Three-dimensional culture models of normal and malignant breast epithelial cells (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 3.
Published in final edited form as: Nat Methods. 2007 Apr;4(4):359–365. doi: 10.1038/nmeth1015
Abstract
Extracellular matrix is a key regulator of normal homeostasis and tissue phenotype1. Important signals are lost when cells are cultured ex vivo on two-dimensional plastic substrata. Many of these crucial microenvironmental cues may be restored using three-dimensional (3D) cultures of laminin-rich extracellular matrix (lrECM)2. These 3D culture assays allow phenotypic discrimination between nonmalignant and malignant mammary cells, as the former grown in a 3D context form polarized, growth-arrested acinus-like colonies whereas the latter form disorganized, proliferative and nonpolar colonies3. Signaling pathways that function in parallel in cells cultured on plastic become reciprocally integrated when the cells are exposed to basement membrane–like gels4–7. Appropriate 3D culture thus provides a more physiologically relevant approach to the analysis of gene function and cell phenotype ex vivo. We describe here a robust and generalized method for the culturing of various human breast cell lines in three dimensions and describe the preparation of cellular extracts from these cultures for molecular analyses. The procedure below describes the 3D ‘embedded’ assay, in which cells are cultured embedded in an lrECM gel8 (Fig. 1). By lrECM, we refer to the solubilized extract derived from the Engelbreth-Holm-Swarm mouse sarcoma cells9. For a discussion of user options regarding 3D matrices, see Box 1. Alternatively, the 3D ‘on-top’ assay, in which cells are cultured on top of a thin lrECM gel overlaid with a dilute solution of lrECM, may be used as described in Box 2 (Fig. 1 and Fig. 2).
Materials
Reagents
- Engelbreth-Holm-Swarm extracellular matrix extract (EHS), growth factor–reduced (Matrigel, BD Biosciences or Cultrex BME, Trevigen)
- Diaminophenylindole (DAPI)
- Hard-set mounting medium (VECTASHIELD HardSet, Vector Laboratories or ProLong Gold, Invitrogen)
- Immunofluorescence (IF) buffer: 0.2% Triton X-100, 0.1% BSA (radioimmunoassay grade), 0.05% Tween 20 in PBS (pH 7.4; sterilized, 0.22 μm filter); for long-term storage, add 7.7 mM NaN3
- IF blocking solution: 10% goat serum, 1% goat F(ab')2 anti-mouse immunoglobulin G (IgG; Caltag) in IF buffer
- PBS: 130 mM NaCl, 13 mM Na2HPO4, 3.5 mM NaH2PO4 (pH 7.4)
- PBS-EDTA: 5 mM EDTA, 1 mM NaVO4, 1.5 mM NaF in PBS
- PBS-glycine: 100 mM glycine in PBS
Procedure
Culturing cells in 3D
- 1
Thaw EHS at 4 °C overnight. - 2
Coat prechilled culture surface (for example, dish or well) with a thin layer of EHS. Slowly pipette the appropriate volume of “EHS coat” (Table 1) directly onto surface and spread evenly with a pipette tip or plunger of a 1-ml syringe for smaller areas, or cell lifter for larger areas. Incubate for 15–30 min at 37 °C to allow the EHS to gel (but do not let it overdry).
- 3
Trypsinize cells from a monolayer to a single-cell suspension.
Use cells that are healthy and not more than 75% confluent. - 4
Aliquot cells to be plated into a 1.5 ml microcentrifuge tube.
To perform drug response assays in 3D cultures7,10,11 (Fig. 3 and Supplementary Videos 1 and 2 online), compounds may be added to the culture in one of two manners:- Small-molecule inhibitors: add to medium when cells are plated in Step 6 of the main protocol and Step b of the 3D on-top protocol (Box 2). Include the compound in all media changes for the duration of the culture.
- Blocking antibodies: mix with cells before plating in Step 5 of the main protocol and Step b of the 3D on-top protocol (Box 2) to ensure complete exposure of the cells to antibodies. Include the antibody in all media changes for the duration of the culture.
