DNA Viruses: The Really Big Ones (Giruses) (original) (raw)
. Author manuscript; available in PMC: 2010 Oct 1.
Abstract
Viruses with genomes greater than 300 kb and up to 1200 kb are being discovered with increasing frequency. These large viruses (often called giruses) can encode up to 900 proteins and also many tRNAs. Consequently, these viruses have more protein-encoding genes than many bacteria, and the concept of small particle/small genome that once defined viruses is no longer valid. Giruses infect bacteria and animals although most of the recently discovered ones infect protists. Thus, genome gigantism is not restricted to a specific host or phylogenetic clade. To date, most of the giruses are associated with aqueous environments. Many of these large viruses (phycodnaviruses and Mimiviruses) probably have a common evolutionary ancestor with the poxviruses, iridoviruses, asfarviruses, ascoviruses, and a recently discovered Marseillevirus. One issue that is perhaps not appreciated by the microbiology community is that large viruses, even ones classified in the same family, can differ significantly in morphology, lifestyle, and genome structure. This review focuses on some of these differences rather than provides extensive details about individual viruses.
Keywords: algal virus, phycodnavirus, Mimivirus, White spot shrimp virus, jumbo phage, NCLDVs
Introduction
Typically, one views viruses as small particles that readily pass through 0.2-µm filters and contain small genomes with a few protein-encoding genes. However, huge viruses with large dsDNA genomes that encode hundreds of proteins, often called giruses, are now being discovered with increasing frequency. This review concentrates on viruses with genomes in excess of 300 kb and focuses on partially characterized viruses with annotated genomes (Table 1; annotated genomes in the public domain have an accession number). Most of these viruses inhabit aquatic environments and infect protists. Examples include Mimivirus, which infects amoebae and has the largest genome (~1.2 Mb); viruses that infect algae (phycodnaviruses) and have genomes up to ~560 kb; viruses that infect bacteria and have genomes up to ~670 kb; and White spot shrimp virus (WSSV), which has a genome of ~305 kb. At least one member of the poxvirus family has a genome larger than 300 kb (canarypox virus – 360 kb); however, most poxviruses have genomes ranging from 180 to 290 kb and therefore we have not discussed canarypox virus in this review. The polydnaviruses are enigmatic with respect to genes and particle structure, means of replication, and transmission (see sidebar, Polydnaviruses); these are not considered further. Other large, dsDNA-containing viruses have genomes ranging from 100 to 280 kb, including herpesviruses, asfarviruses, baculoviruses, iridoviruses, and ascoviruses, and also are not discussed in this review.
Table 1.
Giruses and their properties
| Classification | Genome | Virion | Host | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virusfamily | Type member | Accessionnumber(NCBI) orReferencenumber | Size(kb) | G+C% | %coding | Shape | PredictedCDS | tRNAgenes | Nucleocapsidsymmetry | Nucleocapsiddiameter(nm) | Lipids | Kingdom |
| Mimiviridae | _Acanthamoeba polyphaga_Mimivirus (APMV) | NC_006450 | 1181 | 27 | 86 | Circular | 911 | 1 | Isometric | 500 | Yes | Protozoa |
| Mamavirus | 34 | 1200 | 28 | N/A | Circular | N/A | N/A | Isometric | N/A | Yes | Protozoa | |
| Myoviridae | Pseudomonas chlororaphis phage 201φ2-1 | NC_010821 | 317 | 45 | 93 | Linear | 461 | 1 | Isometric | 122 | None | Proteobacteria |
| Bacillus megaterium phage G | 13 | 670 | N/A | N/A | N/A | N/A | N/A | Isometric | N/A | N/A | Firmicutes | |
| Nimaviridae | White spot shrimp virus 1 (WSSV1) | NC_003225 | 305 | 41 | 92 | Circular | 531 | None | Helical | N/A | Yes | Animalia |
| Phycodnaviridae | Chlorovirus PBCV-1 | NC_000852 | 331 | 39 | 90 | Linear | 366 | 11 | Isometric | 190 | Yes (I)a | Plantae |
| Chlorovirus NY2A | NC_009898 | 369 | 40 | 92 | Linear | 404 | 7 | Isometric | N/A | Yes (I) | Plantae | |
| Chlorovirus AR158 | NC_009899 | 345 | 40 | 92 | Linear | 360 | 6 | Isometric | N/A | Yes (I) | Plantae | |
| Chlorovirus FR483 | NC_008603 | 321 | 44 | 93 | Linear | 335 | 9 | Isometric | N/A | Yes (I) | Plantae | |
| Chlorovirus MT325 | DQ491001 | 321 | 45 | N/A | Linear | 331 | 10 | Isometric | N/A | Yes (I) | Plantae | |
| Ectocarpus siliculosus virus 1 (EsV-1) | NC_002687 | 336 | 51 | 70 | Circular | 240 | None | Isometric | N/A | N/A | Plantae | |
| Emiliania huxleyi virus 86 (EhV86) | NC_007346 | 407 | 40 | 90 | Circular | 472 | 6 | Isometric | N/A | N/A | Chromista | |
| _Chrysochromulina ericina_virus 1(CeV01) | 53 | 510 | N/A | N/A | N/A | N/A | N/A | Isometric | 160 | N/A | Chromista | |
| Pyramimonas orientalis virus 1 (PoV-01) | 53 | 560 | N/A | N/A | N/A | N/A | N/A | Isometric | 222×180 | N/A | Plantae | |
| Phaeocystis globosa virus 1 (PgV group 1) | 2 | 466 | 40–52 | N/A | Linear | N/A | N/A | Isometric | 150–190 | Yes (I) | Chromista | |
| Phaeocystis pouchetii virus 01 (PpV-01) | 69 | 485 | N/A | N/A | N/A | N/A | N/A | Isometric | 220 | Yes (I) | Chromista | |
| Cafeteria roenbergensis virus 1 (CroV 1) | M. Fischer,personalcommunication | 618 | 23 | 90 N/A | N/A | 543 N/A | 22 N/A | Isometric | 300 N/A | Yes (I)N/A | Chromista | |
| Heterocapsa circularisquama virus (HcDNAV) | 47 | 356 | N/A | N/A | N/A | N/A | N/A | Isometric | 197 | N/A | Protozoa | |
| Ectocarpus fasciculatus virus a (EfasV) | 28 | 340 | N/A | N/A | N/A | N/A | N/A | Isometric | N/A | N/A | Chromista | |
| Myriotrichia clavaeformis virus a (MclaV) | 28 | 340 | N/A | N/A | N/A | N/A | N/A | Isometric | N/A | N/A | Chromista | |
| Polydnaviridae | Cotesia congregata bracovirus (CcBv) | NC_006633b | 568 | 34 | 27 | Circularc | 156 | 7 | Helical multipartite | N/A | Yes | Animalia |
| Poxviridae | Canarypox virus (CNPV) | NC_005309 | 360 | 30 | 90 | Linear | 328 | None | Isometric | 160–190 | Yes | Animalia |
| N/A | Marseillevirus | 6 | 368 | 45 | 89 | Circular | 457 | N/A | Isometric | 250 | N/A | Protozoa |
To put the size of these large viral genomes into perspective, the smallest free-living bacterium, Mycoplasma genitalium, encodes 470 proteins (18). Although estimates of the minimum genome size required to support life are ~250 protein-encoding genes (45), a symbiotic bacterium (Candidatus Hodgkinia cicadicola) in the cicada Diciceroprocta semicincta has a 145-kb genome (37). Thus, giruses have more protein-encoding genes (CDSs) than some single-celled organisms.
