Role of Methionine Sulfoxide Reductases A and B of Enterococcus faecalis in Oxidative Stress and Virulence (original) (raw)
Abstract
Methionine sulfoxide reductases A and B are antioxidant repair enzymes that reduce the _S_- and _R_-diastereomers of methionine sulfoxides back to methionine, respectively. Enterococcus faecalis, an important nosocomial pathogen, has one msrA gene and one msrB gene situated in different parts of the chromosome. Promoters have been mapped and mutants have been constructed in two E. faecalis strains (strains JH2-2 and V583) and characterized. For both backgrounds, the mutants are more sensitive than the wild-type parents to exposure to H2O2, and in combination the mutations seem to be additive. The virulence of the mutants has been analyzed in four different models. Survival of the mutants inside mouse peritoneal macrophages stimulated with recombinant gamma interferon plus lipopolysaccharide but not in naïve phagocytes is significantly affected. The msrA mutant is attenuated in the Galleria mellonella insect model. Deficiency in either Msr enzyme reduced the level of virulence in a systemic and urinary tract infection model. Virulence was reconstituted in the complemented strains. The combined results show that Msr repair enzymes are important for the oxidative stress response, macrophage survival, and persistent infection with E. faecalis.
Enterococci are rather harmless commensals of the human biliary and gastrointestinal tract which belong to the group of lactic acid bacteria. Even probiotic effects are claimed for some strains. On the other hand, enterococci have emerged as important nosocomial pathogens causing wound, bloodstream, and urinary tract infections. Due to their general robustness and intrinsic and acquired resistance to antibiotics, enterococci are well equipped to survive and colonize hospital environments. Upon room colonization, patients with low levels of immunity may become colonized by these hospital strains, and it is likely that blood infections are the result of dissemination from the intestine (10).
Concerning enterococcal virulence, a few virulence factors have been characterized (see reference 13 and references therein). Esp, Ace, and pilus-like structures are thought to mediate attachment to host tissues. Aggregation substance promotes aggregation between cells, and altogether, this leads to the formation of a biofilm. Quorum sensing-based mechanisms then lead to expression of the metalloprotease gelatinase and the hemolytic cytolysin, provoking cell lysis and the spread of infection. However, the importance of these virulence factors does not always seem to be supported by the findings of clinical studies, demonstrating that we still have a lack of knowledge concerning the virulence of enterococci.
Phagocytic cells are among the first line of defense against invading pathogens. After phagocytosis, the phagocytes initiate a high-output production of reactive oxygen intermediates (ROIs) and reactive nitrogen intermediates (RNIs) with the aim to destroy the pathogens. The primary target is thought to be DNA, but ROIs and RNIs also react with other macromolecules, including proteins. Pathogens respond to this attack by synthesizing molecules with antioxidant activities, such as superoxide dismutases, catalases, and peroxidases, in order to neutralize these radicals, but they have also evolved systems which repair oxidative damage. The methionine (Met) residues of proteins are particularly vulnerable to oxidation, forming methionine sulfoxide (MetSO). The oxidation of Met residues actually leads to the formation of equimolar amounts of two epimers of MetSO, Met-_S_-SO and Met-_R_-SO (reaction 1): methionine + ROI (RNI) → Met-_S_-SO + Met-_R_-SO.
These MetSO residues can be reduced by methionine sulfoxide reductases (Msr), evolutionarily highly conserved enzymes able to reduce MetSO back to Met using electrons derived from thioredoxin, thioredoxin reductase, and NADPH (6, 7). Two nonhomologous Msr enzymes, named MsrA and MsrB, have been identified, and as has been shown with Msr proteins of different Gram-positive and Gram-negative bacteria, the two diastereoisomers are specifically reduced by MsrA and MsrB enzymes, according to reactions 2A and 2B (6, 20): Met-_S_-SO + MsrA → methionine (reaction 2A) and Met-_R_-SO + MsrA → methionine (reaction 2B).
The chemical mechanism involved in the reduction of MetSO to Met catalyzed by the Msr enzymes is the same and can be divided into three steps: (i) formation of a sulfenic acid intermediate on a catalytic cysteine (Cys) residue of the enzymes, leading to the release of methionine, (ii) formation of an intramonomeric disulfide bond between the catalytic Cys and a recycling Cys, and (iii) reduction of the resulting disulfide bond by the NADPH/thioredoxin reductase/thioredoxin system (6). Since msrA and msrB rank among the best-conserved genes in nature, it is suggested that their products have very important functions for cellular life, most impressively demonstrated by the finding that Msr deficiency affects longevity in yeast and mammals (15, 22, 23, 25). With one exception (39), the bacterial msrA mutants tested were more sensitive to exposure to oxidants, such as H2O2, organic hydroperoxides, and/or nitric oxide (26, 30). Hitherto, only a few studies have addressed the role of MsrB in bacterial resistance to oxidative stress. While a Helicobacter pylori msrB mutant was more sensitive to three different oxidants, this mutation had no effect on oxidative stress resistance in Mycobacterium tuberculosis (1, 20). Besides that, the copy number and genetic organization of msr genes vary widely among the different organisms. The majority of microorganisms contain one copy of each gene in two different transcription units (12, 30). In other bacteria, the msrA and msrB genes are part of the same operon, and the two genes are sometimes translationally fused. Some bacteria have more than one msrA and/or msrB paralog, which are most frequently present on the chromosome but which are also sometimes present on a plasmid (12, 30).