- 5
Gently pellet the cells by centrifugation at ∼115_g_ and, on ice, resuspend the cells into the appropriate volume of EHS (Table 1).
The number of cells to be plated per milliliter of EHS depends on the growth properties of the cell line and may need some optimization, but we recommend the following ranges: for nonmalignant cells, 0.85 × 106 cells/ml; for malignant cells, 0.50–0.60 × 106 cells/ml.
- 6
Pipette the mixture of cells and EHS onto the precoated surface. Incubate 30 min at 37 °C to allow EHS to gel. Add the appropriate volume of medium (Table 1).
- 7
Maintain the culture for 10 d, changing medium every 2 d.
Table 1.
Suggested volumes for 3D culture
| 3D embedded | 3D on-top | ||||||
|---|---|---|---|---|---|---|---|
| Number of wells | Diameter (mm) | Area (cm2) | Medium volume (μl) | EHS coat (μl) | EHS plate (μl) | EHS coat (μl) | |
| Dish | NA | 60 | 28.3 | 5,000 | 250 | 3,600 | 850 |
| Plates | 6 | 35 | 9.6 | 2,000 | 120 | 1,200 | 500 |
| 24 | 16 | 2.0 | 500 | 50 | 300 | 120 | |
| 48 | 10 | 0.75 | 200 | 30 | 150 | 80 | |
| 96 | 6 | 0.26 | 60 | 5 | 75 | 15 | |
| Chamber slides | 4 | NA | 1.8 | 500 | 50 | 300 | 120 |
| 8 | NA | 0.8 | 200 | 30 | 150 | 90 |
Figure 3.
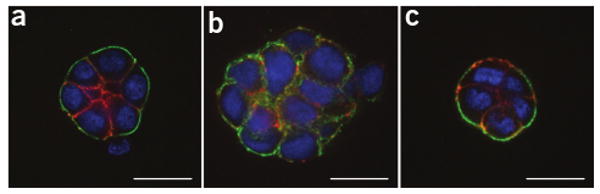
3D drug response assay. (a–c) HMT-3522 S1 (a), HMT-3522 T4-2 (b) and HMT-3522 T4-2 treated with an EGFR inhibitor, AG1478 (c) were cultured in the 3D on-top assay for 4 d. Colonies were then extracted and immunostained against α6 integrin (green) and β-catenin (red). Nuclei were counterstained with DAPI (blue). Confocal sections through the centers of the colonies are shown. Scale bar, 20 μm.
BOX 1 EHS USER'S GUIDE
EHS is available commercially from several sources, including BD Biosciences (Matrigel) and Trevigen (Cultrex Basement Membrane Extract). EHS can also be prepared directly from EHS tumors grown as xenografts22. As EHS is a biological product, its components and properties, including ECM protein and growth factor concentrations, endotoxin levels and stiffness, vary between lots. It is therefore desirable to perform a series of experiments using the same lot number to minimize variation introduced by slight differences in the properties of the EHS. It is also important, when a new lot is obtained, to test whether it is appropriate for culture by performing a side-by-side comparison with cells grown in EHS from a previous lot. We routinely evaluate new lots for the typical and appropriate morphogenesis of nonmalignant and malignant cells along with the expression of several markers of interest. We also exclusively use growth factor–reduced EHS as much of our work is done in the absence of serum and under defined medium conditions. Depending on the nature of the cell type and parameters to be measured, you may develop your own strategy for validation and arrive at your own EHS preferences.
BOX 2 3D ON-TOP ASSAY
As an alternative to the 3D embedded assay, we developed the 3D on-top assay, which requires a shorter amount of time, a decreased amount of EHS, and facilitates imaging as colonies are in a single plane. Therefore, the on-top assay is ideal for time-lapse imaging and also for in situ immunostaining of cell lines that form invasive stellate structures in 3D (see Step 8, Option C). Because less EHS is required, it is also a more cost-effective approach.
- Follow Steps 1–4 of the main protocol to prepare the surface and cells for plating.
