An Allosteric Mechanism of Rho-dependent Transcription Termination (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 28.
Published in final edited form as: Nature. 2010 Jan 14;463(7278):245–249. doi: 10.1038/nature08669
Abstract
Rho is the essential RNA helicase that sets the borders between transcription units and adjusts transcriptional yield to translational needs in bacteria1–3. Although Rho is the first termination factor to be discovered4, the actual mechanism by which it reaches and disrupts the elongation complex (EC) is unknown. Here we show that the termination-committed Rho molecule associates with RNA polymerase (RNAP) throughout the transcription cycle, i.e. it does not require the nascent transcript for initial binding. Moreover, the formation of the RNAP-Rho complex is crucial for termination. We show further that Rho-dependent termination is a two-step process that involves rapid EC inactivation (trap) and a relatively slow dissociation. Inactivation is the critical rate-limiting step that establishes the position of the termination site. The trap mechanism depends on the allosterically-induced rearrangement of the RNAP catalytic center via the evolutionary conserved mobile trigger loop (TL) domain, which is also required for EC dissociation. The key structural/functional similarities, which we found between Rho-dependent and intrinsic (Rho-independent) termination pathways, argue that the allosteric mechanism of termination is general and likely to be preserved for all cellular RNAPs throughout evolution.
Rho is the general transcription termination factor in bacteria. Concerted cycles of ATP binding, hydrolysis and release induce conformational changes that pull the nascent transcript through the cavity of the hexamer and induce Rho to translocate along the RNA in the 5′ → 3′ direction5–8. Classical tracking models postulate that, once loaded onto the nascent RNA, Rho catches up to the EC and pulls the transcript away from RNAP and the DNA template9–11. Extensive biochemical and structural analysis of Rho over the past two decades has explained the molecular details of RNA loading and translocation5–11, but not the actual mechanism of termination. Different models that address this issue have been proposed9,10,12 (Supplementary Fig. 1). The majority of these models envisioned RNAP as passive in the termination process. Indeed, simple kinetics, i.e. the speed at which RNAP elongates, is thought to be the major determinant of Rho termination sites, many of which coincide with RNAP pause sites. Although kinetic coupling is certainly an important parameter in controlling the efficiency of Rho termination13,14, it does not define the conformational state of the enzyme, which renders the EC susceptible to Rho.
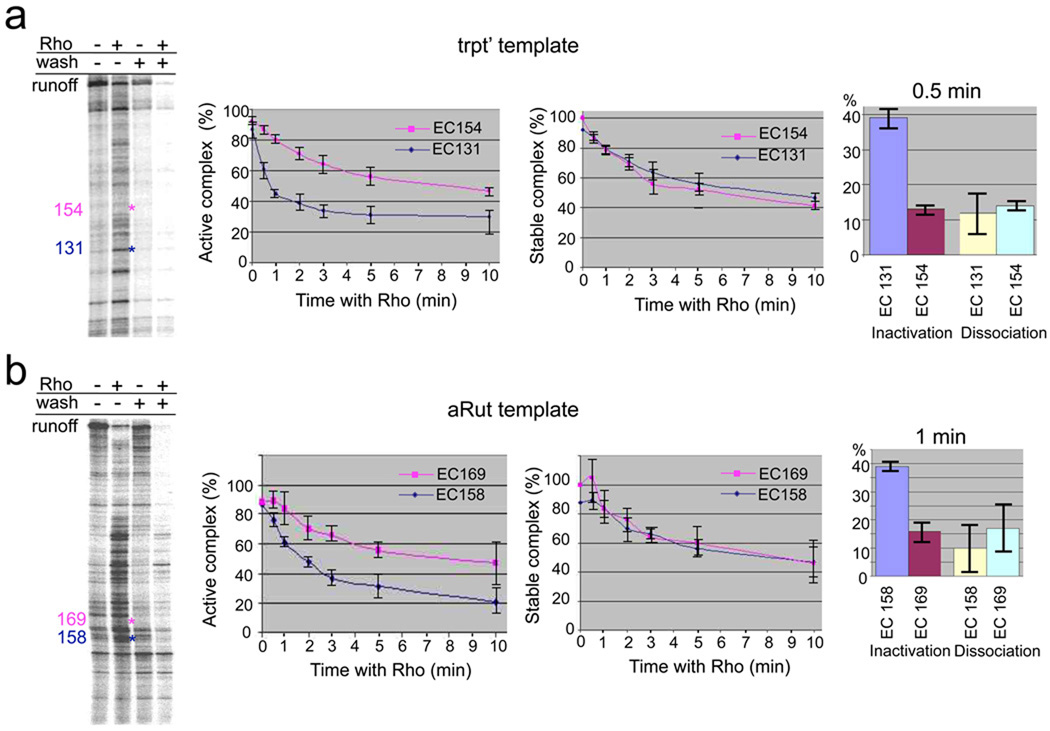
To study Rho termination in vitro we constructed two templates containing the T7A1 promoter fused to well-characterized Rho termination sequences: trpt’ and aRut14–16. We also utilized His6-tagged RNAP and biotinylated DNA adsorbed to metal chelating and NeutrAvidin beads, respectively, to permit “walking” of the EC along the template to a terminator site (see Methods). Under specified chase conditions termination occurs at many distinct positions within the trpt’ and aRut-lacZ sequences (Fig. 1a and b, left panels). To determine the parameters of the EC that define the termination site, we selected two representative complexes on each template for detailed biochemical analyses: EC131 (termination-prone) and EC154 (termination resistant) on the trpt’ template (Fig. 1a); EC158 (termination-prone) and EC169 (termination-resistant) on the aRut template (Fig. 1b). Each individual complex was prepared by walking of the EC followed by treating with Rho (Supplementary Fig. 2 and 3). We monitored inactivation (shown by the inability of the EC to extend the nascent RNA after chasing with a limited set of NTPs) and dissociation (shown by RNA release after washing the beads). Whereas the rate of dissociation was similar between EC131/EC158 and EC154/EC169, the rate of inactivation was significantly faster for EC131/EC158 (Fig. 1a and b). The inactivation and dissociation rates were identical for termination-resistant EC154 or EC169. These rates were similar to the dissociation rates of EC131 and EC158 (Fig. 1, middle and right panels). Similar results were seen for other complexes (Supplementary Fig. 2 and 3). Thus, Rho termination can be viewed as a two-phase process: a fast inactivation (irreversible trap) followed by slow dissociation with inactivation as the rate-limiting step, which determines the position of natural termination sites.
