Global Dynamics of Proteins: Bridging Between Structure and Function (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 13.
Abstract
Biomolecular systems possess unique, structure-encoded dynamic properties that underlie their biological functions. Recent studies indicate that these dynamic properties are determined to a large extent by the topology of native contacts. In recent years, elastic network models used in conjunction with normal mode analyses have proven to be useful for elucidating the collective dynamics intrinsically accessible under native state conditions, including in particular the global modes of motions that are robustly defined by the overall architecture. With increasing availability of structural data for well-studied proteins in different forms (liganded, complexed, or free), there is increasing evidence in support of the correspondence between functional changes in structures observed in experiments and the global motions predicted by these coarse-grained analyses. These observed correlations suggest that computational methods may be advantageously employed for assessing functional changes in structure and allosteric mechanisms intrinsically favored by the native fold.
Keywords: elastic network models, normal modes, principal component analysis, collective motions, allosteric changes in conformation, closed/open conformations
INTRODUCTION
Biomolecular structures enjoy several degrees of freedom under equilibrium conditions, ranging from small fluctuations in atomic positions to collective movements of entire domains, subunits, or molecules. These motions are not random. They are dominated, if not fully determined (owing to environmental perturbations), by intra- and intermolecular interactions, which in turn depend on the three-dimensional (3D) structure, i.e., they are structure-encoded. Structure-encoded dynamics are defined at a minimum by the geometry or topology of native contacts, apart from specific interactions that distinguish between different types of amino acids. Recent years have witnessed an explosion in the number of studies that use elastic network models (ENMs) and normal mode analysis (NMA) for exploring the structure-encoded dynamics that depends exclusively on native contact topology (8, 17). Several servers are now available (1, 21, 48, 63, 64), which permit one to readily compute and visualize a range of collective motions; identify residues that play critical roles in mediating these cooperative movements; or take advantage of predicted modes for structure refinement, for improving docking algorithms, or for steering molecular dynamics (MD) simulations to explore longer timescales and larger length scales.
These studies are now inviting attention to the functional significance of structural dynamics, as also suggested by experimental data (13, 30, 61). There is now an ever-increasing volume of computational studies aimed at gaining insights into the mechanisms of function and allostery, or the links bridging 3D structure and function, with the help of ENM-based methodologies (5, 7, 65). Recent studies raise additional fundamental questions concerning the possible evolution/selection of structures to lend themselves to structure-specific dynamics relevant to their biological function (see, for example, Reference 71).
Given the extensive use of ENMs in conjunction with NMAs and other approaches based on principal component analysis (PCA), as well as graph theoretical methods, in a broad variety of applications, a critical assessment of the underlying assumptions, theory, and methods is in order. The purpose of this review is to give a brief summary of these theoretical foundations, along with illustrative examples of the use and limitations of ENM-based examination of protein dynamics. We begin by presenting an overview of the theoretical foundations and basic features of ENMs, focusing in particular on two ENMs that have been widely used in recent years, the Gaussian network model (GNM) (4, 27) and the anisotropic network model (ANM) (2, 21, 67). We then summarize the extensive studies performed for benchmarking ENMs, or optimizing their parameters, against experimental data on the equilibrium fluctuations of residues observed in structurally resolved proteins. We expose and discuss the relevance of predictions to functional changes in the structures of proteins. We conclude with recent extensions and future prospects.
ELASTIC NETWORK MODELS: THEORY AND ASSUMPTIONS
Why ENMs?
Three basic features have made ENMs attractive for exploring biomolecular dynamics in recent years. First is their simplicity, both conceptual and methodological. In an ENM, the complex structures of biomolecules are reduced to a network of nodes and springs. The network topology is endowed by the native state contact topology. The positions of the nodes are known from X-ray or NMR structures, and those residues that are close to each other are connected by uniform springs. The distribution of interactions is thus a major determinant of structural dynamics. This simple model lends itself to an easily calculated and unique solution for the collective dynamics of each examined system.
A second feature of ENMs is the robustness of the predictions. The global dynamics predicted by the network are almost indistinguishable from those that would otherwise be obtained by full-fledged atomic force fields. Global dynamics refer here to collective motions that cooperatively involve large substructures, usually at the lowest frequency end of the normal mode spectrum, as opposed to local motions associated with high-frequency vibrations. Starting from the pioneering work of Tirion (69), numerous applications have demonstrated that these modes are insensitive to detailed structure and energetics. They are defined, instead, by the overall shape of the biomolecule (e.g., by the fold of the protein), as recently reviewed (5, 50, 65, 76).
The robustness of ENM predictions entails another important feature: scalability. The nodes may indeed be selected at various levels of resolution, provided that the network accurately preserves the gross features of the molecular structure. The most broadly used approach has been to represent each residue as a single node, usually coincident with the Cα-atom for proteins, but models that use only a single node per 40 residues, for example, also predict global dynamics and allosteric communication properties surprisingly well (14, 19). The ENM also lends itself to use in mixed models, in which specific regions of the molecule are modeled in detail and the remainder at low resolution (43), and to rigid blocking schemes as in the rotating-translating blocks model (66). Clearly, the scalability of ENMs makes them particularly useful for investigating large complexes/assemblies or supramolecular systems (79), or even organelles such as the nuclear pore complex (46) or intact viruses (47), i.e., bimolecular systems that are well beyond the scope of exploration via conventional molecular models and simulations.
Simplicity, scalability, and robustness are attractive features; but ENMs would not have found such broad use in molecular and structural biology if it were not for the physical insights they provided into the intrinsically accessible motions of biomolecules, the same motions that are essential to biological function. This review presents examples of the relevance of ENM predictions to such functional changes. Mainly, structures favor well-defined collective modes, and a few cooperative modes underlie the passage between the alternative functional forms of proteins, as illustrated for a few cases in Figure 1 and further elaborated below.
Figure 1.
