BioXSD: the common data-exchange format for everyday bioinformatics web services (original) (raw)
Abstract
Motivation: The world-wide community of life scientists has access to a large number of public bioinformatics databases and tools, which are developed and deployed using diverse technologies and designs. More and more of the resources offer programmatic web-service interface. However, efficient use of the resources is hampered by the lack of widely used, standard data-exchange formats for the basic, everyday bioinformatics data types.
Results: BioXSD has been developed as a candidate for standard, canonical exchange format for basic bioinformatics data. BioXSD is represented by a dedicated XML Schema and defines syntax for biological sequences, sequence annotations, alignments and references to resources. We have adapted a set of web services to use BioXSD as the input and output format, and implemented a test-case workflow. This demonstrates that the approach is feasible and provides smooth interoperability. Semantics for BioXSD is provided by annotation with the EDAM ontology. We discuss in a separate section how BioXSD relates to other initiatives and approaches, including existing standards and the Semantic Web.
Availability: The BioXSD 1.0 XML Schema is freely available at http://www.bioxsd.org/BioXSD-1.0.xsd under the Creative Commons BY-ND 3.0 license. The http://bioxsd.org web page offers documentation, examples of data in BioXSD format, example workflows with source codes in common programming languages, an updated list of compatible web services and tools and a repository of feature requests from the community.
Contact: matus.kalas@bccs.uib.no; developers@bioxsd.org; support@bioxsd.org
1 INTRODUCTION
The bioinformatics community shares a common feature with a global business corporation, namely the diversity of IT systems. A global corporation owns a myriad of IT solutions belonging to its smaller or bigger sub-companies, implemented in diverse ways and covering diverse aspects and business areas of the corporation. To achieve efficiency within the corporation or consortium, all the IT systems must be able to work together: to inter-operate.
In bioinformatics, we do not have any corporate management to force standards for interoperability. But it is clear that the bioinformatics community would benefit greatly from an IT infrastructure that allows more efficient use of biological data and computational resources, in order to support new exploration and discoveries. Since the scientific community lacks a centralized authority, the community must develop its standards within collaborative efforts.
In his visionary comment, Lincoln Stein called for standardization in bioinformatics, suggesting web services (http://www.w3.org/standards/webofservices) as the unifying platform for programmatic interfaces to tools and data sources (Stein, 2002). Nowadays, the ELIXIR project chooses SOAP web services for programmatic access to all considered bioinformatics databases and tools (http://www.elixir-europe.org/page.php?page=wp7). The Web Service Interoperability Organisation (WS-I, http://ws-i.org), supported by the main IT companies, constrains even more strictly the W3C's SOAP-service standards in order to maximize interoperability among the web services and the web-service programmatic libraries.
The EMBRACE project (European Model for Bioinformatics Research and Community Education) has developed guidelines for providing data sources and computational tools that are globally interoperable on the web of services. The guidelines recommend WS-I compliant web services with document/literal wrapped SOAP binding (Pettifer et al., 2010; Stockinger et al., 2008; technical details in http://www.embracegrid.info/page.php?page=tech_documents).
Even while following the W3C and WS-I standards, the practical interoperability within the field of bioinformatics web services is compromised by the incompatibility or inconsistency of input and output formats of different services (Hull et al., 2006). Standard exchange formats have been identified as a necessary key to global interoperation by the ELIXIR and EMBRACE projects, and by the BioHackathon jamboree (http://hackathon.dbcls.jp). Developing an XML Schema of standard data-exchange formats, called a canonical data model, is typical within IT-system integration in industry, business and public administration. The standard formats of the exchanged data enable web-service developers to use them as the input and output data formats, eliminating the need to define their own formats and thus saving development and maintenance costs. This is especially useful when services are developed in a WSDL-centric fashion, as recommended by EMBRACE.