Given the coding capacity of these large viruses, it is not surprising that they encode many proteins that are atypical of or novel for a virus. However, the majority of their CDSs do not match anything in the databases. Some of these viruses also encode introns and inteins, which are uncommon in viruses. A type IB intron exists in several phycodnaviruses (66). Mimivirus has six self-excising introns (9). Because introns are often detected when they interrupt coding sequences of known proteins, additional introns located within anonymous virus CDSs will probably be discovered. Inteins are protein-splicing domains encoded by mobile intervening sequences, and they catalyze their own excision from the host protein. Although found in all domains of life, their distribution is sporadic. Mimivirus and phycodnaviruses NY-2A and CeV01 are among the few intein-containing viruses.
The morphogenesis of large viruses is also interesting because presumably they are too large to self-assemble. Furthermore, structures of giruses vary significantly (Figure 1). Therefore, these viruses must encode proteins that measure size and other proteins that serve as assembly catalysts or scaffolds.
Figure 1.
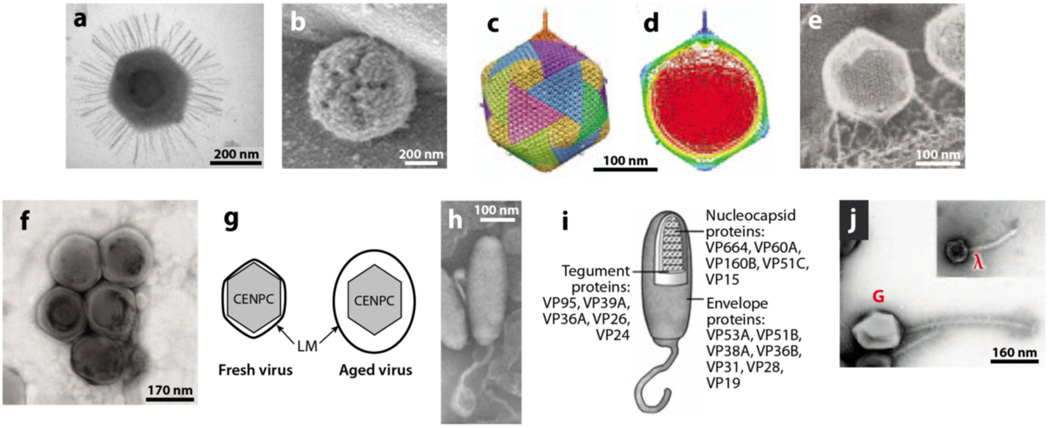
(a) Transmission electron micrograph of Mimivirus. (b) Atomic force microscopy of defibered Mimivirus. The unique star-faced vertex is clearly visible. (c) Fivefold averaged cryo-electron micrographs of virus PBCV-1 reveal a long, thin, cylindrical spike structure at one vertex and protrusions (fibers) extending from one unique capsomer per trisymmetron. (d) Central cross section of panel c. Note the gap between the unique vertex and the membrane enclosing the DNA. Also the unique vertex contains a portal-like protein. (e) PBCV-1 attached to the cell wall as viewed by the quick-freeze, deep-etch procedure. Note fibers attach the virus to the wall. ( f ) Transmission electron micrograph of EhV. (g) Schematic of freshly isolated EhV (left) and stored EhV (right). Note the external membrane swells with age. (h, i) Morphology of the White spot shrimp virus (WSSV) virion. (h) Negative contrast electron micrograph of intact WSSV virion with its tail-like extension. (i ) Schematic based on panel h showing the layered structures of a WSSV virion, i.e., envelope, tegument, and nucleocapsid. ( j ) Electron micrograph of bacteriophage G. The insert shows coliphage lambda to the same scale. Panel a from Reference 10, b from Reference 72, c and d from Reference 7, e from Reference 61, f and g from Reference 36, h and i from Reference 35, and j from Reference 24—all published with permission.
One issue that is perhaps not appreciated by the microbiology community is that large viruses, even ones classified in the same family, can differ significantly in morphology, lifestyle, and genome structure. This review focuses on some of these differences and only provides brief details about individual viruses. See noted comprehensive reviews for more information about specific viruses.
Discovery of Large Viruses
Most large viruses have been discovered and characterized in the last few years. Two exceptions are bacteriophage G, initially described about 40 years ago (13) and largely ignored until recently, and the chlorella virus Paramecium bursaria chlorella virus 1 (PBCV-1), which was first described in 1982 (62) and has since been studied continuously.
Large viruses went undetected for many years for several reasons. First, there were technical problems. For example, classical virus isolation procedures include filtration through 0.2-µm-pore filters to remove bacteria and protists. However, these filters also often exclude large viruses. Standard methods for plaquing bacteriophage missed large phages because the high soft agar concentrations prevented formation of visible plaques (54). In addition, large viruses may grow more slowly than smaller viruses and have lower burst sizes. Large viruses have larger surface areas and are thus more likely to aggregate and/or adsorb to extraneous material. None of these issues is a problem as long as one is aware of them. Second, the hosts for some of these large viruses were not examined for virus infections until recently. Finally, the discovery of some of these large viruses was serendipitous; e.g., Mimivirus was initially believed to be a parasitic bacterium (9).
It is now obvious that many large viruses await discovery. For example, Monier et al. (41) recently conducted a metagenomic study using samples collected on the Sorcerer II Global Ocean Sampling Expedition to determine the relative abundance of DNA polymerase fragments that could be assigned to virus groups. In 86% of sample sites, Mimivirus relatives were the most abundant, after bacteriophages. This high abundance suggests that Mimivirus-like viruses may infect other marine protists (11). In another metagenomics study using three other proteins as the queries, phycodnaviruses were commonly found (31).
Large DNA Virus Families
Many of the viruses listed in Table 1 probably have a common evolutionary ancestor, perhaps arising before the divergence of the major eukaryotic kingdoms (26, 51, 64, 71). These viruses, which include the phycodnaviruses, poxviruses, asfarviruses, iridoviruses, ascoviruses, and the Mimiviruses, are referred to as nucleocytoplasmic large dsDNA viruses (NCLDVs) (25, 26). Recently, another large NCLDV, named Marseillevirus, that is distantly related to the iridoviruses and ascoviruses was isolated from an amoeba (6). NCLDVs contain 9 common genes, and 177 additional genes are present in at least two of the virus families (71).
Although the hypothesis of a common ancestor for the NCLDVs is generally accepted, there is disagreement on the size and morphology of its ancestral virus and how it diverged into the different virus families. A recent maximum-likelihood reconstruction of NCLDV evolution produced a set of 47 conserved genes, which are considered the minimum genome for the common ancestor; NCLDVs then evolved by losing some of these common genes and acquiring new genes from their hosts and bacterial endosymbionts as well as by gene duplications (71). Another scenario suggests the ancestral NCLDV was a huge virus or even a cellular organism that evolved primarily via genome contraction (51). Finally, Filee et al. (15) proposed that NCLDVs evolved from a small DNA virus by gene acquisition from cells.