Convincing data indicating a close relationship between Msr and bacterial virulence has accumulated in recent years. For different pathogenic bacteria, the reduced virulence of _msr_-deficient strains as well as effects on bacterial adherence, biofilm formation, intracellular survival, and colonization capabilities has been reported (4, 9, 37). In the present work we address the question of whether the msrA and msrB genes of Enterococcus faecalis are important in the oxidative stress response and virulence of this opportunistic pathogen.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. Cultures of E. faecalis strains were performed on M17 medium (33) supplemented with 0.5% (wt/vol) glucose (GM17). Overnight cultures of the E. faecalis strains were grown at 37°C without shaking in 30-ml glass tubes containing 10 ml GM17. When appropriate, erythromycin (150 μg ml−1) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 80 μg ml−1) were added to the medium. Escherichia coli strains were cultivated under vigorous agitation at 37°C in Luria-Bertani (LB) medium (29) with ampicillin, when it was required. Growth was followed by measuring the optical density at 600 nm (OD600) on a Biophotometer (Eppendorf).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. faecalis strains | ||
| JH2-2 | Fusr Rifr, plasmid-free wild-type strain | 38 |
| JHD_msrA_ | JH2-2 isogenic msrA deletion mutant | This study |
| JHD_msrB_ | JH2-2 isogenic msrB deletion mutant | This study |
| JHD_msrA/D_msrB | JH2-2 isogenic double mutant with deletions in the aforementioned genes | This study |
| JH_msrA_ complemented strain | Δ_msrA_ strain complemented by allelic exchange | This study |
| JH_msrB_ complemented strain | Δ_msrB_ strain complemented by allelic exchange | This study |
| V19 | Derivative of E. faecalis strain V583 (27) cured of its plasmids, Vanr | This study |
| V19_msrA_::Tet | V19 with an integration of plasmid pUCB30 in the msrA gene | This study |
| V19_msrB_::Tet | V19 with an integration of plasmid pUCB30 in the msrB gene | This study |
| E. coli Top10F′ | F′ [lacI_q Tn_1(TetR)] mcrA Δ(mrr-hsdRMS_-mcrBC) φ80d_lacZ_ΔM15 Δ_lacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pUCB30 | oripMB1 lacZ′ Ampr Emr | 5 |
| pMAD | oripE194ts Emr Ampr_bgaB_ | 3 |
| pVE14218 | oripWV01 RepA− Tetr | L. Rigottier-Gois and P. Serror, unpublished data |
| pGhost3 | oripWV01 RepAts Cmr | 21 |
Survival experiments.
Overnight cultures of E. faecalis strains were prepared. The cells were harvested by centrifugation of 2 ml of the overnight culture. The cells were resuspended in 10 ml of M17 medium. A 96-well microtiter plate was prepared as follows: lane 1 to lane 8 contained 20 μl of 0 mM, 5 mM, 6 mM, 7 mM, 7 mM, 6 mM, 5 mM, and 0 mM H2O2, respectively. A total of 180 μl of the resuspended cultures was added to each well using an eight-channel pipette (Eppendorf Research Plus). This setup allows testing of 12 strains per microtiter plate. The plates were incubated for 2 h at 37°C without agitation. Then, 20 μl was recovered and used to inoculate another plate containing 180 μl of GM17 in each well. The plates were put into a microtiter plate reader (model 680; Bio-Rad). Before reading of the OD600, the plates were agitated for 10 s. The incubation temperature was 37°C, and readings were performed every 30 min for 24 h.
Construction of insertion and unmarked deletion mutants and of complemented strains.
Primers were purchased from Operon (Cologne, Germany). The general protocol for the construction of deletion mutants using plasmids pUCB30 and pMAD is detailed elsewhere (16). Briefly, a DNA chromosomal fragment of 3.5 kbp comprising the msrA or msrB gene to be deleted was amplified by PCR using Pfu ultraproofreading DNA polymerase (Stratagene) and corresponding oligonucleotides deduced from the E. faecalis V583 sequence (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=gef) with overhanging 5′ ends containing recognition sites for restriction endonucleases (REs). Each PCR fragment was then purified with a NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany), cut with the corresponding RE, and ligated into cloning vector pUCB30 and, for the complementation experiments, cloning vector pMAD (see below). Recombinant plasmids were purified, diluted (1 ng/μl), and used as templates for inverse PCR with Pfu ultrapolymerase in order to introduce the deletion. This introduced central deletions of 300 bp and 328 bp in the msrA and msrB genes, respectively. After cloning of the sequences into the pMAD vector (3), msrA and msrB deletion mutants were obtained following the procedures described previously (16). The single Δ_msrA_ mutant was subsequently used to construct the double Δ_msrA_ Δ_msrB_ mutant using the same protocol. All mutants were finally verified to have deletions of the copies of the msrA and msrB genes as well as to lack plasmid pMAD by PCR analysis. The complementation of each single mutant was performed by knocking in the wild-type allele into the corresponding mutant using pMAD, as described previously (36).
Single-crossing-over insertion-duplication mutagenesis was based on a two-vector system and was performed essentially as described previously (18) using plasmid pG+host (RepAts [temperature sensitive {ts}]) and integrative plasmid pVE14218 (Tetr).
Animal studies.
Galleria mellonella larvae were reared on beeswax and pollen at 37°C in darkness. The E. faecalis strains used for infection were grown for 24 h in GM17. After centrifugation of 4 ml of each culture, cells were washed twice in 1 ml of 0.9% NaCl. The bacterial cells were then resuspended in 0.9% NaCl to get a suspension with an OD600 of 1.2. For each strain, 15 G. mellonella caterpillars of about the same size (body weight, 200 to 300 mg) were infected, and each experiment was repeated at least three times. Ten microliters of the cell suspension was injected into the second hindmost proleg of each G. mellonella larva using an automatic syringe pump (KD Scientific, Holliston, MA). After injection, the caterpillars were incubated at 37°C in petri dishes, and the number of survivor caterpillars was scored every 2 h after 15 h of infection. Caterpillars were considered dead when they displayed no movement in response to touch and had turned black.
