Phosphotyrosine Signaling: Evolving a New Cellular Communication System (original) (raw)
. Author manuscript; available in PMC: 2011 Mar 3.
Abstract
Tyrosine phosphorylation controls many cellular functions. Yet the three-part toolkit that regulates phosphotyrosine signaling -- tyrosine kinases, phosphotyrosine phosphatases, and Src Homology 2 (SH2) domains -- is a relatively new innovation. Genomic analyses reveal how this revolutionary signaling system may have originated and why it rapidly became critical to metazoans.
Throughout human history, new technologies and technological platforms have constantly been invented. Only a small fraction of these technologies go on to be widely adopted, but these can ultimately have transformational consequences. In the evolutionary history of living organisms, we know that innovative molecular systems have appeared at key points in time, and these are thought to have played a transformative role in major evolutionary transitions in the tree of life. But how do such innovative molecular systems emerge, and how and why do some proliferate and become stably adopted by subsequent lineages?
An example of such an innovative molecular system is phosphotyrosine (pTyr)-based signal transduction. This molecular system for transmitting cellular regulatory information is estimated to have appeared relatively recently in the history of life ---~600 million years ago, just prior to the emergence of multicellular animals (King et al., 2004; Pincus et al., 2008; Manning et al., 2008). The pTyr signaling system has become an essential part of metazoan biology. For example, pTyr signaling plays a central role in many cell-to-cell communication pathways, including those that regulate proliferation, differentiation, adhesion, hormone responses, and immune defense (Hunter, 2009).
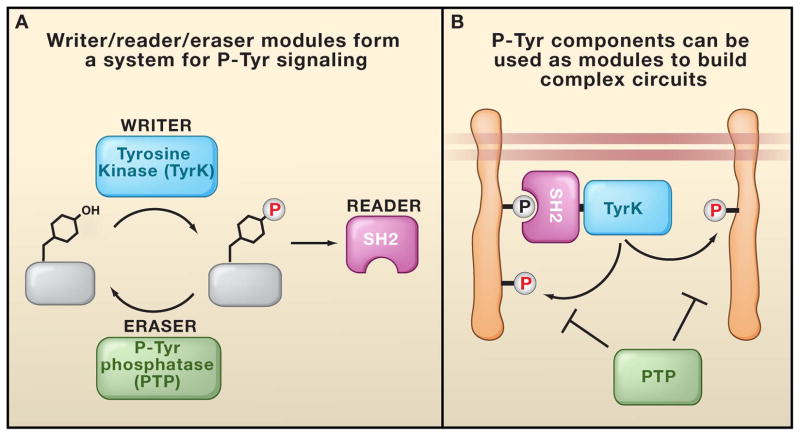
In modern metazoans, pTyr signaling is mediated by a toolkit of three distinct functional modules:tyrosine kinases (TyrK) phosphorylate specific target tyrosine residues, phosphotyrosine phosphatases (PTP) remove the phosphates, and Src Homology 2 (SH2) domains recognize the modifications (Pawson 1995). Together, these three modules form the “writer,” “eraser,” and “reader” toolkit that is common to many diverse cellular information processing platforms (Figure 1A). A rich array of diverse and complex regulatory schemes can be achieved through the dynamic interplay of these three modular functions (Pawson 1995; Bhattacharyya et al., 2006; Kholodenko, 2006). A combination of these modules can lead to higher order functions (Figure 1B). For example, there are several proteins containing a combination of SH2 and kinase domains that can generate positive feedback (phosphorylation of tyrosine sites leads to SH2-mediated recruitment of the kinase, and subsequently, more extensive phosphorylation) (Pawson, 2004). Similarly SH2-phosphatase domain combinations can generate negative feedback (Tonks et al., 2001).
Figure 1. The Writer, Reader, Eraser pTyr Toolkit.
(A) In pTyr signaling, the tyrosine kinase (TyrK), Src Homology 2 (SH2), and phosphotyrosine phosphatase (PTP) domains form a highly interdependent signaling platform. This platform serves as the writer, reader, and eraser modules, respectively, for processing pTyr marks. (B) Components of pTyr signaling can be used to build complex circuits. For example, recruitment of an SH2-TyrK protein to an initiating pTyr site can lead to amplification of tyrosine phosphorylation through a positive feedback loop.