The origin of NCLDVs is controversial. For example, some researchers have suggested that NCLDVs should be considered the fourth kingdom of life (12, 51), others have suggested that NCLDV genes arose from the original gene pool that led to prokaryotes and eukaryotes (26), and still others have suggested that horizontal gene transfer has driven the evolution of their genomes (39). These suggestions have stimulated controversy over whether the tree of life should include these viruses.1 Another interesting hypothesis is that primitive NCLDVs gave rise to the eukaryotic nucleus or vice versa (5). Regardless, some viruses, including the NCLDVs, have a long evolutionary history, and viruses probably contributed to the emergence and subsequent structure of modern cellular life forms (29).
It is becoming difficult to classify some of these large viruses into distinct families. Recent phylogenetic analysis of the DNA polymerase protein from four putative phycodnaviruses illustrates this problem. (a) The DNA polymerase from three phycodnaviruses, CeV01, PpV01, and PoV01 (Table 1), is more similar to Mimivirus than to the other phycodnaviruses (41). (b) The HcDNAV polymerase indicates its closest relative is African swine fever virus (50). Therefore, it is clear that giruses, like the DNA phages (23), have exchanged genes for eons.
Contributing to the uncertainty about NCLDV evolution is the discovery that the structure of the PBCV-1 major capsid protein (MCP) resembles MCPs from other smaller dsDNA viruses with hosts in all three domains of life, including human adenoviruses, bacteriophage PRD1, and a virus infecting an archaeon, Sulfolobus solfataricus. This similarity suggests that these three viruses might have a common evolutionary ancestor with the NCLDVs, despite the lack of amino acid sequence similarity among their MCPs (32).
All NCLDVs are assembled in virus factories located in the cytoplasm. The role of the nucleus in the replication of NCLDVs varies. For example, poxviruses (43) and Mimivirus (46) carry out their entire life cycle in the cytoplasm. In contrast, the nucleus is probably essential for replication of the phycodnaviruses and other NCLDVs. However, the nuclear role in virus replication probably differs, even among the phycodnaviruses.
The four viruses in Table 1 that are not NCLDVs are a polydnavirus, WSSV, and two bacteriophages, PhageG and 201φ2-1. WSSV, which causes huge economic losses to the shrimp industry, is an enigma because it is not obviously related to known viruses. Large bacteriophages, referred to as jumbo phages, resemble smaller phages that may have acquired increased genome functions over evolutionary time.
Brief Descriptions of Some Large Viruses
Mimiviridae
Mimivirus, Mamavirus, and Marseillevirus all infect amoebae; the first two are the largest viruses ever reported (9). Mamavirus has an 18.3-kb DNA satellite virus (called Sputnik) that can only replicate in the presence of Mamavirus (34).
Mimivirus virions have an icosahedral core capsid with a diameter of ~500 nm. The capsid is uniformly covered with a 140-nm-thick layer of closely packed fibers, forming an ~750-nm spherical object (Figure 1_a_) (67). This peripheral fiber layer, which is absent in other NCLDVs, might be linked to the heterotrophic nature of the host amoeba; i.e., Mimivirus particles might mimic the bacteria on which amoebae prey. For amoebae to initiate phagocytosis, individual particles have to be larger than 600 nm in diameter or aggregate before being engulfed (30). The external fiber layer might also serve another role. After engulfing bacteria, bacterial surface lipopolysaccharides (LPSs) stimulate endocytic vesicle formation in amoebae. Therefore, the outer layer of Mimivirus particles, which stains gram positive (51), might resemble the gram-positive bacterial surface LPSs. Consistent with this hypothesis, Mimivirus encodes several sugar-manipulating enzymes and some of them are directly related to surface LPS-specific sugars, such as perosamine (9).
Electron tomography and volume-reconstruction analyses of viral particles inside infected amoeba cells at final infection stages establish that the Mimivirus capsid is composed of two superimposed shells with different densities (72). The inside of the virus particle has a membranous sac enveloping the viral genome. In addition to the two shells, a prominent fivefold star-shaped structure is located at one icosahedral vertex and extends along the entire length of the five icosahedral edges that center around this unique vertex (Figure 1_b_) (46, 56, 67, 72). Mimivirus initiates infection by phagocytosis followed by lysosome fusion with the phagosomes. This lysosomal activity is predicted to open the viral capsid at the stargate portal. The fusion of the particle’s internal membrane with the endocytic vacuole membrane forms a large membrane conduit through which the genome-containing Mimivirus core enters the cytoplasm. Although initial studies suggested the Mimivirus genome moved into the nucleus and shuttled back to the cytoplasm following a few rounds of replication (56), a more recent study indicates its genome remains in the cytoplasm and that, like the poxviruses, the entire replication cycle takes place in the cytoplasm (9, 46). Like poxviruses, Mimivirus possesses its own transcription machinery and it packages 12 transcription proteins in Mimivirus particles. Transcription of early Mimivirus genes, in conjunction with a conserved promoter element AAAATTGA, is believed to occur in the core particles. The cores release virus DNA, forming cytoplasmic replication factories where virus DNA replication begins, followed by transcription of late genes. The replication factories form around the viral cores and expand until they occupy a large fraction of the amoeba cell volume at 6 h post infection (p.i.).
Later stages of the Mimivirus replication cycle occur from 6 to 9 h p.i., when empty fiberless procapsids, which are only partially assembled, as well as icosahedral procapsids undergoing DNA packaging, appear at the periphery of the large replication factories. A statistical survey of particles undergoing DNA packaging indicated that 60% of the particles package DNA through a face-centered rather than a vertex-centered aperture. Thus, Mimivirus DNA exit and packaging proceeds through different portals, which is a unique feature among viruses (72).
The Mimivirus linear ~1.2-Mb genome encodes ~910 predicted CDSs and six tRNA genes. Despite its much larger genome, Mimivirus has a high coding density (90.5%) similar to that of other NCLDVs. Adjacent open reading frames (ORFs) are separated by an average of 157 bp. In contrast to some other NCLDV members, Mimivirus genomic termini lack large inverted repeats. Instead the Mimivirus genome has a 617-bp inverted repeat beginning at nucleotide position 22,515; its unique complementary counterpart begins at nucleotide position 1,180,529 (9). The extreme conservation of these intergenic regions suggests they serve an important role in Mimivirus replication. Pairing these regions produces a putative Q-like form in the genome, with a long (22,514 bp) and a short (259 bp) tail. The short tail region has no CDSs. The long tail region has a lower coding density than the rest of the genome (75% versus 90.5%) and larger intergenic spacers (435 bp versus 157 bp on average). This region encodes 12 proteins, 7 of which are involved in DNA replication.
Thirty-three percent of the Mimivirus genes are related to at least one other Mimivirus gene because of gene duplications (55). Thirty-six of the Mimivirus 910 CDSs are associated with functions not previously found in a virus (9). For instance, Mimivirus possesses a complete set of DNA repair enzymes capable of correcting nucleotide mismatches as well as errors induced by oxidation, UV irradiation, and alkylating agents. Mimivirus is also the only virus to encode three topoisomerases. In addition, Mimivirus encodes a variety of polysaccharide-, amino acid–, and lipid-manipulating enzymes. Such metabolic capabilities, although covering a broader biochemical spectrum in Mimivirus, also exist in other NCLDVs, especially the phycodnaviruses (66), where they often vary among isolates. Probably the most unexpected discovery in the Mimivirus genome was finding homologs to 10 translation-related proteins (9). Finally, Mimivirus, like the chlorella viruses, encodes several putative glycosyltransferases that might help glycosylate its MCP. Proteomic analysis of the Mimivirus virion identified 114 virus-encoded proteins, including the transcription proteins mentioned above.