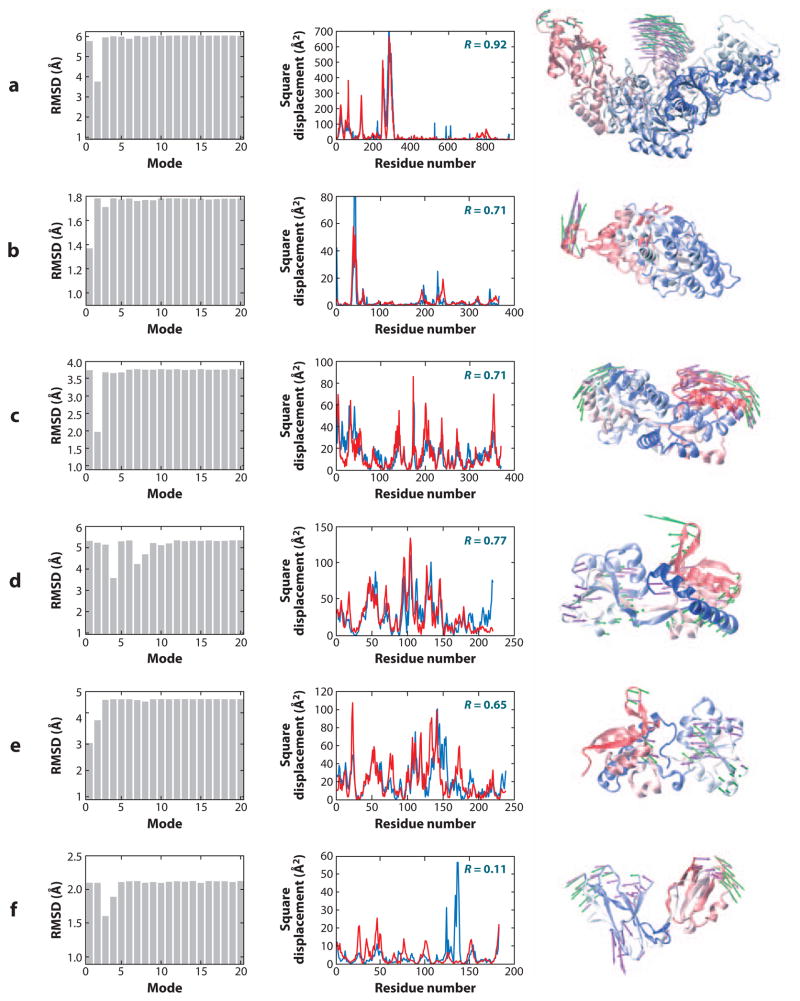
Correlation between collective motions predicted by the anisotropic network model (ANM) and experimentally observed structural changes. The bar plots in the left column illustrate the decrease in root-mean-square deviation (RMSD) between the two endpoints, which can be achieved by moving along particular ANM modes. The original RMSDs are reflected by the plateau values in each plot. There is usually a single or a few low-frequency modes that can be used to deform one of the conformations to get significantly closer to the other. The middle column compares the square displacements of residues calculated for this mode (red) to those experimentally observed between the two conformers (blue). Calculations were performed with Rc = 15 Å. Correlation coefficients between the theoretical and experimental curves appear in the upper-right corner of each graph. The right column shows one structure for each pair, as well as the largest components of the experimentally observed displacement vector (purple arrows) and the relevant ANM mode (green arrows). Panels refer to structural changes between (a) HIV-1 reverse transcriptase complexed with a non-nucleoside inhibitor (PDB code: 1vrt) and its unliganded (with K103N mutation) form (1hqe), using mode 2 of 1vrt; (b) actin in its free form (1j6z) and bound to DNase (1atn), using mode 1 of 1j6z; (c) two forms of a maltodextrin-binding protein (1anf and 1omp), using mode 2 of 1anf; (d) glutamine-binding protein in the unbound (1ggg) and glutamine-bound (1wdn) forms, using mode 4 of 1ggg; (e) LAO in the unbound (2lao) and lysine-bound (1lst) forms, using mode 1 of 2lao; and (f) LIR-1 in its unbound (1g0x) and HLA-A2-bound (1p7q) forms, using mode 3 of 1g0x.
Anisotropic Network Model
The ANM is the most broadly used ENM. It was inspired by the original work of Tirion (69) in which a quadratic potential with a uniform force constant for all atomic interactions within a cutoff distance reproduced almost identically the low-frequency modes of motion obtained with a detailed force field. The ANM is essentially a coarse-grained (CG) version of Tirion’s model, in which the interaction sites, or the network nodes, are the individual residues rather than atoms (2, 21, 67). The corresponding potential reads
| EANM=∑i,j=1Nγ2(Rij−Rij0)2Θ(Rc−Rij0), | 1 |
|---|
where Rij and Rij0 are the instantaneous and equilibrium separations between residues i and j; γ is the uniform force constant, also called stiffness; and Θ(x) designates the Heaviside step function, equal to 1 if x > 0, and zero otherwise. In Equation 1, Θ(x) selects all ij pairs that are within a cutoff distance, Rc, while eliminating farther neighbors. N is the number of nodes/residues in the network. The spatial locations of residues are usually identified by the coordinates of the α-carbons (in proteins), or those of selected nucleotide atoms (in DNA or RNA oligomers). We note that Hinsen et al. (32) introduced a slightly different CG ENM, where the term γ/2Θ(Rc−Rij0) is replaced by a semiempirical function, the form and parameters of which optimally fit the effective force constants obtained (for crambin) with a realistic force field. This distance-weighted form has the advantage of eliminating the parameter Rc and providing a physical description of inter-residue interactions. Jernigan and coworkers also proposed a force constant that decreases with inter-residue separation, which gives a satisfactory description of equilibrium motions (75).
The expansion of the ANM potential near the equilibrium state reads
| EANM=EANM0+∑i∂EANM∂αi|R0(αi−αi0)+12∑i,j∂EANM∂αi∂αj|R0(αi−αi0)(αj−αj0)+⋯, | 2 | | -------------------------------------------------------------------------- | - |
where α is x, y, or z; EANM0=0 according to Equation 1; and ∂EANM/∂αi|R0 vanishes at equilibrium. The (remaining) second order term may be written in compact notation as 1
Here Δ**R** is the 3_N_-dimensional vector of fluctuations Δ**Ri = (Δ_xi_ Δ_yi_ Δ_zi_)T of all residues (1 ≤ i ≤ N) organized as ΔR**i = (Δ_x1_ Δ_y1_ Δ_z1_…Δ_zN_)T, and H is the Hessian, a 3_N_ × 3_N_ matrix composed of the second derivatives of the potential with respect to the components of the position vectors. The ANM yields a concise, closed form expression for the elements of H, e.g.,
| [Hij]xy=∂2EANM∂xi∂yj|R0=−γ(xj0−xi0)(yj0−yi0)(Rij0)2 | 4 |
|---|
for the xy element of the ijth off-diagonal block Hij with a size of 3 × 3. The diagonal blocks are the negative sum over all off-diagonal blocks in the same row/column (H is symmetrical). In NMA using a conventional force field, an expensive initial energy minimization is required prior to calculating H, which becomes a challenge as the size of the system increases. Conversely, ANM requires no such minimization and is readily applicable to supramolecular systems.
The 3_N_ × 3_N_ covariance matrix, C(3_N_) for residue fluctuations reduces to
| C(3N)=〈ΔRΔRT〉=∫−∞∞ΔRΔRTexp(−EANM/kBT)dΔR∫−∞∞exp(−EANM/kBT)dΔR=kBTγH−1 | 5 |
|---|
using Gaussian integrals ∫−∞∞e−ax2dx=(πa) and ∫−∞∞x2e−ax2dx=π2a−3/2 to evaluate the above ensemble. H has six zero eigenvalues, corresponding to the external rotational and translational degrees of freedom of the molecule. As a result, the inversion of H is not straightforward and H−1 is a pseudoinverse expressed in terms of its nonzero eigenvalues (λ_k_, 1 ≤ k ≤ 3_N_-6) and corresponding eigenvectors uk, as
The implication of Equation 6 is that the positional covariance is contributed by a set of normal modes (uk). The eigenvalues serve as the weights of these contributions; they are the squared frequencies of vibrational modes and also the stiffness (curvature of the energy landscape) as the molecule reconfigures along the mode directions (3, 27). Clearly, lower-frequency modes make a larger contribution to the covariance. The modes with the lowest frequencies are generally of interest, as they entail the most cooperative and largest amplitude motions and are the softest modes favored by the overall architecture.
Gaussian Network Model
The GNM is rooted in the statistical mechanical theory of elasticity originally introduced by Flory and coworkers (23, 36) for polymer networks. It uses the same assumption underlying the original theory put forward three decades ago, that of independent, Gaussian fluctuations of network nodes given by the distribution (3, 4)
| W(ΔRi)∼exp{−3(ΔRi)2/2〈(ΔRi)2〉}, | 7 |
|---|
which, in turn, implies Gaussian fluctuations in internode distances. In the GNM, each node is identified by a residue (of the protein or oligonucleotide), and each connector by the interaction that stabilizes the native contact topology. In the language used above for describing ANM, the isotropic Gaussian distributions of fluctuations map to harmonic potentials of entropic origin (4)
| EGNM=γ2ΔRT(Γ⊗I)ΔR=γ2(ΔXTΓΔX+ΔYTΓΔY+ΔZTΓΔZ), | 8 |
|---|
where ΔXT = (Δ_x_1 Δ_x_2… Δ_xN_), and similarly forΔYT and ΔZT. Γ is the N × N Kirchhoff matrix, fully controlling the dynamics. Its ijth off-diagonal element is Γ_ij_ = −1 if node i is within a cutoff distance Rc from node j, and Γ_ij_ = 0; otherwise, the diagonal elements are evaluated from Γii=−∑j=1,j≠iNΓij and represent the residue coordination number. No information on the directions of fluctuations can be obtained from the GNM. The theory provides _N_-dimensional predictions on the mean-square fluctuations (MSFs) of residues and the cross-correlations between their fluctuations given by the respective diagonal and off-diagonal elements of an N × N covariance matrix C(N),
| C(N)=〈ΔXTΔX〉+〈ΔYTΔY〉+〈ΔZTΔZ〉=3kBTγΓ−1, | 9 |
|---|
along with the collective modes in an _N_-dimensional space obtained by the eigenvalue decomposition of Γ. Note that Equation 8 is simply the counterpart of Equation 5 upon substitution of EGNM. It implies, upon substitution from Equation 7, an entropic cost of the form (9)
| ΔSi(ΔRi)=kBlnW(ΔRi)∼−γ(ΔRi)2/(2T[Γ−1]ii) | 10 |
|---|
for the deformation of residue i away from its equilibrium position. Residues subject to large amplitude MSFs thus incur lower entropic cost for a given deformation.