Fig. 1. Uncoupling EC inactivation from dissociation at Rho termination sites.
a, Comparison of naturally terminating and non-terminating ECs formed on the trpt’ template. Left panel shows the distribution of Rho termination sites on the trpt’ template. The initial radiolabeled EC21 was immobilized on metal-chelating beads and chased in the presence (lane 2) or absence (lane 1) of Rho. The locations of EC131 (a representative terminating EC) and EC154 (a representative non-terminating EC) are indicated in blue and pink, respectively. “Wash” lanes demonstrate disappearance of released RNA from immobilized terminated complexes. The kinetics of Rho-mediated inactivation and dissociation of EC131 and EC154 were measured by walking the EC to the indicated position, incubating the corresponding ECs for the indicated time intervals with or without Rho, followed by either a chase (GTP, UTP, ATP) or a wash with transcription buffer (TB) (Supplementary Fig. 2). Data are shown as mean ±S.E. from four independent experiments. The Y-axis indicates either the proportion of EC that remained active (active complex %) or that is retained RNA (stable complex %); The X-axis indicates the time of incubation with Rho. The right panel shows the extent of EC inactivation and dissociation after 30 sec of incubation with Rho. A similar analysis was performed for several other ECs on this template (Supplementary Fig. 2). b, Comparison of the naturally terminating and non-terminating ECs obtained at the aRut template. The left panel shows the distribution of Rho termination sites at the aRut template. The initial radiolabeled EC43 was chased in the presence (lane 2) or absence (lane 1) of Rho. The locations of EC158 (a representative terminating EC) and EC169 (a representative non-terminating EC) are indicated in blue and pink, respectively. “Wash” lanes demonstrate disappearance of released RNA from immobilized terminated complexes. The kinetics of Rho-mediated inactivation and dissociation of EC158 and EC169 were measured by walking the EC to the indicated position, incubating the corresponding ECs for the indicated time intervals with or without Rho, followed by either a chase (GTP, UTP, CTP) or a wash with transcription buffer (TB) (Supplementary Fig. 3). Data are shown as mean ±S.E. from four independent experiments. The Y-axis indicates either the proportion of EC that remained active (active complex %) or that is retained RNA (stable complex %); The X-axis indicates the time of incubation with Rho. The right panel shows the extent of EC inactivation and dissociation after 1 min of incubation with Rho. A similar analysis was performed for several other ECs on this template (Supplementary Fig. 3).
Taken together, the results indicate that rapid EC inactivation is the determining event at natural Rho termination sites.
To determine parameters of EC inactivation by Rho, systematic alterations of the nucleic acid scaffold have been made so that the RNA:DNA hybrid, catalytic site base pair (i site), and downstream DNA duplex of EC158 were modified (Supplementary Fig. 4). Changes in the DNA downstream sequence had minor effect on the rate of EC158 termination: inactivation and dissociation rates changed little, regardless of whether AT- or GC-rich sequences were present downstream of the “i” site. In contrast, an AT-rich RNA:DNA hybrid had a protective effect against inactivation (Supplementary Fig. 4). These data are inconsistent with a shearing, spooling, or forward translocation models that predict an inverse relationship between the termination efficiency and the strength of the hybrid and/or downstream DNA duplex (Supplementary Fig. 1). Two TG substitutions at the 3’ terminus of the hybrid strongly reduced the inactivation rate, whereas shifting the 3’ terminal position upstream by one nucleotide substantially increased the inactivation rate (Supplementary Fig. 4). We conclude that Rho termination is most sensitive to sequence alterations in the vicinity of the catalytic site.