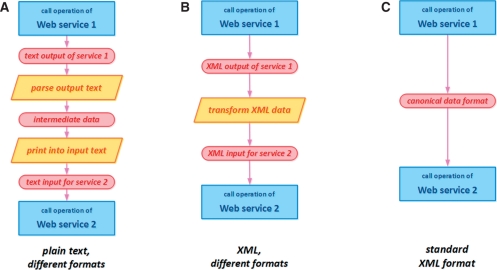
The biggest advantage of the common standard formats is however for the users of services. The common, canonical data model gives users the ability to easily mix-and-match diverse web services into custom analysis pipelines: workflows. Workflows are developed using either ordinary programming or scripting languages, or specialized workflow tools like Taverna (Hull et al., 2006) or workflow languages like BPEL (http://docs.oasis-open.org/wsbpel/2.0/wsbpel-v2.0.html). The use of standard canonical formats diminishes the need to translate data between different formats of the output and input of the various tools. Such translations normally require custom parsers or development of predefined transformers, referred to as shims (Hull et al., 2004). The various scenarios of mix-and-matching different web services are illustrated in Figure 1. A standard, canonical format of the exchanged data simplifies workflow development, and reduces the effort of developing and tailoring analysis pipelines. Standard exchange formats aim to save time and resources, and decrease the needs for specialized personnel with advanced programming skills.
Fig. 1.
Three different scenarios of sending data from an output of one web service to an input of another web service. (A) Plain textual data inside SOAP messages. Proprietary parsing and serialization, or shims are necessary. (B) Different XML formats of output of the first service and input of the second. Translation of data is in general easier but still necessary. (C) Both services using the same standardized exchange format. No transformation is necessary and data flow smoothly from one service to another.
Data formats can be defined in several alternative ways. XML Schema (XSD, http://www.w3.org/standards/techs/xmlschema) is a formal language for defining data types and their formats. XSD defines the structure of data objects, restricts the allowed values of atomic data and defines specialization and inclusion relations on the data types. XML Schema is primarily used to define the structure of XML documents, SOAP messages, or, from the web-service perspective, the data objects. Not limited to define XML formats, XSD can also be used to define object data models for object-oriented programming languages. The source code needed to define the data objects in the chosen programming language can be automatically generated from the particular Schema using ordinary tools. When used for so-called marshalling and unmarshalling of objects between their XML-serialized representation and the computer-memory representation, such a framework is called a data binding. Due to a Schema and the data binding, we can abstract from the textual XML appearance of the exchanged data, and regard it as a medium for directly transferring data objects.
A number of industry-supported tools are available for parsing, validating or translating XML data corresponding to a defined XML Schema. If a Schema describes the data format at an appropriately detailed level, these widely available frameworks provide us with useful functionality. This includes automatic validation of the data ensuring that our tools receive only valid inputs, thus improving the security for the providers and eliminating senseless invocations and the need for validating the input in the source code. Data validation thus lowers the burden for the tool provider and increases the usability for the user. Detailed Schemas eliminate the need for writing and maintaining special parsers for each type and format of data, making the inputs and outputs ‘parsed on arrival’, unmarshalled into the data-binding objects. Selection and projection, or translation among different formats is simplified. A detailed XSD makes it possible to semantically annotate details of data formats using the SAWSDL standard (Semantic Annotation for WSDL and XML Schema, http://www.w3.org/standards/techs/sawsdl). The detailed XSD can also help develop or even generate comprehensive graphical user interfaces for editing data and invoking services. In addition, very efficient compression of data can be achieved by Schema-aware compression tools which use the constraining XML Schema to provide the minimum binary encoding of the data. Examples of such frameworks are emerging (Augeri et al., 2007), and their adoption will be crucial for the increasingly data-intensive bioinformatics, for instance related to high-throughput sequencing.
Initiated by the EMBRACE project partners, we have developed BioXSD. BioXSD is the candidate for a reasonably lightweight, but formal and detailed, standard XML exchange format of commonly used, everyday bioinformatics data. BioXSD makes web services easily interoperable and markedly simplifies the construction of workflows.
The following section briefly summarizes related work and approaches, and discusses their relationship with BioXSD. Section 3 describes the BioXSD development and summarizes the design principles. Section 4 describes the developed BioXSD formats and their highlights. It also presents a feasibility test of BioXSD adoption by web services and a case-study client workflow. Section 5 concludes the article.
2 RELATED EFFORTS AND APPROACHES
In this section, we introduce previous efforts, related and alternative approaches and discuss their relationship with BioXSD.