For comparative purposes with the ANM potential, Equation 8 can be rewritten as
| EGNM=∑i,j=1Nγ2(ΔRij•ΔRij)Θ(Rc−∣Rij0∣). | 11 |
|---|
The difference with respect to Equation 1 is therefore the replacement of (Rij−Rij0)2 by (ΔRij•ΔRij)=(Rij−Rij0)•(Rij−Rij0), where Δ**Rij is the vectorial difference between the instantaneous and equilibrium distance vectors Rij and Rij0. The term (Rij−Rij0)2 in Equation 1 becomes zero if the instantaneous distance vector maintains its magnitude while changing its orientation. Its counterpart in the GNM, on the other hand, penalizes the change in orientation (including both internal and external rotation) in addition to the changes in distance (magnitude). In fact, Γ has one zero eigenvalue (trivial mode), and consequently, Γ⊗I** has three, instead of six, trivial modes, which correspond exclusively to the translational invariance of the results (68). Notably, the GNM analysis shares much in common with spectral graph-theoretical methods based on network connectivity that have found wide applications in other disciplines (15, 16).
Benchmarking ENM Predictions Against Experimental Data on Equilibrium Fluctuations
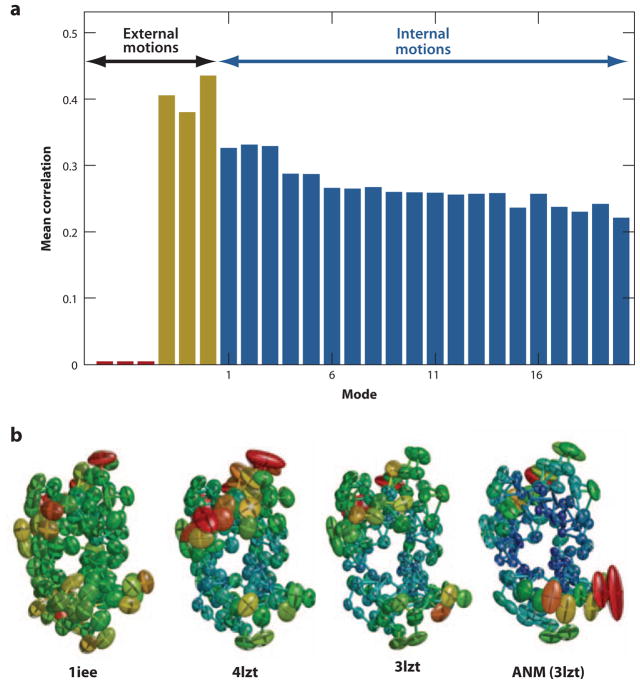
The most abundant data for the equilibrium fluctuations of residues near their folded state are the X-ray crystallographic temperature factors, also known as _B_-factors or Debye-Waller factors, reported with the majority of Protein Data Bank (PDB)-deposited X-ray structures. The _B_-factors are not direct experimental measurements, however, but refined values that reflect more than pure internal motion—including rigid-body motion and static disorder in the crystal. In some structures, rigid-body motions and uniform static disorder make even larger contributions to observed _B_-factors than do thermal fluctuations (31, 62), as indicated by the separation of internal and external degrees of freedom in NMA-based refinement of structures (35, 57, 62). Figure 2_a_ illustrates the results from a systematic examination of a set of 90 high-resolution structures. No correlation with rigid-body translation is observed as these modes are constants for all residues, while rigid-body rotations exhibit significant correlations.
Figure 2.
Analysis of X-ray crystallographic _B_-factors observed in experiments. (a) Decomposition of _B_-factors into external and internal contributions. The bars indicate the correlation between experimentally observed _B_-factors and external (translational, red; rotational; yellow) and internal [top-ranking 20 anisotropic network model (ANM) modes; _blue_] motions computed for a representative set of 90 high-resolution structures. By definition, no net contribution from rigid-body translation is detectable in this comparison because the corresponding eigenvectors are constants for all residues, whereas observed _B_-factors can be compared with, and appear to be affected by, rigid-body rotations. (b) Anisotropic displacements computed and observed for hen egg white lysozyme. Anisotropic _B_-factor data are displayed as color-coded (from red to blue, with decreasing size of fluctuations) ellipsoids for residues whose fluctuation volumes are at least one standard deviation away from mean values. The left three diagrams refer to experimental values reported in the PDB files 1iee, 4lzt, and 3lzt, and the right diagram to theoretical values predicted by ANM using the mean coordinates in 3lzt. 3lzt and 4lzt have the same crystal form; 1iee is different. Figure created using Rastep (53). For more details see Reference 20.
Despite these limitations, B_-factors have been used broadly for exploring whether the equilibrium fluctuations in residue positions predicted by ENM-based NMA exhibit any correlation with experimental data, and for assessing the optimal parameters for these CG studies (37, 38, 42, 62, 78, 80), given that the B_-factors theoretically scale with the MSFs of individual atoms around their equilibrium positions, as Bi = (8_π 2/3) 〈Δ**Ri)2〉. Note that in the ENM analysis, 〈(ΔR**i)2〉 is simply the ith diagonal element of C (Equation 9) or the trace of the ith super-element of C(3_N) (Equation 5). The MSFs computed by the GNM usually yield a correlation with _B_-factors of about 0.60 (20, 37, 42, 78) and increase to 0.66 when the contacts made with neighboring molecules in the crystal environment are taken into consideration (31, 37, 42, 62). For ANM, the mean correlation is about 0.55 and increases to 0.60 upon consideration of crystal contacts (21); the use of distance-weighted spring constants further increases the average correlation with experimental _B_-factors to 0.67 (60). The better correlation of GNM predictions with _B_-factors has been attributed to the energetic penalty on internal and external rotations inherent to the GNM. Recent studies show that correlations higher than 0.8 can be achieved in individual cases (49, 62, 81) upon optimizing ANM parameters against experimental data.
In high-resolution crystallographic structures, the diffraction data are sufficiently detailed to refine six anisotropic displacement parameters (ADPs), instead of a single isotropic parameter, per atom. The ADPs for the ith atom are simply the six distinctive elements of the ith diagonal superelement (a 3 × 3 symmetric matrix) of C(3_N_). Three of these, 〈(Δ_xi_)2〉, 〈(Δ_yi_)2〉, and 〈(Δ_zi_)2〉, provide information on the sizes of motions along different directions. The remaining three, 〈Δ_xi_ Δ_yi_〉, 〈Δ_xi_ Δ_zi_〉, and 〈Δ_yi_ Δ_zi_〉, are cross-correlations between fluctuations along different axes. Systematic calculations (20, 38, 49) showed that the MSFs, 〈(Δ**R**i)2〉, are predicted with higher accuracy than the individual components 〈(Δ_xi_)2〉, 〈(Δ_yi_)2〉, and 〈(Δ_zi_)2〉; that these components are, in turn, more accurately predicted than their cross-correlations (20); and that use of detailed force fields like CHARMM slightly improves the predictions (12, 38). ADPs suffer from the same limitations as the isotropic _B_-factors, and should be considered with caution. Figure 2_b_ shows, for example, that the ADPs predicted by the ANM for hen egg white lysozyme may agree better with experimental data than do two sets of experimental data obtained for the same protein from crystals with different symmetries (20), illustrating the sensitivity of ADPs to experimental conditions.