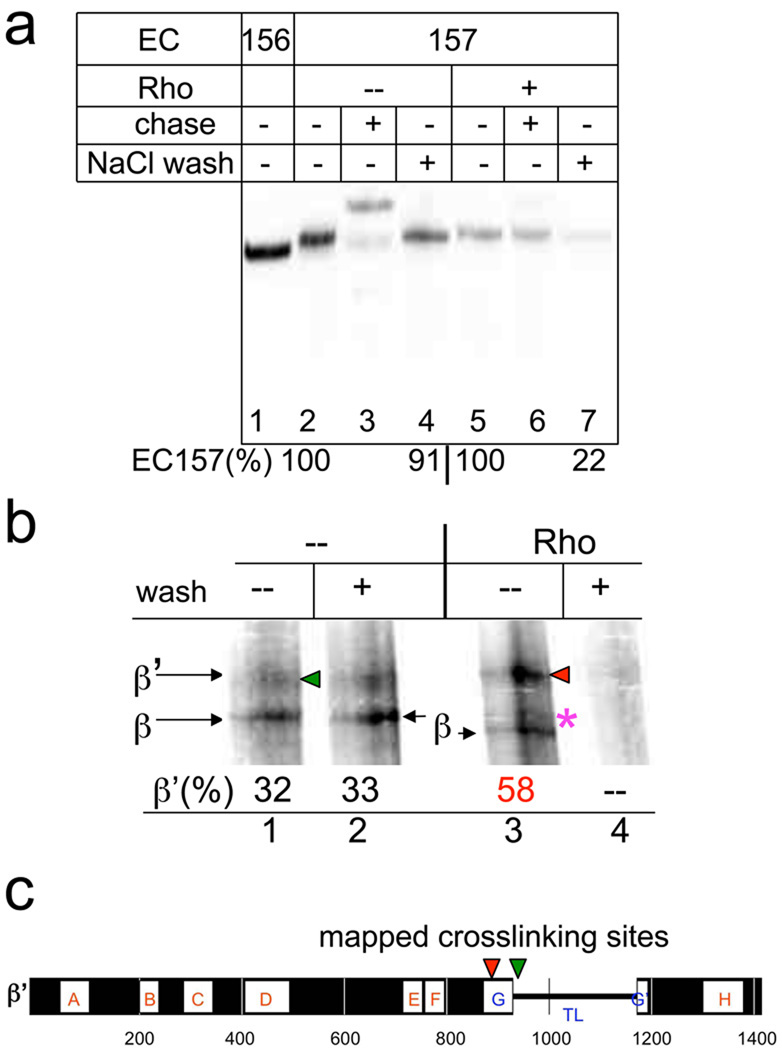
We used an RNA crosslinkable probe to monitor changes in protein-RNA interactions in the catalytic site in response to Rho. We incorporated the cross-linkable reagent 4-thio-uridine-5’-monophosphate (sU) specifically at the Rho termination position (U157 position, Fig. 2a). To avoid EC157sU dissociation, the Rho reaction was conducted under low salt conditions, which did not affect the rate of trapped complex formation. At the time of the crosslinking, almost all of the Rho-treated EC157sU had been inactivated (trapped). In both the active and trapped EC157sU, the sU probe generated two major cross-linked species corresponding to the β’ and β subunit (Fig. 2b). However, the ratio between these adducts changed dramatically in the trapped complex. Furthermore, the electrophoretic mobility of the β crosslinking adduct differed between active and trapped ECs, indicating a change of the crosslinking site within the β subunit. We mapped the major crosslinking sites to the β’ trigger-loop (TL) domain in both the trapped and active complexes (Supplementary Fig. 5–7). However, the precise locations of adducts within the TL differed between the two types of complexes (Fig. 2c). We conclude that: (i) Rho-induced EC inactivation (trap) is accompanied by alterations of the contacts in the catalytic center; (ii) the conformational change in the TL may function in the termination process; (iii) the presence of the RNA 3’ terminus in the catalytic center of the termination (trapped) complex provides direct evidence contrary to both the forward translocation and shearing/spooling models of Rho action.
Fig. 2. EC inactivation (trap) by Rho is accompanied by a rearrangement of protein-RNA contacts in the catalytic center.
a, EC157 carries the crosslinking probe (4-thioU) at the RNA 3’-terminus. EC157 was incubated with Rho for 3 min and either chased (with GTP, ATP, CTP) or washed with TB containing 0.7 M NaCl (NaCl wash). The panel shows that Rho traps and destabilizes EC157 (renders it salt sensitive). “%” indicates the relative amount of RNA retained in the immobilized EC157 after the beads were washed. b, RNA-protein crosslinking in the trapped (lanes 3, 4) and intact (lanes 1, 2) EC157. A 4% SDS gel showing β’ and β subunits derivatized by UV-induced RNA crosslinking. Numbers indicate the fraction of radiolabeled β’ in relation to the total amount of derivatized subunits. An asterisk indicates the change in β mobility. The control (lane 4) demonstrates that all crosslinking takes place in the terminating (trapped) EC, because all label was eliminated by washing. c, Summary of RNA crosslink mapping results (see Supplementary Fig. 5–7 for mapping details). The horizontal bar represents the E.coli β’ subunit. Lettered boxes show evolutionarily conserved regions. G helix-loop-helix (TL) domain is indicated. The locations of crosslinking sites from derivatized β’ probes obtained with and without Rho are indicated by red and green arrowheads, respectively.