- Pellet the cells by centrifugation at ∼115 g, resuspend in half the “medium volume” (Table 1) and plate onto the coated surface. Allow the cells to settle and attach to the EHS for 10–30 min at 37 °C. The number of cells to be plated per square centimeter of culture surface area may need some optimization depending on the growth properties of the cell line, but we recommend the following ranges: for nonmalignant cells, 0.25 × 105 cells/cm2; for malignant cells, 0.175–0.20 × 105 cells/cm2. Cells of some lines tend to aggregate with one another and may not adhere as quickly to the EHS, resulting in cells not resting singly on the layer of EHS or concentrating in the center of the well. Agitation of the plate in the x-y plane at intervals during incubation at 37 °C may assist with preventing cell concentration in the center of the well (do not apply a swirling motion as cells will then accumulate around the edge of the well).
- Chill the remaining medium on ice and add EHS to 10% volume. Gently add the EHS-medium mixture to the plated culture.
Medium must be thoroughly chilled before addition of EHS to ensure homogenous mixing and even deposition of EHS onto cells in culture. Pipette the EHS-medium mixture down the side of the well to avoid disturbance of the cells or EHS gel. - Maintain culture for 4 d, replacing EHS-medium mixture every 2 d.
- To perform drug response assays in the 3D on-top assay, see Step 8 of the main protocol.
Extracting cells from 3D cultures
- 8
Colonies may be fully extracted from their 3D culture environment for further analyses, for example, immunostaining, DNA, RNA and protein extraction (Option A, complete extraction). If only an immunostaining endpoint is desired, a complete extraction is often not necessary. An abbreviated procedure may be performed in which the gel is partially broken down, allowing for sufficient dissolution of polymerized EHS in the sample for immunostaining (Option B, in-well extraction). Alternatively, colonies may be fixed and then immunostained directly in the gel (Option C, whole-culture fixation; proceeding to Box 3, whole-culture immunostaining). This latter method is generally only preferable if a 3D colony you would like to visualize would be disrupted by extraction (as in Fig. 2c) because the immunostaining procedure then requires a much larger amount of antibody per sample and is less flexible as a single culture may only be stained against one set of markers. Frozen sections for subsequent immunostaining may also be obtained after embedding of whole cultures in Optimal Cutting Temperature (OCT) compound. If only RNA extraction is desired, the culture may be directly solubilized by addition of Trizol.
Perform all steps on ice.
Figure 2.
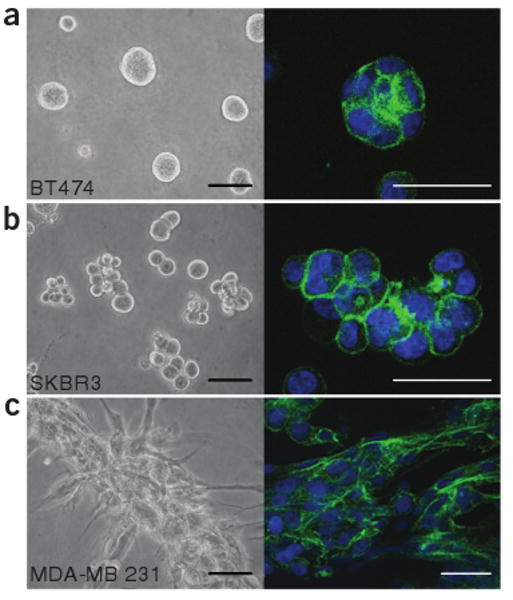
Breast cancer cell lines in 3D culture. (a–c) Phase-contrast images (left) and confocal cross-sections of Phalloidin staining of F-actin (right) of BT-474 (a), SK-BR-3 (b) and MDA-MB-231 (c) cell lines grown for 4 d in the 3D on-top assay. In a and b, colonies were completely extracted from the gel for immunostaining; in c, colonies were immunostained in the gel. Scale bars, 100 μm (left) and 50 μm (right).
Option A. Complete extraction
- Aspirate the medium and rinse 2× with 1 medium-volume equivalent of ice-cold PBS.