In addition to X-ray crystallographic data, GNM predictions correlate well with H/D exchange protection factors (9), folding nuclei, (28, 59), NMR order parameters (26), and root-mean-square deviations (RMSDs) between NMR models (78). Notably, the RMSDs from NMR ensembles agree better with ENM results than do the crystallographic _B_-factors. This difference has been explained by the more accurate sampling of low-frequency/large-amplitude movements in solution, reflected in NMR models, as opposed to their suppression in the crystal environment (78).
FUNCTIONAL SIGNIFICANCE OF STRUCTURAL DYNAMICS
The comparative studies summarized above focus on equilibrium fluctuations. More significant, however, is the relevance of ENM predictions to functional changes in structure, which are elucidated by focusing on the global modes.
Normal Modes and Passages Between Substates
In principle, there is always a combination of normal modes that accounts for a given conformational change, since the normal modes form an orthonormal basis set that spans the complete space of internal motions. However, the question is whether a single mode or a small subset of low-frequency modes, predictable by ENMs, can largely explain the functional changes. This ability of ENMs could then assist in structural refinement, molecular docking, and development of drugs that target particular functional motions.
The existence of a correlation between GNM predictions and structural changes inferred from the structures of a given protein resolved in different forms (e.g., closed or open forms, or inhibitor-bound, DNA-bound, or unbound forms) was pointed out a decade ago for HIV-1 reverse transcriptase (6) and other proteins (3), along with the implications of observed domain motions on biomolecular function. We note in particular a pioneering survey by Tama & Sanejouand (67) in which 20 different proteins, all resolved in both open and closed states, were examined using the ANM to demonstrate the relevance of predicted slow modes to observed (functional) changes in structure. Since then, this important feature has been exploited for many systems (5). The idea is the following: Given two conformations R(A) and R(B) for a given protein, the question is whether one or more modes, uk, accessible to R(A) closely approximate the 3_N_-dimensional difference vector Δ**R**AB = R(B) − R(A) between the two conformers. In mathematical terms, the goal is to determine the displacement of size sk along eigenvector uk that will minimize the RMSD,
| RMSDk=1N∣(R(A)+skuk)−R(B)∣2, | 12 |
|---|
between the end points. The sk value that minimizes RMSD_k_ is readily found by differentiating Equation 12 with respect to sk, as the projection of Δ**RAB onto uk, (note that uk is normalized), i.e., sk = ΔR**AB • uk, which substituted into Equation 12, leads to the smallest RMSD_i_ that can be achieved upon moving along the kth mode axis:
| min(RMSD)k=1N(∣R(B)−R(A)∣2−sk2). | 13 |
|---|
Figure 1 illustrates some typical results obtained for six proteins, each resolved in at least two conformers, R(A) and R(B). We see in each case at least one mode uk, among those in the lowest frequency regime (e.g., k ≤ 20), that makes a significant contribution to the structural change Δ**RAB between two conformers. This property is evidenced by (a) the decrease in the RMSD between the two conformers upon reconfiguration of one of them along that particular mode, and (b) the correlation between the square displacement profiles of residues derived from experimental (ΔRAB) and theoretically predicted (u**k) changes in coordinates. For example, in Figure 1_c_, the second slowest mode alone allows for a reduction from 3.80 to 1.95 Å in the RMSD between the open and closed structures of the maltodextrin-binding protein. The square displacements of residues along this particular ANM mode exhibit a correlation of 0.71 with the changes experimentally observed between the two conformers. Yet, this particular mode represents just one of the 3_N_-6-accessible directions of reconfiguration (at the residue-level description) in the conformational space. Notably, this motion is predicted by the ANM to be one of the most readily accessible (entropically favorable) modes of motion and, when observed experimentally, exhibits a remarkable correlation with the global change in structure.
Similar features are observed in five of the proteins shown in Figure 1. However, not all conformational changes can be described by low-frequency modes, especially if the transitions involve rearrangements on a local scale. In some proteins such as calmodulin, the conformational change can be almost fully accounted for by a few slow modes; in others, such as NtrC, this is not the case (25). Among motor proteins, the functional motions of myosin and F1-ATPase can be accurately described by one or two modes, but this is not the case for the evolutionarily related kinesin (83). For the ligand-binding domain of the leukocyte-immunoglobulin-like receptor (LIR-1) (Figure 1_f_), mode 3 helps to reduce the RMSD between the open and closed forms, presumably due to the correctly predicted anti-correlated movements of the two domains, but the overall mobility profile exhibits poor correlation with experiments owing to inadequate description of a solvent-exposed loop region. The RMSDs and correlation coefficients thus provide complementary information.
From another perspective, we can see that correlation cosines of ≥0.90 between the theoretical and experimental mobility profiles can be obtained upon adding up the contributions of only a handful of slow modes. Gerstein and coworkers (1) have now constructed a database of more than 3800 molecular motions, which shows that the slow modes often exhibit maximum overlap with the experimentally observed directions of reconfiguration. The slow modes can be readily evaluated and visualized for any structure deposited into the PDB, or any model written in PDB format, using the ANM server (21).
We note that the proteins illustrated in Figure 1 exhibit RMSDs of 1.8–6.0 Å between their different conformers, with the largest changes occurring in the case of reverse transcriptase (Figure 1_a_). Reverse transcriptase is composed of two subunits (p66 and p51), each of which is composed of multiple subdomains. The large structural change observed between its inhibitor-bound and inhibitor-free forms presumably falls within the same global energy minimum represented at a CG scale. In a sense, coarse-graining of structure and energetics allows for overlooking the ruggedness of the energy landscape—which otherwise arises from specific, highly nonlinear interactions at atomic scale. Motivated by such findings, and presumably inspired by the examination of folding kinetics using Gō-models, transitions between substates have been explored, using ENMs for the endpoints (51, 54, 81). A recent study performed for the bacterial chaperonin GroEL along these lines (79) revealed that a small subset of modes is conducive to the next functional state, but there is also a need to gradually sample steps along higher mode directions as the structure moves away from the original state. Calculations demonstrated that more than half of the 12 Å RMSD undergone by the complex during its allosteric cycle was achieved upon moving along the slowest two to three modes, with minimal perturbation, if any, in the distribution of native contacts (79). The concurrence between the naturally selected (or entropically favored) modes of motion and those required for achieving its chaperonin function provides a nice example of the evolutionary optimization of structure to intrinsically favor functional modes.