2.1 Specialized standard formats
Standardized XML data-exchange formats do exist for specialized sub-domains of bioinformatics and the data types in focus. Examples are ‘SBML’ for systems-biology models (Hucka et al., 2003), ‘PDBML’ for structural bioinformatics (Westbrook et al., 2005), The HUPO PSI's Molecular Interaction format (‘MIF′; Hermjakob et al., 2004), or ‘phyloXML’ (Han and Zmasek, 2009) and ‘NeXML’ (http://www.nexml.org) for phylogenetic data. The ‘Minimum Information’ standards for different fields of experimental molecular biology often include an XML Schema of the data-exchange format: for example ‘MAGE-ML’ in MIAME (Spellman et al., 2002) or ‘GCDML’ in MIGS/MIMS (Kottmann et al., 2008). The scope of BioXSD is to offer standard exchange formats for the common bioinformatics data not covered by these specialized, mostly heavyweight standards. We encourage using the ‘big’ standards for data exchange always when applicable, and BioXSD for the exchange of common, everyday bioinformatics data like sequences, alignments, references and unified generic sequence annotations.
2.2 XML formats for common bioinformatics
‘DAS’ (Distributed Annotation System; Prlić et al., 2007) and ‘BioMoby’ (Wilkinson and Links, 2002) are web-based, service-oriented bioinformatics infrastructures, enabling interoperability across distributed resources. To ensure the interoperability within the infrastructure, they include a set of common XML formats. In BioMoby, it is an open library of data types and formats, and in DAS it is a set of lightweight formats for sequences and annotation data. The family of ‘HOBIT XML’ Schemas developed by the Helmholz Open BioInformatics Technology project (HOBIT) has defined common XML formats of everyday bioinformatics data types. These are used among web services within a set of German bioinformatics research institutes (Seibel et al., 2006). HOBIT XML defines a substantial number of types spread over multiple XSDs, which are programmatically accessible using a dedicated Java library (BioDOM). The ‘CBS Common Data Types’ have been in use among web services at the Center for Biological Sequence analysis (CBS) at the Technical University of Denmark and at EMBL, Heidelberg (http://www.cbs.dtu.dk/ws/doc/datatypes.php). They constitute a couple of lightweight XML data formats including ones corresponding to the FASTA format and a subset of General Feature Format (GFF, http://www.sanger.ac.uk/resources/software/gff). The CBS-EMBL data model has served as one of the starting points for the development of BioXSD.
2.3 Semantic web
Semantic web standards offer alternative languages to define a data format in a formal way. For example ‘BioPAX’ (http://www.biopax.org), the common exchange format for biological pathways, is defined in OWL (http://www.w3.org/standards/techs/owl). Some semantic web-service-oriented architectures propose using a canonical reference ontology instead of a canonical XML Schema, and perform lifting and lowering of the output and input data through data individuals of the reference ontology. However, to achieve the main goal of BioXSD and EMBRACE, namely interoperable programmatic access, we have opted for a combined approach of a pure XML Schema annotated by a data-type ontology using SAWSDL. This ensures the practical usability by presently mature common tools proven by an extensive industrial use, including the simple interoperability with programming languages using the ordinary SOAP libraries. For similar reasons, we do not model data annotations and resources using RDF (http://www.w3.org/standards/techs/rdf). RDF use will be considered for future versions of BioXSD after evaluating the progressed maturity and user-friendliness of the necessary frameworks. References to resources in BioXSD are modeled using pure XML-Schema elements, though in a formalized and at the same time versatile way. Strict formalization of the references enables traceability within the semantic web of data, using URI and preferably dereferenceable URL or REST-service links. Versatility of representation in BioXSD enables also formal references to resources that are volatile, do not offer stable URIs, or offer data only through a SOAP-service interface. This way, BioXSD supports formal links to any database, public or private, taxonomies and ontologies, allowing any cross-references and annotations by controlled vocabularies, for example the Sequence (Eilbeck et al., 2005), Gene (Ashburner et al., 2000) or BioSapiens Protein Feature Ontology (Reeves et al., 2008) or any future ontologies.