Phycodnaviridae
Three viruses, PBCV-1, EhV, and EsV, are selected to represent the Phycodnaviridae family. These three viruses group into a single family (14, 66) and at first glance appear to be more similar than they actually are. As noted below, the apparent long evolutionary history of these three viruses has led to major differences in propagation strategies (lytic versus lysogenic), virus release (lytic versus budding), and virus structure (unique vertex with a spike versus probably no spike). The fact that these three viruses only have 14 common genes provides additional evidence of their long evolutionary history. Thus, over 1000 different genes exist just among these three phycodnaviruses!
Chlorella viruses
The chlorella viruses (genus Chlorovirus) infect symbiotic chlorella, often called zoochlorellae, which are associated with the protozoan Paramecium bursaria, the coelenterate Hydra viridis, and the heliozoon Acanthocystis turfacea (58, 68). Paramecium bursaria chlorella virus (PBCV-1) is the type member of the genus (61). The zoochlorellae are resistant to virus infection in the symbiotic state. Fortunately, some zoochlorellae can be grown independently of their hosts, permitting plaque assay of the viruses and synchronous infection of their hosts. Therefore, one can study the virus replication cycle in detail. The 46.2-Mb genome of the PBCV-1 host Chlorella NC64A was sequenced recently by the Department of Energy Joint Genome Institute and its genome annotation is publicly available. Availability of both host and virus sequences makes chlorella viruses a favorable model system.
Freshwater throughout the world contains chlorella viruses with titers as high as 100,000 plaque-forming units (PFUs) per milliliter of native water, although typically, virus titers are 1–100 PFU ml−1. The titers fluctuate during the year, with the highest titers occurring in the spring. Although chlorella viruses are ubiquitous in freshwater, little is known about their natural history. For example, do they have another host?
Cryo-electron microscopy and 3D image reconstruction of PBCV-1 indicate the outer capsid is icosahedral and covers a single lipid bilayered membrane, which is required for infection. The capsid shell consists of 1680 donut-shaped trimeric capsomers plus 12 pentameric capsomers, one at each icosahedral vertex. The trimeric capsomers are arranged into 20 triangular facets (trisymmetrons, each containing 66 trimers) and 12 pentagonal facets (pentasymmetrons, each containing 30 trimers and one pentamer at the icosahedral vertices) (Figure 1_c_). PBCV-1 has a triangulation number of 169d quasi-equivalent lattice (66).
Recent fivefold symmetry averaging 3D reconstruction experiments revealed that one of the PBCV-1 vertices has a cylindrical spike, 250 °A long and 50 °A wide (Figure 1_c_) (7). A pocket exists between the inside of the unique vertex and the enveloped nucleocapsid; i.e., the internal virus membrane departs from icosahedral symmetry adjacent to the unique vertex (Figure 1_d_). Consequently, the virus DNA located inside the envelope is packaged nonuniformly in the particle. The PBCV-1 MCP is a glycoprotein and comprises ~40% of the total virus protein. The MCP consists of two eight-stranded, antiparallel β-barrel jelly-roll domains related by a pseudo-sixfold rotation (48).
External fibers extend from some of the trisymmetron capsomers (probably one per trisymmetron) and may facilitate attachment to the host (Figure 1_c, e_). The spike at the unique vertex is too thin to deliver DNA and so it probably aids in penetration of the wall. PBCV-1 initiates infection by attaching rapidly and specifically to the Chlorella NC64A cell wall (58), probably by the fibers mentioned above (7). Following host cell wall degradation by virus-packaged enzyme(s), the viral internal membrane presumably fuses with the host membrane, facilitating entry of the viral DNA and virion-associated proteins into the cell, leaving an empty capsid attached to the surface (57). This fusion process initiates rapid depolarization of the host membrane and the rapid release of K+ from the cell. The rapid loss of K+ and associated water fluxes from the host reduce its turgor pressure, which may aid ejection of viral DNA and virion-associated proteins into the host. Depolarization may also prevent infection by a second virus (22).
PBCV-1 lacks a recognizable RNA polymerase gene, and so circumstantial evidence suggests PBCV-1 DNA and DNA-associated proteins quickly move to the nucleus, where early transcription begins 5 to 10 min p.i. (66). In this immediate-early phase of infection (5– 10 min p.i.), host transcription machinery is reprogrammed to transcribe viral DNA. Details of reprogramming are unknown, but host chromatin remodeling is probably involved. PBCV- 1 encodes a SET domain–containing protein (referred to as vSET) that methylates Lys-27 in histone 3. vSET is packaged in the PBCV- 1 virion, and circumstantial evidence indicates vSET helps to repress host transcription following PBCV-1 infection (44). In addition, host chromosomal DNA degradation begins within minutes after infection, presumably by PBCV- 1-encoded and packaged DNA restriction endonuclease(s) (1). This degradation also inhibits host transcription and facilitates recycling of nucleotides for viral DNA replication.
Viral DNA replication begins 60 to 90 min p.i. and is followed by transcription of late genes (58). Approximately 2 to 3 h p.i., assembly of virus capsids begins in localized regions in the cytoplasm, which become prominent 3 to 4 h p.i. Five to 6 h p.i. the cytoplasm fills with infectious progeny virus particles, and localized lysis of the host cell releases progeny at 6–8 h p.i. Each cell releases ~1000 particles, of which ~30% are infectious.
Global transcription of PBCV-1 genes during virus replication (70) indicate that (a) 98% of the 365 PBCV-1 protein-encoding genes are expressed in laboratory conditions, (b) 63% of the genes are expressed before 60 min p.i. (classified as early genes), (c)37% of the genes are expressed after 60 min p.i. (classified as late genes), and (d) 43% of the early gene transcripts are also detected at late times following infection (classified as early/late genes).
The PBCV-1 genome is a linear, ~334-kb, nonpermuted dsDNA molecule with covalently closed hairpin termini. Identical ~2.2-kb inverted repeats flank each hairpin end. The remainder of the PBCV-1 genome contains primarily single-copy DNA. Of the 365 predicted PBCV-1 CDSs, ~35% resemble proteins of known function, including many that are novel for a virus (e.g., hyaluronan synthase, K+ channel protein, and four polyamine biosynthetic enzymes). PBCV-1 CDSs are evenly distributed on both DNA strands with minimal intergenic spaces. Exceptions to this rule include a 1788-nucleotide sequence in the middle of the PBCV-1 genome that encodes 11 tRNAs [cotranscribed as a large precursor and then processed to mature tRNAs (68)].