Allosteric Changes Exploit Conformations Accessible via Global Modes
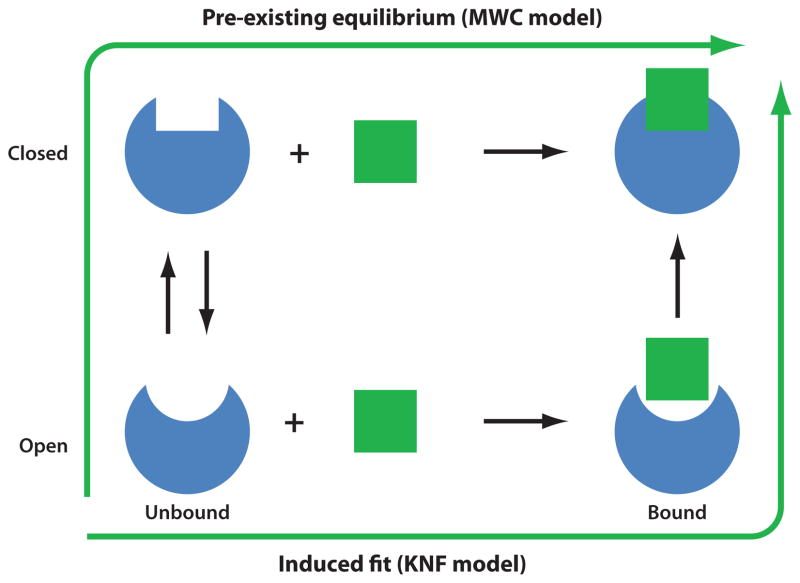
Two models are broadly used in the literature for explaining allosteric changes elicited by ligand binding (see Figure 3): (a) induced fit, originally proposed by Koshland (39), which confers an active role to the ligand as the determinant of the conformational change undergone by the protein upon binding it, and (b) pre-existing equilibrium (55), in which the substrate-bound conformer results from the selection of an already existing (albeit at low proportion) conformation of the protein. The transition is an all-or-none change, cooperatively involving the entire molecule, similar to slow modes predicted by NMA. The Koshland-Néméthy-Filmer (KNF) model (40), on the other hand, proposes a sequential transition that is initiated locally and gradually engages other subunits.
Figure 3.
Two models of structural change observed upon ligand binding, induced fit versus selection of pre-existing conformer. In the elastic network model description of structural dynamics, pre-existing conformers are those readily accessible via movements along low-frequency modes. KNF, Koshland-Néméthy-Filmer; MWC, Monod-Wyman-Changeux.
An important result from ENM-based studies, with implications for molecular docking, is that the low-frequency modes calculated for the unbound protein can account for the conformational changes observed upon substrate binding (70). This observation reveals the important property of intrinsic accessibility of functional movements, endowed by the equilibrium structure, per se, prior to substrate binding. Recent analyses further suggest that this property holds not only for protein-protein interactions, but even for protein-ligand (small molecule) interactions (10, 73), and that it enables allosteric regulation in general.
A classical example is hemoglobin, the T → R transition of which has been demonstrated in both atomic (56) and ANM-based (72) NMAs to be achieved by one slow mode (second in that case) intrinsically accessible to the T state. The transition is cooperative, i.e., all subunits concertedly undergo a conformational switch, the mechanism of which is defined by the overall quaternary structure. Another prime example is the bacterial chaperonin, GroEL, an ATP-regulated machine. The conformational transitions undergone by GroEL during its allosteric cycle are highly dominated by a few low-frequency modes (79, 82). Zheng et al. (82) demonstrated that not only are ligand-induced conformational changes dominated by few low-frequency modes, but that these modes are robust to sequence variations.
An alternative way of examining the correspondence between experiments and computations is to compare existing structural data for a given protein in the presence of different ligands with snapshots from its MD simulations. This type of comparison may be particularly useful for local changes at the ligand-binding pocket that fall within the range of observation of MD simulations. Such an analysis recently performed for acetylcholine esterase showed that the side chain conformational changes upon ligand binding correspond to pre-existing states observable from MD (73). In summary, increasing experimental and computational evidence supports the dominance of intrinsic, as opposed to induced, dynamics in controlling the conformational switches and allosteric signals of proteins (5, 10, 13, 22, 25, 70, 73); but the bound conformations understandably undergo further stabilizing rearrangements induced upon substrate binding (70) consistent with an alteration in the energy landscape in favor of the bound state (61).
Principal Changes in Structure Agree with ENM Dynamics
Perhaps the most striking evidence of consistency between the ensembles of structures observed in experiments and the structural changes predicted by the ANM is provided by PCA of structures deposited into the PDB for a given protein. The comparison is straightforward: PCA extracts the dominant modes of structural variation in a given ensemble, permitting us to directly compare these principal modes, based on experimental structural data, with the lowest-frequency normal modes predicted (for one conformation in the ensemble) by the ANM.
With increasing structural data on the same protein in different conformers, we are now in a position to make such comparisons and assess the realism of the global modes predicted by the ANM. We may also take a closer look at the differences between the structures determined by NMR and X ray for a given protein and see to what extent these differences comply with the intrinsic dynamics of the protein. Calculations recently performed (77) along those lines showed that in 20 of 24 proteins examined, the conformational differences between the NMR structures and their X-ray counterparts were consistent with the principal modes identified by PCA of NMR models and supported by ANM. This study unambiguously shows that X-ray and NMR structures simply reflect the reconfigurations of the protein along its intrinsically accessible global modes of motions (77).
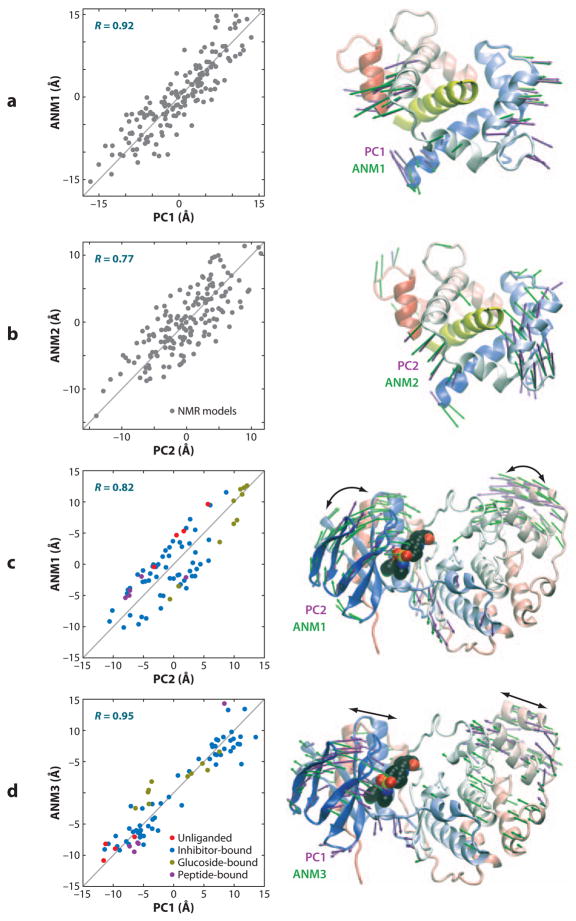
Notably, global modes can be deduced from the ANM analysis of NMR models. Figure 4_a,b_ illustrates the results from such an ANM investigation of an ensemble of NMR models determined for calmodulin complexed with myosin light chain kinase (10). The PCA of the ensemble yields two principal modes of structural variations, PC1 and PC2. ANM analysis of one representative structure, on the other hand, yields the top-ranking modes, designated ANM1 and ANM2. The plots display the ensemble of models dispersed along the top-ranking ANM and PCA directions, demonstrating the close correspondence between the experimentally determined structural variations (PCA) and theoretically predicted global modes (ANM). Precisely, ANM modes 1 and 2 exhibit respective correlations of 0.88 and 0.77 with PC1 and PC2.
Figure 4.
Comparison of principal changes in structure observed in experiments and predicted by the anisotropic network model (ANM). Results are displayed for calmodulin (CaM) complexed with myosin light chain kinase (MLCK) resolved by NMR in panels a and b and for p38 kinase in multiple forms in panels c and d. Both sets of experimental data (160 NMR models for CaM-MLCK, 74 PDB structures for p38) were subjected to principal component analysis (PCA) to obtain the two dominant changes in structures, PC1 and PC2, in each case. A representative structure [the apo structure (1p38) for p38 kinase, and the average model with the lowest root-mean-square deviation (RMSD) from all others in NMR ensemble for CaM-MLCK] from each set was analyzed by ANM to determine the global modes ANM1–ANM3. The plots on the left display the dispersion of the examined models/structures along these top-ranking mode axes derived from experiments (PC1–2) and theory (ANM1–3). Correlations in the range 0.77–0.95 are observed. The colored dots in the left plots of panels c and d refer to 4 unliganded (red), 56 inhibitor-bound (blue), 10 glucoside-bound (yellow), and 4 peptide-bound (purple) p38 structures; the gray dots in panels a and b refer to NMR models. The ribbon diagrams (right) illustrate the global movements predicted by theory (green arrows) and exhibited by experiments (mauve arrows). The MLCK is displayed in yellow in the ribbon diagrams, and a bound inhibitor is shown in space-filling representation in p38 kinase structures. See the text and Reference 10 for more details.