The three-part pTyr signaling toolkit thus raises a classic question in evolutionary biology: How do complex, interdependent systems arise? It is clear why a new system encompassing a writer, eraser, and reader might be extremely useful. But given their interdependence, how could these individual components arise in a stepwise fashion consistent with an evolutionary process? Proteins that bind or remove a post-translational modification would seem useless without an enzyme to generate the modification, and, in principle, would not provide a fitness advantage leading to its retention and spread. The pTyr signaling platform provides a case study to look for plausible stepwise pathways of the evolution of a multipart system.
Here we reconstruct a possible history for the evolution of pTyr signaling. This reconstruction is based on the recent sequencing of the genomes of a number of organisms that originated both before and after the emergence of metazoans from single-celled eukaryotic ancestors (King et al., 2008). The genome sequence of the choanoflagellate, Monosiga brevicollis, has been particularly illuminating as choanoflagellates are thought to be one of the closest single-celled relatives of metazoans. We present a model for how this three-part signaling system could have plausibly evolved in a stepwise manner. We propose that once the complete three-part system was in place, it may have rapidly taken hold in subsequent lineages because it could generate new regulatory behaviors without significant cross-interference with existing regulatory circuits. We also discuss the possible role of this new communication system in facilitating the transformative evolutionary shift to multicellularity.
Given the incomplete record, however, such an evolutionary reconstruction is highly speculative. For example, we cannot rule out more complex paths involving cycles of evolutionary gain and loss of components, nor the possibility that similar components in distinct lineages have independent origins. Nonetheless, this model may provide a useful framework for focusing studies of pTyr signaling origins and the origins of analogous multi-component signaling platforms.
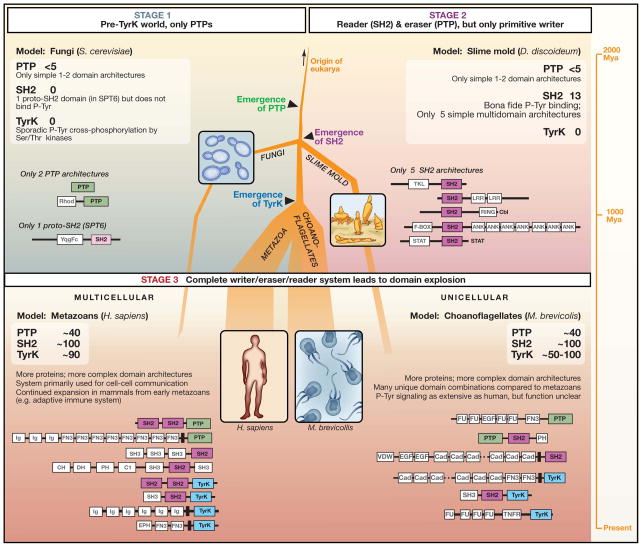
We describe three possible stages in the emergence of the modern pTyr signaling toolkit, each represented by an extant model organism (Figure 2). These stages are representative; we do not claim to define the exact path of evolution, but rather focus on identifying the dominant classes of stable intermediates than can exist in the broader evolutionary landscape.
Figure 2. Evolution of pTyr signaling.
Shown is a possible path for the emergence of phosphotyrosine (pTyr) signaling. . We postulate three successive stages, each represented by what is observed in a modern organism. The thickness of the tree reflects the approximate degree of usage of pTyr signaling (thicker lines means more usage). Stage 1 (exemplified by the budding yeast Saccharomyces cerevisiae) reflects the situation in early eukaryotes, in which PTPs emerged but were limited in number and complexity. They were most likely used to reverse or process sporadic cross-phosphorylation of tyrosine residues by Ser/Thr kinases. S. cerevisiae has <5 PTP proteins and no functional SH2 or TyrK domains. Stage 2 reflects systems in which functional SH2 domains emerged that were able to bind to pTyr motifs. Together with Ser/Thr kinases with increased cross-reactivity for Tyr (such as tyrosine kinase-like or dual specificity Ser/Thr kinases), these systems may reflect the most primitive of pTyr writer/reader/eraser systems. However, the lack of a dedicated Tyr kinase may have limited the utility and expansion of this toolkit. This stage is potentially represented by the slime mold, Dictyostelium discoideum. Stage 3 reflects systems that evolved after the emergence of the modern TyrK domain. We postulate that the full writer/reader/eraser system was of so much greater utility, that its use expanded dramatically. This likely resulted in many more proteins in these families, as well as much more complex, multi-domain architectures than those seen in the earlier stages. This stage is represented by both the multicellular metazoan and unicellular choanoflagellate lineages.