Mouse experiments were performed with the approval of an institutional animal use committee. Female BALB/c mice (weight, 20 to 25 g; Harlan Italy S.r.l) were housed in filter-top cages at the Catholic University Unit for Laboratory Animal Medicine and had free access to food and water. Two peritoneal macrophage models were used to test the survival of E. faecalis single and double mutants and of the complemented mutants. The first is a previously described in vivo/in vitro infection model (14). In the second model, peritoneal macrophages were isolated from mice 4 days after intraperitoneal injection of 2 ml of a sterile 10% thioglycolate solution and cultured as described previously (8, 31). Macrophages were plated in 24-well (2 × 106 cells/well) tissue culture plates (Corning). After overnight incubation, the cells were washed with phosphate-buffered saline (PBS) to remove nonadherent cells and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10 mM HEPES, 2 mM glutamine, 10% bovine fetal serum, and 1× nonessential amino acids (DMEM complete medium) for 24 h before treatment. The macrophages (>99%, as evaluated by anti-mouse F4/80 antibody staining) were stimulated with medium alone or recombinant gamma interferon (rIFN-γ; 20 ng/ml; Sigma-Aldrich, Milan, Italy) plus lipopolysaccharide (LPS; 10 ng/ml; Sigma-Aldrich) for 24 h and then infected with E. faecalis cells (multiplicity of infection = 10) in DMEM complete medium and incubated for 2 h at 37°C to permit phagocytosis of the E. faecalis cells. The macrophages were washed with PBS and placed in DMEM complete medium with vancomycin (10 μg/ml) and gentamicin (10 μg/ml) to kill extracellular bacteria. To quantify the number of intracellular E. faecalis cells, the macrophages were washed and then lysed with detergent at different time points (2, 5, 8, 24, 48, and 72 h). After dilution with PBS, the lysates were plated onto Enterococcus selective agar (ESA; Fluka Analytical, Switzerland) to determine the numbers of CFU of viable intracellular bacteria. All experiments were performed at least three times, and the results were analyzed by one-way analysis of variance (ANOVA) with a Bonferroni correction posttest.
In addition, to assess the virulence of the E. faecalis single and double mutants and the complemented mutants for the JH2-2 wild-type strain, two mouse models were used. In the intravenous infection model, experiments were performed as described previously (14). Briefly, overnight cultures of the strains grown in brain heart infusion (BHI) broth supplemented with 40% heat-inactivated horse serum were centrifuged, and the resulting pellets were resuspended in sterile PBS to achieve final concentrations of 1 × 109 bacteria/ml. Aliquots of 100 μl from each strain suspension were used to inject the tail veins of groups of 10 mice each. The infection experiments were repeated three times. The mice were monitored with twice-daily inspections, and at 7 days after infection they were killed using CO2 inhalation. The kidneys and livers were then removed aseptically, weighed, and homogenized in 5 ml of PBS for 120 s at high speed in a Stomacher 80 apparatus (Pbi International, Milan, Italy). Serial homogenate dilutions were plated onto ESA medium for determination of the numbers of CFU.
In the urinary tract infection (UTI) model, we followed a previously described protocol (19). Briefly, each bacterial strain was grown in 10 ml of BHI broth supplemented with 40% heat-inactivated horse serum for 10 h at 37°C with shaking. The cells were pelleted, resuspended in 10 ml of sterile PBS, and adjusted to reach a concentration of 1 × 107 bacteria/ml. Groups of five isoflurane-anesthetized mice per bacterial inoculum (102 to 106 CFU) were first infected with 200 μl of each strain suspension via intraurethral catheterization (polyethylene catheter ∼4 cm long; outer diameter, 0.61 mm; Becton Dickinson, Sparks, MD). Additionally, groups of 15 mice each were infected with a sole inoculum of 104 CFU. The mice were killed 48 h after transurethral challenge, and the bladders and kidney pairs were processed as described above. For each strain, the 50% infective dose (ID50) was determined as described previously (28). The bacterial detection limits were 50 and 10 CFU/ml for the kidney and bladder homogenates, respectively. Differences between the total numbers of infected kidney pairs or bladders, obtained by combining the data for all inoculum (102 to 106 CFU) groups, were analyzed by Fisher's exact test. For both models, CFU counts were analyzed by the unpaired t test. All statistical analyses were performed using Prism software (version 5.00) for Windows (GraphPad Software, San Diego, CA). For all comparisons, a P value of less than 0.05 was considered significant.
RESULTS
Definition of msrA and msrB operon structures and promoter localizations.
Analysis of the genome sequences of different E. faecalis strains available at The Institute for Genome Research (TIGR; strain V583) and at the Human Genome Sequencing Center at Baylor College of Medicine (strains OG1RF, TX0104, and HH22) revealed that all strains harbor one msrA gene and one msrB gene, located on different parts of the chromosome. Sequence analysis of the MsrA and MsrB proteins of E. faecalis revealed that they show significant identity with several prokaryotic Msr proteins (data not shown) and that they are part of the third and first subclasses of MsrA and MsrB enzymes, respectively (6). The third class of MsrA enzymes, represented by the Bacillus subtilis protein, has an N-terminal CFWC thioredoxin-like signature, also present in the enterococcal homologue, which is supposed to correspond to the catalytic center of the enzyme (24). The first subclass of MsrB enzymes is represented by the Neisseria meningitidis protein, in which C-117 (C-118 in E. faecalis) corresponds to the catalytic Cys and C-63 (the position is conserved in the E. faecalis enzyme) corresponds to the regeneration Cys (6).