- Add 2–3 volumes of ice-cold PBS-EDTA. Detach EHS from the bottom of culture surface using a cell lifter (for dishes of diameter ≥35 mm) or by gently scraping the bottom with a pipette tip. Shake gently for 15–30 min.
The 3D-embedded cultures will require a larger volume of PBS-EDTA than 3D on-top cultures and will also take longer to break down. - Transfer solution to a conical tube. Rinse culture surface once with 0.5 volume of PBS-EDTA and transfer the rinse to the conical tube. Gently shake tube on ice for 15–30 min.
- Inspect the tube to check that EHS has dissolved completely (invert tube gently and look for a homogeneous suspension of cell colonies without visible EHS gel fragments). If not, wait longer and/or add more PBS-EDTA.
To collect colonies for immunostaining, follow (v). Otherwise, proceed to (vi). - Centrifuge the solution at ∼115_g_ for 1–2 min such that cell colonies collect at the bottom of the tube but do not form a tight pellet. Then carefully aspirate the majority of the supernatant. Gently resuspend the colonies in the remaining supernatant. Pipette approximately 15 μl of the colony suspension onto a slide, allow cells to settle and adhere to the glass, and fix the cells using a fixative appropriate for the antigen of interest.

 Colonies fixed on slides may be stored at −20 °C for several months.
Colonies fixed on slides may be stored at −20 °C for several months. - Centrifuge the colonies at ∼115_g_ for 5 min into a pellet. Aspirate the supernatant, lyse the cells with an appropriate extraction buffer and process using standard procedures12.
For protein extraction an additional wash with PBS-EDTA will minimize the amount of EHS in the final extract. Extracts may be stored at −80 °C for several months.
Extracts may be stored at −80 °C for several months.
Option B. In-well extraction
- Follow Step 8 Option A (i–ii).
- Check under microscope to verify that majority of EHS has broken apart and colonies have settled to bottom of the well. If not, wait longer and/or add more PBS-EDTA.
- Carefully aspirate the majority of the supernatant. Pipette approximately 15 μl of the colonies in solution onto a slide, allow cells to settle and adhere to the glass, and fix them.

 Cultures fixed on slides may be stored at −20 °C for several months.
Cultures fixed on slides may be stored at −20 °C for several months.
Option C. Whole-culture fixation
- Aspirate the medium and rinse 2× with 1 medium-volume equivalent of ice-cold PBS.
- Fix culture with 4% paraformaldehyde at room temperature (18–23 °C) for 10 min.
- Stop fixation by aspirating 4% paraformaldehyde and adding at least 2× medium-volume PBS-glycine for 10 min; wash once with PBS and store in PBS.
 Fixed cultures may be stored at 4 °C for up to 4 d.
Fixed cultures may be stored at 4 °C for up to 4 d.
Immunostaining of 3D cultures
- 9
To immunostain the 3D cultures, first wash slides 3× with PBS-glycine for 10 min at room temperature.
This immunostaining procedure applies to cultures fixed on slides generated in Step 8 Option A or B (Figs. 2a,b and 3) and, with modifications indicated in Box 3, also to fixed whole cultures generated in Step 8 Option C (Fig. 2c).
Box 3 Whole-Culture Immunostaining
When performing whole-culture immunostaining in a well, the general procedure for immunostaining may be applied (see Steps 9–18), with some slight modifications:
Washes are performed in the well and require careful pipetting and aspiration; if cultures are not treated gently, colonies may detach from the EHS and be lost. It is advisable to check cultures frequently under a microscope to ensure they are still attached. We suggest using 2× medium-volume equivalents for washes; for antibody incubations, 0.5× volume is sufficient.
Samples immunostained in a chamber slide may be mounted directly after removal of the chamber and aspiration of excess fluid.
Samples immunostained in a tissue-culture well should be removed and mounted on a slide for long-term storage and higher-quality imaging.