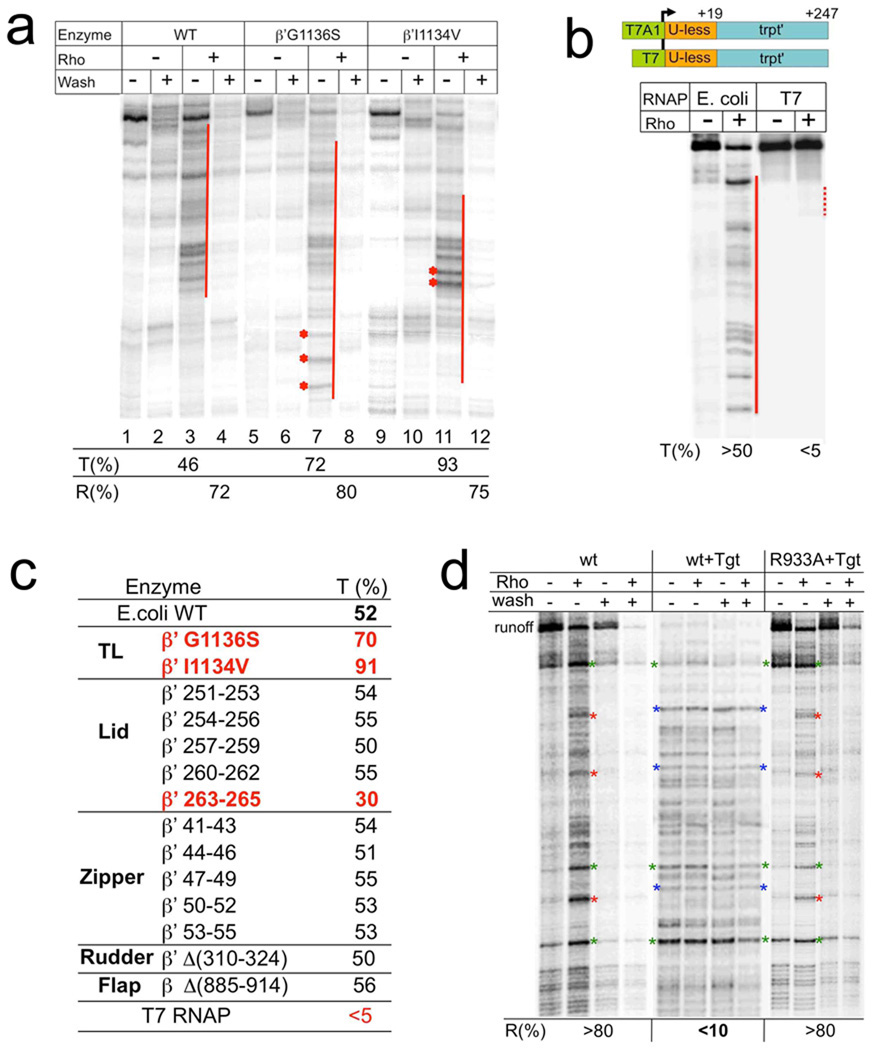
To identify RNAP domains involved in Rho termination we examined the effect of RNAP mutations (Fig. 3). The majority of the mutants had no significant effect on Rho termination. However, a mutation in the lid domain (β’ 263–265) strongly decreased the termination efficiency (Fig. 3c) without altering the rate of transcription elongation (Supplementary Fig. 8b). Interestingly, mutations in other parts of the lid had little effect on termination (Fig. 3c). Thus, β’ lid plays a specific mechanistic role in the termination process.
Fig. 3. Effect of RNAP mutations, heterologous (T7) RNAP, and tagetitoxin (Tgt) on Rho termination.
a, A representative runoff assay. Wild type and TL mutant ECs were immobilized on beads and chased in the presence or absence of Rho. The termination efficiency (%T) was calculated as described in Methods. %R indicates the fraction of released RNA at the termination zones after washing the beads. Red lines show the termination zones. Asterisks show the termination sites, which are particularly strong in the case of “fast” G1136S and “slow” I1134V mutants as compared to wt. b, Rho is not effective against heterologous RNAP. The initial EC19 was prepared with either E.coli or T7 RNAP using identical DNA templates except for the corresponding promoters (schematically shown on top). The initial ECs were then chased in the presence of equal amount of Rho and dATP. NTP concentration was selected to ensure the slower elongation rate of T7 RNAP as compared to that of E. coli RNAP (Supplementary Fig. 9). Red solid and dotted lines indicate the robust and weak termination zones, respectively. c, The table summarizes the effects of E.coli RNAP mutations and T7 RNAP on Rho termination. %T represents the average values from at least two independent experiments. Mutations leading to profound changes in Rho termination are highlighted in red. d, Suppression of Rho termination by Tgt. Rho and Tgt were added as indicated to the initial EC immobilized on beads. After the completion of the chase reaction, the beads were washed (as indicated) to monitor the RNA release. %R indicates the amount of released (washed out) RNA due to Rho action. Red asterisks indicate representative major Rho termination sites, which were absent in Tgt probes. Blue asterisks indicate representative major EC stop sites caused by Tgt. These stalled complexes became resistant to Rho termination, i.e. remained mostly intact after incubation with Rho. Green asterisks indicate representative major pause/arrest sites present in all probes, and serve as reference points. Tgt also inhibited Rho-mediated dissociation of these ECs. WT represents the wild type RNAP. R933A represents the point mutation in the TL, which prevents Tgt binding to RNAP. This control demonstrates that all the negative effects of Tgt on Rho termination are due to alterations in the properties of the RNAP catalytic center.
Two point mutations (G1136S and I1134V) of the TL stimulated Rho termination (Fig. 3a). The effect of slowly elongating I1134V could in principle be explained in terms of the kinetic coupling model. However, the G1136S mutant exhibited an increased elongation rate17, arguing against a simple kinetic coupling model. Furthermore, the antibiotic tagetitoxin (Tgt) abolished both Rho-mediated inactivation and EC dissociation (Fig. 3d). Tgt binds RNAP in the vicinity of the catalytic site18 and stabilizes the TL in its folded (closed) state. In combination with the crosslinking results described above, these findings implicate the conformational state of the mobile TL as a key element of the termination process.
To examine further the specificity of Rho-RNAP interaction, the validity of the kinetic coupling model, and the role of RNAP domains in termination we compared the termination efficiency for E. coli RNAP (natural Rho target) and structurally unrelated phage T7 RNAP on the same template (Fig. 3b). Even though we caused the elongation rate of T7 RNAP to be much slower than that of E. coli RNAP (Supplementary Fig. 9), we detected very weak termination of T7 RNAP, but robust termination of E.coli RNAP (Fig. 3b). Since T7 ECs are less stable than E.coli ECs19, we conclude that specific interactions between Rho and E.coli RNAP determine natural termination sites and their efficiency.