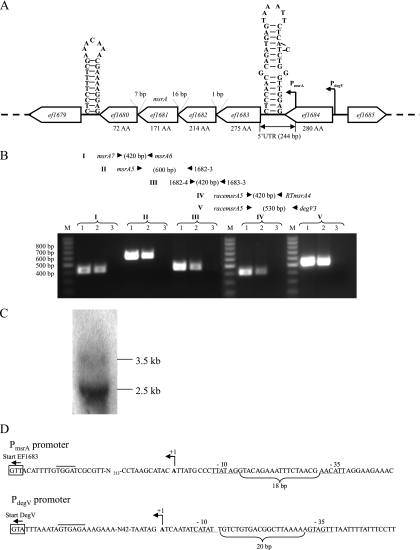
The genetic context of each msr gene of E. faecalis strain V583 is shown in Fig. 1 and 2; this organization is also conserved in E. faecalis strains OG1RF, TX0104, and HH22. The E. faecalis msrA gene (ef1681) is in tandem organization with three other genes (ef1683, ef1682, and ef1680) transcribed counterclockwise and before the last gene in this structure. The four genes are separated by short intergenic regions (1 bp, 16 bp, and 7 bp) and are flanked by putative transcription terminators (Fig. 1A). These characteristics suggested that the four genes form an operon, named the msrA operon throughout this article. The ef1683 gene encodes a putative lipase/acylhydrolase of unknown function, and the ef1682 and ef1680 genes encode conserved hypothetical proteins. In order to experimentally verify the presumed operon structure, RT-PCR experiments were performed (Fig. 1B). This confirmed that the msrA gene is cotranscribed with ef1683, ef1682, and ef1680 but also revealed the presence of transcripts containing sequences upstream of ef1683 and the putative transcription terminator present in the ef1684-ef1683 intergenic region. This suggested the existence of multiple promoters, with one also mapping upstream of the stem-and-loop structure. The promoters were mapped by 5′ rapid amplification of cDNA ends-PCR (RACE-PCR), and the oligonucleotides used were designed to be able to identify transcriptional start sites situated downstream as well as upstream of the putative terminator. These experiments showed that the only promoter mapped, named P_msrA_ (Fig. 1A and D), was situated upstream of the transcriptional terminator in the 3′ end of the ef1684 gene, leading to the synthesis of a transcript with a 244-bp untranslated region (UTR). P_msrA_ displays −10 (GATATT) and −35 (TTACAA) regions separated by 18 bp whose sequences differ from the consensus sequences (TATAAT and TTGACA) of the bacterial promoters at two and three positions, respectively. However, in some electropherograms the sequence seemed to continue after the poly(G) tail added at the 5′ ends during the RACE-PCR protocol, which suggested the existence of a longer transcript as well. Another RT-PCR experiment using oligonucleotides racemsrA5 and degV3 (mapping upstream of P_msrA_) indeed revealed a PCR product, strengthening our hypothesis (Fig. 1B). For mapping the second supposed promoter for expression of the msrA operon, another round of 5′ RACE-PCR was conducted. The only promoter which was identified is situated upstream of the ef1684 gene. This last gene encodes a putative DegV family protein of unknown function, and the promoter identified was named P_degV_ (Fig. 1D). This promoter has characteristics similar to those of the consensus sequences in the −10 element (TATACT) and −35 element (TTGATG), which are separated by 20 bp. However, as judged by Northern blot analysis (Fig. 1C), the main promoter for the expression of the msrA operon seems to be P_msrA_, since the main transcript of approximately 2.5 kbp is compatible if the msrA operon is expressed from this degV internal promoter. Nevertheless, a minor transcript with a size of approximately 3.5 kbp also seems to exist on the Northern blot and may correspond to a pentacistronic transcript containing the msrA operon and degV expressed from P_degV_.
FIG. 1.
(A) Genetic context of the msrA gene of E. faecalis. The open reading frames are represented by open arrows, and their orientation indicates the transcriptional direction. Numbers above the intergenic regions indicate the gene distances. The numbers below the genes indicate the sizes of the gene products in amino acids (AA). The size of the 5′ UTR is given in bp. The nucleotide sequences of the putative rho_-independent terminators, located 4 and 8 nucleotides downstream of the ef1680 and ef1684 stop codons, respectively, are shown. P_msrA and P_degV_ indicate the positions of two promoters mapped by 5′ RACE-PCR (see panel D) (B) Positions of hybridization of 5 oligonucleotide pairs (I to V) used for RT-PCR. The expected amplimer sizes are indicated in bp. The results of the RT-PCRs with the five primer pairs are shown in the electropherogram. Lanes 1, control PCR with chromosomal DNA; lanes 2, RT-PCR; lanes 3, PCR with RNA extraction without prior reverse transcriptase reaction (negative control); lanes M, molecular size standard, with sizes given at the left of the gel. (C) Northern blot analysis using RNA extracted from exponentially growing cells. Hybridization was performed with an [α-32P]dATP-labeled single-strand probe complementary to the msrA mRNA. The size of the transcript was estimated by comparison with the sizes on an RNA ladder. (D) Mapping of promoters P_msrA_ and P_degV_ by 5′ RACE-PCR. The transcriptional initiation sites (+1) are indicated, the putative −35 and −10 motifs are underlined, and the distances between the two boxes are given in bp. The putative ribosome binding sites are overlined, and the start codons are boxed.
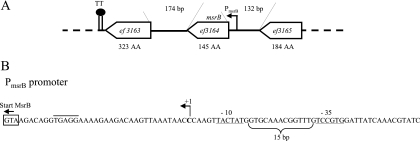
The E. faecalis msrB gene (ef3164), annotated as a PilB family protein in the TIGR database, was already the subject of a previous work (17). In that study, it has been shown that msrB expression is induced by heavy metals, and it was suggested from the results of Northern blot experiments using total RNA preparations from cadmium-induced cultures that msrB forms an operon with the downstream end of the ef3163 gene encoding a ribose-phosphate pyrophosphokinase implicated in purine ribonucleotide biosynthesis. Both genes are transcribed counterclockwise and are separated by a relatively long intergenic region of 174 bp in which no putative transcription terminator could be detected (Fig. 2A). In the present work, we mapped the transcriptional start site by 5′ RACE-PCR, which showed that it is located 34 bp upstream of the ATG translational start codon of the msrB gene. Upstream of this point, regions similar to consensus −10 and −35 boxes separated by 15 bp are found (Fig. 2B). This experimentally defined promoter is different from that proposed in the previous study (17).
FIG. 2.
(A) Genetic context of the msrB gene of E. faecalis. For detailed information, see the legend for Fig. 1A. TT, position of a putative _rho_-independent transcriptional terminator located 8 nucleotides downstream of the ef3163 stop codon. (B) The transcription start site of the msrB operon has been mapped using 5′ RACE-PCR (for details, refer to the legend for Fig. 1D).
Effect of msrA and msrB gene inactivation on E. faecalis resistance to oxidative stress.