Most chlorella virus genomes contain methylated bases. For example, genomes from 37 sampled chlorella viruses have 5- methylcytosine (5 mC) in amounts ranging from 0.12 to 47.5% of the total cytosine. In addition, 24 of the 37 viral DNAs contain N6- methyladenine (6 mA) in amounts ranging from 1.5 to 37% of the total adenine (61). The methylated bases occur in specific DNA sequences, which led to the discovery that the chlorella viruses encode multiple 5 mC and 6 mA DNA methyltransferases. About 25% of the virus-encoded DNA methyltransferases have companion DNA site-specific (restriction) endonucleases, including some with unique cleavage specificities (61).2
Five additional chlorella viruses have been sequenced, including two more viruses (NY-2A and AR158) that infect the same host as PBCV- 1, Chlorella NC64A; two (MT325 and FR483) that infect Chlorella Pbi; and one (ATCV-1) that infects Chlorella SAG 3.83 (66). Approximately 80% of the genes are common to all six sequenced chlorella viruses, suggesting they are essential for virus replication. However, the number of chlorella-virus-encoded genes is much larger than those present in any one virus. Not surprisingly, orthologs from viruses infecting the same host are the most similar; the average amino acid identity between orthologs from PBCV-1 and NY-2A or AR158 is ~73%. PBCV-1 and MT325 or FR483 orthologs have ~50% amino acid identity, and PBCV-1 and ATCV-1 orthologs have ~49% amino acid identity. Using PBCV-1 as a model, there is high synteny between the three viruses that infect Chlorella NC64A. In contrast, PBCV-1 has only slight synteny with the two Pbi viruses and the SAG virus (16).
Many PBCV-1-encoded enzymes are either the smallest or among the smallest proteins in their family. Phylogenetic analyses indicate some of these minimalist proteins are potential evolutionary precursors of more complex cellular proteins. Despite their small size, the virus enzymes typically have all the catalytic properties of larger enzymes. Their small size and the fact that they are often laboratory friendly have made them excellent models for mechanistic and structural studies (66).
The chloroviruses are also unusual because they encode enzymes involved in sugar metabolism. For example, two PBCV-1- encoded enzymes synthesize GDP-l-fucose from GDP-d-mannose (19), and three enzymes contribute to the synthesis of hyaluronan, a linear polysaccharide typically found in vertebrates (68). All three genes are transcribed early during PBCV-1 infection and hyaluronan accumulates on the external surface of the infected chlorella cells. In addition, PBCV-1 encodes at least five putative glycosyltransferases that likely participate in glycosylating the virus MCP (60).3
Emiliania huxleyi virus
The coccolithophore Emiliania huxleyi is a globally important unicellular marine phytoplankton. The alga forms huge blooms that extend over 100,000 km2 and it is important in ocean carbon and sulfur cycles, as well as influencing the climate (20). It is now generally accepted that the Emiliania huxleyi virus (EhV) contributes to the collapse of these blooms (66).
E. huxleyi has two phenotypes in its haplodiploid lifestyle. The diploid calcified phase forms algal blooms; this form is infected by EhV (genus Coccolithovirus). In contrast, the ecological status of the noncalcified haploid phase is largely unknown. However, haploid cells are resistant to EhV (17).
Currently, no detailed structural studies exist for the icosahedral EhV virion (Figure 1_f, g_). However, the initial assumption that it is structurally similar to PBCV-1 is probably incorrect because the EhV capsid is surrounded by an external lipid membrane and it infects its host by fusion with the host plasma membrane and enters by endocytosis (36). In contrast, PBCV-1 uncoats at the surface of the cell wall.
EhV has a different propagation strategy than either the lytic chlorella viruses or the latent EsV-1 viruses. The host alga E. huxleyi is covered with a calcium carbonate shell that would appear to create a physical barrier to virus adsorption. However, despite this barrier, virus adsorption to the host membrane is rapid and intrinsically linked to the host cell cycle (36). Real-time fluorescence microscopy revealed that EhV-86 rapidly enters its host intact via either an endocytotic or an envelope fusion mechanism where it rapidly disassembles.
Whereas both the chlorella viruses and EsV- 1 depend on host transcription machinery, EhV is unique among the phycodnaviruses because it has six RNA polymerase-encoding genes (65). These genes suggest some virus independence from the host nucleus. Viral transcription begins immediately after infection, but it is limited to a specific 100-kb region, containing ~150 CDSs, of the virus genome. This 100- kb region contains a unique promoter element, and only the genes transcribed during the first hour p.i. contain this element. Thus, these CDSs undoubtedly play a crucial and integral role early during virus infection. However, none of these CDSs matches anything in the databases.
Intriguingly, proteomic analysis did not detect any transcriptional proteins in mature EhV-86 virions; therefore, host nuclear RNA polymerase(s) is presumably responsible for early transcription (66). Between 1 and 2 h p.i., a second transcription phase begins with gene expression occurring from the remainder of the genome. Because viral RNA polymerase components are expressed in this second phase, viral replication may no longer be nuclear dependent and transcription may occur in the cytoplasm. At ~4.5 h p.i., virus progeny begin to be released via budding, during which EhV-86 virions become enveloped with host plasma membrane. Therefore, unlike chlorella viruses, for which nascent infectious virions accumulate in the cytoplasm prior to release by cell lysis, EhV virions are released gradually (36).
The EhV-86 407-kb genome, encoding 472 CDSs, was originally thought to be linear. PCR amplification over the termini revealed a random A/T single nucleotide overhang (50% A, 50% T), suggesting the virus genome has both linear and circular phases. EhV-86 has three repeat families (none of which is located at the ends of the genome); one repeat family is postulated to act as a replication origin (suggesting a circular form of DNA replication), another family is postulated to contain immediate-early promoter elements, and the last family has a large repetitive proline-rich domain that may bind calcium (66).
EhV-86 also has some unusual CDSs, including an entire metabolic pathway of seven genes encoding sphingolipid metabolic enzymes (65). The host also contains genes encoding this entire pathway and it is clear that horizontal gene transfer occurred between EhV and E. huxleyi (42). However, the direction of the transfer is not obvious. This biosynthetic pathway appears to function during lytic infection and the glycosphingolipids (GSLs) produced induce programmed cell death (PCD) with corresponding activation of an algal metacaspase, an essential activity for EhV-86 replication. Susceptible hosts accumulate both algal and viral derived GSLs that may coordinate virus maturation, whereas resistant cells accumulate only algal derived GSLs. The viral GSLs accumulate in the viral envelope, and it is hypothesized that this is a mechanism to activate virus release and subsequently induce PCD in surrounding algal cells during natural blooms as a type of quorum-sensing device, terminating the bloom (63). This example of cell signaling by the E. huxleyi/EhV interaction suggests that aquatic viruses are very much in control of their environment in ways virologists and ecologists are only just beginning to fathom.
Ectocarpus siliculosus virus
Ectocarpus siliculosus virus 1 (EsV-1) is the type species for the genus Phaeovirus and its infection strategy is regarded as typical for the genus (59, 66). Collectively, Phaeovirus members infect freeswimming, wall-less gametes or spores of filamentous marine brown macroalgae (order Ectocapales, class Phaeophyceae) by fusing with the host plasma membrane. Their hosts are members of benthic communities in near-shore coastal environments in all the world’s oceans. Phaeovirus DNAs are integrated into the host genome and are passed to daughter cells during cell division. The EsV-1 genome persists as a latent infection in vegetative cells, and infected algae show no obvious growth or developmental defects, except for partial or total inhibition of their reproductive organs. The viral genome is only expressed in sporangia and gametangia cells, where the cellular organelles disintegrate and are replaced with densely packed viral particles. Environmental stimuli, such as temperature and light, cause lysis of reproductive organs, synchronously releasing spores or gametes as well as viruses. Phaeoviruses are the only known phycodnaviruses to infect members of more than one algal family.