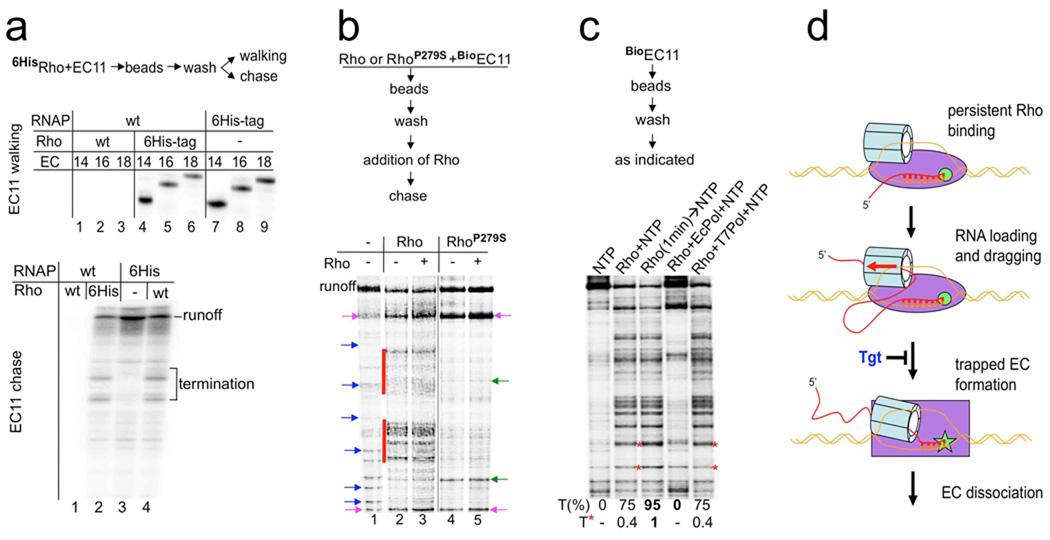
The classical tracking model postulates that Rho loads onto the nascent RNA prior to its contact with RNAP, i.e. once loaded, Rho moves along the transcript in pursuit of elongating RNAP (Supplementary Fig. 1). To test this model directly we performed a transcription walking reaction in the presence of His6-tagged Rho (6HisRho) as an affinity tag. Surprisingly, the EC containing RNA only 11 nt long (EC11) was retained on the Rho-beads under normal salt conditions (Fig. 4a), indicating that Rho binds the EC tightly in the absence of any available RNA. Moreover, such a complex between Rho and the initially transcribed EC could be then walked along the template without loss (Fig. 4a, top panel), demonstrating persistent Rho binding. Chasing the washed 6HisRho-EC11 resulted in efficient Rho termination at predictable downstream sites (Fig. 4a, bottom panel), indicating that the same hexameric Rho acted in the elongation and termination events. Furthermore, pre-incubation of RNAP with a catalytically inactive Rho mutant (P279S)16 resulted in defective Rho termination, even after addition of wt Rho (Fig. 4b). We conclude that initial binding of Rho to RNAP is strong enough to preclude exchange of Rho molecules. Interestingly, RhoP279S significantly affected the RNAP pausing profile. A similar effect was observed with wt Rho, although it was masked considerably by termination.
Fig. 4. Functional Rho binds RNAP throughout the transcription cycle.
a, Rho binds to the EC at the start of transcription and terminates the complex at downstream sites. The flowchart describes the experimental design. The initial EC11-Rho6His and 6HisEC11-Rho were prepared on the aRut template, immobilized on Co2+-chelating sepharose beads, and then washed multiple times with TBGlu (100 mM KGlu, 10 mM MgGlu). Radioautograms show [32P] RNA isolated from the EC immobilized via 6HisRho or 6HisRNAP that was walked three steps in TB to the indicated positions (top panel; lanes 4–9) or chased with all four NTPs (bottom panel, lanes 2–4). Major termination sites are indicated. Without the 6His tag no RNA could be recovered on the beads after the washes (top panel, lanes 1–3; bottom panel, lane 1). b, The same Rho molecule that binds the EC at the start of transcription terminates transcription at downstream sites. The flowchart describes the experimental design. Wild type (Rho) and mutant Rho (RhoP279S), the ATPase activity of which was abolished, were used in the experiment. Biotinilated RNAP was used to prepare the startup BioEC11 on the trpt’ template that carried [32P] RNA. Rho or RhoP279S were added to BioEC11. Next, BioEC11-Rho and BioEC11-RhoP279S were immobilized on NeutrAvidin beads, washed multiple times with TBGlu (100 mM KGlu, 10 mM MgGlu), and then chased in the presence of excess Rho as indicated. The radioautogram shows [32P] RNA isolated from the chased reactions. Lane 1 shows the chased reaction without Rho. Blue arrows indicate natural pause/arrest sites. Major Rho termination signals are indicated by red stripes (lanes 2, 3). No increase in Rho termination was observed if excess Rho was added to the washed BioEC11-Rho (lane 3), indicating that the initially-bound Rho was responsible for the majority of the termination signals. Initially-bound RhoP279S did not cause any termination (lane 4), and it also prevented termination by subsequently-added wild type Rho (lanes 5). Note that RhoP279S dramatically altered the pause distribution profile: the majority of pauses disappeared, some new pauses appeared (indicated by green arrows) and some pauses became much stronger (indicated by pink arrows). A similar effect on elongation can be observed with wild type Rho, although they are masked by strong termination signals. c, Early binding of Rho to the EC is crucial for termination. The initial radiolabeled EC11 was chased by the addition of NTPs without Rho (lane 1), together with Rho (lanes 2, 4, and 5), or after a 1 min preincubated with Rho (lane 3). In lanes 4 and 5, Rho was premixed with a 2-fold molar excess of E.coli or T7 RNAP, respectively. Rifampicin was added to ensue a single round of transcription. The overall termination efficiency (%T) was calculated as described in Methods. Red asterisks indicate early Rho termination sites, which became particularly strong if Rho preincubation occurred (lane 3). T* indicates the relative amount of these terminated products. d, The molecular pathway to Rho termination. Schematic shows the newly identified critical steps in the Rho termination process. Rho binds to RNAP at the start of transcription. Once RNA becomes long enough, Rho has an opportunity to load onto it (forming the RNA loop) and to thread the transcript through its cavity using energy derived from ATP. Eventually a topological strain results in the inactivation of RNAP (trapped complex formation), which involves a conformational change of the TL domain at the catalytic center. The trapped EC is sensitive to salt, indicating that its hybrid is partially unwound and/or that the clamp is opened due to the motion of the TL. Unwinding of the hybrid results from the direct action of Rho-mediated RNA:DNA helicase activity and/or to the clash caused by Rho between the lid domain and the hybrid. The unstable, trapped complex eventually dissociates. Rho, however, remains bound to RNAP for the next cycle of transcription. The antibiotic tagetitoxin (Tgt) abolishes Rho termination by interfering with the formation of the trapped complex.