In order to define the physiological roles of the E. faecalis Msr proteins, mutants with markerless single and double deletions have been constructed in strain JH2-2. Furthermore, insertional msr mutants have been constructed in a plasmid-cured strain (V19) of E. faecalis V583. The growth of all mutant strains either with or without agitation or in the presence of different concentrations of H2O2 (1.5 mM, 2 mM, 2.5 mM, and 3 mM) was comparable to that of the corresponding wild-type strains (data not shown). The msr mutants were then tested for resistance to H2O2 (Table 2). In this test system, cells are exposed to a given H2O2 concentration for 2 h. After the treatment, an aliquot is used to inoculate fresh medium and the time needed by the culture to reach an OD of 0.2 is determined. This showed that in both E. faecalis strains, the time of outgrowth of the msr mutants was always longer than that for their wild-type counterparts, indicating that they are more sensitive to the peroxide treatments. The Δ_msrA_ Δ_msrB_ double mutant constructed in strain JH2-2 is more sensitive than the single mutants, suggesting an additive effect of the mutations. Comparing the results between both strains showed that in the JH2-2 background the msrA mutant was less affected than the msrB mutant, whereas the corresponding msr mutants constructed in the plasmid-cured derivative of strain V583 demonstrated comparable sensitivities.
TABLE 2.
Sensitivities of Msr-deficient isolates of two E. faecalis strains toward exposure to three different H2O2 concentrations
| Strain | Time (h) to reach OD of 0.2 with H2O2 concn ofa: | |||
|---|---|---|---|---|
| 0 mM | 5 mM | 6 mM | 7 mM | |
| JH2-2 | 0.6 ± 0.1 | 5.2 ± 0.8 | 6.4 ± 0.8 | 8.2 ± 0.8 |
| JHΔ_msrA_ | 0.6 ± 0.1 | 7.3 ± 0.9 (P = 0.037) | 8.2 ± 0.8 (P = 0.067) | 10.0 ± 0.8 (P = 0.18) |
| JH_msrA_compb | 0.6 ± 0.1 | 5.9 ± 0.7 | 7.0 ± 0.6 | 8.5 ± 0.9 |
| JHΔ_msrB_ | 0.6 ± 0.1 | 10.0 ± 0.5 (P = 0.001) | 12.9 ± 0.9 (P = 0.001) | 15.6 ± 1.4 (P = 0.002) |
| JH_msrB_compb | 0.6 ± 0.1 | 5.0 ± 0.5 | 6.2 ± 0.5 | 7.9 ± 0.9 |
| JHΔ_msrA/B_ | 0.6 ± 0.1 | 12.2 ± 0.6 (P < 0.001) | 14.2 ± 1.5 (P = 0.002) | 17.6 ± 1.5 (P = 0.001) |
| V19c | 1.4 ± 0.1 | 7.4 ± 0.7 | 8.3 ± 1.0 | 9.4 ± 1.6 |
| V19_msrA_::Tet | 1.5 ± 0.1 | 12.7 ± 2.0 (P = 0.001) | 14.5 ± 2.5 (P = 0.002) | 16.5 ± 2.5 (P = 0.002) |
| V19_msrB_::Tet | 1.4 ± 0.15 | 13.3 ± 1.3 (P < 0.001) | 14.3 ± 1.0 (P < 0.001) | 15.7 ± 1.6 (P = 0.001) |
Effects of msrA and msrB gene deletion on E. faecalis virulence.
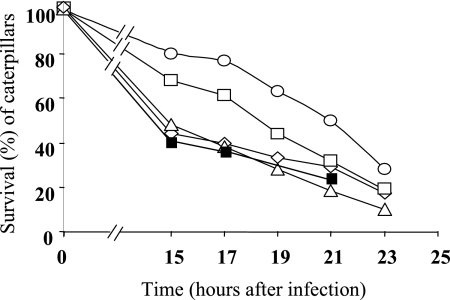
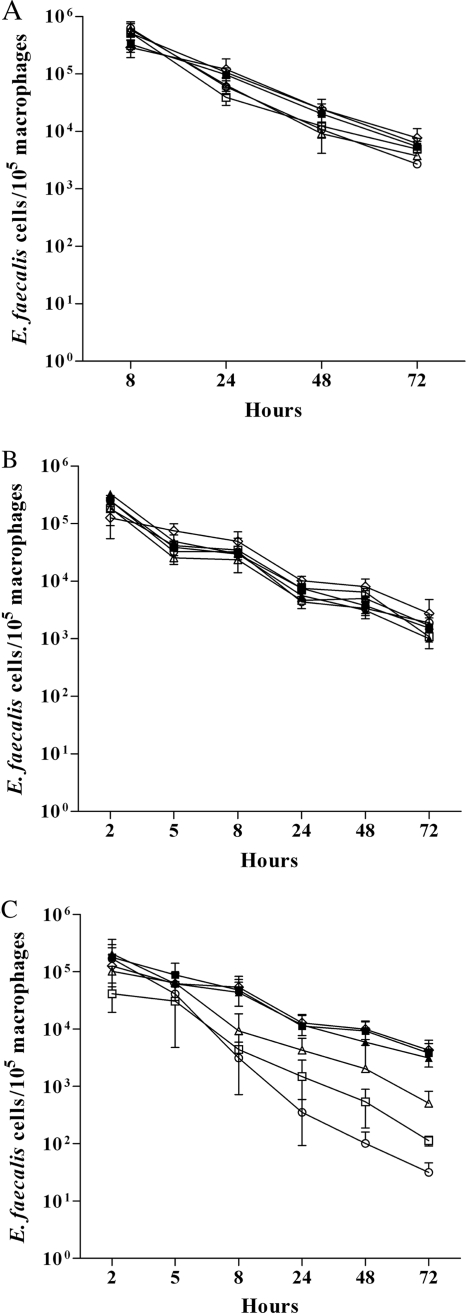
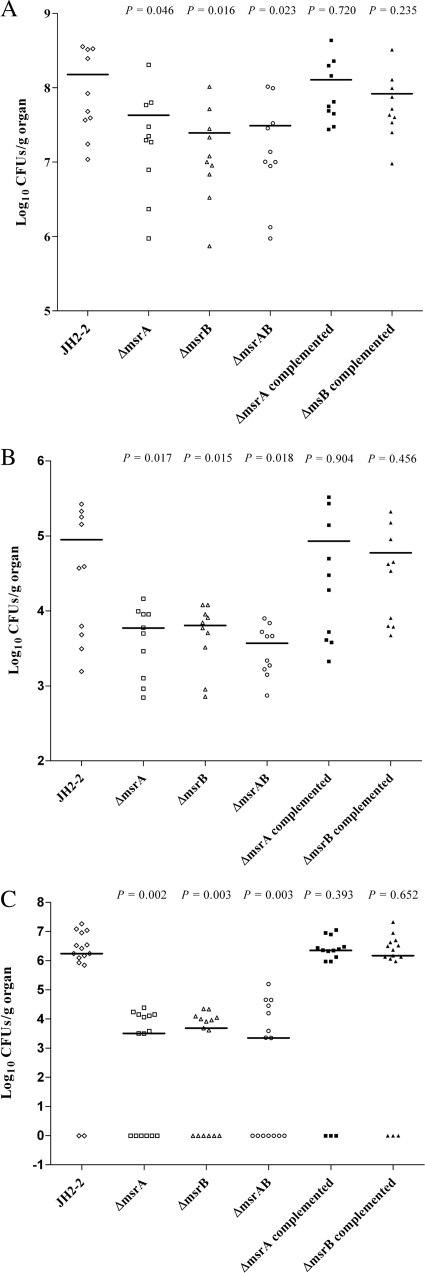
In order to assess the effects of the Msr enzymes on the virulence of E. faecalis, different animal models were used. To exclude the stability problems frequently observed with insertional mutants, only the stable deletion mutants constructed in strain JH2-2 were tested. Moreover, complemented strains were constructed for the Δ_msrA_ and Δ_msrB_ mutants. We started our analysis on the impact of Msr deficiencies on virulence in the simple Galleria mellonella insect model (Fig. 3), which has proved useful for the screening of virulence factors of pathogens (13). The level of killing of the insect larvae by the Δ_msrB_ mutant was comparable to that of the wild-type control. The virulence of the Δ_msrA_ mutant seems attenuated in this model, at least at 15 and 17 h after infection (P = 0.045). However, the combination of both mutations in one strain led to the most significant attenuation of virulence in this model (P = 0.039). The survival capacities of the different strains inside mouse peritoneal macrophages were then measured using an in vivo/in vitro model described previously (16). All strains survived equally