PTPs in a pre-tyrosine kinase world
What came first, TyrK, PTP, or SH2 domains? Sequence analysis suggests it was PTP domains. The genome of a simple single-celled eukaryote like the budding yeast Saccharomyces cerevisiae shows no TyrK proteins and one proto-SH2 domain, but a handful of PTP proteins (Pincus et al., 2008) (Figure 2, Stage 1). Most fungi have ≤5 PTP proteins, and several of these have tyrosine phosphatase activity. We refer to the single putative “SH2” domain in yeast, found within the gene Sup6T, as a proto-SH2 domain because it does not show pTyr binding (the domain has been reported to show phospho-Ser/Thr binding; Dengl et al., 2009). Thus, functionally, it cannot be considered a “reader” domain that is part of a pTyr regulatory system. These observations suggest a simple model: the first step in the evolution of the three-part pTyr signaling machinery was likely to have been the emergence of a functional tyrosine phosphatase. But why would PTPs arise in the pre-tyrosine kinase world? What functional use and fitness advantage would this eraser domain provide in organisms lacking a writer domain?
The answer may lie in the fact that some Ser/Thr kinase domains, which are more ancient than tyrosine kinases (dating back close to the origins of eukaryotes), can carry out sporadic but functionally important phosphorylation of tyrosines. Phosphoamino acid analysis of yeast reveals a small but significant population of pTyr (Schieven et al., 1986). Moreover, certain events, such as the activation of mitogen activated protein kinases (MAPKs) and inhibition of the cell cycle kinase Cdk1, are known to involve phosphorylation of tyrosine residues (for activation, a MAPK must be phosphorylated by an upstream Ser/Thr kinase on both a Thr and Tyr residue within its activation loop; Cdk1 is phosphorylated on Tyr14 by the inhibitory kinase Wee1). These tyrosine modifications are clearly not recognized by SH2 domains, but exert direct allosteric effects within the proteins in which they occur. Thus, PTP domains may have provided a fitness benefit by negatively modulating these rare but functionally important phosphorylation events. Consistent with this model, the proteins PTP2 and PTP3 in yeast clearly have a functionally important role in downregulating MAPK-mediated signaling in response to pheromones or osmolarity changes, explaining their fitness benefit (Pincus et al., 2008). In addition, PTPs may have played a general role buffering against the occasional harmful stray phosphorylation of functionally important tyrosines.
Where did these PTPs come from? PTPs are likely to have arisen from a common ancestor of the related dual-specificity phosphatases, which are also found in most single-celled eukaryotes (Kennelly et al., 2001; Alonso et al., 2004). Dual-specificity phosphatases are catalytic domains that can dephosphorylate both pSer/Thr and pTyr substrates. The PTP and dual-specificity phosphatase catalytic domains are distinct, but are evolutionarily related. They share a common fold and the core catalytic motif HC(X)5R in which a phospho-cysteine enzyme intermediate is generated during catalysis. (Sometimes both dual-specificity phosphatases and classical PTPs are referred to as PTPs; here, we use this nomenclature only for the classical PTP domains that act only on pTyr). The domains of dual-specificity phosphatases have a shallower active site than classical PTPs, which may explain why they can dephosphorylate either Tyr or Thr/Ser residues. In some lineages, dual-specificity phosphatases have functionally diverged further, giving rise to members that can act on lipid substrates, such as the phosphoinositide phosphatases PTEN and the myotubularins (Alonso et al., 2004). Thus, the PTPs appear to have arisen from a somewhat promiscuous class of multi-functional phosphatases.
Despite the presence of PTP proteins in fungi, there are striking differences between these proteins and those found in metazoans (Figure 2). For example, there are far fewer PTPs in fungi (~5/genome versus ~40/genome in metazoans) and they are considerably less complex in domain architecture (Pincus et al., 2008). Metazoan PTP proteins tend to be large multi-domain proteins in which the PTP module has been functionally recombined with multiple other signaling modules. In contrast, in fungi, the PTP domains are all either in simple single domain proteins, or in combination with a single rhodanase-like domain (a putative regulatory domain that is homologous to a class of sulfur transfer enzymes; Bordo and Bork, 2002). Thus, fungal PTP proteins are very simple (1–2 domains) and lack the combinatorial complexity of metazoan PTP proteins. The simplicity and low number of PTPs in yeast suggests that in early single-celled eukaryotes, PTP domains had fairly limited functional utility, especially when compared to their broad and complex usage in metazoans.