Rho associates with RNAP independently of DNA, RNA or other proteins. It binds free RNAP and RNAP:DNA in open complex in vitro (Supplementary Fig. 10). To address the functional role of early Rho binding to RNAP, we determined the effect of delayed or no Rho binding on termination (Fig. 4c). Termination efficiency was greater if Rho was pre-bound to the early EC. Indeed, persistent association of Rho with RNAP should dramatically increase its local concentration, thereby facilitating the initial loading of Rho onto RNA. Note, that due to rapid Rho binding to RNAP and short time window for RNAP+NTPs to move without Rho, the most profound difference could be seen only at the early termination sites. We also showed that preincubation of Rho with free E. coli RNAP, but not T7 RNAP, completely abolished termination (Fig. 4c). Thus, E.coli RNAP molecules that have not been engaged in transcription sequestered all Rho and prevented it from acting on active ECs. Taking together these results demonstrate that: (i) initial and specific Rho binding to RNAP is a critical step in the termination pathway. (ii) RNAP does not move along the RNA towards RNAP. Rather, Rho association with RNAP results in an RNA loop that tightens as Rho drags the transcript through itself (Fig. 4d). (iii) Rho association with RNAP alters its elongation properties (Fig. 4b), suggesting a role for Rho during elongation.
In this work we identify two crucial steps in the Rho termination pathway: specific Rho binding to RNAP early in elongation and formation of the trapped complex at the termination sites (Fig. 4d). Mapping of the 3’ RNA contacts in the trapped ECs showed that it interacts with a different part of the mobile TL domain than it does in the active EC (Fig. 2, Supplementary Fig. 5–7), suggesting that Rho induces a conformational change in the TL leading to EC inactivation. Recent structural and biochemical studies revealed that the TL assumes different conformations, serving as an ultimate sensor and effector of the elongation process17,20–23. Consistent with these observations, point mutations in the TL17 change the efficiency of Rho termination (Fig. 3). Inhibition of Rho termination by Tgt (Fig. 3d) directly implicates TL in both Rho-induced formation of the trapped complex and in EC dissociation. The latter implies that Rho-induced unfolding of the TL, which comprises the bottom of the clamp, results in clamp opening and loss of all major interactions between RNAP and nucleic acids, thus causing EC destabilization.
Our data exclude alternative models of Rho termination. Neither the forward translocation model, nor the shearing/spooling models of Rho termination are consistent with the experimental data presented here (Fig. 2 and 3, Supplementary Fig. 4 and 11). We propose that instead of pushing RNAP or pulling RNA, Rho invades the main channel of the EC to unwind the hybrid from its upstream edge (Fig. 4d), using either its powerful RNA:DNA helicase activity24,25 or steric interference, similar to the mechanism of intrinsic termination, in which the hairpin invades the main channel, causing the upstream portion of the hybrid to melt26–28. The lid domain has been implicated in the separation of the nascent RNA from the DNA template strand at the upstream edge of the hybrid29,30. Mutation of the lid strongly inhibited Rho termination (Fig. 3c). It is therefore possible that by pushing against the lid Rho facilitates hybrid melting. Simultaneously, the lid is connected to the TL via rigid domains of β’, which could allosterically transmit the inhibitory signal to the catalytic center. Formation of the trapped complex is the rate-limiting step in the Rho termination pathway that determines the position of the termination site. Because the dismantling of the stable EC requires more time and energy than its inactivation, such a “spider” strategy (trap first, kill later) must be highly effective. The structural organization and principles by which RNA and DNA are retained in the EC are conserved among all cellular RNAPs, arguing that the basic structural pathway leading to termination is universal.
METHODS SUMMARY
The full method section describes DNA and enzyme preparations, transcription reactions protocols, the Rho-binding assay, and protein-RNA crosslinking and mapping techniques.