2.4 Ontology of bioinformatics data types
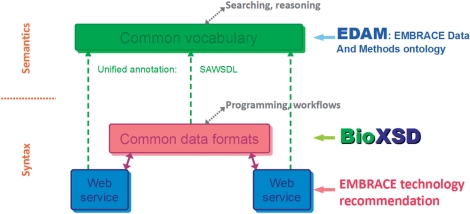
The ‘EMBRACE Data And Methods’ ontology (EDAM; Pettifer et al., 2010, http://edamontology.sourceforge.net), developed within the EMBRACE project, is a comprehensive controlled vocabulary of bioinformatics-specific data types, computational methods and data sources. BioXSD is closely coordinated with the EDAM initiative. BioXSD types and applicable local elements are annotated by EDAM terms using the SAWSDL standard. Model references to EDAM serve as formal semantics of the syntactic BioXSD types, in addition to the detailed, but only human-understandable semantics in the documentation. BioXSD thus constitutes a collection of ready-made, semantically annotated building blocks for the web-service interfaces. The architecture of the EMBRACE standards is shown in Figure 2. When EDAM is used to annotate multiple XML Schemas defining different formats, it can help in matching and translating, thus enabling interoperability across formats. In addition to data types, EDAM is used to annotate data and ontology resources and numerical scores within BioXSD, and is recommended to be used to annotate computational methods and other resources within the BioXSD-formatted data. Using SAWSDL, any BioXSD types or BioXSD-typed variables can be in the same way additionally annotated by any other model references to existing data-type ontologies or future models.
Fig. 2.
Strategy to reach maximum interoperability, as recommended by EMBRACE. Strategy comprises the technology for implementing web-service interfaces (WS-I compliant, document/literal wrapped SOAP); the common exchange format for basic bioinformatics data (BioXSD); the semantic annotation format (SAWSDL model reference); and the ontology of bioinformatics-specific computational methods, data types and resources (EDAM). The common data format (BioXSD) is essential for increasing interoperability within the programmatic access and for construction of workflows. The common vocabulary of meanings (EDAM) is essential when discovering and matching services, and doing additional semantic reasoning.
3 METHODS
BioXSD has been designed by analyzing the existing approaches, tools and data. Focused on existing web services and bioinformatics tools, we analyzed their inputs and outputs. Most important for interoperability, usability and maintenance is precise modeling of data that appears both as output and as input of tools. BioXSD models the common, everyday-bioinformatics data types, for which no standard exchange formats have been widely adopted. We have specified formats for biological sequences, sequence annotations and alignments and references to data and resources. These common data types cover inputs and outputs of about 2/3 of web services in the EMBRACE Registry (Pettifer et al., 2009). We analyzed legacy textual formats, modern tabular and XML formats, and related life-scientific ontologies. Some inconveniences of the existing solutions have been taken into account, keeping contact with bench molecular biologists and focusing on the present requirements of their research. These resulted for example in generalization of some data types, formalized handling of volatile resources and provenance and inclusion of structured optional metadata, including the possibility to annotate the metadata by externally controlled meaning (defined by any external taxonomy or ontology).
Technical requirements for BioXSD included WS-I compliance and interoperability with the existing frameworks: the ordinary SOAP and XML/XSD libraries. Compatibility with such libraries for the main programming languages is crucial for successful provider-side development of web services and for client-side usability. We have used a pure XML Schema with a constrained subset of its features, tested with a number of the most common SOAP and XML/XSD frameworks. BioXSD has been designed as a detailed XML Schema, allowing in-depth data validation and semantic annotation. At the same time, a requirement has been to limit the BioXSD to a lightweight to medium-heavy model, and in particular to ensure that its usage will be as easy as possible.
BioXSD has been developed in an iterative fashion, passing through a number of refactorings. Iterations resulted in prototypes and later beta-versions which were revised and tested by a group of service users and providers representing the stakeholders. Future versioning and maintenance process is designed as follows: a necessary major rearrangement that demands changes in the providers' and clients' software will constitute a major release of BioXSD. We hope for as rare major rearrangements as possible, but changes are unavoidable while keeping progress. A major release is identified by two major version numbers, and defines its own namespace and schema location: for example http://bioxsd.org/BioXSD-1.0 and http://bioxsd.org/BioXSD-1.0.xsd. This way, all the services that use the obsolete version will stay fully functional, while their providers will be informed about the new major release. More frequent, intermediate minor releases (updates) will be identified by a third version number (1.0.5) and will be designed not to break the interoperability of the canonical model, and thus not to demand any changes in the software for providers and clients.
To prove the feasibility of our effort, we have adapted a number of web services provided by different institutes, so that they use BioXSD for their inputs and outputs. These services have been developed in Python using the Zolera ZSI library, in Perl using SOAP::Lite and XML::Compile and in Java using Axis2 with XMLBeans data binding. We implemented a client worflow in Java to test the interoperability. As more services will adopt BioXSD, more example client workflows in more diverse programming or workflow languages will appear on the http://bioxsd.org web page. BioXSD is annotated with EDAM ontology terms using SAWSDL. Within the web-service and workflow implementations, we have tested the feasibility of user support and consulting.