10
To block slides, spread 100 μl of IF blocking solution over the fixed cells with a strip of Parafilm, and incubate the slides for 1.5 h, at room temperature in a humid chamber.
The anti-mouse F(ab')2 fragments in the IF blocking solution block against immunoreactive mouse IgG species in EHS.11
Stain cultures with the primary antibody of interest diluted in 100 μl of IF buffer for 2 h at room temperature in a humid chamber.12
Wash slides 3× for 20 min in IF buffer in a Coplin jar at room temperature.13
Incubate slides with a secondary antibody diluted in 100 μl of IF buffer for 45 min at room temperature in a humid chamber.14
Wash slides 1× with IF buffer for 20 min, and then 2× with PBS for 10 min at room temperature.15
Counterstain the nuclei with DAPI (0.5 μg/ml in PBS) for 5 min at room temperature.16
Wash slides in PBS for 10 min at room temperature.17
To mount slides, spread one drop of hard-set mounting medium with a number 1½ coverslip and allow to set overnight at 4 °C.
Take care to avoid forming bubbles when mounting. Mounted slides may be stored at −20 °C for several months.
Mounted slides may be stored at −20 °C for several months.18
Owing to the thickness of the samples, confocal microscopy is ideal for imaging 3D cultures.
TROUBLESHOOTING TABLE
| Problem | Solution |
| Step 6 Cells are not suspended and have settled to the bottom of the EHS gel. | If the cell-EHS mixture in Step 5 is diluted with medium before setting, the cells may settle to the bottom of the gel. Be sure to aspirate the majority of the supernatant to avoid this problem. |
| Step 18 Excess EHS causes immunostaining background. | Colonies fully extracted from the EHS gel as in Step 8 Option A should have little or no background. Partial extraction of colonies by Step 8 Option B may result in background if EHS is not sufficiently broken down, and a haze or cloud of EHS adheres to cells. To avoid this, increase both the volume of PBS-EDTA used and increase the incubation time in Step 8, Option A (ii). |
Critical Steps
Step 2 Culture surfaces must be prechilled and coated on ice to ensure even spreading of EHS. Pipette EHS slowly and directly onto culture surface to avoid formation of bubbles, which may allow cells to come in direct contact with the culture surface and begin to spread as a monolayer beneath the gel. Note that the viscosity of EHS causes it to form a meniscus in the well, the effect of which increases with decreasing well size. Thus, for 3D on-top cultures, the smaller the well size, the less flat the plane of plating will be.
Step 5 After aspiration of the supernatant, gently flick the tube to loosen the cell pellet so that when EHS is added, the cells are in a single cell suspension. Pipette carefully when mixing to avoid bubbles.
Step 8, Option A (v) Centrifugation time will depend on the size of your colonies and may require some optimization. Larger colonies will settle on their own and may only require a pulse to collect at the bottom of the tube whereas smaller colonies may have only just collected in the conical area of the tube after 2 minutes of centrifugation.
The amount of aspiration required to achieve the balance between getting a high number of colonies on the slide in a relatively low volume of liquid may require some practice. If the volume of liquid used to pipette a sufficient number of colonies is too high it may decrease the efficiency of fixation. The slides may sit for some time to allow excess liquid to evaporate, but do not allow the cells to dry out completely at any point as this will alter the cell structure.
Step 8, Option B (iii) See second comment in Critical Steps, Step 8, Option A (v).
Comments
Here we describe a generalized protocol for monotypic 3D breast epithelial cell culture in the presence of lrECM, an approach that has proved to be extremely informative in our laboratory and those of others. For instance, we recently used the 3D on-top assay to analyze the correlations between colony morphology and gene expression in a large panel of breast cancer cell lines13. Other workers have succeeded in culturing cell types from a wide variety of tissues using these techniques (summarized in ref. 14) and informative protocols have been published for several of these systems15–20. Although much of this work has been performed using EHS, other 3D substrata, such as collagen I gels, are excellent for assays of mammary gland branching morphogenesis21, and we are watching with interest the development of additional synthetic and natural 3D substrata. Using these approaches, we hope to develop functional organotypic cultures comprised of multiple cell types—including epithelial, myoepithelial, stromal and endothelial—to more appropriately model signaling and cell-cell interactions in an environment similar to complex breast tissue.