Unlike PTPs, there are no known pTyr-binding SH2 domains in fungi, although there is one clearly homologous domain found in the yeast protein SPT6. This protein, which has a domain with an SH2-like sequence and fold, is involved in the regulation of transcription elongation, and the SH2 domain binds to the Ser/Thr phosphorylated C-terminal tail of RNA polymerase II. The domain does not bind to pTyr (Dengl et al., 2009). Interestingly, a single SPT6 ortholog, with the same overall domain architecture, is found in all eukaryotes, including all fungi and metazoans (but not prokaryotes). This finding suggests that in early eukaryotes, a proto-SH2 domain emerged to perform a highly specialized function — one that was unrelated to the flexible modular pTyr recognition function of the modern SH2 domain. This proto-SH2 domain most likely did not “read” pTyr modifications, but instead recognized a specialized related modification. Thus, although SPT6 is likely to represent an early ancestor or relative that eventually gave rise to modern SH2 domains, it cannot be considered a functional part of a pTyr regulatory toolkit. We therefore postulate that early eukaryotes had only a pTyr eraser function (mediated by PTPs) with no specialized complementary reader or writer functions.
In summary, the PTP domain and a structural ancestor of the SH2 domain appear to have arisen in early single-celled eukaryotes, but are likely to have functional origins that are not directly related to their later function in modern pTyr regulatory systems. These components may have provided a limited but incremental fitness advantage, even in the absence of a specialized tyrosine kinase domain.
Towards a Write/Read/Erase System
In the early days of a more sophisticated pTyr-signaling system, we suggest that a proto-SH2 domain (mostly likely a homologue of the yeast Spt6 protein) in a single-celled organism acquired the new and functionally beneficial ability to bind to pTyr-containing peptide motifs. The slime mold Dictyostelium discoideum has the simplest repertoire of bona fide pTyr-binding SH2 domains, and may therefore provide a living representative of this second evolutionary stage (Figure 2, Stage 2). Dictyostelium is a soil-living amoeba that has a unicellular lifestyle in the presence of bacterial food. However, when food is depleted, individual cells aggregate in response to the chemoattractant cAMP to form a multicellular structure, which then develops into a fruiting body through the differentiation of stalk and spore cells. The rudimentary pTyr-SH2 system in Dictyostelium is important for aspects of this differentiation process, including intracellular responses to both cAMP and the morphogen differentiation inducing factor or DIF (which induces the differentiation of pre-stalk cells), as well as for transcriptional regulation in response to hyperosmotic stress. These observations are consistent with early pTyr-SH2 signaling playing a role in cellular responses to changing environmental conditions.
The Dictyostelium genome specifies 13 proteins with SH2 domains (as well as a single Spt6 homologue). These 13 proteins cluster into five basic domain architectures, two of which are homologous to metazoan SH2 proteins. Notably, Dictyostelium has four STAT (Signal Transducers and Activators of Transcription) proteins that are very similar to metazoan STAT transcription factors (Kay et al., 1997; Kawata et al., 1997). For example, they all have an SH2 domain juxtaposed to a DNA-binding region; they are inducibly phosphorylated on tyrosine residues in response to stress or the extracellular signaling molecule DIF; they undergo pTyr/SH2-mediated dimerization and then translocate to the nucleus to regulate the expression of specific genes. Dictyostelium also has an ortholog of the mammalian E3 ubiquitin ligase Cbl, which uses SH2 and Ring domains to couple pTyr signals to the ubiquitination machinery (Langenick et al., 2008). The remaining three domain architectures of Dictyostelium SH2 proteins are distinct from those found in other sequenced organisms. The LrrB protein has an SH2 domain linked to a leucine-rich repeat domain (Sugden et al., 2010), whereas the FbxB protein has an F-box followed by an SH2 domain and ankyrin repeats. In addition, the Shk proteins have a protein kinase domain followed by an SH2 domain, a domain combination that is somewhat similar to metazoan cytoplasmic tyrosine kinases like Src (Moniakis et al., 2001). The Shk catalytic domain, however, lacks motifs characteristic of bona fide tyrosine kinases, and biochemically displays dual specificity towards serine/threonine and tyrosine residues.
Indeed, Dictyostelium differs from metazoans and choanoflagellates in that its genome does not encode any modern tyrosine-specific protein kinases. For example, metazoan STAT proteins are usually phosphorylated by Janus tyrosine kinases (JAKs), but there are no JAKs in Dictyostelium (Kay et al., 1997). This suggests the possibility that signaling proteins containing SH2 domains such as STATs evolved before the modern tyrosine kinases with which they are associated in metazoans. The identity of the kinase responsible for STAT tyrosine phosphorylation, and the consequent formation of SH2-binding sites, remains mysterious.