4 RESULTS AND DISCUSSION
4.1 BioXSD 1.0
BioXSD 1.0 is the first stable version of the canonical data model for everyday bioinformatics. It defines exchange formats for the most common data types: biological sequences, sequence alignments, sequence annotation and references to data, resources and vocabularies. As supporting definitions, it offers a safe URI format, a recommended set of restricted numeric and string types, formats of the main accession numbers, as well as recommended qualitative values and identifying names, for example for the main public data sources (values for which stable, widely used vocabularies are still missing). In the following paragraphs, we briefly describe features of the most important BioXSD data formats.
4.1.1 Sequence
BioXSD includes type definitions for pure strings of one-letter-coded sequences, sequence records with additional metadata to identify and describe the sequence and a reference to a sequence in an external resource. The schema includes distinguished types for generic ‘biosequence’, nucleotide or amino-acid sequence, unambiguous or general. The optional metadata of a BioXSD sequence record are listed in Table 1. Figure 3A–D shows a diagram of NucleotideSequenceRecord, a defined restriction of GeneralAminoacidSequence and examples of BioXSD-formatted sequence records.
Table 1.
Optional elements for sequence metadata
| Element | Description |
|---|---|
| species | Identifies the biological source of the sequence: typically an organism, but possibly also a sample, tissue, cell line, individual, conditions or a geographic location. The generic species type may formally refer to any taxonomy and supports meta- and individual genomics data |
| customName | A name to identify the sequence for a human user |
| customNote | A textual note for a human user, if necessary. (not to be parsed) |
| formalReference | Identifies the data source of the sequence: typically a public or private database or data set. Can contain an accession, database identification, provenance data (version, date), isoform. Can also identify a position of the sequence in a super-sequence or a genome. May include an explicit super-sequence, necessary in special cases |
| translationData | Element to hold data for forward or backward translation, if necessary. May identify for example a genetic code and translational phase of an incomplete coding sequence |
Fig. 3.
BioXSD format of a sequence record including the sequence and optional metadata. (A) Diagram showing the structure of the sequence record (example type is specialized towards nucleotides). (B) Restricting pattern of the sequence string (example is a general amino-acid sequence type allowing ambiguous and additional residues: Pyl and Sec). (C) Example of a simple sequence record in BioXSD. (D) Example of a BioXSD sequence record with more metadata. Figure highlights which metadata elements are textual and focus purely on human understandability, and which are formally structured allowing more automatic usage by computer applications.
4.1.2 Sequence alignment
BioXSD offers a unified format for global and local, pair-wise and multiple alignments. It enables optimization in case of very long or very many sequences and supports frame shifts and direction. The aligned sequence records remain intact (no gap characters are inserted into the sequences), and can thus be directly used for further computation. This is also important for provenance, together with metadata identifying the methods used to construct the alignment. Any scores can be stated for the alignment and for each single aligned sequence.
4.1.3 Sequence annotation
The AnnotatedSequence is a versatile format for describing any kind of feature annotations of a biological sequence or genome. Nucleotide and protein sequences can be annotated with non-positioned and positioned features. Features can be spread over multiple segments of a sequence, and can be related to each other. Wrapping in blocks with local dependencies enables consistent merging of annotations. Features can contain a wide scale of formalized metadata, including cross-references and inter-feature relations with semantic meaning, sequence variation, alignment, experimental or predictive evidence with generic annotated scores, verdicts, literature citations and reliability. The format supports controlled ontological meanings of types of features, their relations with cross-referenced data and terms and of types of prediction tools and scores. Computational methods can additionally contain references to literature and web services. The BioXSD format for sequence annotations has been designed to fully cover the expressive power of GFF3 (http://www.sequenceontology.org/gff3.shtml) and DAS features. An additional expressiveness enables ‘loss-less’ exchange of for example UniProt features (The UniProt Consortium, 2010), data from specialized feature databases such as, for example, phiSITE (Klucar et al., 2010) or outputs of feature-prediction and similarity-search tools. Examples of diverse feature data modeled in BioXSD can be found at http://bioxsd.org, together with an extensive documentation of all defined types.
