful fragment insertion, and positive clones were sequenced to determine the allelic status of SNP rs6983269 (rs6983267-seq F: TAGACACCAAGAGGGAGGTATCA; rs6983267-seq R: CCAGGTTAAAGGAAACTGAACTG). Clones containing sequence harboring both the risk (G) and non-risk (T) rs6983267 allele were transferred to a Gateway-HSP68-lacZ reporter vector using the LR recombination reaction (Invitrogen) (Poulin et al. 2005). All plasmids were again verified by restriction analysis and direct sequencing prior to pronuclear mouse injections.
Production of transgenic mice
Tn7β-lacZ tagged BAC DNA was purified using the Nucleobond BAC 100 Kit (Macherey-Nagel), rehydrated in injection buffer (10 mM Tris at pH 7.5; 0.1 mM EDTA), and diluted to a concentration of 2 ng/μL. BAC DNA was injected in its circular form.
Plasmid DNA was purified using the Plasmid Maxi Kit (Qiagen), and 50 μg of each plasmid was digested with SalI to excise the vector backbone. Following a gel purification step using the QIAquick Gel Extraction Kit (Qiagen), the DNA to be injected was further purified using a standard ethanol precipitation. The purified DNA was dialyzed for 24 h against injection buffer (10 mM Tris at pH 7.5; 0.1 mM EDTA), and its concentration was determined fluorometrically and by agarose gel electrophoresis. The DNA was diluted to a concentration of 2 ng/μL. Purified BAC and plasmid DNA were then used for pronuclear injections of CD1 mouse embryos in accordance with standard protocols approved by the University of Chicago.
For the Tn7β-lacZ tagged BACs, multiple stable transgenic lines were generated for each construct, and F1 animals were analyzed for each line at multiple postnatal developmental time points. BAC CTD-2506D10 DNA injections yielded 12 independent lines (0/12 positive for prostate beta-galactosidase expression); injections of RP11-124F15 and CTD-2533C10 both resulted in two independent beta-galactosidase-expressing lines.
For the rs6983267-containing enhancer plasmid, a total of three beta-galactosidase-expressing independent transgenics was obtained for rs6983267-G; three beta-galactosidase-expressing independent transgenic animals/lines were also obtained for rs6983267-T. For several of these independent lines, the F0 animals themselves were analyzed at P8; this excluded any analysis of the line at other time points. For the risk allele, rs6983267-G, we obtained two F0 animals positive for beta-galactosidase expression in the prostate. The third independent rs6983267-G transgenic was maintained as a stable line. For the non-risk allele, rs6983267-T, one F0 transgenic animal was obtained; the remaining two independent transgenics were maintained as stable lines.
Mouse in vivo transgenic reporter assay
Prostates and mammary glands were harvested from mice at P0, P8, and P21 and dissected into cold 100 mM phosphate buffer (PBS) (pH 7.3), followed by 30–45 min of incubation with 4% paraformaldehyde at 4°C. E14.5 embryos were incubated in 4% paraformaldehyde for 2 h. Tissues were then washed two times for 20 min with wash buffer (2 mM MgCl2; 0.01% deoxycholate; 0.02% NP-40; 100 mM phosphate buffer at pH 7.3), and stained for 18 h at room temperature with freshly made staining solution (0.8 mg/mL X-gal; 4 mM potassium ferrocyanide; 4 mM potassium ferricyanide; 20 mM Tris at pH 7.5 in wash buffer). After staining, samples were rinsed five times for 20 min in PBS and post-fixed in 4% paraformaldehyde. For each animal analyzed, tail samples were taken at the time of dissection and DNA was isolated through the addition of lysis buffer (100 mM Tris-HCl at pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl, and 1 mg/mL proteinase K) and incubation overnight at 55°C. Genotyping was performed by PCR with primers within the reporter cassette/vector (using beta-globin and lacZ primers for the Tn7β-lacZ tagged BACs, rs6983267-seq primers for the plasmids).
Imaging
All photographs were taken using a Leica MZ16 F stereomicroscope and QCapture Pro software. Settings (lighting, exposure time) were kept constant between structure- and aged-matched samples. Images displayed in the paper were generated using an image processing software package (CombineZM) that allows for the creation of extended depth of field images. Multiple pictures of each structure were taken at varying depth of fields and then computationally integrated; the focus areas are blended to create a composite high-resolution image with an extended depth of field. This allowed for the production of images where all the multiple plains of the urogenital apparatus appear well focused and defined.