How, then, is tyrosine phosphorylation of Dictyostelium proteins such as the STATs controlled? Thus far, genetic analysis has not identified a specific relevant kinase, and it has been proposed that, in contrast to mammalian STATs, there may be basal constitutive phosphorylation of Dictyostelium STAT tyrosine sites, which is regulated by changes in PTP activity in response to extracellular signals (Langenick et al., 2008). One of the PTPs in Dictyostelium, PTP3, binds and dephosphorylates STATc, thereby blocking SH2-mediated dimerization and STATc accumulation in the nucleus. Signaling induced by the DIF morphogen appears to transmit signals by inhibiting PTP3 activity and consequently boosting STATc tyrosine phosphorylation and STATc-dependent gene expression.
Although Dictyostelium lacks true tyrosine kinases, it is noteworthy that its genome has a significant expansion in the number of putative dual-specificity protein kinases (there are ~70, also known as tyrosine kinase-like or TKL kinases) (Manning et al., 2008). This set includes the Shk catalytic domain, described above. It is unlikely that any of these kinases are precursors of modern tyrosine kinases. However, it is plausible that these represent the first evolutionary form of the “writer” function in a prototype pTyr three-part regulatory system. The union of an SH2 domain and a dual specificity kinase domain, as found in the Shk proteins, may be an early example of linking “reader” and “writer” modules to achieve more complex functions such as positive feedback. Nonetheless, the limited functionality of the dual specificity kinases in carrying out tyrosine phosphorylation may have limited the capabilities of this early system. This may explain the very modest expansion of pTyr signaling in organisms such as Dictyostelium.
These observations paint the following picture of Dictyostelium pTyr signaling, and by extension, of an early phase in the evolution of pTyr communication. SH2 domains have acquired pTyr-binding activity, and are found in several distinct combinations with other types of signaling domains. Among these, the STAT and Cbl proteins are shared with metazoans, whereas the LrrB, FbxB, and Shk proteins are unique to Dictyostelium. But no dedicated modern tyrosine kinases have been found, and the dynamic control of tyrosine phosphorylation may be primarily regulated by PTPs. Although functionally important for aggregation and differentiation, the pTyr signaling system has not acquired the pervasive influence evident in M. brevicollis and metazoans, perhaps due to the lack of an efficient tyrosine kinase. Put another way, Dicytostelium has effective pTyr readers and erasers, but the writer is poorly developed.
Invention of TyrK and expansion of the pTyr toolkit
Current analysis suggests that the modern tyrosine kinases arose just prior to the evolution of the metazoans. Aside from metazoans, canonical tyrosine kinases have thus far only been observed in the choanoflagellates, which appear to be the closest known single-celled relatives of metazoans (King et al., 2008). The absence of significant numbers of such tyrosine kinases in any other branch of life, suggests that this new catalytic domain evolved in a recent common ancestor of choanoflagellates and metazoans, most likely as a branch of the older Ser/Thr kinases. Some bacteria do have specialized tyrosine kinases (BY kinases), but these resemble P-loop NTPases (nucleotide triphosphatases) and are structurally unrelated to eukaryotic tyrosine kinases (Lee et al., 2009). It is therefore probable that BY kinases evolved separately from metazoan tyrosine kinases, and operate in a different fashion.
The new eukaryotic tyrosine kinase domain appears to have been a game changing innovation (Figure 2, Stage 3). The total number of tyrosine kinase proteins in both choanoflagellate and metazoan species is in the range of 30–150 per genome (Pincus et al., 2008; Manning et al., 2008). Among sequenced genomes, there is a striking absence of species with only a small number of TyrK proteins. This all-or-none sudden jump in the number of TyrK proteins suggests their importance as they appear to have undergone rapid expansion and subsequent retention.
What is perhaps more striking is the observation that the emergence of the TyrK domain and its rapid expansion correlates with an equally rapid expansion of PTP and SH2 domains within the same genomes (Pincus et al., 2008). Although fungi and Dictyostelium have ~5 PTP proteins, metazoans and choanoflagellates have 30–40 per genome. Similarly, Dictyostelium has ~10 SH2-domain containing proteins (fungi have none), whereas metazoans and choanoflagellates have ~100 each. Thus, both PTP and SH2 proteins undergo a roughly 10-fold increase in number per genome after the emergence of the TyrK domain. Moreover, the proteins containing SH2 and PTP domains become far more complex and varied (Jin et al., 2009). For example, in yeast and Dictyostelium, SH2 and PTP proteins normally are very simple one or two domain proteins. However, in lineages that have modern TyrK proteins, SH2 and PTP proteins almost always comprise 3 to 10 domains.