In a recent study, the ensembles of structures experimentally resolved for well-studied enzymes in the presence of different ligands/substrates, or in the unbound state, were shown to correlate with ANM-predicted conformational changes (10). Figure 4_c,d_ illustrates the results for an ensemble of 74 structures determined for p38 kinase. PCA of this ensemble revealed two principal modes of structural changes, PC1 and PC2, that essentially correspond to a concerted opening/closing of the two lobes (PC1), and a twisting of the N-lobe with respect to the C-lobe (PC2). ANM calculations performed for a representative member from this set revealed that the global modes 1 and 3 closely correlate with PC2 and PC1, respectively, as illustrated in Figure 4_c,d_. Again, the major, largest-scale conformational changes observed between the experimentally resolved structures are essentially reconfigurations along the respective ANM modes 1 and 3 intrinsically accessible to p38 kinase structure, irrespective of ligand binding.
These results demonstrate that the structural changes undergone by the protein in different functional forms conform to a large extent to those intrinsically encoded by the native contacts. In a recent study of HIV-1 protease, Jernigan and coworkers (74) also demonstrated the close correlation between the top-ranking PCA modes derived for experimentally resolved structures and the low-frequency ANM modes computed for a representative structure. Such comparisons are expected to become a frequent benchmarking technique for assessing and consolidating the conformational changes predicted for functional mechanisms.
These observations draw attention to two important concepts. First, functional information may be extracted from ensembles of conformations resolved for a given protein; i.e., the data in the presence of different ligands or in different crystal space groups are not redundant but actually contain valuable information on functional dynamics that may be extracted by PCA, stipulating the utility of depositing into the PDB multiple structures for a given protein. Second, the principal variations in structure are consistent with the top-ranking ANM modes, implying that information on potential changes in structure, needed for assessing protein-protein and protein-ligand interactions, can be advantageously deduced from knowledge of inter-residue contact topology.
Notably, the residues identified to be highly constrained in the PCA or NMA modes are further verified to be evolutionarily conserved, supporting the utility of the PCA of structural ensembles or ANM analysis of known structures for identifying potential functional sites. We have now constructed a portal, PCA NEST (PCA of Native Ensembles of STructures) (77), which uses as input the ensembles of structures known from experiments (or snapshots from simulations), and releases as output the corresponding principal modes of structural changes and potential functional sites via a user-friendly interface.
ENMs in Homology Modeling and Structure Refinement
Despite recent advances in ab initio methods for protein structure prediction, ongoing structural genomics initiatives strive to resolve representative structures for many different folds such that most proteins can be accurately modeled by homology modeling. In this method, target proteins are modeled on the basis of evolutionarily related templates with known structures. Homology modeling methods, however, are often not accurate enough for practical purposes because of our inability to successfully refine the structure of a template protein based on the target sequence (41). Any advances in this refinement mark significant steps toward the goal of exploiting protein models for practical needs, such as drug design, in the structural genomics era.
We are currently in a position to improve the quality of homology models by refining the template structures based on their equilibrium motions using ENM and NMA. The changes in protein topology due to limited sequence variation between family members tend to take place along a set of low-frequency normal modes (44). Therefore, it should be possible to refine the template backbone using the target sequence by effectively restricting the search space to a subspace spanned by a few lower-frequency normal modes. This will increase the chance of finding correct solutions while reducing the chance of getting trapped in false solutions. Implementing these ideas, Qian et al. (58) performed a grid search for refining template structures by PCA of structures available for a given family and selecting conformations based on sensitive energy functions. Given that the principal components from experimental structures correlate well with normal modes (44, 77), it should be possible to use normal modes instead of PCA. The advantage of normal modes is that they can be calculated based on a single template structure. This endows NMA with the advantage of filling the gaps in the sequence-structure space. Han et al. (29), on the other hand, developed a combined method that utilizes both PCA and NMA. It effectively restricts the search space for homology modeling, making it feasible to adopt sensitive optimization procedures that require lower dimensionality. A recent application is the prediction of open and closed conformations of Rat heme-free oxygenase (52). Starting from the heme-bound X-ray structure, and using templates corresponding to human and incomplete rat heme-free structures, models of the rat unbound species with open and closed conformations were generated.
Finally, there is an even larger body of literature on the use of NMAs for structure refinement, beginning with original studies by Diamond (18) and Kidera & Gō (34) soon after the pioneering NMA articles in the 1980s (11, 24, 45). Owing to space limitations, we simply acknowledge here that significant advances have been made in this field, not only for X-ray structure refinement (57), but also fitting high-resolution structures into low-resolution cryo-electron microscopy density maps (33, 63).
CONCLUSION
Despite their simplicity, ENM-based approaches have shown considerable success in improving our understanding of allosteric mechanisms and explained a wealth of experimental data. Why do such simplified models work?
The answer presumably lies in two features. First, the major ingredient of the theory, native contact topology represented by the spatial distribution of network nodes, plays a dominant role in defining the collective motions at the low-frequency end of the mode spectrum, i.e., the most cooperative modes. The types (attraction or repulsion) or grades (strong or weak) of interactions are not as important as their existence or absence defined by the network topology. Alternatively, one can think that entropic effects make significant contributions to the selection of structural changes. In the GNM, the network topology defines the conformational space accessible to the network/structure via Gaussian fluctuations, as originally set forth in the statistical mechanical theory of elasticity developed for polymer networks, allowing us to assess the entropic cost of each residue’s movement (Equation 10). The agreement between GNM predictions and experimental data, which is usually better than that achieved by ANM, has raised the possibility of entropic drive playing a major role in defining optimal directions/modes of structural changes on the free energy landscape in the neighborhood of the native state (72).
Second, the network models lend themselves to analytical treatments and unique solutions for the collective modes of particular architectures. In other words, although the models are approximate, the use of rigorous statistical mechanical theories of solid-state physics and rubber elasticity, and the effective implementation of NMA and/or spectral graph theoretic methods, allow for obtaining exact analytical solutions for these approximate models. In the opposite case, we may predict some approximate behavior (from simulations that suffer from sampling inefficiencies) despite the use of highly precise models (full atomic representation and detailed force field).
Our contention is that the former group of models and methods provides superior results when exploring large-scale, long-time (microseconds or slower) processes of biomolecular systems, or highly cooperative or allosteric responses that involve long-range spatial couplings. The latter type of studies/simulations, on the other hand, appears to be suited, or required, for stochastic processes in the nanoseconds regime, where local structural heterogeneities, side chain rotameric transitions, and specific interactions can make big differences. These are in sharp contrast to the robust processes elucidated by ENM-based methods, examples of which were presented above. There is definitely room and a need for both of these complementary approaches in exploring biomolecular structural dynamics at global and local scales. In the next few years, we will likely see more examples of integrative methodologies that exploit the capabilities of these two approaches.
SUMMARY POINTS.
- Associated with each protein fold is a set of intrinsically accessible global motions that arise solely from the fold geometry.
- ENMs provide robust information on global protein dynamics by using network representation of the structure with uniform springs accounting for native contact topology.
- A growing body of literature emphasizes the importance of ENM-predicted motions in many aspects of protein function, including ligand and substrate binding and allosteric regulation.