EsV-1 has a linear dsDNA genome with almost perfect inverted repeats at each end that allows circularization. Indeed, before sequencing, EsV-1 was thought to have a circular genome. The inverted repeats are proposed to anneal with each other to form a cruciform structure that effectively circularizes the genome. In addition to the terminal repeats, tandem repeats are located throughout the EsV-1 genome and comprise ~12% of the total genome size. The genome also contains several single-stranded regions randomly distributed over its length whose functions are unknown. Another characteristic of the EsV-1 genome is its low gene density, compared with the other phycodnaviruses. The 231 CDSs only occupy 70% of the EsV-1 genome; they are located in islands of densely packed genes that are separated by large regions of DNA repeats and noncoding sequences.
EsV-1 also encodes some unusual CDSs, including six putative hybrid histidine kinases (two-component systems that form part of a stimulus-responsive transduction pathway) that are widespread in archaea and bacteria. The relevance of these genes to EsV-1 infection is unknown.
Nimaviridae
The first reported appearance of WSSV occurred in 1992–1993 in shrimp farms in southern provinces of mainland China and also in northern Taiwan. The virus quickly spread to shrimp farming regions all over the world, including North and South America, Europe, and the Middle East. WSSV is lethal to most commercially cultivated penaeid shrimp species, causing serious economic damage. For example, an acute outbreak of white spot disease in cultured shrimp can result in 100% fatality in 3 to 10 days (35).
Unlike many viruses, WSSV infects a wide range of marine, brackish water, and freshwater crustaceans in addition to penaeid shrimp, including crayfishes, crabs, spiny lobsters, and hermit crabs. However, WSSV infection is usually not lethal to these other crustaceans; consequently, these other crustaceans may serve as virus reservoirs.
Structurally WSSV virions resemble baculoviruses and WSSV was initially classified as a baculovirus. However, WSSV has now been assigned to its own family called Nimaviridae (genus Whispovirus). WSSV virions are enveloped, cylindrical to elliptical in shape (Figure 1_h_). They measure 80–120 nm wide and 250–380 nm long. Some purified virions contain a 279- to 310-nm filamentous tail-like appendage at one end. The nucleocapsid has a segmented appearance, with ring-like segments running perpendicular to the longitudinal axis of the nucleocapsid (Figure 1). Each segment (or ring) is composed of two parallel rows of 12–14 globular subunits, each of which is approximately 8–10 nm in diameter. At least 40 structural proteins have been identified in the virus particles (35).
WSSV infection studies have been hampered by the lack of a cell culture system, although this may be changing (27). Currently, researchers agree that WSSV replicates and assembles in the nucleus, and in an acute infection, its life cycle is completed within 24 h. However, there are conflicting reports on the events associated with morphogenesis. One report suggests that nuclear protein is packaged into a partially enveloped empty capsid, whereas another report suggests that the electron-dense nucleocapsid is assembled first and then enveloped by a viral membrane (35).
The ~305-kb WSSV genome is circular. Most of the WSSV genome sequences are unique and only 3% of the genome consists of repetitive sequences. These repetitive sequences are organized into nine homologous regions containing 47 repeated mini-segments, which are distributed throughout the genome, mainly in intergenic regions.
Annotation of the WSSV genome identified 531 ORFs that consist of at least 60 codons (35). One hundred and eighty-one ORFs are non-overlapping and are classified as CDSs. About 80% of these CDSs have a potential 3′- polyadenylation site (AATAAA). The sizes of the proteins encoded by these CDSs range from 60 to 6077 amino acids. The 6077-amino-acid CDS encodes the extraordinarily large MCP. Only 45 of the CDSs resemble known proteins (>20% amino acid identity) or contain recognizable motifs. Twenty-seven CDSs are classified into 10 WSSV gene families; these families probably arose from gene duplications.
The few WSSV identifiable CDSs primarily encode gene products involved in nucleotide metabolism (35). Surprisingly, only one WSSV CDS, a DNA polymerase, is related to DNA replication. WSSV also encodes a collagen-like protein, which is the first collagen gene to be identified in a virus genome.
Bacteriophage
Large dsDNA bacteriophages are being discovered with increasing frequency (33). However, when this review was written only two phages, 670-kb Phage G and 317-kb 201φ2- 1, had genomes larger than 300 kb (Table 1). Both viruses are members of the tailed family Myoviridae. PhageG infects Bacillus megaterium and phage 201φ2-1 infects Pseudomonas chlororaphis. Although both genomes have been sequenced, annotation of only 201φ2-1 is in the public domain.
The majority of the proteins predicted from the genome sequences of these phages have no database matches, and the genomes themselves are diverse enough to preclude the detailed comparative analysis that has occurred with smaller phages, for which hundreds of genome sequences are available. However, one can extrapolate the better-known genome organizations and mechanisms of evolution seen in the smaller phages to the jumbo phages. Typically, larger phages contain the same core genes (structural and DNA replication genes), plus many additional, generally smaller genes that do not match anything in the databases and can usually be deleted without affecting replication. It is possible that the jumbo phages evolved from smaller-tailed phages, possibly in a process mediated by constraints on genome size by capsid size (24).4
The phage G genome sequence is 498 kb, but the chromosome is ~670 kb. This means terminal redundancy is about 35%. Phage G is predicted to have 682 CDSs and about 10% of these are families of paralogs. Phage G, like some other large viruses, encodes several translation system components, e.g., 17 tRNAs covering 14 codon specificities and a homolog of a serine aminoacyl-tRNA synthetase. The phage 201φ2-1 genome is also circularly permuted and terminally redundant and is predicted to encode 468 CDSs.
Concluding Remarks
Although giruses are probably ancient, they are relatively new to virologists. Even with our limited knowledge, research efforts on large viruses are contributing scientific and economic benefits. For example, chlorella viruses, which encode as many as 400 CDSs, are sources of new and unexpected genes. The genes not only encode commercially important enzymes such as DNA restriction endonucleases, but many viral proteins are the smallest in their class. Consequently, these proteins serve as biochemical models for mechanistic and structural studies (21). The viruses are also a source of genetic elements for genetically engineering other organisms. Examples include (a) promoter elements from chlorella viruses that function well in both monocots and dicots of higher plants, as well as bacteria (38); and (b) a translational enhancer element from a chlorella virus that functions well in Arabidopsis (49).
The hosts for some of these viruses either have been sequenced recently or are in the process of being sequenced. Annotation of these sequences will certainly contribute to studies on giruses. However, one obstacle to studying these viruses is that currently none of the eukaryotic viruses described in this review can be genetically modified by molecular techniques. The development of successful and reproducible host transformation procedures should lead to the genetic analysis of these viruses, which would be a major achievement.
It is obvious that the discovery and characterization of giruses are in their infancy and that many more interesting and unusual members await discovery. For example, metagenomic studies on environmental microbial DNA sequences collected in the Sargasso Sea revealed many homologs of Mimivirus genes. Thus, many Mimivirus relatives certainly exist in nature, some of which probably infect novel protists. Classifying these newly discovered large viruses will be complicated because of horizontal gene swapping.
The origin of giruses is controversial. One interesting suggestion is that amoebae, which harbor many diverse microorganisms, such as viruses, are melting pots for gene mixing, leading to new viruses, including large viruses with complex gene repertoires of various origins (6).
Summary Points
- Really big dsDNA viruses (giruses), with genomes up to 1.2 Mb in size, are rapidly being discovered.