These observations are consistent with the following model. When a far more efficient TyrK domain – or “writer” function – emerged, this dramatically increased the functional utility of the pre-existing PTP (eraser) and SH2 (reader) domains. As a three-part toolkit---a catalytic domain to generate pTyr, an interaction domain to bind to these pTyr sites, and an enzyme to dephosphorylate them---this domain set could be used to encode and execute a far wider and diverse range of regulatory functions, thus leading to the subsequent expansion of the complete set. Although the PTP and SH2 domains had utility in simpler organisms, their much larger functional potential was not unleashed until the emergence of the TyrK domain.
The rapid expansion of the pTyr signaling machinery in the ancestors of choanoflagellates and animals is reminiscent of how technology expands in quantum jumps, especially in situations involving co-dependent technologies. For example, the value of the laser expanded dramatically after the later invention of the complementary technology of fiber optics. This co-dependent technology allowed lasers to be repurposed to rapidly displace electrical transmission via copper wires as the backbone of global communication (Alwayn, 2004). Thus, although lasers had standalone utility, their major application had to await the introduction of complementary technology. The expansion of molecular components in biology is likely to be similar. A toolkit of writer, reader, and eraser functions may be of full use only when all components are present. Thus, it may be common for any system of this type to show a quantum “all-or-none” expansion only when the final piece of the toolkit emerges.
Applying the new pTyr toolkit to different functions
Although both choanoflagellate and metazoan lineages show a large expansion of the three-part pTyr regulatory machinery, the way in which these components are used appears to be quite different. When one examines the domain types that co-occur with TyrK, SH2, or PTP domains one finds many distinct combinations that are unique to each lineage (Pincus et al., 2008; Manning et al., 2008). These differences in domain combinations imply distinct functions for proteins containing these domains in the choanoflagellate and metazoan lineages (Li et al., 2009). Assuming that the evolution of new TyrK, SH2 and PTP proteins occurred by recombination with new accessory domains (Jin et al., 2009; Peisajovich et al., 2010), this observation also implies that the complete signaling toolkit emerged only shortly before the divergence of metazoans and choanoflagellates (i.e. shortly before the evolution of metazoan multicellularity), and that much of the divergent expansion of these domain families occurred after the lineage split.
Thus, earlier assumptions that pTyr signaling is only used in metazoan cell-cell communication are clearly incorrect. Choanoflagellates do not form the complex and permanent cell-cell organization that metazoans do, yet surprisingly they have a comparable (if not greater) number of pTyr signaling proteins (Manning et al., 2008). Sequencing of other organisms that arose near the origins of metazoans is ongoing. Preliminary data also suggest a large number of pTyr signaling proteins in other single-celled relatives of metazoans. Thus, it may be more reasonable to view the pTyr signaling system as an innovative but generic information processing system that could potentially be used for transmitting many different types of information.
Orthogonal Signaling: A Platform For Biological Innovation
When the three-part pTyr system first emerged, it presented a new platform with which to transmit information that was orthogonal to pre-existing signaling systems. Because it was based on a distinct covalent modification, new regulatory circuits could be assembled with these components without significant cross-interference with pre-existing networks. Thus, this brand new signaling apparatus likely had a high encoding potential for evolving dramatically new functions, such as those involved in multicellularity. One possible problem that could be caused by the expansion of the new pTyr signaling enzymes might be excessive general phosphorylation of tyrosine residues throughout the proteome. Interestingly, however, organisms using pTyr signaling may have developed a simple solution to deal with this problem -- a decrease in the tyrosine content of proteins across the proteome is observed to correlate with tyrosine kinase expansion (Tan et al., 2009).
A new orthogonal signaling system like the pTyr signaling platform can be viewed as analogous to a newly opened region in the telecommunications spectrum. New frequencies provide the opportunity for transmitting large amounts of information as there is little interference from existing communication. Because of this valuable high encoding potential, there is extreme pressure to quickly fill this region of the spectrum. Moreover, the exact type of information carried by each region of the spectrum is flexible – for example the same region of the spectrum can be assigned to different functions in different countries. We hypothesize that the new pTyr signaling system that emerged prior to metazoans presented similar new opportunities to transmit more information. This virgin system was rapidly exploited, though the way it has been used appears to be different in the two branches (metazoans and choanoflagellates) that emerged after the complete toolkit was established.