well in this test system (Fig. 4A ). While the same results were also obtained using isolated mouse peritoneal macrophages without stimulation (Fig. 4B), differences were noted when the macrophages were stimulated by rIFN-γ plus LPS before infection. Under these conditions, the rates of survival of the Δ_msrA_, Δ_msrB_, and Δ_msrA_ Δ_msrB_ mutants were significantly lower than those of the wild-type strain at 8 h (P = 0.002, P = 0.0049, and P = 0.0016, respectively), 24 h (P = 0.0011, P = 0.0019, and P = 0.0001, respectively), 48 h (P < 0.0001, P = 0.0014, and P < 0.0001, respectively), and 72 h (P < 0.001, P = 0.0011, and P < 0.0001, respectively) (Fig. 5C). We next compared the burdens in the kidney and liver tissues of groups of infected mice using a well-established intravenous infection model (14). As shown in Fig. 5, all mutants (Δ_msrA_, Δ_msrB_, and the Δ_msrA_ Δ_msrB_ double mutant) showed statistically significant reductions in the burdens in both kidney and liver tissues. In detail, the Δ_msrA_ mutant exhibited reductions of 0.54 log unit in the kidneys (P = 0.046) and 1.18 log units in the livers (P = 0.0179) compared to the burdens of the JH2-2 wild-type strain and reductions similar to those obtained for the Δ_msrB_ mutant (0.78 log unit in kidneys [P = 0.016]; 1.15 log units in livers [P = 0.0159]). Virulence was reconstituted to the wild-type level in the msrA and msrB complemented strains (Fig. 5A and B). The results obtained for the double mutant, showing reductions of 0.69 log unit in the kidneys (P = 0.023) and 1.38 log units in the livers (P = 0.0184), confirmed the involvement of MsrA and MsrB in E. faecalis virulence. Finally, the same strains were also tested in a UTI model described elsewhere (19). The resulting ID50s showed that the Δ_msrA_, Δ_msrB_, and double mutant strains required 0.66, 0.68, and 1.11 log10 more cells (2.5 × 103, 2.6 × 103, and 7.0 × 103, respectively) than wild-type strain JH2-2 (5.4 × 102) to infect 50% of mice. In addition, for all the bacterial inocula used, the proportions of infected kidneys were 84% for JH2-2, 64% for the both Δ_msrA_ and Δ_msrB_ single mutant strains (P = 0.02), and 56% for the double mutant strain (P = 0.002). As a representative example, Fig. 5C shows the log10 numbers of CFU recovered from the kidney pairs of mice infected with 104 cells from JH2-2 or each of the mutant strains. As expected, all three mutant strains exhibited statistically significant reductions (2.85 log units for both Δ_msrA_ and Δ_msrB_ [P = 0.002]; 2.39 log units for the double mutant [P = 0.003]) in kidney tissue burden compared to the reduction for wild-type strain JH2-2. In the UTI model, the msrA and msrB complemented strains also showed a wild-type level of virulence (Fig. 5C).
FIG. 3.
Virulence test of msr mutants using the Galleria mellonella insect model. About 5 × 108 CFU was injected into 1 caterpillar, and 15 caterpillars were used for each strain. Viable caterpillars infected with the JH2-2 wild-type strain (⋄), the Δ_msrA_ mutant (□), the Δ_msrB_ mutant (▵), the Δ_msrA_ Δ_msrB_ double mutant (○), and the Δ_msrA_ (▪) complemented strain were counted after 15, 17, 19, 21, and 23 h after infection.
FIG. 4.
Survival of the E. faecalis JH2-2 wild type (⋄); its isogenic Δ_msrA_ (□), Δ_msrB_ (▵), and Δ_msrA_ Δ_msrB_ (○) mutant strains; and the Δ_msrA_ (▪) and Δ_msrB_ (▴) complemented strains in mouse peritoneal macrophages derived from an in vitro/in vivo infection model (A) or isolated from the animals and cultured in vitro with medium alone (B) or with rIFN-γ plus LPS (C) before infection. The data are expressed as means ± standard deviations for the number of viable intracellular bacteria per 105 macrophages in at least three different experiments.