- Giruses infect a variety of hosts, bacteria, protists, and animals. Thus, their sizes are not restricted to a specific host or phylogenetic clade.
- Giruses are much more diverse than might be expected. For example, they have diverse capsid structures, lifestyles, and genome structures.
- Giruses that infect the same hosts and are members of the same family, e.g., phycodnaviruses, can differ significantly.
- Many giruses are evolutionarily old, possibly going back to the time prokaryotes and eukaryotes diverged.
- The majority of the predicted CDSs in giruses do not match anything in gene databases, indicating these viruses are a rich source of novel biochemical functions yet to be discovered.
Acknowledgments
Research in the Van Etten laboratory was supported in part by Public Health Service grant GM32441 and NIH grant P21RR15635 from the COBRE program of the National Center for Research Resources. We thank Michele Malchow for help with the figures.
Glossary
WSSV
White spot shrimp virus
CDS
protein-encoding gene
Polydnaviruses
Polydnaviruses that infect thousands of species of endoparasitic wasps have complex lifestyles and large, multipartite genomes. Two genera (Bracovirus and Ichnovirus) represent this family, and these viruses manipulate the defenses, development, and physiology of the parasitized lepidopteran larval hosts, where the virus facilitates a symbiotic or mutualistic condition of the wasp larvae with an otherwise resistant lepidopteran host. The viruses are evolutionarily linked to the family Baculoviridae (3). The life cycle of polydnaviruses life cycle may be the most complex known in virology. Upon infection the virus integrates into the genome of specialized cells of the wasp’s ovaries and is carried in the parasitic eggs and larvae as a provirus, as well as circular episomal DNA within virions. In both wasp and lepidopteran cells, the virus has a closed circular DNA that replicates in the nucleus. In the case of bracoviruses, replication and particle production occur only in the ovaries of the wasp and the virus is transmitted vertically, yet the virions contain no bracovirus structural proteins. Rather, the structural components are derived from a baculovirus. Another distinctive feature of the polydnaviruses is their low coding density (~27%) compared to other giruses, which are typically ~90%. In addition, their protein-encoding genes have little relationship to free replicating viruses. Thus, polydnaviruses blur the distinction of viruses as obligate parasites and challenge virologists’ understanding of the role of viruses in nature, where a virus appears to be domesticated by a cellular organism for the sake of obligate mutualism (4).
PBCV-1
Paramecium bursaria chlorella virus 1
NCLDV
nuclear cytoplasmic large DNA virus
MCP
major capsid protein
LPS
lipopolysaccharide
p.i.
post infection
ORF
open reading frame
PFU
plaque forming unit
5 mC
5-methylcytosine
6 mA
N6-methyladenine
GSL
glycosphingolipids
PCD
programmed cell death
Footnotes
1
For example, References 8, 52, 40, and comments by seven others in Nat. Rev. Microbiol. 2009. 7:614–27 relate to this discussion.
2
The chlorella viruses were the first nonbacterial source of DNA restriction endonucleases.
3
PBCV-1 was the first virus reported to encode most, if not all, of the machinery to glycosylate its MCP.
4
In the construction of the virion of all the tailed phages, an empty protein capsid is assembled first and then DNA is pumped into the capsid, presumably by a head-full packaging mechanism. This puts an upper size limit on the genome, and in fact the DNA is usually packed as tightly as physically possible. This agrees with the circularly permuted and terminally redundant structure of the phage DNAs.
Literature Cited
- 1.Agarkova IV, Dunigan DD, Van Etten JL. Virion-associated restriction endonucleases of chloroviruses. J. Virol. 2006;80:8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoux AC, Brussaard CP. Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology. 2005;341:80–90. doi: 10.1016/j.virol.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Bézier A, Annaheim M, Herbinière J, Wetterwald C, Gyapay G, et al. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science. 2009;323:926–930. doi: 10.1126/science.1166788. [DOI] [PubMed] [Google Scholar]
- 4.Bézier A, Herbinière J, Lanzrein B, Drezen JM. Polydnavirus hidden face: The genes producing virus particles of parasitic wasps. J. Invertebr. Pathol. 2009;101:194–203. doi: 10.1016/j.jip.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Bell PJL. Sex and the eukaryotic cell cycles is consistent with a viral ancestry for the eukaryotic nucleus. J. Theoret. Biol. 2006;243:54–63. doi: 10.1016/j.jtbi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, et al. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. Describes a spike structure at a unique vertex in a girus.
- 8.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claverie JM, Abergel C. Mimivirus and its virophage. Annu. Rev. Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 10.Claverie JM, Abergel C, Ogata H. Mimivirus. 2009:89–121. doi: 10.1007/978-3-540-68618-7_3. See Ref. 58a. [DOI] [PubMed] [Google Scholar]
- 11.Claverie JM, Grzela R, Lartigue A, Bernadac A, Nitsche S, et al. Mimivirus and Mimiviridae: Giant viruses with an increasing number of potential hosts, including corals and sponges. J. Invertebr. Pathol. 2009;101:172–180. doi: 10.1016/j.jip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Donelli G, Dore E, Frontali C, Grandolfo ME. Structure and physico-chemical properties of bacteriophage G. III. A homogeneous DNA of molecular weight 5 times 10(8) J. Mol. Biol. 1975;94:555–565. doi: 10.1016/0022-2836(75)90321-6. [DOI] [PubMed] [Google Scholar]
- 14.Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: A peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. Indicates that there are more than 1000 unique genes among three phycodnaviruses.
- 15.Filee J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol. Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald LA, Boucher PT, Yanai-Balser G, Suhre K, Graves MV, Van Etten JL. Putative gene promoter sequences in the chlorella viruses. Virology. 2008;380:388–393. doi: 10.1016/j.virol.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. USA. 2008;105:15944–15949. doi: 10.1073/pnas.0807707105. Establishes that the diploid phase of E. huxleyi is infected by virus EhV, whereas the haploid phase is resistant.
- 18.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 19.Fruscione F, Sturla L, Duncan G, Van Etten JL, Valbuzzi P, et al. Differential role of NADP+ and NADPH in the activity and structure of GDP-D-mannose 4,6-dehydratase from two chlorella viruses. J. Biol. Chem. 2008;283:184–193. doi: 10.1074/jbc.M706614200. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman JA. Microbial community structure and its functional implications. Nature. 1999;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 21.Gazzarrini S, Kang M, Van Etten JL, Tayefeh S, Kast SM, et al. Long distance interactions within the potassium channel pore are revealed by molecular diversity of viral proteins. J. Biol. Chem. 2004;279:28443–28449. doi: 10.1074/jbc.M401184200. First demonstration that orthologs from the chlorella viruses can provide important information on protein structure/mechanistic studies.
- 22.Greiner T, Frohns F, Kang M, Van Etten JL, Kaesmann A, et al. Chlorella viruses prevent multiple infections by depolarizing the host membrane. J. Gen. Virol. 2009;90:2033–2039. doi: 10.1099/vir.0.010629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix RW. Bacteriophage genomics. Curr. Opin. Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix RW. Jumbo bacteriophages. 2009:229–240. doi: 10.1007/978-3-540-68618-7_7. See Ref. 58a. [DOI] [PubMed] [Google Scholar]
- 25.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. Establishes the phylogenetic relationship between the Poxviridae, Iridoviridae, Asfarviridae, and Phycodnaviridae and introduced the term nucleocytoplasmic large DNA viruses.