It is tempting to speculate that the emergence of a new signaling system with high encoding potential may have played a key role in the emergence of a new, complex biological function such as metazoan multicellularity. Such large-scale phenotypic evolutionary innovations may require and coincide with innovations in basic molecular components (King et al., 2004; Rokas et al., 2008).
Indeed, we speculate that pTyr signaling may provide a more general model for the generation of multi-component biological systems, involving first a limited stepwise development of elements that together have a rudimentary biological utility, followed by an explosive spread, once all of the components of the mature system are in place. Exploration of this concept, and further analysis of the evolution of pTyr signaling, will be assisted by the increasing sequence information being gathered for both unicellular and multicellular eukaryotes (Srivastava et al., 2010), which will no doubt yield surprises akin to the discovery of extensive pTyr signaling in M. brevicollis. Moreover, as genomic information bracketing other major evolutionary transitions becomes available, it will be interesting to see if these innovations are also associated with the explosive expansion of new molecular toolkits.
A key point here is that the specific emergence of the pTyr toolkit may not have been essential for the evolution of multicellularity, but rather, any number of new orthogonal signaling toolkits with the same high encoding potential could have served a similar role. Other analogous new molecular information currencies could have, in principle, been able to serve as the substrate for dramatic phenotypic innovation. In this context, plants make extensive use of protein phosphorylation, and have numerous transmembrane receptor Ser/Thr kinases, but they lack conventional tyrosine kinases, indicating that pTyr-based signaling is not the only mechanism of information transfer through which organisms can achieve multicellularity.
Is pTyr Signaling Saturated?
How close is the pTyr signaling system to being saturated? Is there still available encoding potential that could be tapped for the evolution of new pathways and behaviors? It is difficult to answer these questions. However, the fact that new pTyr signaling proteins appear to be associated with advanced processes like adaptive immunity suggests that there was still some remaining encoding potential in the system as late as the evolution of mammals. The evolutionary history reconstructed here begs many questions. Are their new regulatory toolkits evolving now or in the future? Will these new toolkits be the substrate required for the next big evolutionary innovation?
The importance of new molecular toolkits is conversely also very relevant to the emerging field of synthetic biology, in which the goal is to engineer cellular systems with new functions. A major potential limitation is how to build such new functions in a reliable fashion that does not cross-interfere in unanticipated ways with existing systems (Lim, 2010). Can we develop new synthetic molecular signaling currencies that are orthogonal to existing natural ones, and would these systems dramatically facilitate our ability to reliably and predictably endow cells with innovative new functions?
Conclusions
Current data suggest that PTP and SH2 domains evolved before modern TyrK domains, most likely to process pTyr modifications sporadically catalyzed by Ser/Thr kinases. However, the PTP and SH2 domain protein families did not expand dramatically until the emergence of an efficient TyrK. We postulate that only with the complete toolkit of writer (TyrK), reader (SH2), and eraser (PTP) domains, was the full encoding potential of this system unleashed, leading to rapid expansion and elaboration of these domain families. This type of explosive increase in component usage may prove to be common to all multi-part molecular systems. The emergence of the modern TyrK maps just prior to the split between metazoans and choanoflagellates. These two lineages appear to have used this new molecular communication system in distinct ways – multicellular metazoans used it for cell-cell coordination, whereas unicellular choanoflagellates used it for distinct but as yet uncharacterized functions.
Thus, we are able to reconstruct a plausible model by which the tyrosine signaling machinery could have evolved in a relatively simple stepwise manner into what today is a complex and highly interdependent system. In this model, evolution is opportunistic and forward looking, borrowing and repurposing machinery that pre-exists. The first simple PTP proteins likely arose from the more ancient Ser/Thr phosphatase family and may have been maintained initially as a way to reverse the unavoidable occasional tyrosine phosphorylation event catalyzed by a Ser/Thr kinase. In some cases, like the MAPKs, which are present in all eukaryotes, these tyrosine phosphorylation events appear to have become exploited and fixed as actual parts of signal transmission, alongside Ser/Thr phosphorylation events. SH2 domains also likely arose from a pre-existing fold in the SPT6 protein, which is found in all eukaryotes but has no pTyr binding activity. But this fold, once co-opted for this function, began to expand, most likely because of its ability to contribute to a wider range of modular signaling events. But the full utility of these components was only unleashed upon the emergence of the modern TyrK domain, which led to the highly expanded three-part system. One cannot help but wonder what other simple pieces of molecular machinery may be lying around in today’s biological systems, of limited utility now, but awaiting the emergence of some as yet unknown complementary component that will generate a complete toolkit that will help to drive future evolutionary innovation.