4.2 Existing implementations
A number of web services have been adapted to use the common BioXSD model for the formats of their inputs and outputs. Their implementation supplied iterations of BioXSD development with community feedback, and has proven the feasibility of the proposed solution. Experience has shown that as soon as services deal with a detailed, structured XML, it is easy to adapt the format. The first step of moving from plain text to structured XML, if necessary, is the one requiring considerable effort, but has to be done only once. Table 2 lists BioXSD-compatible web services by the time of writing this article. An updated list is maintained on the BioXSD web page.
Table 2.
BioXSD-compatible web services by the time of article submission
| Service | Provider | Function |
|---|---|---|
| BLAST | IBCP, France | Similarity search (Altschul et al., 1990) |
| ClustalW | IBCP, France | Multiple sequence alignment (Thompson et al., 1994) |
| GorIV | IBCP, France | Prediction of secondary structure of proteins (Garnier et al., 1996) |
| BLAST | BCCS, Norway | Similarity search |
| MaxAlign | CBS, Denmark | Optimization of multiple sequence alignment (Gouveia-Oliveira et al., 2007) |
| ProP | CBS, Denmark | Prediction of pro-peptide cleavage sites (Duckert et al., 2004) |
| NetNES | CBS, Denmark | Prediction of nuclear export signals (la Cour et al., 2004) |
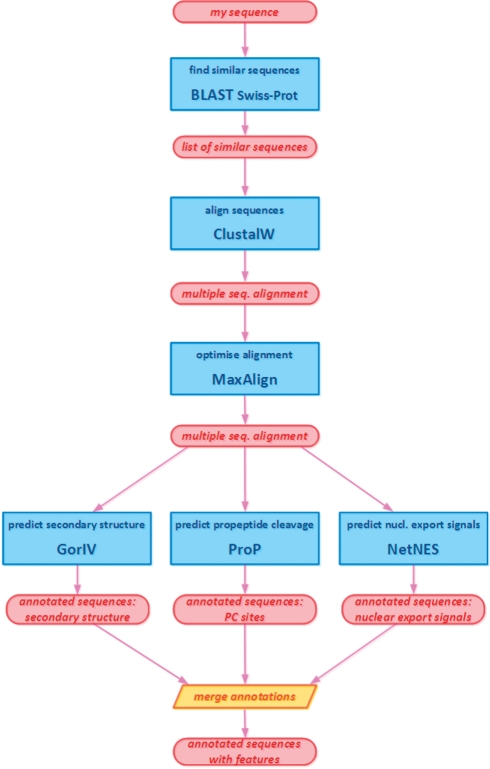
An example workflow, combining diverse web services from different providers, is schematically outlined in Figure 4. The workflow performs a routine bioinformatics analysis. Given a query sequence, similar sequences are searched and fetched from a database. These sequences are then brought together in a multiple sequence alignment. For each of the sequences, annotations with features of interest are predicted or fetched. Thanks to the common, detailed format of the exchanged data, the interoperability has highly improved resulting in smooth orchestration of the services in the workflow. Writing the source code of such a workflow in a common programming language has been markedly simplified. Another advantage when using BioXSD is the easiness with which a service in a workflow can be changed to a different service doing the same task (using a different algorithm, database or being hosted elsewhere). In the example, we can change the BLAST service for another BLAST or a different similarity search. The alignment service can be changed to any other BioXSD-compatible multiple-alignment tool. We can add any additional feature-prediction tools or data sources and merge the annotations without an effort. Changing or adding services that have the same programmatic interface in the form of BioXSD, requires only minimal changes in the script or workflow. Full working source code of the example workflow can be found among ‘example workflows’ on the BioXSD web page (http://bioxsd.org).
Fig. 4.
An example bioinformatics workflow (analysis pipeline). Blue rectangles are web-service calls, red ovals are data. Common, standardized BioXSD format of the exchanged data makes sure that there is no additional parsing and transforming of the data necessary between the service calls. Such web services are smoothly interoperable, allowing users to combine them without any substantial effort.
5 OUTLOOK
BioXSD is a candidate for the standard exchange formats of everyday bioinformatics data. It enables automatic validation and sustainably decreases the amount of necessary programming both on the side of service users and the service providers. Users of BioXSD-compatible web services gain most from the smooth interoperability, allowing them to combine services easily and to focus purely on the scientific aspects of their analysis. Providers can use BioXSD directly as the common format whenever applicable, and BioXSD types can be included, extended or restricted as building blocks of other custom or standard data types. With web services that use and will keep using other formats, BioXSD can serve as a canonical intermediate exchange format, into/from which other formats can be translated. The tool and database providers are encouraged to include BioXSD as one of the supported formats for input and output data.