- Increasing structural data for a given protein in different forms provide a broad perspective of functional changes in structure, which may be further elucidated by PCA and compared with ENM predictions to gain a deeper understanding of the mapping of sequence to dynamics to function.
FUTURE ISSUES.
- The overall architecture of biomolecular systems is now recognized to define accessible changes in conformation, or pre-existing substates, which in turn define functional mechanisms of biomolecular systems. How far can ENMs be applied for exploiting the collective dynamics of networks of protein interactions or organelles?
- Can ENMs be used to accurately predict alternative conformations for proteins? For a novel protein, can we predict the bound form from the unbound form, and vice versa?
- Can ENM global modes be exploited to rationally design drugs, such as allosteric inhibitors?
- How well can ENMs capture the global response of a protein to specific stimuli, such as conformational changes induced by ligand binding?
- Is there a well-defined set of evolutionarily selected motions intrinsic to proteins, and essential for life, that can be elucidated by ENM analysis of protein families?
- Can ENM be applied consistently to successfully refine template structures during homology modeling?
- Can proteins be classified with respect to their intrinsic motions? Does this classification overlap with functional classification or other classification based on annotation?
- Is there a simple way of combining evolutionary data with results from ENM to predict the mode/modes that are critical for biological function?
Acknowledgments
Support from NIH grant 1R01GM086238-01 is gratefully acknowledged by I.B. We also thank Ahmet Bakan for his help in preparing Figure 4.
ENM
elastic network model
NMA
normal mode analysis
MD
molecular dynamics
PCA
principal component analysis
GNM
Gaussian network model
ANM
anisotropic network model
CG
coarse-grained
MSF
mean-square fluctuations
PDB
Protein Data Bank
ADP
anisotropic displacement parameter
RMSD
root-mean-square deviation
LIR
leukocyte-immunoglobulin-like receptor
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alexandrov V, Lehnert U, Echols N, Milburn D, Engelman D, Gerstein M. Normal modes for predicting protein motions: a comprehensive database assessment and associated Web tool. Protein Sci. 2005;14:633–43. doi: 10.1110/ps.04882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–15. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahar I, Atilgan AR, Demirel MC, Erman B. Vibrational dynamics of folded proteins: significance of slow and fast motions in relation to function and stability. Phys Rev Lett. 1998;80:2733–36. [Google Scholar]
- 4.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–81. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 5.Bahar I, Chennubhotla C, Tobi D. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr Opin Struct Biol. 2007;17:633–40. doi: 10.1016/j.sbi.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahar I, Erman B, Jernigan RL, Atilgan AR, Covell DG. Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J Mol Biol. 1999;285:1023–37. doi: 10.1006/jmbi.1998.2371. [DOI] [PubMed] [Google Scholar]
- 7.Bahar I, Lezon TR, Bakan A, Shrivastava IH. Normal mode analysis of biomolecular structures: intrinsic motions of membrane proteins. Chem Rev. 2010;110:1463–97. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–92. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahar I, Wallqvist A, Covell DG, Jernigan RL. Correlation between native-state hydrogen exchange and cooperative residue fluctuations from a simple model. Biochemistry. 1998;37:1067–75. doi: 10.1021/bi9720641. [DOI] [PubMed] [Google Scholar]
- 10.Bakan A, Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci USA. 2009;106:14349–54. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks B, Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci USA. 1983;80:6571–75. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–28. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 14.Chennubhotla C, Bahar I. Markov propagation of allosteric effects in biomolecular systems: application to GroEL-GroES. Mol Syst Biol. 2006;2:36. doi: 10.1038/msb4100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chennubhotla C, Bahar I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS Comput Biol. 2007;3:1716–26. doi: 10.1371/journal.pcbi.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci USA. 2005;102:7426–31. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Q, Bahar IE. Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. Boca Raton, FL: Chapman & Hall/CRC; 2006. [Google Scholar]
- 18.Diamond R. On the use of normal modes in thermal parameter refinement: theory and application to the bovine pancreatic trypsin inhibitor. Acta Crystallogr A. 1990;46(Pt. 6):425–35. doi: 10.1107/s0108767390002082. [DOI] [PubMed] [Google Scholar]
- 19.Doruker P, Jernigan RL, Bahar I. Dynamics of large proteins through hierarchical levels of coarse-grained structures. J Comput Chem. 2002;23:119–27. doi: 10.1002/jcc.1160. [DOI] [PubMed] [Google Scholar]
- 20.Eyal E, Chennubhotla C, Yang LW, Bahar I. Anisotropic fluctuations of amino acids in protein structures: insights from X-ray crystallography and elastic network models. Bioinformatics. 2007;23:i175–84. doi: 10.1093/bioinformatics/btm186. [DOI] [PubMed] [Google Scholar]
- 21.Eyal E, Yang LW, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22:2619–27. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 22.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc Natl Acad Sci USA. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flory P. Statistical thermodynamics of random networks. Proc R Soc London Ser A. 1976;351:351–80. [Google Scholar]
- 24.Go N, Noguti T, Nishikawa T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc Natl Acad Sci USA. 1983;80:3696–700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr Opin Struct Biol. 2004;14:104–9. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Haliloglu T, Bahar I. Structure-based analysis of protein dynamics: comparison of theoretical results for hen lysozyme with X-ray diffraction and NMR relaxation data. Proteins. 1999;37:654–67. doi: 10.1002/(sici)1097-0134(19991201)37:4<654::aid-prot15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 27.Haliloglu T, Bahar I, Erman B. Gaussian dynamics of folded proteins. Phys Rev Lett. 1997;79:3090–93. [Google Scholar]
- 28.Haliloglu T, Keskin O, Ma B, Nussinov R. How similar are protein folding and protein binding nuclei? Examination of vibrational motions of energy hot spots and conserved residues. Biophys J. 2005;88:1552–59. doi: 10.1529/biophysj.104.051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han R, Leo-Macias A, Zerbino D, Bastolla U, Contreras-Moreira B, Ortiz AR. An efficient conformational sampling method for homology modeling. Proteins. 2008;71:175–88. doi: 10.1002/prot.21672. [DOI] [PubMed] [Google Scholar]
- 30.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–72. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 31.Hinsen K. Structural flexibility in proteins: impact of the crystal environment. Bioinformatics. 2008;24:521–28. doi: 10.1093/bioinformatics/btm625. [DOI] [PubMed] [Google Scholar]
- 32.Hinsen K, Petrescu A-J, Dellerue S. Harmonicity in slow protein dynamics. Chem Phys. 2000;261:25–37. [Google Scholar]
- 33.Hinsen K, Reuter N, Navaza J, Stokes DL, Lacapere JJ. Normal mode-based fitting of atomic structure into electron density maps: application to sarcoplasmic reticulum Ca-ATPase. Biophys J. 2005;88:818–27. doi: 10.1529/biophysj.104.050716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidera A, Gō N. Refinement of protein dynamic structure: normal mode refinement. Proc Natl Acad Sci USA. 1990;87:3718–22. doi: 10.1073/pnas.87.10.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidera A, Inaka K, Matsushima M, Go N. Normal mode refinement: crystallographic refinement of protein dynamic structure applied to human lysozyme. Biopolymers. 1992;32:315–19. doi: 10.1002/bip.360320404. [DOI] [PubMed] [Google Scholar]
- 36.Kloczkowski A, Mark JE, Erman B. Chain dimensions and fluctuations in random elastomeric networks. Macromolecules. 1989;10:1426–32. [Google Scholar]
- 37.Kondrashov DA, Cui Q, Phillips GN., Jr Optimization and evaluation of a coarse-grained model of protein motion using X-ray crystal data. Biophys J. 2006;91:2760–67. doi: 10.1529/biophysj.106.085894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondrashov DA, Van Wynsberghe AW, Bannen RM, Cui Q, Phillips GN., Jr Protein structural variation in computational models and crystallographic data. Structure. 2007;15:169–77. doi: 10.1016/j.str.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshland DE., Jr Enzyme flexibility and enzyme action. J Cell Comp Physiol. 1959;54:245–58. doi: 10.1002/jcp.1030540420. [DOI] [PubMed] [Google Scholar]