Acknowledgments
We thank D. Pincus, B. Mayer, P. Beltrao, O. Hoeller, R.Linding, G. Superti-Furga, N.King, N.Helman, L. Holt, A. Horwitz, T. Miller, G.Manning, T. Hunter, D.Morgan, J. Williams and H. Bourne for helpful comments. This work was supported by the Howard Hughes Medical Institute (WL), the NIH (GM55040, GM62583 and EY016546 – WL), the Packard Foundation (WL), the NSF Synthetic Biology Engineering Research Center (WL), the Canadian Institutes for Health Research (TP - MOP – 6849), Genome Canada (TP) and the Canadian Cancer Society Research Institute (TP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Alwayn V. Optical Network Design and Implementation. Cisco Press; 2004. [Google Scholar]
- Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Annu Rev Biochem. 2006;75:655–80. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Bordo D, Bork P. EMBO reports 3. 2002;8:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengl S, Mayer A, Sun M, Cramer P. J Mol Biol. 2009;389(1):211–25. doi: 10.1016/j.jmb.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Hunter T. Curr Opin Cell Biol. 2009;21(2):140–6. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Xie X, Chen C, Park JG, Stark C, James DA, Olhovsky M, Linding R, Mao Y, Pawson T. Sci Signal. 2009;2(98):ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG. Cell. 1997;89(6):909–16. doi: 10.1016/s0092-8674(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Kay RR. Curr Biol. 1997;7(11):R723–5. doi: 10.1016/s0960-9822(06)00366-6. [DOI] [PubMed] [Google Scholar]
- Kennelly PJ. Chem Rev. 2001;101(8):2291–312. doi: 10.1021/cr0002543. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN. Nat Rev Mol Cell Biol. 2006 Mar;7(3):165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N. Science. 2004;301(5631):361–3. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- King N. Dev Cell. 2004 Sep;7(3):313–25. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- King N, et al. Nature. 2008;451(7180):783–8. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenick J, Araki T, Yamada Y, Williams JG. J Cell Sci. 2008;121(Pt 21):3524–30. doi: 10.1242/jcs.036798. [DOI] [PubMed] [Google Scholar]
- Lee DC, Jia Z. Trends Biochem Sci. 2009;34(7):351–7. doi: 10.1016/j.tibs.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Li W, Scarlata S, Miller WT. Biochemistry. 2009 Jun 16;48(23):5180–6. doi: 10.1021/bi9000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA. Nat Rev Mol Cell Biol. 2010 Jun;11(6):393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y. Proc Natl Acad Sci U S A. 2008;105(28):9674–9. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniakis J, Funamoto S, Fukuzawa M, Meisenhelder J, Araki T, Abe T, Meili R, Hunter T, Williams J, Firtel RA. Genes Dev. 2001;15(6):687–98. doi: 10.1101/gad.871001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich SG, Garbarino E, Wei P, Lim WA. Science. 328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Letunic I, Bork P, Lim WA. Proc Natl Acad Sci U S A. 2009;105:9680–4. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Olivier P, Rozakis-Adcock M, McGlade J, Henkemeyer M. Philos Trans R Soc Lond B Biol Sci. 1993;340(1293):279–85. doi: 10.1098/rstb.1993.0069. [DOI] [PubMed] [Google Scholar]
- Pawson T. Nature. 1995;373(6515):573–80. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pawson T. Cell. 2004;116(2):191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- Rokas A. Annu Rev Genet. 2008;42:235–51. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- Schieven G, Thorner J, Martin GS. Science. 1986;231(4736):390–3. doi: 10.1126/science.2417318. [DOI] [PubMed] [Google Scholar]
- Sugden C, Ross S, Bloomfield G, Ivens A, Skelton J, Mueller-Taubenberger A, Williams JG. J Biol Chem. 2010 Jul 23;285(30):22927–35. doi: 10.1074/jbc.M110.139733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CS, Pasculescu A, Lim WA, Pawson T, Bader GD, Linding R. Science. 2009;325(5948):1686–8. doi: 10.1126/science.1174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK, Neel BG. Curr Opin Cell Biol. 2001;13(2):182–95. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]