In situ hybridization
In situ hybridization analysis on whole P8 prostates using digoxigenin-labeled Myc antisense and sense riboprobes was performed according to standard protocols (Wilkinson and Nieto 1993). The probes were generated from a full-length mouse Myc cDNA clone (IMAGE ID 3962047). Staining was performed for 48 h, and the stained prostates were then transferred to 10% buffered formalin phosphate prior to imaging.
Acknowledgments
We thank François Spitz for kindly providing us with the beta-globin-Tn7 vector and Linda Degenstein for assistance in generating transgenic animals. We also thank James Noonan, Rick Kittles, and Gail Prins for their consultation and support. The urogenital apparatus and mammary gland cartoons in Figure 1, B and D, were kindly drawn by John Westlund. This work was partially supported by grant HG004428 (M.A.N.). N.F.W. is supported by a DoD Prostate Cancer Training Award (PC094251).
Footnotes
References
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, et al. 2009. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 41: 1058–1060 [DOI] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, et al. 2006. A common variant associated with prostate cancer in European and African populations. Nat Genet 38: 652–658 [DOI] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI 2003. Pathological and molecular aspects of prostate cancer. Lancet 361: 955–964 [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. 2007. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, et al. 2007. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 39: 631–637 [DOI] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, et al. 2007. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet 39: 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW 2005. Information networks in the mammary gland. Nat Rev Mol Cell Biol 6: 715–725 [DOI] [PubMed] [Google Scholar]
- Hens JR, Wysolmerski JJ 2005. Key stages of mammary gland development: Molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res 7: 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, et al. 2008. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 40: 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ 2009. Cancer statistics, 2009. CA Cancer J Clin 59: 225–249 [DOI] [PubMed] [Google Scholar]
- Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, et al. 2009. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5: e1000597 doi: 10.1371/journal.pgen.1000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, et al. 2008. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 40: 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J 1989. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105: 707–714 [DOI] [PubMed] [Google Scholar]
- Krajnc-Franken MA, van Disseldorp AJ, Koenders JE, Mosselman S, van Duin M, Gossen JA 2004. Impaired nipple development and parturition in LGR7 knockout mice. Mol Cell Biol 24: 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR 2007. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res 67: 8504–8510 [DOI] [PubMed] [Google Scholar]
- Liu Y, Blackwood DH, Caesar S, de Geus EJ, Farmer A, Ferreira MA, Ferrier IN, Fraser C, Gordon-Smith K, Green EK, et al. 2010. Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry doi: 10.1038/mp.2009.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Oakes SR, Hilton HN, Ormandy CJ 2006. The alveolar switch: Coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res 8: 207 doi: 10.1186/bcr1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009a. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, Hawksworth DJ, Schumacher FR, Mucci L, Penney KL, et al. 2009b. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res 69: 5568–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin F, Nobrega MA, Plajzer-Frick I, Holt A, Afzal V, Rubin EM, Pennacchio LA 2005. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85: 774–781 [DOI] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, et al. 2009. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet 41: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X 2010. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci 107: 3001–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405–417 [DOI] [PubMed] [Google Scholar]
- Sternlicht MD 2006. Key stages in mammary gland development: The cues that regulate ductal branching morphogenesis. Breast Cancer Res 8: 201 doi: 10.1186/bcr1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura Y, Cunha GR, Donjacour AA 1986. Morphogenesis of ductal networks in the mouse prostate. Biol Reprod 34: 961–971 [DOI] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. 2007. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39: 984–988 [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. 2009. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 41: 885–890 [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA 2009. Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newton DC, Miller TL, Teichert AM, Phillips MJ, Davidoff MS, Marsden PA 2002. An alternative promoter of the human neuronal nitric oxide synthase gene is expressed specifically in Leydig cells. Am J Pathol 160: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA 1993. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225: 361–373 [DOI] [PubMed] [Google Scholar]
- Williams K, Fernandez S, Stien X, Ishii K, Love HD, Lau YF, Roberts RL, Hayward SW 2005. Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. Prostate 63: 369–384 [DOI] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD 2010. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 30: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Yang W, Pei D, Cao X, Fan Y, Pounds SB, Neale G, Trevino LR, French D, et al. 2009. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA 301: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, et al. 2007. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 39: 989–994 [DOI] [PubMed] [Google Scholar]