Compared to the previous attempts of standardizing everyday bioinformatics formats for web-service data exchange, BioXSD offers more expressive power thanks to the extensive structured metadata including formal annotation by external biological ontologies throughout the format. It enables provenance, and modeling of data from emerging research fields like meta- and individual genomics. BioXSD itself is annotated by the EDAM ontology for discovery and interoperability of web services and data formats. Of highest importance for practical adoption within the community, BioXSD is compatible with the widely available, ordinary tools for programmatic access to web services.
The standardization of exchange-data format for basic bioinformatics data types is an initiative coming from within the scientific community. It can reach its goal of becoming the standard only with active participation of the community itself. BioXSD development has been, and should further be done, in form of an open but organized collaboration. We have been and are further trying to develop a model that is sufficiently expressive yet easy to use. BioXSD is welcoming new features and change requests from the community, for which a submission system has been designed. If the formats in their current form do not fit some providers or users, they are encouraged to submit their need for a change or addition. Changes and additions that do not demand rearrangement may be included in an instant update. Requests that lead to rearrangements of the model will be considered for the next major release.
To successfully establish the World Wide Web of bioinformatics services that use common exchange-data standard is desired by many projects. Highly important for reaching this goal is to offer service providers the necessary user support and consulting from the consortium responsible for maintenance of the standard. We offer full user support and consulting, and hope to establish a sustainable consortium which will guarantee the future maintenance of BioXSD.
ACKNOWLEDGEMENTS
We thank Kjell Petersen, Peter Fischer Hallin and Torbjørn Lium for knowledgeable advice and extensive technical support.
Funding: European Commission within FP6 (grant LHSG-CT-2004-512092, the EMBRACE project); Research Council of Norway (within eVITA) (grant 178885/V30, eSysbio project, to M.K., P.P., A.T., P.V., I.J.); l'Agence Nationale de la Recherche (grant ANR-06-CIS6-005, HIPCAL project, to A.J.); Villum Foundation (grant for the Center of Disease Systems Biology, to E.B. and K.R.).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augeri CJ. Proceedings of the 2007 Workshop on Experimental Computer Science. New York, USA: ACM; 2007. An analysis of XML compression efficiency. San Diego, CA. [Google Scholar]
- Duckert P, et al. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Eilbeck K, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, et al. GOR secondary structure prediction method version IV. Meth Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- Gouveia-Oliveira R, et al. MaxAlign: maximizing usable data in an alignment. BMC Bioinformatics. 2007;8:312. doi: 10.1186/1471-2105-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, et al. The HUPO PSI's Molecular Interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Hull D, et al. Proceedings of First Advanced Knowledge Technologies Workshop on Semantic Web Services (AKT-SWS04) KMi. Milton Keynes,UK: The Open University; 2004. Treating shimantic web syndrome with ontologies. [Google Scholar]
- Hull D, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucar L, et al. phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res. 2010;38:D366–D370. doi: 10.1093/nar/gkp911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottmann R, et al. A standard MIGS/MIMS compliant XML schema: toward the development of the Genomic Contextual Data Markup Language (GCDML) OMICS. 2008;12:115–121. doi: 10.1089/omi.2008.0A10. [DOI] [PubMed] [Google Scholar]
- la Cour T, et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Pettifer S, et al. An active registry for bioinformatics web services. Bioinformatics. 2009;25:2090–2091. doi: 10.1093/bioinformatics/btp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettifer S, et al. The EMBRACE Web service collection. Nucleic Acids Res. 2010;38:W683–W688. doi: 10.1093/nar/gkq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlić A, et al. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GA, et al. The Protein Feature Ontology: a tool for the unification of protein feature annotations. Bioinformatics. 2008;24:2767–2772. doi: 10.1093/bioinformatics/btn528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, et al. XML schemas for common bioinformatic data types and their application in workflow systems. BMC Bioinformatics. 2006;7:490. doi: 10.1186/1471-2105-7-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0046. research0046.1-0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. Creating a bioinformatics nation. Nature. 2002;417:119–120. doi: 10.1038/417119a. [DOI] [PubMed] [Google Scholar]
- Stockinger H, et al. Experience using web services for biological sequence analysis. Brief. Bioinform. 2008;9:493–505. doi: 10.1093/bib/bbn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, et al. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J, et al. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief. Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]