Supplementary Material
Supplementary Data, Video 1
Supplementary Data, Video 2
Figure 1.
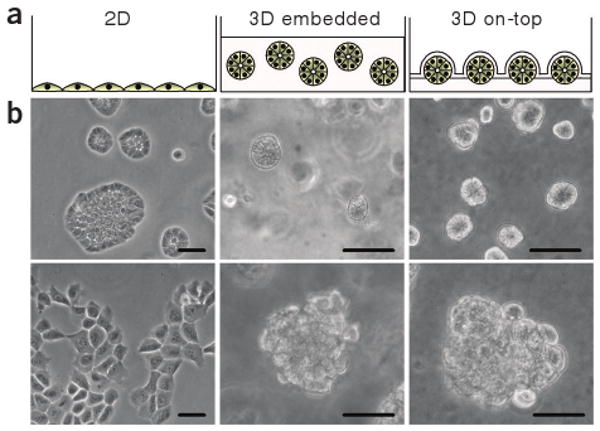
Breast epithelial cell morphology in different culture conditions. (a) Schematic of nonmalignant breast epithelial cells grown as a monolayer on tissue-culture plastic (left), in the 3D embedded assay (middle) and in the 3D on-top assay (right). (b) Phase-contrast images of nonmalignant HMT-3522 S1 cells grown in the three different culture conditions (top) and malignant HMT-3522 T4-2 cells grown in the same conditions (bottom). Scale bars, 50 μm.
Acknowledgments
The protocol described here has been the work of many members of the Bissell laboratory over many years. We apologize to those whose work could not be cited owing to space limitations and have cited reviews where possible. This work was supported by grants from the Office of Biological and Environmental Research of the US Department of Energy (DE-AC03-76SF00098 and a Distinguished Fellow Award to M.J.B.), the US National Cancer Institute (CA64786 to M.J.B.; CA57621 to Zena Werb and M.J.B.) and the Breast Cancer Research Program of the US Department of Defense (Innovator Award DAMD17-02-1-438 to M.J.B).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
Competing Interests Statement: The authors declare no competing financial interests.
References
- 1.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anders M, et al. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the coxsackievirus and adenovirus receptor. Proc Natl Acad Sci USA. 2003;100:1943–1948. doi: 10.1073/pnas.0337599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinman HK, et al. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry. 1982;21:6188–6193. doi: 10.1021/bi00267a025. [DOI] [PubMed] [Google Scholar]
- 10.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver VM, et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 13.Kenny PA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. doi: 10.1016/j.molonc.2007.02.004. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Auerbach R, Lewis R, Shinners B, Kubai L, Akhtar N. Angiogenesis assays: a critical overview. Clin Chem. 2003;49:32–40. doi: 10.1373/49.1.32. [DOI] [PubMed] [Google Scholar]
- 16.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 17.Toda S, et al. A new organotypic culture of thyroid tissue maintains three-dimensional follicles with C cells for a long term. Biochem Biophys Res Commun. 2002;294:906–911. doi: 10.1016/S0006-291X(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 18.Tonge D, Edstrom A, Ekstrom P. Use of explant cultures of peripheral nerves of adult vertebrates to study axonal regeneration in vitro. Prog Neurobiol. 1998;54:459–480. doi: 10.1016/s0301-0082(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan MB, Ramirez RD, Wright WE, Minna JD, Shay JW. A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006;74:141–148. doi: 10.1111/j.1432-0436.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, et al. Three-dimensional co-culture models to study prostate cancer growth, progression, and metastasis to bone. Semin Cancer Biol. 2005;15:353–364. doi: 10.1016/j.semcancer.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 2004;6:1–11. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kibbey MC. Maintenance of the EHS sarcoma and matrigel preparation. Methods Cell Sci. 1994;16:227–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data, Video 1
Supplementary Data, Video 2