Methods
DNA templates and proteins
The T7A1-trpt’ template was a 550 bp PCR-amplified fragment carrying the T7A1 promoter fused to the trpt’ terminator. Its transcribed sequence is: ATCGAGAGGG agcaaccgct ggagttccaa cttacgtcat ttttccgccc aacagtaata taatcaaaca aattaatccc gcaacataac accagtaaat caataatttt ctctaagtca cttattcctc aggtaattgt taatatatcc agaatgttaa tcaaaatata ttttccctct atcttctcgt tgcgcttaat ttgactaatt ctcattagcg actaatttta atgagtgtgg gatcc. The transcribed portion of T7A1 is shown in capital letters; rutA and rutB sites are underlined. The T7A1-aRut-LacZ template was a 744 bp PCR-amplified fragment carrying the T7A1 promoter. Its transcribed sequence is: ATCGAGAGGG CCACGGCGAA CAGCCAACCC AACCGAACAG GCCTGCTGGT gacttctcct ctgtctcctt cttccttctc ctctgtctcc ttcttcctcc gtcgttttac aacgtcgtga ctgggaaaac cctggcgtta cccaacttaa tcgccttgca gcacatcccc ctttcgccag ctggcgtaat agcgaagagg cccgcaccga tcgcccttcc caacagttgc gcagcctgaa tggcgaatg. The transcribed portion of T7A1 is shown in capital letters; the artificial rut (aRut) site (underlined) is followed by a part of the lacZ gene. All other templates were prepared from this template by PCR-mediated mutagenesis. All modifications are shown in the Figures. His6-tagged wild type, Zipper, Lid, Zn-finger mutant RNAP were purified and immobilized on Talon™ Co++-beads (Clontech) as described31. A plasmid expressing RpoC lacking entire Zn-finger was provided by R. Weisberg (NIH, Bethesda). His6-tagged Δflap RNAP was provided by K. Severinov (Rutgers University), TL R933A RNAP was provided by I. Artsimovitch (Ohio State University); T7 RNAP was from Roche; EcoRQ111 was expressed and purified as described32. The σ70 subunit was purified as in ref. 33. Rho and 6HISRho proteins were expressed from pET21a (+) vectors and purified as described in ref. 16. RhoP279S was obtained by site directed mutagenesis and purified in the same way.
Transcription reactions
All NTP substrates were purified by ion exchange chromatography1. The preparation of start-up ECs and solid phase transcription reactions (walking) were performed as described1. Briefly, His6-RNAP (~2 pmol) was mixed with a 2-fold molar excess of DNA in 20 µl of TB50 (10 mM MgCl2, 40 mM Tris-HCl, pH 7.9, 50 mM KCl) for 5 min at 37° followed by addition of ApUpC (10 µM, Oligos Etc.), GTP and ATP (25 µM) for 10 min. Next, 5 µl TB100-equilibrated Talon Co++ affinity bead suspension (Clontech) were added for 5 min at room temperature. 1 µl [α-32P] CTP (3000Ci/mmol; NEN Life Sciences Products) was added for 5 min, followed by washing of the beads in 3x1 ml TB 1000 (as TB 50 but 1000 mM NaCl) and 2x1 ml TBGlu (100 mM KGlu [pH=8.0]; 10 mM MgGlu). For run-off experiments the start-up EC was chased with 1 mM ATP, CTP, UTP and 25 µM GTP for 10 minutes with or without 0.4 µM Rho at 37°C. To prepare trapped complexes, 0.1 µg EcoRQ111 was added to the start-up EC for 5 min at 23°C, followed by chasing with 100 µM NTPs for 2 min. The resulting EC156 was washed in 3x1 ml TB100, 2x1 ml TB1000 (to remove the roadblocking EcoRQ111), and 2x1 ml TBGlu. For walking to downstream positions 5 µM NTPs were used. The final complexes were washed in 3x1 ml TBGlu and divided into 10 µl aliquots. Rho and dATP were added to 0.4 µM and 1 mM, respectively. Complexes were incubated at 37°C before chasing with a limited NTP set (100 µM) or washed with TB700. For the experiment of Fig 3d, 0.5 unit tagetin (Epicenter) was used.
For the experiment of Fig. 4c, 3 pmol template DNA (A1-trpt’) were mixed with 6 pmol E.coli RNAP holoenzyme in 20 µL TB Glu [100 mM K Glutamate pH=8.0; 10 mM Mg Glutamate] and incubated 5 minutes at 37°C. AUC primer (10 µM), GTP (25 µM), ATP (25 µM) were added to the mixture and incubated for 10 minutes at 37°C. The mixture was transferred to room temperature, 2 µL [α-P32]CTP were added and incubation was continued for another 5 minutes. Resulting EC11 was transferred to 10 µl TB Glu-equilibrated Talon Co++ affinity bead suspension (Clontech) and incubated for 5 min at room temperature. The beads then were washed 4x1 ml TBGlu. 30 µL TB Glu were added to the EC11 and the mixture divided into 10 µL aliquots. Rho was added to the final concentration of 100 nM either alone or together with 200 µM NTPs. 200 nM E.coli RNAP core enzyme or 200 nM T7 RNAP were added to the chase reaction as indicated. One sample was chased without Rho, but the equal amount of mock solution (50% glycerol, 50% TB Glu) was added instead. Chase continued for 5 minutes at 37°C in the presence of 10 µM rifampicin. One sample was preincubated with Rho for 5 minutes at room temperature before the chase. Reactions were stopped by the addition of 10 µL Stop Buffer [1xTBE; 8M Urea; 20 mM EDTA], samples were phenol/chlorophorm extracted, ethanol precipitated and re-dissolved in 10 µL water plus 10 µL Stop Buffer.
For the experiment of Fig. 3b, 3 pmol template DNA were mixed with 6 pmol T7 RNAP or E.coli RNAP holoenzyme in 20 µL TB Glu [100 mM K Glutamate pH=8.0; 10 mM Mg Glutamate] and incubated for 5 minutes at 37°C. GTP (200 µM), ATP (100 µM) were added to the mixture and incubated for 10 minutes at 37°C. The mixture was transferred to room temperature, 2 µL [α-P32]CTP were added and incubation was continued for another 5 minutes. The resulting EC19 was divided into 10 µL aliquots and Rho was added to a final concentration of 0.4 µM to one sample and an equal amount of the mock solution (50% glycerol, 50% TB Glu) was added to another. After a 5 minute incubation at room temperature the samples were chased in the presence of 100 µM ATP, GTP and 3 µM CTP, UTP (T7 RNAP) or 100 µM ATP, GTP, CTP, UTP (E.coli RNAP) for 5 min at 37°C. Reactions were stopped by the addition of 10 µL Stop Buffer [1xTBE; 8M Urea; 20 mM EDTA], samples were phenol/chrolophorm extracted, ethanol precipitated and re-dissolved in 10 mL water plus 10 mL Stop Buffer.