- 40.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–85. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 41.Kryshtafovych A, Fidelis K, Moult J. Progress from CASP6 to CASP7. Proteins. 2007;69(Suppl 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- 42.Kundu S, Melton JS, Sorensen DC, Phillips GN., Jr Dynamics of proteins in crystals: comparison of experiment with simple models. Biophys J. 2002;83:723–32. doi: 10.1016/S0006-3495(02)75203-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurkcuoglu O, Turgut OT, Cansu S, Jernigan RL, Doruker P. Focused functional dynamics of supramolecules by use of a mixed-resolution elastic network model. Biophys J. 2009;97:1178–87. doi: 10.1016/j.bpj.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leo-Macias A, Lopez-Romero P, Lupyan D, Zerbino D, Ortiz AR. An analysis of core deformations in protein superfamilies. Biophys J. 2005;88:1291–99. doi: 10.1529/biophysj.104.052449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levitt M, Sander C, Stern PS. Protein normal-mode dynamics: trypsin inhibitor, crambin, ribonuclease and lysozyme. J Mol Biol. 1985;181:423–47. doi: 10.1016/0022-2836(85)90230-x. [DOI] [PubMed] [Google Scholar]
- 46.Lezon TR, Sali A, Bahar I. Global motions of the nuclear pore complex: insights from elastic network models. PLoS Comput Biol. 2009;5(9):e1000496. doi: 10.1371/journal.pcbi.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lezon TR, Srivastava I, Zheng Y, Bahar I. Elastic network models for biomolecular dynamics: theory and application to membrane proteins and viruses. In: Boccaletti S, Latora V, Moreno Y, editors. Handbook on Biological Networks. Hackensack, NJ: World Scientific; 2009. pp. 129–58. [Google Scholar]
- 48.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu M, Ma J. A minimalist network model for coarse-grained normal mode analysis and its application to biomolecular X-ray crystallography. Proc Natl Acad Sci USA. 2008;105:15358–63. doi: 10.1073/pnas.0806072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–80. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Maragakis P, Karplus M. Large amplitude conformational change in proteins explored with a plastic network model: adenylate kinase. J Mol Biol. 2005;352:807–22. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 52.Marechal JD, Perahia D. Use of normal modes for structural modeling of proteins: the case study of rat heme oxygenase 1. Eur Biophys J. 2008;37:1157–65. doi: 10.1007/s00249-008-0279-y. [DOI] [PubMed] [Google Scholar]
- 53.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–24. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 54.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100:12570–75. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 56.Mouawad L, Perahia D. Motions in hemoglobin studied by normal mode analysis and energy minimization: evidence for the existence of tertiary T-like, quaternary R-like intermediate structures. J Mol Biol. 1996;258:393–410. doi: 10.1006/jmbi.1996.0257. [DOI] [PubMed] [Google Scholar]
- 57.Poon BK, Chen X, Lu M, Vyas NK, Quiocho FA, et al. Normal mode refinement of anisotropic thermal parameters for a supramolecular complex at 3.42-Å crystallographic resolution. Proc Natl Acad Sci USA. 2007;104:7869–74. doi: 10.1073/pnas.0701204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian B, Ortiz AR, Baker D. Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation. Proc Natl Acad Sci USA. 2004;101:15346–51. doi: 10.1073/pnas.0404703101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rader AJ, Bahar I. Folding core predictions from network models of proteins. Polymer. 2004;45:659–68. [Google Scholar]
- 60.Riccardi D, Cui Q, Phillips GN., Jr Application of elastic network models to proteins in the crystalline state. Biophys J. 2009;96:464–75. doi: 10.1016/j.bpj.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song G, Jernigan RL. vGNM: a better model for understanding the dynamics of proteins in crystals. J Mol Biol. 2007;369:880–93. doi: 10.1016/j.jmb.2007.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suhre K, Navaza J, Sanejouand YH. NORMA: a tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Crystallogr D. 2006;62:1098–100. doi: 10.1107/S090744490602244X. [DOI] [PubMed] [Google Scholar]
- 64.Suhre K, Sanejouand YH. ElNémo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–14. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tama F, Brooks CL. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu Rev Biophys Biomol Struct. 2006;35:115–33. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- 66.Tama F, Gadea FX, Marques O, Sanejouand YH. Building-block approach for determining low-frequency normal modes of macromolecules. Proteins. 2000;41:1–7. doi: 10.1002/1097-0134(20001001)41:1<1::aid-prot10>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 67.Tama F, Sanejouand YH. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 68.Thorpe MF. Comment on elastic network models and proteins. Phys Biol. 2007;4:60–63. doi: 10.1088/1478-3975/4/1/N01. [DOI] [PubMed] [Google Scholar]
- 69.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77:1905–8. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 70.Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc Natl Acad Sci USA. 2005;102:18908–13. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–7. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 72.Xu C, Tobi D, Bahar I. Allosteric changes in protein structure computed by a simple mechanical model: hemoglobin T → R2 transition. J Mol Biol. 2003;333:153–68. doi: 10.1016/j.jmb.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 73.Xu Y, Colletier JP, Jiang H, Silman I, Sussman JL, Weik M. Induced-fit or preexisting equilibrium dynamics? Lessons from protein crystallography and MD simulations on acetylcholinesterase and implications for structure-based drug design. Protein Sci. 2008;17:601–5. doi: 10.1110/ps.083453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang L, Song G, Carriquiry A, Jernigan RL. Close correspondence between the motions from principal component analysis of multiple HIV-1 protease structures and elastic network modes. Structure. 2008;16:321–30. doi: 10.1016/j.str.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Song G, Jernigan RL. Protein elastic network models and the ranges of cooperativity. Proc Natl Acad Sci USA. 2009;106:12347–52. doi: 10.1073/pnas.0902159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang LW, Chng C-P. Coarse-grained models reveal functional dynamics—I. elastic network models—theories, comparisons and perspectives. Bioinform Biol Insights. 2008;2:25–45. doi: 10.4137/bbi.s460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang LW, Eyal E, Bahar I, Kitao A. Principal component analysis of native ensembles of biomolecular structures (PCA NEST): insights into functional dynamics. Bioinformatics. 2009;25:606–14. doi: 10.1093/bioinformatics/btp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang LW, Eyal E, Chennubhotla C, Jee J, Gronenborn AM, Bahar I. Insights into equilibrium dynamics of proteins from comparison of NMR and X-ray data with computational predictions. Structure. 2007;15:741–49. doi: 10.1016/j.str.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Z, Majek P, Bahar I. Allosteric transitions of supramolecular systems explored by network models: application to chaperonin GroEL. PLoS Comput Biol. 2009;5:e1000360. doi: 10.1371/journal.pcbi.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng W. A unification of the elastic network model and the Gaussian network model for optimal description of protein conformational motions and fluctuations. Biophys J. 2008;94:3853–57. doi: 10.1529/biophysj.107.125831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng W, Brooks BR, Hummer G. Protein conformational transitions explored by mixed elastic network models. Proteins. 2007;69:43–57. doi: 10.1002/prot.21465. [DOI] [PubMed] [Google Scholar]
- 82.Zheng W, Brooks BR, Thirumalai D. Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations. Proc Natl Acad Sci USA. 2006;103:7664–69. doi: 10.1073/pnas.0510426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng W, Doniach S. A comparative study of motor-protein motions by using a simple elastic-network model. Proc Natl Acad Sci USA. 2003;100:13253–58. doi: 10.1073/pnas.2235686100. [DOI] [PMC free article] [PubMed] [Google Scholar]