- 26.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Jiravanichpaisal P, Söderhäll K, Söderhäll I. Characterization of White spot syndrome virus replication in in vitro-cultured haematopoietic stem cells of freshwater crayfish, Pacifastacus leniusculus. J. Gen. Virol. 2006;87:847–854. doi: 10.1099/vir.0.81758-0. [DOI] [PubMed] [Google Scholar]
- 28.Kapp M. Viruses infecting marine brown algae. Virus Genes. 1998;16:111–117. doi: 10.1023/a:1007962112756. [DOI] [PubMed] [Google Scholar]
- 29.Koonin EV, Senkevich TG, Dolga VV. The ancient virus world and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korn ED, Weisman RA. Phagocytosis of latex beads by Acanthamoeba. II. Electron microscopic study of the initial events. J. Cell Biol. 1967;34:219–227. doi: 10.1083/jcb.34.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristensen DM, Muchegian AR, Dolga VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krupovic M, Bamford D. Virus evolution: How far does the double β-barrel viral lineage extend? Nat. Rev. Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 33.Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann HW. “PhiKZ-like viruses,” a proposed new genus of myovirus bacteriophages. Arch. Virol. 2007;152:1955–1959. doi: 10.1007/s00705-007-1037-7. [DOI] [PubMed] [Google Scholar]
- 34.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 35.Leu JH, Yang F, Zhang X, Xu X, Kou GH, Lo CF. Whispovirus. See Ref. 58a. 2009:197–227. doi: 10.1007/978-3-540-68618-7_6. [DOI] [PubMed] [Google Scholar]
- 36.Mackinder LC, Worthy CA, Biggi G, Hall M, Ryan KP, et al. A unicellular algal virus, Emiliania huxleyi virus 86, exploits an animal-like infection strategy. J. Gen. Virol. 2009;90:2306–2316. doi: 10.1099/vir.0.011635-0. [DOI] [PubMed] [Google Scholar]
- 37.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra A, Higgins DW, Rohe NJ. A chlorella virus gene promoter functions as a strong promoter both in plants and bacteria. Biochem. Biophys. Res. Comm. 1994;204:187–194. doi: 10.1006/bbrc.1994.2443. [DOI] [PubMed] [Google Scholar]
- 39.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: The multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreira D, López-García P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 41.Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. Marine Mimivirus relatives are probably large algal viruses. Virol. J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monier A, Pagarete A, de Vargas C, Allen MJ, Read B, et al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19:1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2945. [Google Scholar]
- 44.Mujtaba S, Manzur KL, Gurnon JR, Kang M, Van Etten JL, Kang MM. Epigenetic transcription repression of cellular genes by a viral SET protein. Nat. Cell Biol. 2008;10:1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mutsafi Y, Zauberman N, Sabanay I, Minsky A. Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl. Acad. Sci. USA. 2010;107:5978–5982. doi: 10.1073/pnas.0912737107. Demonstrates that, like poxviruses, the entire replication cycle of Mimivirus occurs in the cytoplasm.
- 47.Nagasaki K. Dinoflagellates, diatoms, and their viruses. J. Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- 48.Nandhagopal N, Simpson A, Gurnon JR, Yan X, Baker TS, et al. The structure and evolution of the major capsid protein of a large, lipid-containing, DNA virus. Proc. Natl. Acad. Sci. USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen PS, Falcone DL, Graves MV. The A312L 5′-UTR of chlorella virus PBCV-1 is a translational enhancer in Arabidopsis thaliana. Virus Res. 2009;140:138–146. doi: 10.1016/j.virusres.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Ogata H, Toyoda K, Tomaru Y, Nakayama N, Shirai Y, et al. Remarkable sequence similarity between the dinoflagellate-infecting marine girus and the terrestrial pathogen African swine fever virus. Virol. J. 2009;6:178. doi: 10.1186/1743-422X-6-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raoult D, Audic S, Robert C, Abergel C, Regesto P, et al. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. Gives the complete genome sequence analysis of Mimivirus and the evolutionary implications of its large gene content.
- 52.Raoult D, Forterre P. Redefining viruses: Lessons from Mimivirus. Nat. Rev. Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 53.Sandaa RA, Heldal M, Castberg T, Thyrhaug R, Bratbak G. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae) Virology. 2001;290:272–280. doi: 10.1006/viro.2001.1161. [DOI] [PubMed] [Google Scholar]
- 54.Serwer P, Hayes SJ, Thomas JA, Hardies SC. Propagating the missing bacteriophages: A large bacteriophage in a new class. Virol. J. 2007;4:21. doi: 10.1186/1743-422X-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J. Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thiel G, Moroni A, Dunigan D, Van Etten JL. Initial events associated with virus PBCV-1 infection of Chlorella NC64A. In: Lüttge U, Beyschlag W, Büdel B, editors. Progress in Botany. Berlin: Springer-Verlag; 2010. pp. 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Etten JL. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 58a.Van Etten J, editor. Lesser Known Large dsDNA Viruses. Berlin: Springer; 2009. [PubMed] [Google Scholar]
- 59.Van Etten JL, Graves MV, Muller DG, Boland W, Delaroque N. Phycodnaviridae —large DNA algal viruses. Arch. Virol. 2002;147:1479–1516. doi: 10.1007/s00705-002-0822-6. [DOI] [PubMed] [Google Scholar]
- 60.Van Etten JL, Gurnon J, Yanai-Balser G, Dunigan D, Graves MV. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta. 2010;1800:152–159. doi: 10.1016/j.bbagen.2009.07.024. Summarizes the evidence that, unlike all other viruses, the chlorella viruses encode most if not all of the machinery to glycosylate their major capsid protein.
- 61.Van Etten JL, Lane LC, Meints RH. Viruses and virus-like particles of eukaryotic algae. Microbiol. Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Etten JL, Meints RH, Kuczmarski D, Burbank DE, Lee K. Viruses of symbiotic chlorella-like algae isolated from Paramecium bursaria and Hydra viridis. Proc. Natl. Acad. Sci. USA. 1982;79:3867–3871. doi: 10.1073/pnas.79.12.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326:861–865. doi: 10.1126/science.1177322. [DOI] [PubMed] [Google Scholar]
- 64.Villarreal LP. Viruses and the Evolution of Life. Washington, DC: Am. Soc. Microbiol. Press; 2005. p. 395. [Google Scholar]
- 65.Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 66.Wilson WH, Van Etten JL, Allen MJ. The Phycodnaviridae: The story of how tiny giants rule the world. See Ref. 58a. 2009:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:958–966. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv. Virus Res. 2006;66:293–366. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan X, Chipman PR, Castberg T, Bratbak G, Baker TS. The marine algal virus PpV01 has an icosahedral capsid with T = 219 quasisymmetry. J. Virol. 2005;79:9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanai-Balser GM, Duncan GA, Eudy JD, Wang D, Li X, Agarkova IV, et al. Microarray analysis of Paramecium bursaria chlorella virus 1 transcription. J. Virol. 2010;84:532–542. doi: 10.1128/JVI.01698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: Clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. Plos Biol. 2008;6:1104–1114. doi: 10.1371/journal.pbio.0060114. Provides an ultrastructural study of Mimivirus virions describing a new DNA delivery system (stargate) distinct from the DNA packaging site.