Efficiencies of Rho-dependent termination (%T) were calculated by dividing the amount of radioactivity in all the termination bands (sensitive to washings) by the total radioactivity present in those and all read-through bands. Efficiency of RNA release (%R) was calculated by dividing the amount of radioactivity in major termination bands remaining after washing the beads by the total radioactivity present in those and corresponding pre-washed bands. When calculating %R, the values are also normalized to the mild background radioactivity loss, which occurs due to the inadvertent loss of beads after multiple washings. All the numbers are statistically significant and represent the average values from at least 3 independent experiments.
Rho binding assays
Conditions for Rho binding to the RNAP holoenzyme and open promoter complex are described in the legend to Supplementary Fig. 10.
Crosslinking and mapping of the cross-linked sites
EC156 was obtained as described above. 20 µM 4-thio-uridine-5’-triphosphate (sUTP; TriLink Inc.) was added for 5 min at 23°C. The resulting EC157 was washed in 4x1ml TBGlu and incubated with 1mM dATP (with or without 0.4 µM Rho) for 10 min at 37°C. The resulting complexes were washed in 4x1ml TB10 (10 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl [pH=8.0]). The crosslink in the resulting trapped complex was induced by UV irradiation at 308 nM for 10 min with a UV hand-held lamp (Cole-Palmer 9815 series lamp) placed above an Eppendorf tube held in ice. The samples were treated with imidazole at 150 mM to remove the complexes from the Co++-beads, and the supernatant material was fractionated by SDS-PAGE, followed by radioautographic visualization. Mapping of crosslinked sites was performed by limited chemical degradation. β’ and β subunits carrying the radioactive adduct were excised from the gel and eluted with 3 vol of 0.2% SDS at 37°C for 1h. The eluate was freeze-dried with a SpeedVac and redissolved in water to a final concentration of 1–2% SDS. Protein degradation effected by CNBr (at Met residues) and NTCBA (at Cys residues) was performed according to ref. 4. Gels containing radioactive materials were scanned on a PhosphoImager.
Supplementary Material
Epshteinsuppl
Acknowledgments
We thank Irina Artsimovitch, Sergei Borukhov, Konstantin Severinov, Barbara Stitt, Dmitry Vassylyev, and Robert Weisberg for materials and discussions. This work was supported by a grant from the NIH R01GM58750 (E.N.).
Footnotes
Author Contributions V.E., D.D., and J.W. conducted the experimental work, discussed the results, and commented on the manuscript. E.N. designed the study and wrote the paper.
References
- 1.Adhya S, Gottesman M. Control of transcription termination. Annu. Rev. Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 2.Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- 3.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 5.Geiselmann J, Yager TD, Gill SC, Calmettes P, von Hippel PH. Physical properties of the Escherichia coli transcription termination factor Rho. 1. Association states and geometry of the Rho hexamer. Biochemistry. 1992;31:111–121. doi: 10.1021/bi00116a017. [DOI] [PubMed] [Google Scholar]
- 6.Browne RJ, Barr EW, Stitt BL. Catalytic cooperativity among subunits of Escherichia coli transcription termination factor Rho. Kinetics and substrate structural requirements. J Biol Chem. 2005;280:13292–13299. doi: 10.1074/jbc.M500221200. [DOI] [PubMed] [Google Scholar]
- 7.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 8.Adelman JL, et al. Mechanochemistry of transcription termination factor Rho. Mol Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: more questions than answers. J Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 10.Platt T. Rho and RNA: models for recognition and response. Mol Microbiol. 1994;11:983–990. doi: 10.1111/j.1365-2958.1994.tb00376.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim DE, Patel SS. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J Biol Chem. 2001;276:13902–13910. doi: 10.1074/jbc.M011043200. [DOI] [PubMed] [Google Scholar]
- 12.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at Rho-dependent terminators depends on kinetic coupling between RNA polymerase and Rho. Proc Natl Acad Sci USA. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu AQ, von Hippel PH. Rho-dependent termination within the trp t' terminator. II. Effects of kinetic competition and rho processivity. Biochemistry. 1998;37:11215–11222. doi: 10.1021/bi972912s. [DOI] [PubMed] [Google Scholar]
- 15.Guerin M, Robichon N, Geiselmann J, Rahmouni AR. A simple polypyrimidine repeat acts as an artificial rho-dependent terminator in vivo and in vitro. Nucleic Acids Res. 1998;26:4895–4900. doi: 10.1093/nar/26.21.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalissery J, Banerjee S, Bandey I, Sen R. Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex. J Mol Biol. 2007;371:855–872. doi: 10.1016/j.jmb.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Nahum G, Epshtein V, Ruckenstein AE, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 18.Vassylyev DG, et al. Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mentesana PE, Chin-Bow ST, Sousa R, McAllister WT. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J Mol Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- 20.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 23.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz A, Margeat E, Rahmouni AR, Boudvillain M. Transcription termination factor rho can displace streptavidin from biotinylated RNA. J Biol Chem. 2007;282:1469–1476. doi: 10.1074/jbc.M706935200. [DOI] [PubMed] [Google Scholar]
- 26.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 27.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 28.Epshtein V, Cardinale C, Ruckenstein A, Borukhov S. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 30.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J Mol Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Nudler E, Gusarov I, Bar-Nahum G. Methods of walking with the RNA polymerase. Methods Enzymol. 2003;371:160–169. doi: 10.1016/S0076-6879(03)71011-8. [DOI] [PubMed] [Google Scholar]
- 32.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989;264:11816–11821. [PubMed] [Google Scholar]
- 33.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr Purif. 1993;4:503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 34.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Epshteinsuppl