FIG. 5.
Enterococcal tissue burdens in kidneys (A) and livers (B) of BALB/c mice infected intravenously with 5 × 108 cells of the E. faecalis JH2-2 wild type (⋄); its isogenic Δ_msrA_ (□), Δ_msrB_ (▵), and Δ_msrA_ Δ_msrB_ (○) mutant strains; and the Δ_msrA_ (▪) and Δ_msrB_ (▴) complemented strains. Groups of 10 mice were killed and necropsied at day 7 postinfection. (C) Enterococcal burdens of the kidneys of BALB/c mice infected transurethrally with 104 cells of the E. faecalis JH2-2 wild type (⋄); its isogenic Δ_msrA_ (□), Δ_msrB_ (▵), and Δ_msrAB_ (○) mutant strains; and the msrA (▪) and msrB (▴) complemented strains. Kidney pair homogenates were obtained from groups of 15 mice that were killed and necropsied 48 h after the transurethral challenge. The results, expressed as log10 CFU per gram of tissue, represent the values recorded separately for each mouse. Horizontal bars represent the geometric means. A value of 0 was assigned to uninfected kidneys.
DISCUSSION
E. faecalis is part of those bacteria in which the msrA and msrB genes constitute two separate transcription units located in different parts of the genome. The msrA gene of E. faecalis is embedded in a complex operon structure and seems to be expressed from two different promoters, resulting in the synthesis of four-gene and five-gene polycistronic messengers. Expression from both promoters led to the synthesis of a transcript with a long 5′ UTR containing a structure resembling a transcriptional terminator. This structure might be implicated in the regulation of expression of the msrA operon. In some bacteria, msrA expression is induced by oxidants (2, 35), although in most bacteria investigated so far, this was not the case (30). In E. coli and Staphylococcus aureus, a modest induction of msrA in stationary phase has also been reported (26, 32). Our preliminary results obtained by reverse transcription-quantitative PCR and from microarrays using RNAs extracted from, respectively, cultures treated for 30 min with 2.5 and 1.5 mM H2O2suggests that msrA transcription is not induced under these conditions (data not shown). However, in order to get closer insight into the regulation of this gene, more sophisticated approaches based on specific anti-MsrA antibodies or use of a reporter gene assay are necessary.
Comparison of the msrA genome region with the regions of other bacteria using the region comparison tool of the TIGR database revealed that the most closely related genetic context is present in Lactobacillus casei ATCC 334. Indeed, it has an identical succession of the five genes which are close homologues of the corresponding E. faecalis genes. Furthermore, as in E. faecalis, the degV gene is separated from the gene encoding the putative lipase by a long intergenic region containing a transcription terminator. The corresponding genomic region in Listeria monocytogenes differs from the organization in E. faecalis by the insertion of the msrB gene in conjunction with a gene encoding a putative dehydrogenase between the msrA gene (ef1681) and the last gene (ef1680). However, in L. monocytogenes the long intergenic region is absent between the degV gene and the following gene, and it has recently been found that the degV gene is an integral part of the _msrA_-msrB operon (34).
Our physiological studies showed that both the msrA and the msrB genes are important to protect cells against exposure to H2O2 concentrations. However, relatively high oxidant concentrations are needed to see differences that seem beyond physiological relevance. A survey of the literature indicated that most bacteria lacking MsrA investigated so far displayed sensitivity to this oxidant (30). Only a few studies concerned the role of msrB in the resistance to H2O2. Whereas the Helicobacter pylori msrB mutant was sensitive (1), the corresponding mutant of Mycobacterium smegmatis had no role in the defense to this oxidant (30).
No difference in survival inside macrophages of the msr mutants was evidenced using the in vitro/in vivo model (14). This was in apparent contrast to other findings showing that mutants deficient for well-known antioxidant activities of E. faecalis, like superoxide dismutase and thiol peroxidase, were hypersensitive in this test system (16, 36). However, when the msr mutants were tested in IFN-γ-activated macrophages, their rate of survival was lower than that of the parent strain. This was in agreement with the concept that oxidative killing is normally regulated by IFN-γ in macrophages, thus implying that the msr mutants displayed a phenotype only under intense oxidative stress conditions. This is consistent with the growth and survival experiments in the presence of H2O2. Differences were seen only when H2O2 concentrations of >5 mM were used. Of note, the reduction in the rate of intracellular survival of a msrA mutant strain of M. smegmatis (a msrB mutant was not tested in that study) compared to that of the wild-type strain was also more marked in IFN-γ-activated macrophages than in unactivated ones (11).
A clear relationship between E. faecalis virulence and Msr enzymes has been demonstrated in two animal models. Whereas the contribution of Msr deficiencies to virulence in the Galleria model was modest, a significant decrease in virulence was evident in a systemic and urinary infection model. Since evidence in support of a role of Msr in bacterial adherence and biofilm formation has been presented (4, 9, 37), one might suggest that defects in these processes in the msr mutants could be the basis for the observed reduction in virulence in these models. However, no difference in adherence in comparison to that of the wild-type strain was observed for the msrA and msrB mutants using CaCo2 cells as a model (unpublished results). Nevertheless, further experiments on adherence using other cell lines and construction of msr mutants in a strong biofilm-forming strain of E. faecalis are needed to examine the influence of Msr enzymes on these processes in more detail.
Acknowledgments
The technical assistance of Annick Blandin was greatly appreciated. We thank P. Serror and L. Rigottier-Gois from the INRA Institute in Jouy-en-Josas (France) for the generous gift of plasmid pVE14218 and E. coli strain VE14188.
This study was partly supported by grants from the Agence Nationale de la Recherche in the framework of a transnational ERA-NET PathoGenoMics program (grant ANR-06-PATHO-008-01).
Footnotes
▿
Published ahead of print on 21 June 2010.
REFERENCES
- 1.Alamuri, P., and R. J. Maier. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53**:**1397-1406. [DOI] [PubMed] [Google Scholar]
- 2.Alamuri, P., and R. J. Maier. 2006. Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress-induced expression. J. Bacteriol. 188**:**5839-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70**:**6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51**:**659-674. [DOI] [PubMed] [Google Scholar]
- 5.Benachour, A., Y. Auffray, and A. Hartke. 2007. Construction of plasmid vectors for screening replicons from gram-positive bacteria and their use as shuttle cloning vectors. Curr. Microbiol. 54**:**342-347. [DOI] [PubMed] [Google Scholar]
- 6.Boschi-Muller, S., A. Olry, M. Antoine, and G. Branlant. 2005. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim. Biophys. Acta 1703**:**231-238. [DOI] [PubMed] [Google Scholar]
- 7.Boschi-Muller, S., A. Gand, and G. Branlant. 2008. The methionine sulfoxide reductases: catalysis and substrate specificities. Arch. Biochem. Biophys. 474**:**266-273. [DOI] [PubMed] [Google Scholar]
- 8.Cornacchione, P., L. Scaringi, K. Fettucciari, E. Rosati, R. Sabatini, G. Orefici, C. von Hunolstein, A. Modesti, A. Modica, F. Minelli, and P. Marconi. 1998. Group B streptococci persist inside macrophages. Immunology 93**:**86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183**:**5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donskey, C. J. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39**:**219-226. [DOI] [PubMed] [Google Scholar]
- 11.Douglas, T., D. S. Daniel, B. K. Parida, C. Jagannath, and S. Dhandayuthapani. 2004. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J. Bacteriol. 186**:**3590-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezraty, B., L. Aussel, and F. Barras. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta 1703**:**221-229. [DOI] [PubMed] [Google Scholar]
- 13.Gaspar, F., N. Teixeira, L. Rigottier-Gois, P. Marujo, C. Nielsen-Leroux, M. T. Barreto Crespo, M. D. Lopes, and P. Serror. 2009. Virulence of Enterococcus faecalis dairy strains in an insect model: role of fsrB and gelE genes. Microbiology 155**:**3564-3571. [DOI] [PubMed] [Google Scholar]
- 14.Gentry-Weeks, C., M. Estay, C. Loui, and D. Baker. 2003. Intravenous mouse infection model for studying the pathology of Enterococcus faecalis infections. Infect. Immun. 71**:**1434-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koc, A., A. P. Gasch, J. C. Rutherford, H. Kim, and V. N. Gladyshev. 2004. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. U. S. A. 101**:**7999-8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Carbona, S., N. Sauvageot, J. C. Giard, A. Benachour, B. Posteraro, Y. Auffray, M. Sanguinetti, and A. Hartke. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66**:**1148-1163. [DOI] [PubMed] [Google Scholar]
- 17.Laplace, J. M., A. Hartke, J. C. Giard, and Y. Auffray. 2000. Cloning, characterization and expression of an Enterococcus faecalis gene responsive to heavy metals. Appl. Microbiol. Biotechnol. 53**:**685-689. [DOI] [PubMed] [Google Scholar]
- 18.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177**:**7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebreton, F., E. Riboulet-Bisson, P. Serror, M. Sanguinetti, B. Posteraro, R. Torelli, A. Hartke, Y. Auffray, and J. C. Giard. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77**:**2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, W. L., B. Gold, C. Darby, N. Brot, X. Jiang, L. P. S. de Carvalho, D. Wellner, G. St. John, W. R. Jacobs, and C. Nathan. 2009. Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol. Microbiol. 71**:**583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174**:**5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskovitz, J., S. B. Berlett, M. Poston, and E. R. Stadtman. 1997. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in-vivo. Proc. Natl. Acad. Sci. U. S. A. 94**:**9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskovitz, J., E. Flescher, S. B. Berlett, J. A. Azare, M. Poston, and E. R. Stadtman. 1998. Overexpression of peptide-methionine sulfoxide reductase (MsrA) in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 95**:**14071-14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskovitz, J., M. Poston, B. S. Berlett, J. N. Nosworthy, R. Szczepanowski, and E. R. Stadtman. 2000. Identification and characterization of a putative active site for peptide-methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 275**:**14167-14172. [DOI] [PubMed] [Google Scholar]
- 25.Moskovitz, J., S. Bar-Noy, W. M. Williams, J. Requena, B. S. Berlett, and E. R. Stadtman. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. U. S. A. 98**:**12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177**:**502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299**:**2071-2074. [DOI] [PubMed] [Google Scholar]
- 28.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. (Lond.) 27**:**493-497. [Google Scholar]
- 29.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sasindran, S. J., S. Saikolappan, and S. Dhandayuthapani. 2007. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2**:**619-630. [DOI] [PubMed] [Google Scholar]
- 31.Shirey, K. A., L. E. Cole, A. D. Keegan, and S. N. Vogel. 2008. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J. Immunol. 181**:**4159-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, V. K., and J. Moskovitz. 2003. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149**:**2739-2747. [DOI] [PubMed] [Google Scholar]
- 33.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic Streptococci and their bacteriophages. Appl. Microbiol. 29**:**807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toledo-Arana, A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M. A. Nahori, G. Soubigou, B. Régnault, J. Y. Coppée, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459**:**950-956. [DOI] [PubMed] [Google Scholar]
- 35.Vattanaviboon, P., C. Seeanukun, W. Whangsuk, S. Utamapongchai, and S. Mongkolsuk. 2005. Important role for methionine sulfoxide reductase in the oxidative stress response of Xanthomonas campestris pv. phaseoli. J. Bacteriol. 187**:**5831-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verneuil, N., A. Mazé, M. Sanguinetti, J. M. Laplace, A. Benachour, Y. Auffray, J. C. Giard, and A. Hartke. 2006. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology 152**:**2579-2589. [DOI] [PubMed] [Google Scholar]
- 37.Wizemann, T. M., J. Moskovitz, B. J. Pearce, D. Cundell, C. G. Arvidson, M. So, H. Weissbach, N. Brot, and H. R. Masure. 1996. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc. Natl. Acad. Sci. U. S. A. 93**:**7985-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143**:**966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You, C., A. Sekowska, O. Francetic, I. Martin-Verstraete, Y. Wang, and A. Danchin. 2008. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 8**:**128. [DOI] [PMC free article] [PubMed] [Google Scholar]